Abstract
The predisposition to neuropsychiatric disease involves a complex, polygenic, and pleiotropic genetic architecture. However, little is known about how genetic variants impart brain dysfunction or pathology. We use transcriptomic profiling as a quantitative readout of molecular brain-based phenotypes across 5 major psychiatric disorders, including autism (ASD), schizophrenia (SCZ), bipolar disorder (BD), depression (MDD), and alcoholism (AAD), compared with matched controls. We identify patterns of shared and distinct gene-expression perturbations across these conditions. Notably, the degree of sharing of transcriptional dysregulation is related to polygenic (SNP-based) overlap across disorders, suggesting a significant causal genetic component. This comprehensive systems-level view of the neurobiological architecture of major neuropsychiatric illness demonstrates pathways of molecular convergence and specificity.
Despite remarkable success identifying genetic risk factors for major psychiatric disorders, it remains unknown how genetic variants interact with environmental and epigenetic risk factors in the brain to impart risk for clinically distinct disorders (1, 2). We reasoned that brain transcriptomes, a quantitative, genome-wide molecular phenotype (3), would allow us to determine whether disease-related signatures are shared across major neuropsychiatric disorders with distinct symptoms and whether these patterns reflect genetic risk.
We first analyzed published gene-expression microarray studies of cerebral cortex across five major neuropsychiatric disorders (3–11) using 700 cerebral cortical samples from subjects with ASD (n=50 samples), SCZ (n=159), BD (n= 94), MDD (n=87), AAD (n=17), and matched controls (n=293) (12). These disorders are prevalent and disabling, contributing substantially to global disease burden. Inflammatory bowel disease (IBD, n=197) was included as a non-neural comparison.
Individual datasets underwent stringent quality control and normalization (Fig. 1; (12)), including re-balancing to alleviate confounding between diagnosis and biological (e.g., age, sex) or technical (e.g., post-mortem interval, pH, RIN, batch, 3′ bias) covariates (Figs. S1, S2). Transcriptome summary statistics for each disorder were computed with a linear mixed-effects model to account for any sample overlap across studies (12). Comparison of differential gene expression (DGE) log2 fold change (log2FC) signatures revealed a significant overlap among ASD, SCZ, and BD and SCZ, BD, and MDD (Fig. 2A; all Spearman’s ρ ≥ 0.23, P < 0.05, 40,000 permutations). The regression slopes between ASD, BD, and MDD log2-FC effect sizes compared to SCZ (5.08, 0.99, and 0.37) indicate a gradient of transcriptomic severity with ASD > SCZ ≈ BD > MDD (Fig. 2B). To ensure robustness, we compared multiple methods for batch correction, probe summarization, and feature selection, including use of integrative correlations, none of which changed the qualitative observations (Fig. S3; (12)). Results were also unaltered after first regressing gene-level RNA degradation metrics, suggesting that systematic sample quality issues were unlikely to drive these correlations (Fig. S3). Further, the lack of (or negative) overlap between AAD and other disorders suggests that similarities are less likely due to comorbid substance abuse, poor overall general health, or general brain-related post-mortem artefacts.
Fig. 1.
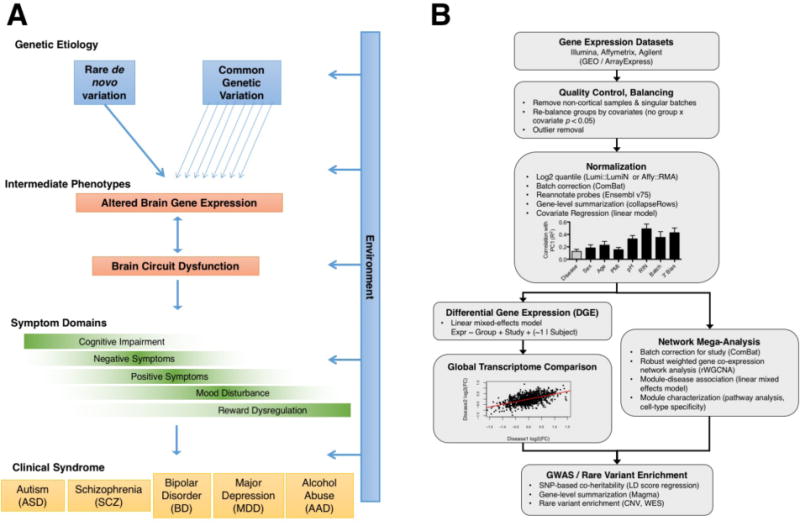
(A) Model of psychiatric disease pathogenesis. (B) Flowchart of the cross-disorder transcriptome analysis pipeline (12). Cortical gene expression datasets were compiled from cases of ASD (n=50 samples), SCZ (n=159), BD (n=94), MDD (n=87), AAD (n=17), and matched non-psychiatric controls (n=293) (12) (see Table S1).
