Abstract
Heterotrophic marine nanoflagellates are important grazers on bacteria in the water column. Some marine bacteria appear more resistant to grazing than do others. Marine nanoflagellates can be grown in the laboratory in batch cultures fed specific bacterial isolates. In some cultures, the flagellates appear unable to completely deplete the bacterial prey even when the bacterial strain otherwise is an excellent prey. This may indicate that some marine bacteria are able to induce defence mechanisms if they are grazed by nanoflagellates. Four morphologically distinct marine heterotrophic nanoflagellates, of which 3 were still identified as Procryptobia sorokini (Kinetoplastea) and one as Paraphysomonas imperforata (Chrysophyceae) were isolated from a coastal location along with 3 isolates of the marine bacterium Pseudoalteromonas sp. Flagellate growth and grazing on bacterial prey were analysed in batch cultures. Pseudoalteromonas was a suitable prey for all 4 flagellate isolates. They grazed and grew on Pseudoalteromonas as sole prey with maximal cell-specific growth rates of 0.1–0.25 h-1 and gross growth efficiencies of 38–61%. Exposure to dense flagellate cultures or their supernatants did, however, cause a fraction of the Pseudoalteromonas cells to aggregate and the bacterium became apparently resistant to grazing. Concentrations of suspended Pseudoalteromonas cells were therefore not decreased below 1,700–7,500 cells μL-1 by any of the flagellate isolates. These results indicate that Pseudoalteromonas sp. can be an excellent prey to marine nanoflagellates but also that is in possession of inducible mechanisms that protect against flagellate grazing.
Introduction
Heterotrophic nanoflagellates play important roles in marine environments as bacterial grazers. Pelagic nanoflagellates control the concentration of bacteria in the water column [1, 2] and mediate the transfer of bacterial biomass to higher trophic levels of aquatic food webs [3]. Not all bacteria are captured and consumed equally efficient by all flagellates. Most interception feeding nanoflagellates (e.g. kinetoplastids and chrysophytes) seem to prefer larger, non-motile bacterial cells while smaller bacterial cells are preferred by e.g. the filter feeding choanoflagellates [4–6]. Bacteria may use a range of defence mechanisms to protect themselves from flagellate grazing, such as cell surface masking, digestional resistance, toxin production, aggregation, and microcolony formation [7]. Bacteria that are resistant to grazing may gain competitive advantages over other bacteria when grazed by nanoflagellate. In chemostat and mesocosm experiments, grazing resistant bacterial species [8, 9] or grazing resistant phenotypes of otherwise grazing sensitive bacteria [10–12] have dominated bacterial populations when nanoflagellates were also present. Evolutionary adaptations improving grazing resistance of bacterial strains have also been described [13]. It has furthermore been observed that some marine bacteria change physiology in the presence of heterotrophic nanoflagellates, grow in microcolonies, and secrete mucus to protect against grazing [14–16]. Flagellates are recognized by chemosensory mechanisms and the protective responses can be induced by cell free medium or filtrates from flagellate cultures [15, 16]. Grazing resistance may have a metabolic cost and grazing sensitive bacteria tends to become dominating in the absence of flagellate predators [9–11, 13].
Growth and bioenergetics have been investigated in several marine nanoflagellates [17– 29]. Maximal specific growth rates are commonly between 0.1–0.25 h-1 at 10–20°C with net growth efficiencies of 15–61% (measured either in units of biomass or cell carbon). The relationship between specific growth rate and concentration of bacterial cells can be described by Monod type saturation kinetics with half saturation constants of 1,300–45,000 bacterial cells per μL [17, 19–21, 23, 29]. These half-saturation constants are higher than the bacterial concentrations (1,000 bacterial cells per μL or less) normally found in marine waters [30] and bacterivorous marine nanoflagellates generally live under food limited conditions. In several studies, however, where flagellates have been grown in batch cultures, have the flagellates stopped taking up bacteria at concentrations of 2–20,000 bacterial cells per μL [19, 23, 28]. We observed the same phenomenon in preliminary batch cultures of marine nanoflagellates fed an isolated marine bacterium, Pseudoalteromonas sp. while there seemed not to be a clear relationship between initial and final Pseudoalteromonas concentrations. This could suggest that Pseudoalteromonas sp. may be able to activate growth independent responses that protect against grazing. Bacteria of the genus Pseudoalteromonas are widespread in marine environments [31] and among the bacteria that can readily be isolated from seawater and grown in axenic cultures in the laboratory. These bacteria are believed to be suitable prey for marine nanoflagellates since flagellate grazing apparently maintains the concentration of Pseudoalteromonas and other large gram-negative bacteria low in the sea [32]. Pseudoalteromonas species are known to secrete anti-bacterial compounds, toxins, extracellular enzymes, and exopolysaccharides, and surface dwelling strains can inhibit growth and settlement of other organisms [31].
If grazing protective mechanisms can be induced also in non-growing marine bacteria, such mechanisms may be found in e.g. Pseudoalteromonas species because these bacteria are heavily grazed [32] and in other connections have shown diverse metabolic capabilities [31]. In this paper, we have quantified growth of 4 heterotrophic, interception feeding, marine nanoflagellates feeding on 3 Pseudoalteromonas isolates, verified that Pseudoalteromonas sp. is excellent prey to these marine flagellates, and observed that Pseudoalteromonas indeed appears to be in possession of defence mechanisms that can be induced to protect them from being grazed by the flagellates.
Materials and methods
Sampling, isolation, and cultivation of marine nanoflagellates
Seawater (temperature = 8°C, salinity = 22‰) was sampled at a coastal location (public beach area) at Dokkedal, Denmark (56°54'21.2"N 10°16'9.2"E) 5–40 cm below the surface and brought to the laboratory and incubated at 15°C in the dark. Seawater for preparation of growth media was collected at the same location, filtered through 0.22 μm filters and autoclaved after enrichment by 0.01–1 g L-1 yeast extract.
Nanoflagellates were propagated by mixing 80 mL of freshly sampled sea water with 20 mL of medium, reaching a final concentration of yeast extract of 0.01 g L-1. After 2 weeks of incubation at 15°C in the dark, the cultures were examined for the presence of heterotrophic nanoflagellates under the microscope, diluted in sterile seawater until a flagellate concentration of 1 cell per μL, and 1 μL was transferred into a well in a microtiter plate containing 200 μL of sterile medium containing 0.01 g L-1 of yeast extract. This left on average 1 flagellate cell in each well. After 4–5 days if incubation at 15°C, each well was examined for the presence of flagellates under the microscope. Water from wells containing nanoflagellates were again diluted to a flagellate concentration of 1 cell per μL and transferred into a new well repeating the procedure described above. This procedure was repeated at least twice until all flagellates in one well appeared morphologically identical under the microscope (phase contrast, 40x and 60x objectives). Biweekly, the flagellate isolates were transferred into new batch cultures at 15°C in 22‰ autoclaved seawater supplemented by 0.01 g L-1 of yeast extract.
Isolation and cultivation of Pseudoalteromonas sp.
Seawater was sampled at Dokkedal, Denmark and diluted in sterile sea water until a bacterial concentration of approximately 10 cells per μL where after 25 μL was spread on petri dishes containing seawater enriched by 1 g L-1 of yeast extract and solidified by 10 g L-1 agar. The plates were incubated at 15°C for 4–5 days, and individual colonies were transferred to 0.22 μm filtered and autoclaved seawater enriched by 1 g L-1 of yeast extract and grown in batch cultures at 15°C. Three of these clonal isolates supported growth of clonal nanoflagellate cultures and were maintained in liquid culture and used as flagellate feed.
Molecular identification of nanoflagellate and bacterial isolates
Four supposedly clonal nanoflagellate isolates and three clonal bacterial isolates that supported growth of the flagellates were identified by partial 18S rDNA or 16S rDNA sequencing, respectively. Approximately 25 mg of lyophilized biomass was subjected to bead-beating (6000 rpm for 3×5 seconds using 1.4 mm ceramic spheres, 0.1 mm silica spheres, and one 4 mm glass bead) on a Precellys tissue homogenizer (Bertin, France). Samples were cooled on ice before and after the bead beating. Thereafter genomic DNA was extracted by the Qiagen DNeasy Plant Mini Kit (Qiagen, Germany) or the FastDNA™ SPIN kit for soil (MP Biomedicals, USA). The DNA regions encoding part of the 18S and 16S rDNA regions were amplified from 25–50 ng genomic DNA template using primer pairs F-566/R-1200, cryso240/ cryso651, and 27F/1492R, respectively (Table B in S1 File). PCR products were generated in 50 μL reaction volumes containing 0,02 U Phusion™ High-Fidelity DNA Polymerase (Thermo Scientific, USA), 1× HF Green Buffer, 200 μM dNTP mix, and 0.5 μM forward and 0.5 μM reverse primer. PCR products were subsequently purified with either the QIAquick PCR Purification Kit (Qiagen, Germany) or the QIAquick Gel Extraction Kit (Qiagen, Germany) and outsourced for sequencing at Eurofins Genomics (Eurofins, Germany). A blastN analysis was performed against the non-redundant database at NCBI to identify the relevant taxa [33].
Grazing experiments
Nanoflagellate isolates were grown in 100 mL batch cultures in conical flasks incubated at 15°C in an orbital shaking incubator operated at 100 RPM. Pseudoalteromonas sp. harvested from 1-day old batch cultures that had entered stationary phase served as feed. Initial concentrations of flagellates were 30–70 cells per μL while initial concentrations of Pseudoalteromonas sp. were 7,500–30,000 cells per μL. The bacteria were harvested by centrifugation and resuspended in 1 mL 0.22 μm sterile filtered seawater before added to flagellate cultures in order to minimize potential transfer of left-over components from the bacterial growth medium to the flagellate cultures. Flagellate and bacterial cells were simultaneously counted in samples taken from the cultures and used to quantify growth and grazing (see below).
Determination of cell densities, cell dry weights, yield coefficients, and gross growth efficiencies
Concentrations of nanoflagellates, cf and bacteria, cb were determined from microscopically counts in a 0.0025 mm3 hemocytometer (Thoma). The concentration of grazing resistant bacterial cells, cb,end was estimated as bacterial concentrations remaining in each culture after flagellate growth has stopped. In bacterial cultures cell concentrations were also followed indirectly from measurements of the optical density at 600 nm, OD600 after dilution to OD600 values below 0.3.
The concentration of dry biomass of nanoflagellate or bacterial cultures were measured after filtration of culture onto pre-dried and pre-weighed 0.22 μm MF membrane filters (Millipore) and drying at 105°C overnight. Dry weights of individual bacterial cells, mb were estimated by comparing dry biomass concentrations to bacterial cell concentrations. Dry weights of individual flagellate cells, mf were estimated the same way, but after subtraction of the dry bacterial biomass concentrations in the cultures.
The yield coefficient, Yf/b was estimated from slopes of linear regressions of plots of increase of flagellate concentration, cf−cf,0. vs. decrease of bacterial cell concentration, cb,0 –cb. The number of bacterial cells needed to produce one nanoflagellate cell, Yb/f corresponds to Yf/b-1. Gross growth efficiency, GGE was calculated by multiplying Yf/b values by the ratio between dry flagellate and bacterial cell weights.