Fig. 2.
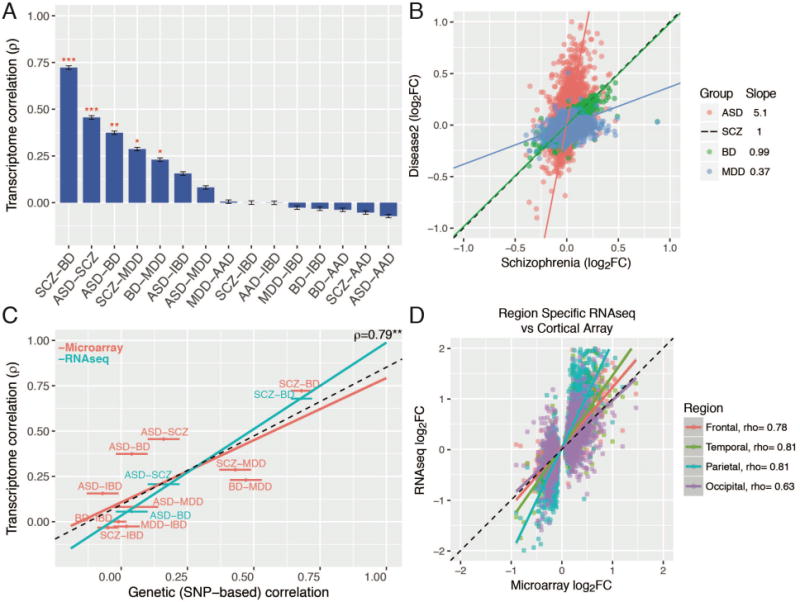
Cortical gene expression patterns overlap. (A) Rank order of microarray transcriptome similarity for all disease pairs, as measured by Spearman’s correlation of differential expression log2FC values. (B) Comparison of the slopes among significantly associated disease pairs indicates a gradient of transcriptomic severity, with ASD > SCZ ~ BD > MDD. (C) Overlapping gene expression patterns across diseases are correlated with shared common genetic variation, as measured by SNP co-heritability (22). The Y-axis shows transcriptome correlations using microarray-based (discovery, red) and RNAseq (replication, blue) datasets. (D) RNAseq across all cortical lobes in ASD replicates microarray results and demonstrates a consistent transcriptomic pattern. Spearman’s ρ is shown for comparison between microarray and region-specific RNAseq replication datasets (all P’s < 10−14). Plots show mean +/− SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Disease-specific DGE summary statistics (Data Table S1) provide human in vivo benchmarks for determining the relevance of model organisms, in vitro systems, or drug effects (13, 14). We identified a set of concordantly down- and upregulated genes across disorders (Fig. S4), as well as those with more specific effects. Complement component 4A (C4A), the top GWAS-implicated SCZ disease gene (15), was significantly upregulated in SCZ (log2FC=0.23, P=6.9×10−6) and in ASD (RNAseq; log2FC=0.91, P=0.014; Data Table S1) but not BD, MDD, or AAD. To investigate potential confounding by psychiatric medications, we compared disease signatures with those from non-human primates treated with acute or chronic dosing of antipsychotic medications. Significant negative overlap (Fig. S5; (12)) was observed, indicating that antipsychotics are unlikely to drive, but rather may partially normalize, these transcriptomic alterations, whereas the psychotomimetic PCP partially recapitulates disease signatures.
To validate that these transcriptomic relationships are generalizable, we generated independent RNAseq datasets for replication for 3 out of the 5 disorders (Fig. S6; (12)). We identify 1099 genes whose DGE is replicated in ASD (OR 6.4, P=3.3×10−236, Fisher’s exact test; Table S2), 890 genes for SCZ (BrainGVEX; OR 4.5, P=7.6×10−155), and 112 genes for BD (BrainGVEX; OR 3.9, P=4.6×10−26), which is likely due to the relatively smaller RNAseq sample size for BD (12). We observed similarly high levels of transcriptomic overlap among ASD, SCZ, and BD, and a similar gradient of transcriptomic severity (Figs. 2C; S7). The SCZ and BD patterns were further replicated in the CommonMind dataset, although gene-level overlap was lower (12, 16) (Fig. S7). The ASD signature was largely consistent across the four major cortical lobules, indicating that this pattern is not caused by regional differences (Fig. 2D).