Modelling of cell concentrations and estimation of flagellate performance
Grazing and growth of flagellates were described using the growth model shown in S1 File. The individual equations in the model were discretized and solved numerically using Euler’s solution at time intervals, Δt = 0.2 h, starting at t = 0, and in the order shown in Table A in S1 File. Maximal clearance rate, Clmax and ingestion rate, Imax were estimated by fitting Eqs. A-F in S1 File to experimentally determined concentrations of flagellate and bacterial cells in 3 flagellate batch cultures grown on different initial concentrations of non-growing bacterial cells. The numerical differences between measured and calculated cell concentrations were simultaneously minimized in all 3 cultures. The half saturation constant, kb was found from Eq. H, while the maximal specific growth rate, μmax of the flagellates was estimated from Eq. E in S1 File setting I = Imax.
Results
Isolation and identification of nanoflagellates and bacterial prey
Four morphologically distinct nanoflagellates (Fig A in S1 File) were isolated and grown in batch cultures. For 3 of the nanoflagellates, blast analyses of their 18S rDNA sequences showed closest sequence similarity (99%) to GenBank ID KF479401.13 identifying them as Procryptobia sorokini (Table D in S1 File). These 3 isolates were named P. sorokini strains G5, B11, and A5, respectively. P. sorokini strain A5 had a hook shaped flagellum (Fig A in S1 File) and resembled the morphotype Neobodo [34]. Procryptobia sorokini (syn. Bodo sorokini) as well as Neobodo sp. belong to the Kinetoplastea, order Bodonida [35], which is a group of motile, interception feeding nanoflagellates that are common members of the heterotrophic picoplankton. The last of the isolates was identified as Paraphysomonas imperforata and named strain A2 (99% sequence similarity to GenBank ID KX431470.1). P. imperforata belongs to the Chrysophyceae [35] and is also a motile bacterivorous predator. The nanoflagellates had dried cell masses between 96 and 219 pg cell-1 during growth phases (Table 1).
Table 1. Characteristic values of batch cultures of Procryptobia sorokini G5, B11, A5 and Paraphysomonas imperforata A2, feeding on Pseudoalteromonas sp. B2, B3 or B4.
Cell dry weights of flagellates, mf, and bacteria, mb, measured during growth and stationary phase, respectively. Yield of flagellates per bacterium taken up, Yf/b, number of bacterial cells needed to produce one flagellate cell Yb/f, and gross growth efficiency, GGE were evaluated from Figs 1C, 1D, 2C and 2D, respectively (data from batch cultures of P. imperforata A2 feeding on Pseudoalteromonas sp. B2 in Fig B in S1 File. Maximal clearance, Clmax, and ingestion rates, Imax, are estimated by fitting numerical solutions to Eqs. A and D in S1 File to experimentally determined concentrations of flagellate and bacterial cells (Figs 1A, 1b, 2A and 2B). Maximal specific growth rate, μmax, and half saturation constant, Kb, are calculated from Eqs. E and H, respectively. Final bacterial concentration, Cb.end. Three batch cultures were carried out for each combination of flagellate and bacterial isolate.
| Flagellate isolate |
Bacterial isolate |
mf | mb | Yf/b | Yb/f | GGE | Clmax | Imax | μmax | Kb | Cb.end |
|---|---|---|---|---|---|---|---|---|---|---|---|
| pg per cell | pg per cell | μL cell-1 h-1 | h-1 | h-1 | cells per μL | cells per μL | |||||
| G5 | B2 | 116 | 4.0 | 0.021 | 48 | 0.61 | 0.0041 | 11.7 | 0.25 | 2,854 | 1,700 |
| B11 | B4 | 124 | 3.6 | 0.016 | 61 | 0.57 | 0.0064 | 6.7 | 0.11 | 1,127 | 4,800 |
| A5 | B4 | 219 | 3.6 | 0.010 | 102 | 0.60 | 0.0025 | 20.7 | 0.20 | 8,350 | 4,600 |
| A2 | B3 | 96 | 3.1 | 0.015 | 67 | 0.46 | 0.0013 | 12.6 | 0.19 | 9,705 | 3,400 |
| A2 | B2 | 96 | 4.0 | 0.016 | 64 | 0.38 | 0.0017 | 6.3 | 0.10 | 3,615 | 7,500 |
At the location at Dokkedal from where the nanoflagellates were isolated, bacterial numbers in the water varied between 200 and 900 cells μL-1 between October 2015 and April 2016. Three bacterial isolates, isolated from the same water samples as the nanoflagellates, were identified through partial 16S rDNA sequencing as Pseudoalteromonas sp. and named strain B2, B3, and B4, respectively (showing 99% sequence identity to GenBank ID MF061255.1, Table D in S1 File). Pseudoalteromonas sp. grew as solitary rod shaped cells, approximately 2 μm in length and with a diameter of 1 μm. Few aggregates of 10 or more cells were, however, also present in growing cultures. Cells harvested from stationary phase of batch cultures grown in 22‰ seawater supplemented with yeast extract had cell masses of 3–4 pg cell-1 (Table 1). All 3 Pseudoalteromonas sp. isolates were grazed by the 4 nanoflagellates and supported their growth when used as the only feed.
Growth and grazing
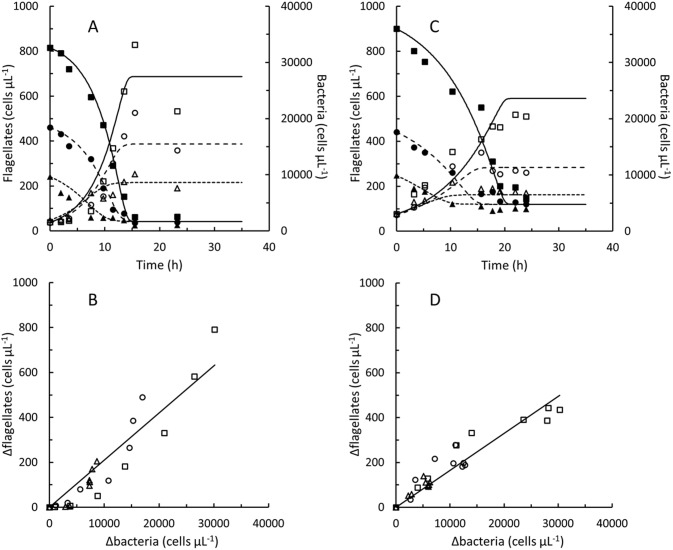
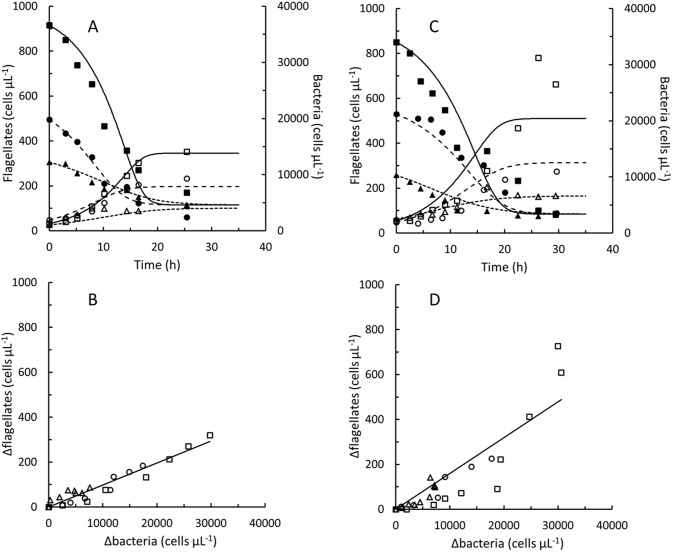
The 4 nanoflagellates, P. sorokini G5, B11, A5 and P. imperforata A2 were grown in batch cultures on different concentrations of Pseudoalteromonas sp. (Figs 1 and 2). In control experiments without flagellates, bacterial concentrations decreased by 0–10% during 14 h periods (corresponding to the growth phase of flagellate cultures). The seawater used was therefore too low in nutritional compounds to supported additional cell divisions in Pseudoalteromonas sp. (under nutrient sufficient conditions the doubling time of Pseudoalteromonas sp. was approximately 1.5 h). In control experiments without addition of Pseudoalteromonas sp., flagellate concentrations stayed constant or decreased by up to 30% during 24 h periods. Flagellate growth and decreasing bacterial concentrations were therefore linked to grazing in these experiments. Flagellate concentrations increased in proportion to the decrease in bacterial concentrations (Figs 1 and 2). The yield, Yf/b varied between 0.010 and 0.021 flagellate cell per ingested Pseudoalteromonas cell (Table 1), meaning that between 48 and 102 bacterial cells were ingested to produce one flagellate cell. The estimated gross growth efficiency, GGE taken into account the masses of the flagellate as well as the Pseudoalteromonas cells were between 38 and 61%. Fairly similar GGE’s (24–50%) were found when cell masses were predicted from rough estimates of cell volumes (Table C in S1 File), although cell masses estimated from the cell volumes were lower than the measured ones.
Fig 1. Procryptobia sorokini.
Batch cultures of Procryptobia sorokini G5 feeding on Pseudoalteromonas sp. B2 (A and B) and P. sorokini B11 feeding on Pseudoalteromonas sp. B4 (C and D). Concentrations of flagellate cells (open symbols) and bacterial cells (solid symbols) in cultures inoculated at approximately 7,500 (△, ▲), 15,000 (○, ●), and 30,000 (□, ■) Pseudoalteromonas sp. μL-1, respectively. Curves (A and C) drawn by fitting Eqs. A-F in S1 File to measured concentrations of Procryptobia sorokini and Pseudoalteromonas sp. Data in S1 Dataset.
Fig 2. Procryptobia sorokini and Paraphysomonas imperforata.
Batch cultures of Procryptobia sorokini A5 feeding on Pseudoalteromonas sp. B4 (A and B) and Paraphysomonas imperforata A2 feeding on Pseudoalteromonas sp. B3 (C and D). Concentrations of flagellate cells (open symbols) and bacterial cells (solid symbols) in cultures inoculated at approximately 7,500 (△, ▲), 15,000 (○, ●), and 30,000 (□, ■) Pseudoalteromonas sp. μL-1, respectively. Curves (A and C) drawn by fitting Eqs. A-F in S1 File to measured concentrations of flagellates and Pseudoalteromonas sp. Data in S1 Dataset.
None of the nanoflagellate cultures were able to decrease the concentration of bacterial cells to zero. Irrespectively of the initial bacterial density, 2–5,000 bacteria per μL remained in the cultures when the flagellates entered stationary phase (Figs 1 and 2). Flagellate and bacterial concentrations were modelled by Eqs. A-H in S1 File and maximal clearance rates, Clmax and ingestion rates, Imax are listed in Table 1, along with maximal specific growth rates, μmax and half saturation constants, kb. Clearance and ingestion rates predicted from Eqs. B and C in S1 File as function of bacterial concentration are shown in Fig C in S1 File.
Responses in Pseudoalteromonas sp.