To more specifically characterize the biological pathways involved, we performed robust weighted gene co-expression network analysis (rWGCNA; (12, 17)), identifying several shared and disorder-specific co-expression modules (Fig. 3). Modules were stable (Fig. S8), showed greater association with disease than other biological or technical covariates (Fig. S9), and were not dependent on corrections for covariates or batch effects (Fig. S10). Moreover, each module was enriched for protein-protein interactions (Fig. S8) and brain enhancer-RNA co-regulation (Fig. S11) derived from independent data, which provides anchors for dissecting protein complexes and regulatory relationships.
Fig. 3. Network analysis identifies modules of co-expressed genes across disease.
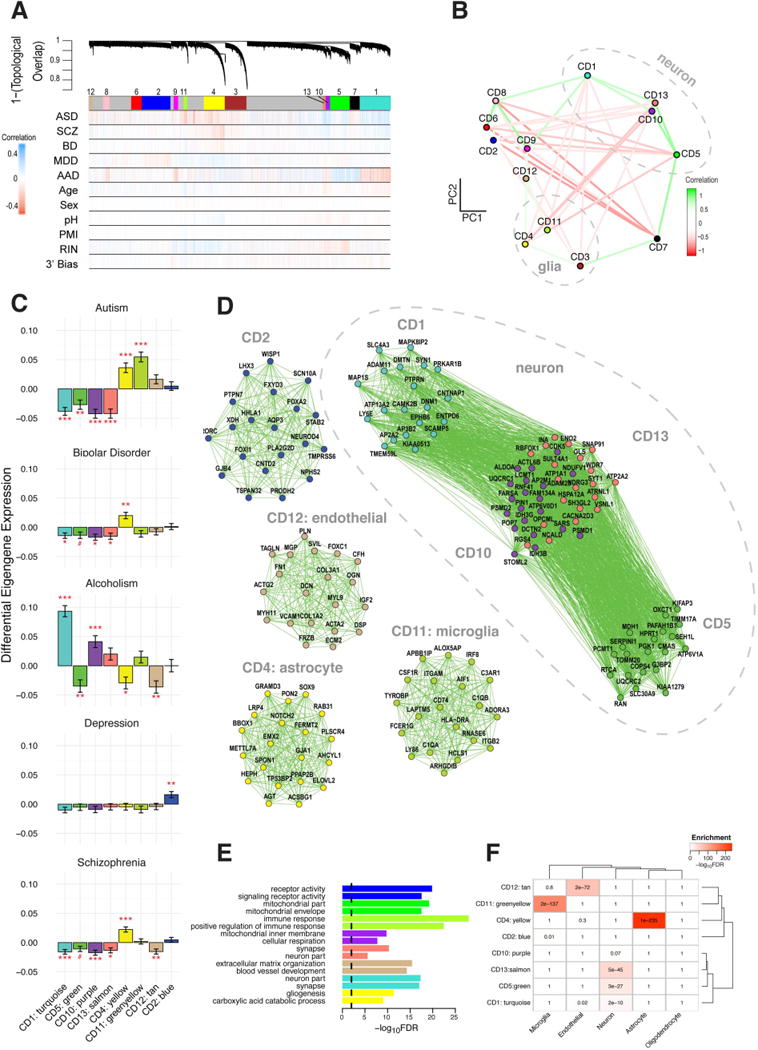
(A) Network dendrogram from co-expression topological overlap of genes across disorders. Color bars show correlation of gene expression with disease status, biological, and technical covariates. (B) Multidimensional scaling plot demonstrates relationship between modules and clustering by cell-type relationship. (C) Module-level differential expression is perturbed across disease states. Plots show beta values from linear mixed-effect model of module eigengene association with disease status (FDR-corrected #P<0.1, *P<0.05, **P<0.01, ***P<0.001). D) The top twenty hub genes are plotted for modules most disrupted in disease. See Data Table S2 for a complete list of genes’ module membership (kME). Edges are weighted by the strength of correlation between genes. Modules are characterized by (E) Gene Ontology enrichment (top two pathways shown for each module) and (F) cell-type specificity, on the basis of RNAseq of purified cell populations from healthy human brain samples (25).