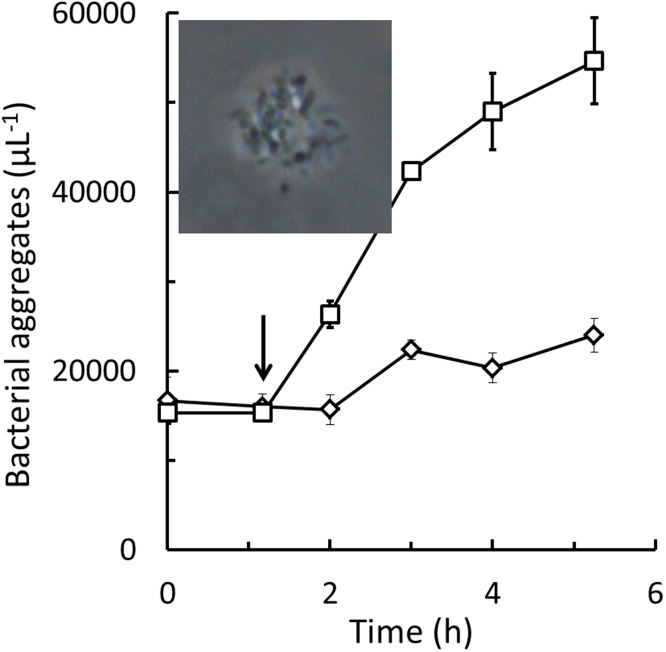
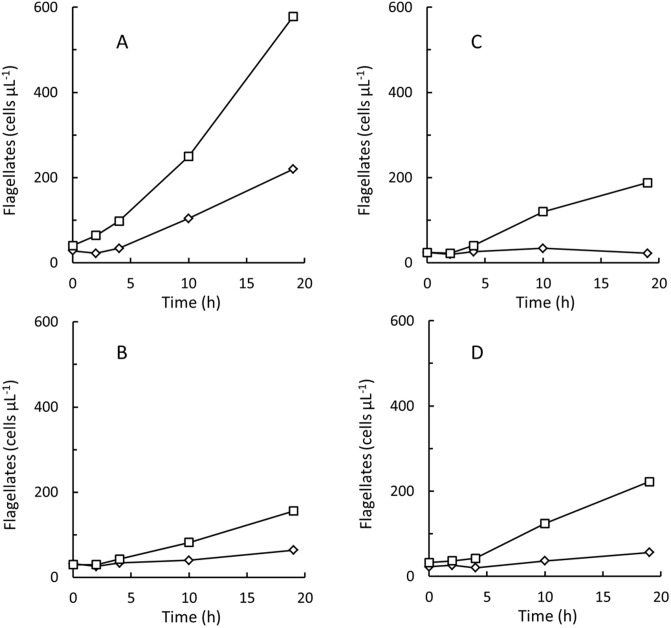
The major fraction of the Pseudoalteromonas sp. cells in flagellate cultures remained solitary until the cultures were terminated (bacterial concentrations shown in Figs 1 and 2 represent solitary cells) but at the end of the experiments, a fraction also formed aggregates of 10 or more bacterial cells. Increased aggregation of Pseudoalteromonas sp. could be induced also by cell free culture supernatant from the nanoflagellate cultures. Fig 3 shows numbers of bacterial aggregates in Pseudoalteromonas sp. B2 diluted in seawater with and without addition of 10% spent supernatant from a stationary phase P. sorokini G5 culture, sterile filtered through a 0.22 μm syringe filter. After 1 h of incubation, higher numbers of bacterial aggregates were observed after the spent culture supernatant had been added. Increased cell aggregation was likewise observed in Pseudoalteromonas sp. B3 and B4 upon addition of 0.22 μm sterile filtered supernatant from cultures of all 4 nanoflagellates. When cell free culture supernatant taken from stationary phase of flagellate batch cultures was added to newly inoculated flagellate cultures, also flagellate growth was inhibited. Fig 4 compares growth of the 4 nanoflagellates in batch cultures with and without addition of 5% 0.22 μm sterile filtered supernatant taken from stationary phase cultures of one of the other flagellate isolates. Bacterial aggregates appeared in all the flagellate cultures to which spent culture supernatant had been added. Finally was it observed, that 0.22 μm sterile filtered supernatant taken from flagellate batch cultures also effected growth of freshly inoculated batch cultures of Pseudoalteromonas sp. (Fig D in S1 File).
Fig 3. Pseudoalteromonas.
Concentration of aggregates >10 cells of Pseudoalteromonas sp. B2. Arrow marks addition of 10% 0.22 μm sterile filtered culture supernatant from a stationary phase culture of Procryptobia sorokini G5 (□) or 10% 0.22 μm sterile filtered seawater (◊). Initial concentrations were 1,000,000 solitary Pseudoalteromonas sp. B2 μL-1. Inset is micrograph showing aggregated Pseudoalteromonas sp. B2 cells viewed under phase contrast.
Fig 4. Procryptobia sorokini and Paraphysomonas imperforata.
Batch cultures of Procryptobia sorokini G5 fed Pseudoalteromonas sp. B2 (A), P. sorokini B11 fed Pseudoalteromonas sp. B4 (B), P. sorokini sp. A5 fed Pseudoalteromonas sp. B4 (C), and Paraphysomonas imperforata A2 fed Pseudoalteromonas sp. B3 (D). Concentrations of flagellates in cultures added 5% 0.22 μm sterile filtered culture supernatant from a stationary phase culture of a different flagellate isolate (◊) or 5% 0.22 μm sterile filtered seawater (□).
Discussion
The 4 nanoflagellates isolated in this study, Procryptobia sorokini G5, B11, A5 and Paraphysomonas imperforata A2 were all able to graze and grow on Pseudoalteromonas sp. as their sole prey. This bacterium seems, however, also able to become resistant to flagellate grazing and was never completely depleted despite the high concentrations of flagellates. Cell aggregation may be part of the mechanism that protects Pseudoalteromonas sp. against flagellate grazing but since most of the Pseudoalteromonas cells remained solitary, cell aggregation may not be the only protective mechanism in their possession. Since addition of cell free culture supernatant from flagellate cultures was sufficient to induce aggregation of Pseudoalteromonas cells, this bacterium must be able to detect flagellates by a chemosensory mechanism. Our data suggest that Pseudoalteromonas can be excellent prey for flagellates but also that the presence of flagellates rapidly can induce growth independently defence mechanisms against grazing in Pseudoalteromonas sp.
Pseudoalteromonas are gram-negative, rod shaped bacteria. They are large bacterial cells [32] with dry weights of 3–4 pg cell-1 (Table 1). They therefore seem to fulfil the several criteria for being suitable prey to interception feeding nanoflagellates [4–6, 17, 18, 22, 27]. Specific growth rates ranged from 0.1–0.25 h-1 (Table 1), which is within the expected range for heterotrophic nanoflagellates [17, 19, 22, 24, 29]. In P. imperforata A2, the maximal clearance rates of 1.3 and 1.7 nL per cell per h agreed excellently with earlier measurement of 1.55 nL per cell per h [36] while higher Clmax values have been found in other heterotrophic nanoflagellates [17]. Maximal ingestion rates ranged from 6 to 21 bacterial cells per cell per h. These values are low compared to what have been found in earlier studies. E.g. has Imax previously been estimated to 62–103 cells per cell per h in P. imperforata [21, 24] while 6 different nanoflagellates had Imax values of 27–254 cells per cell per h [17]. Clearance and ingestion rates show, however, considerable variations among nanoflagellates [22] and the low ingestion rates were probably related to the large cell size of Pseudoalteromonas sp. Gross growth efficiencies (GGE’s) may therefore represent at better foundation for comparing the performances of different flagellates feeding on different prey. The GGE’s of the 4 nanoflagellates feeding on Pseudoalteromonas sp. were between 38 and 61% (Table 1). These GGE estimates are high but within the range observed also for other heterotrophic nanoflagellates [17–20, 22, 23, 25–27]. The GGE’s of P. imperforata A2 of 36–46% (Table 1) were also within the range of 15–54% (based on either mass or bio volume) previously reported for this species [21, 24, 29]. Changes in flagellate cell size during the final cell generation [17, 20] or cryptic bacterial growth stimulated by re-mineralising of nutrients during the experiments [23] may, however, potentially have resulted in too high GGE estimates. Still, the high GGE’s in combination with the high μmax indicate that Pseudoalteromonas can be an excellent prey to all 4 nanoflagellate isolates. The 4 nanoflagellates and the 3 Pseudoalteromonas strains were isolated from the same waters. Potentially, it may therefore be advantageous for Pseudoalteromonas sp. to be in possession of inducible defensive mechanisms against flagellates, although the defensive reactions observed in this study appeared at artificially high concentrations of flagellate and bacterial cells.
When flagellate grazing had decreased the concentration of Pseudoalteromonas sp. to 2–5,000 cells per μL grazing stopped and bacterial and flagellate concentrations became stable (Figs 1 and 2). The decrease in concentrations of P. sorokini G5 after 15 h (Fig 1A) is maybe a result of cannibalism [37]. Grazing and growth did not resume if additional Pseudoalteromonas sp. was added (data not shown). Inhibition of newly inoculated flagellate cultures by spent culture supernatant (Fig 4) demonstrates that it was not the high flagellate concentrations that were growth-inhibiting. Because grazing stopped at fairly high bacterial concentrations, Kb values could only be estimated indirectly from Eq. H. The accuracy of these estimates may be hampered by the fact that the grazing-resistant bacterial sub-population, cb,end was modelled as being constant (Eq. F in S1 File) despite the protective mechanisms in Pseudoalteromonas sp. rather appeared to be inducible (Figs 3 and 4). Still, the estimated Kb values of 1,100–9,700 bacterial cells per μL (Table 1) are within the range of Kb values found also in other marine nanoflagellates [17, 19, 20, 22, 24, 29].
Bacterial concentrations of 2–5,000 cells per μL at which flagellate growth stopped cannot be expected to represent lower limits for nanoflagellate growth and grazing, also the same has been observed also in cultures of other heterotrophic marine nanoflagellates [19, 23, 28], but may rather be a result of the induction of protective mechanisms in the bacterial prey. The Kb values (Table 1) were of the same order of magnitude as the lowest bacterial cell concentrations in the cultures (Figs 1 and 2). If no protective mechanisms had been induced in Pseudoalteromonas sp., ingestion and specific growth rates of the flagellates should expectedly have proceeded at half their maximal values at the bacterial concentrations at which grazing and growth actually stopped. In comparison to bacterial concentrations in seawater [30], 2–5,000 bacteria per μL are a high. We found e.g. only 200–900 bacterial cells per μL at the location where the nanoflagellates were isolated during the winter period from October to March while a second study counted 2–4,000 bacterial cells per μL at a nearby location during August and September [1]. Data from earlier studies do also show that nanoflagellate batch cultures in some cases have been able to decrease bacterial numbers to levels at or below 1,000 cells per μL [17, 29, 38].
Aggregation could be one mechanism used by Pseudoalteromonas sp. to protect against flagellate grazing [7] as partial aggregation of non-growing cells could be induced in all 3 Pseudoalteromonas by supernatant from all the isolated nanoflagellates, despite P. imperforata taxonomically is unrelated to P. sorokini. Aggregation is probably not the only protective mechanism in Pseudoalteromonas sp. since the concentration of solitary bacterial cells remained above the Kb values after flagellates entered stationary phase (Figs 1 and 2). Aggregation may also not be particularly efficient in natural waters if cell concentrations are low. Although bacterial aggregates are avoided by some nanoflagellates they may actually be preferred by others [39]. This could be one reason for at bacterium to use more than one protective mechanism. Protective mechanisms impose a metabolic load [9–11, 13]. Inducible grazing mechanisms may therefore be way to preserve the ability of Pseudoalteromonas sp. to compete with other bacteria when grazing pressures are low. The results of this study therefore suggest that Pseudoalteromonas sp. may indeed be an excellent prey to marine nanoflagellates but also that inducible defence mechanisms can protect it against flagellate grazing.