An astrocyte-specific module (CD4, hubs GJA1, SOX9) was broadly upregulated in ASD, BD, and SCZ (FDR-corrected P’s < 0.05, Fig. 3C, Data Table S2; (12)) and enriched for glial cell differentiation and fatty-acid metabolism pathways. In contrast, a module strongly enriched for microglial markers (CD11) was upregulated specifically in ASD (two-sided t-test, FDR-corrected P=4×10−9). Hubs include canonical microglial markers (HLA-DRA, AIF1), major components of the complement system (C1QA, C1QB) and TYROBP, a microglial signalling adapter protein (18). Results fit with convergent evidence for microglial upregulation in ASD and an emerging understanding that microglia play a critical role regulating synaptic function during neurodevelopment (19).
One module was upregulated specifically in MDD (CD2, FDR-corrected P=0.009; Data Table S2) and was enriched for G-protein coupled receptors, cytokine-cytokine interactions, and hormone activity pathways, suggesting a link between inflammation and dysregulation of the hypothalamic-pituitary (HPA) axis, consistent with current models of MDD pathophysiology (20). Several modules annotated as neuronal/mitochondrial were downregulated across ASD, SCZ, and BD (CD1, CD10, CD13; Fig. 3C, Data Table S2; (12)). The overlap of CD10 with a mitochondrial gene-enriched module previously associated with neuronal firing rate (21) links energetic balance, synaptic transmission, and psychiatric disease (Data Table S2).
The transcriptome may reflect the cause or the consequence of a disorder. To refine potential causal links, we compared SNP-based genetic correlations between disease pairs (22) with their corresponding transcriptome overlap. SNP co-heritability was significantly correlated with transcriptome overlap across the same disease pairs (Fig. 2C, Spearman’s ρ =0.79, 95% confidence interval [0.43–0.93], P=0.0013), suggesting that a major component of these gene-expression patterns reflects biological processes coupled to underlying genetic variation.
To determine how disease-associated variants may influence specific biological processes, we investigated whether any modules harbor genetic susceptibility for specific disorders or for relevant cognitive or behavioral traits (12). We identified significant enrichment among several of the downregulated, neuronal co-expression modules (CD1, CD10, CD13) for GWAS signal from SCZ and BD, as well as for educational attainment and neuroticism (Fig. 4A; FDR-corrected P’s < 0.05, Spearman; (12)). We also observe enrichment for the three downregulated neuronal co-expression modules in the iPSYCH Consortium (23) ASD GWAS cohort (Fig 4A; Table S3; (12)). In contrast, these modules showed no enrichment for MDD, AAD, or IBD. Further, none of the microglial- or astrocyte-specific modules showed psychiatric GWAS enrichment. Extending this analysis to disease-associated rare variants (Data Table S3; (2, 12)), we found that the CD1 neuronal module was enriched for genes harbouring rare, non-synonymous de novo mutations identified in ASD (OR 1.36, FDR-corrected P=0.03, logistic regression) and SCZ cases (OR 1.82, FDR-corrected P=0.014) but not unaffected controls (Fig. 4B). A similar CD1-enrichment was observed for genes affected by rare, recurrent copy-number variation (CNV) in ASD (OR 2.52, FDR-corrected P=0.008) and SCZ (OR 2.46, FDR-corrected P=0.014). These results suggest convergence of common and rare genetic variation acting to downregulate synaptic function in ASD and SCZ.
Fig. 4. Downregulated neuronal modules are enriched for common and rare genetic risk factors.
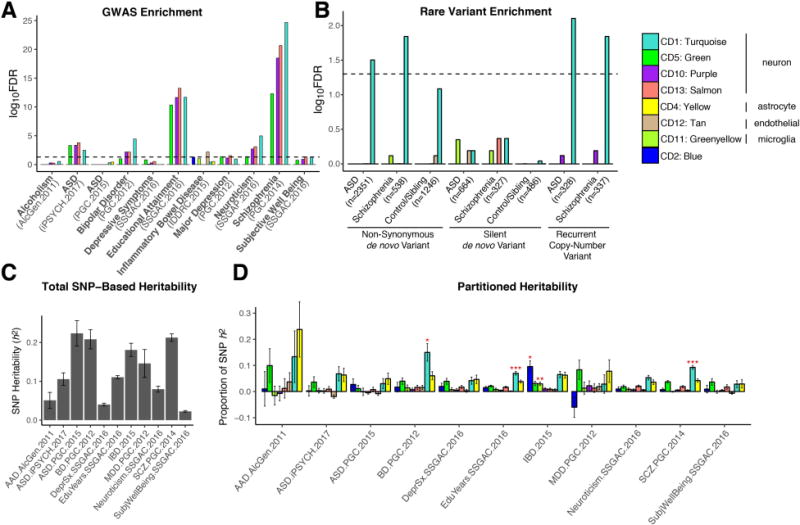
(A) Significant enrichment is observed for SCZ-, ASD-, and BD-associated common variants from GWAS among neuron/synapse & mitochondrial modules (12). GWAS datasets are listed in Table S3. (B) The CD1 neuronal module shows significant enrichment for ASD- and SCZ-associated non-synonymous de novo variants from whole exome sequencing. The number of genes affected by different classes of rare variants is shown in parentheses. Significance was calculated using logistic regression, correcting for gene length. P-values are FDR corrected. (C) Total SNP-based heritability (liability scale for psychiatric diagnoses) calculated from GWAS using LD-score regression. (D) Proportion of heritability for each disorder or trait that can be attributed to individual co-expression modules. Significance (FDR-corrected *P<0.05, **P<0.01, ***P<0.001) is from enrichment statistics comparing the proportion of SNP heritability within the module divided by the proportion of total SNPs represented. The CD1 module shows significant enrichment in SCZ, BD, and educational attainment.