Supporting information
(XLSX)
Table A The order in which Eqs. A-E in the growth model are evaluated to model the concentrations of flagellates and bacterial prey cells. Table B DNA primers used in this study of the identification of the nanoflagellates and the bacterium Pseudoalteromonas sp. Table C Gross growth efficiencies estimated from cell dimensions. Table D Partial 18S rDNA or 16S rDNA sequences used to identify flagellates and Pseudoalteromonas sp. Fig A Micrographs of nanoflagellate isolates fixed in Lugol’s solution. Fig B Batch cultures of Paraphysomonas imperforata A2 feeding on Pseudoalteromonas sp. B2. Fig C Predicted clearance and ingestion rates as function of bacterial prey concentration. Fig D Batch cultures of Pseudoalteromonas sp. grown with and without addition of sterile supernatant from a Procryptobia sorokini culture.
(PDF)
Acknowledgments
We thank Per Andersen, Aarhus University for his help morphotyping Neobodo sp.
Data Availability
Data are included in S1 Dataset.
Funding Statement
This work was supported by a research grant (9278) from Villum Fonden to NTE. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fenchel T. Ecology of heterotrophic microflagellates. IV. Quantitative occurrence and importance as bacterial consumers. Mar Ecol Prog Ser. 1982; 9: 35–42. [Google Scholar]
- 2.Massana R, Jürgens K. Composition and population dynamics of planktonic bacteria and bacterivorous flagellates in seawater chemostat cultures. Aquat Microb Ecol. 2003; 32: 11–22. [Google Scholar]
- 3.Azam F, Fenchel T, Field JG, Gray JS, Meyer-Reil LA, Thingstad F. The Ecological role of water-column microbes in the sea. Mar Ecol Prog Ser. 1983; 10: 257–263. [Google Scholar]
- 4.González JM, Sherr EB, Sherr BG. Size-selective grazing on bacteria by natural assemblages of estuarine flagellates and ciliates. Appl Environ Microbiol. 1990; 56: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.González JM. Efficient size-selective bacterivory by phagotrophic nanoflagellates in aquatic ecosystems. Mar Biol. 1996; 126: 785–789. [Google Scholar]
- 6.Boenigk J, Arndt H. Comparative studies on the feeding behavior of two heterotrophic nanoflagellates: the filterfeeding choanoflagellate Monosiga ovata and the raptorial-feeding kinetoplastid Rhynchomonas nasuta. Mar Ecol Prog Ser. 2000; 22: 243–249. [Google Scholar]
- 7.Matz C, Kjelleberg S. Off the hook–how bacteria survive protozoan grazing. TRENDS Microbiol. 2005; 13: 302–307. doi: 10.1016/j.tim.2005.05.009 [DOI] [PubMed] [Google Scholar]
- 8.Corno G, Caravati E, Callieri C, Bertoni R. Effects of predation pressure on bacterial abundance, diversity, and size-structure distribution in an oligotrophic system. J Limnol. 2008; 67: 107–119. [Google Scholar]
- 9.Corno G, Jürgens K. Structural and functional patterns of bacterial communities in response to protist predation along an experimental productivity gradient. Environ Microbiol. 2008; 10: 2857–2871. doi: 10.1111/j.1462-2920.2008.01713.x [DOI] [PubMed] [Google Scholar]
- 10.Hahn MW, Höfle MG. Flagellate predation on a bacterial model community: Interplay of size-selective grazing, specific bacterial cell size, and bacterial community composition. Appl Environ Microbiol. 1999; 65: 4863–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matz C, Deines P, Jürgens K. Phenotypic variation in Pseudomonas sp. CM10 determines microcolony formation and survival under protozoan grazing. FEMS Microbiol Ecol. 2002; 39: 57–65. doi: 10.1111/j.1574-6941.2002.tb00906.x [DOI] [PubMed] [Google Scholar]
- 12.Boenigk J, Stadler P, Wiedlroither A, Hahn MW. Strain-specific differences in the grazing sensitivities of closely related ultramicrobacteria affiliated with the Polynucleobacter cluster. Appl Environ Microbiol. 2004; 70: 5787–5793. doi: 10.1128/AEM.70.10.5787-5793.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baumgartner M, Neu TR, Blom JF, Pernthaler J. Protistan predation interferes with bacterial long-term adaptation to substrate restriction by selecting for defence morphotypes. J Evol Biol. 2016; 29: 2297–2310. doi: 10.1111/jeb.12957 [DOI] [PubMed] [Google Scholar]
- 14.Corno G, Jürgens K. Direct and indirect effects of protist predation on population size structure of a bacterial strain with high phenotypic plasticity. Appl Environ Microbiol. 2006; 72: 78–86. doi: 10.1128/AEM.72.1.78-86.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blom JF, Hornák K, Šimek K, Pernthaler J. Aggregate formation in a freshwater bacterial strain induced by growth state and conspecific chemical cues. Environl Microbiol. 2010; 12: 2486–2495. [DOI] [PubMed] [Google Scholar]
- 16.Blom JF, Zimmermann YS, Ammann T, Pernthaler J. Scent of danger: Floc formation by a freshwater bacterium is induced by supernatants from a predator-prey coculture. Appl Envrion Microbiol. 2010; 76: 6156–6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fenchel T. Ecology of heterotrophic microflagellates. II. Bioenergetics and growth. Mar Ecol Prog Ser. 1982; 8: 225–231. [Google Scholar]
- 18.Sherr BF, Sherr EB, Berman T. Grazing, growth, and ammonium excretion rates of a heterotrophic microflagellate fed with four species of bacteria. Appl Environ Microbiol. 1983; 45: 1196–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geider RJ, Leadbeater BSC. Kinetics and energetics of growth of the marine choanoflagellate Stephanoeca diplocostata. Mar Ecol Prog Ser. 1988; 47: 169–177. [Google Scholar]
- 20.Andersen P. Functional biology of the choanoflagellate Diaphanoeca grandis Ellis. Mar Microb Food Webs. 1988/1989; 3: 35–50. [Google Scholar]
- 21.Choi JW, Peters F. Effects of temperature on two psychrophilic ecotypes of a heterotrophic nanoflagellate, Paraphysomonas imperforata. Appl Environ Microbiol. (1992; 58: 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eccleston-Parry JD, Leadbeater BS. A comparison of the growth kinetics of six marine heterotrophic nanoflagellates fed with one bacterial species. Mar Ecol Prog Ser. 1994; 105: 167–177. [Google Scholar]
- 23.Snyder RA, Hoch MP. Consequences of protist-stimulated bacterial production for estimating protist growth efficiencies. Hydrobiologia. 1996; 341: 113–123. [Google Scholar]
- 24.Peters F, Choi JW, Gross T. Paraphysomonas imperforata (Protista, Chrysomonadida) under different turbulence levels: feeding, physiology and energetics. Mar Ecol Prog Ser. 1996; 134: 235–245. [Google Scholar]
- 25.Sleigh MA, Zubkov MV. Methods of estimating bacterivory by protozoa. Europ J Protistol. 1998; 34: 273–280. [Google Scholar]
- 26.Zubkov MV, Sleigh MA. Comparison of growth efficiencies of protozoa growing on bacteria deposited on surfaces and in suspension. J Eukaryot Microbiol. 2000; 47: 62–69. [DOI] [PubMed] [Google Scholar]
- 27.Frolov AO, Karpov SA, Mylnikov AP. The ultrastructure of Procryptobia sorokini (Zhukov) comb.nov. and rootlet homology in kinetoplastids. Protistology. 2001; 2: 85–95. [Google Scholar]
- 28.Ishigaki T, Sleigh MS. Grazing characteristics and growth efficiencies at two different temperatures for three nanoflagellates fed with Vibrio bacteria at three different concentrations. Microb Ecol. 2001; 41: 264–271. doi: 10.1007/s002480000060 [DOI] [PubMed] [Google Scholar]
- 29.Rose JM, Vora NM, Countway PD, Gast RJ, Caron DA. Effects of temperature on growth rate and gross growth efficiency of an Antarctic bacterivorous protist. ISME J. 2009; 3: 252–260. doi: 10.1038/ismej.2008.96 [DOI] [PubMed] [Google Scholar]
- 30.Ferguson RL, Rublee P. Contribution of bacteria to standing crop of coastal plankton. Limnol Oceanogr. 1976; 21: 141–145. [Google Scholar]
- 31.Holmström C, Kjelleberg S. Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol Ecol. 1999; 30: 285–293. [DOI] [PubMed] [Google Scholar]
- 32.Beardsley C, Pernthaler J, Wosniok W, Amann R. Are readily culturable bacteria in coastal North Sea waters suppressed by selective grazing mortality? Appl Environ Microbiol. 2003; 69: 2624–2630. doi: 10.1128/AEM.69.5.2624-2630.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990; 215: 403–10. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 34.Jeuck A, Arndt H. A short guide to common heterotrophic flagellates of freshwater habitats based on the morphology of living organisms. Protist. 2013; 164: 842–860. doi: 10.1016/j.protis.2013.08.003 [DOI] [PubMed] [Google Scholar]
- 35.Guiry MD, Guiry GM. AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. http://www.algaebase.org. 2018.
- 36.Fu Y, O’Kelly C, Sieracki M, Distel DL. Protistan grazing analysis by flow cytometry using prey labeled by in vivo expression of fluorescent proteins. Appl Environ Microbiol. 2003; 69: 6848–6855. doi: 10.1128/AEM.69.11.6848-6855.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martel CM, Flynn KJ. Morphological controls on cannibalism in a planktonic marine phagotroph. Protist. 2008; 159: 41–51 doi: 10.1016/j.protis.2007.05.003 [DOI] [PubMed] [Google Scholar]
- 38.Pelegrí SP, Christaki U, Dolan J, Rassoulzadegan F. Particulate and dissolved organic carbon production by the heterotrophic nanoflagellate Pteridomonas danica Patterson and Fenchel. Microb Ecol. 1999; 37: 276–284. [DOI] [PubMed] [Google Scholar]
- 39.Sibbald MJ, Albright LJ. Aggregated and free bacteria as food sources for heterotrophic microflagellates. Appl Environ Microbiol. 1988; 54: 613–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Table A The order in which Eqs. A-E in the growth model are evaluated to model the concentrations of flagellates and bacterial prey cells. Table B DNA primers used in this study of the identification of the nanoflagellates and the bacterium Pseudoalteromonas sp. Table C Gross growth efficiencies estimated from cell dimensions. Table D Partial 18S rDNA or 16S rDNA sequences used to identify flagellates and Pseudoalteromonas sp. Fig A Micrographs of nanoflagellate isolates fixed in Lugol’s solution. Fig B Batch cultures of Paraphysomonas imperforata A2 feeding on Pseudoalteromonas sp. B2. Fig C Predicted clearance and ingestion rates as function of bacterial prey concentration. Fig D Batch cultures of Pseudoalteromonas sp. grown with and without addition of sterile supernatant from a Procryptobia sorokini culture.
(PDF)
Data Availability Statement
Data are included in S1 Dataset.