We next used LD score regression (24) to partition GWAS heritability (Fig. 4C; Data Table S4) into the contribution from SNPs located within genes from each module ((12); Fig. 4D). CD1 again showed significant enrichment for SCZ (2.5 fold, FDR-corrected P=8.9×10−11), BD (3.9 fold, FDR-corrected P<0.014), and educational attainment (1.9 fold, FDR-corrected P<0.0008; χ2) GWAS, accounting for ~10% of SNP-based heritability within each dataset, despite containing only 3% of the SNPs. This illustrates how gene network analysis can begin to parse complex patterns of common variants, each of small effect size, to implicate specific biological roles for common variant risk across neuropsychiatric disorders.
These data provide a quantitative, genome-wide characterization of the cortical pathology across five major neuropsychiatric disorders, providing a framework for identifying the responsible molecular signalling pathways and interpreting genetic variants implicated in neuropsychiatric disease risk. We observe a gradient of synaptic gene down-regulation, with ASD > SZ ≈ BD. BD and SCZ appear most similar in terms of synaptic dysfunction and astroglial gene up-regulation, which may represent astrocytosis, activation, or both. ASD, an early-onset disorder, shows a distinct upregulated microglial signature, which may reflect the role for microglia in regulation of synaptic connectivity during neurodevelopment (19). MDD shows neither the synaptic nor astroglial pathology, but does exhibit dysregulation of HPA-axis and hormonal signalling not observed in the other disorders.
Our data suggest that shared genetic factors underlie a substantial proportion of cross disorder expression overlap. Given that a minority of these relationships represent eQTL (Fig. S12), most of the genetic effects are likely acting indirectly, through a cascade of developmental and cell-cell signalling events rooted in genetic risk. Genetic variation is also not the only driver of expression variation; there is undoubtedly a contribution from environmental effects. Hidden confounders could introduce a correlation structure that matches SNP-level genetic correlations, but parsimony and hidden covariate correction suggests that this is unlikely. Diagnostic misclassification could artificially elevate shared signals, but the results are robust to disorder removal (Fig. S13), and misclassification would not account for the substantial overlap we observe with ASD, which has a highly distinct phenotypic trajectory from later onset disorders. Finally, we have replicated broad transcriptomic and cell-type specific patterns independently for ASD, SCZ and BD, providing an organizing pathological framework for future investigation of the mechanisms underlying specific gene and isoform-level transcriptomic alterations in psychiatric disease.
Supplementary Material
One Sentence Summary.
Autism, schizophrenia, and bipolar disorder share specific global gene expression patterns, characterized by astrocyte activation and disrupted synaptic processes.
Acknowledgments
The work is funded by the US National Institute of Mental Health (P50-MH106438, DHG; R01-MH094714, DHG; U01-MH103339, DHG; R01-MH110927, DHG; R01-MH100027, DHG; R01-MH110920, CL; U01-MH103340, CL), the Simons Foundation for Autism Research (SFARI 206733, DHG; SFARI Bridge to Independence Award, MJG), and the Stephen R. Mallory schizophrenia research award at UCLA (MJG). Published microarray datasets analyzed in this study are available on GEO (accession numbers GSE28521, GSE28475, GSE35978, GSE53987, GSE17612, GSE12649, GSE21138, GSE54567, GSE54568, GSE54571, GSE54572, GSE29555, GSE11223), ArrayExpress (accession E-MTAB-184), or directly from the study authors (4). New RNAseq data (available on Synapse with accession numbers syn4590909 and syn4587609, with access governed by NIMH RGR) were generated as part of the PsychENCODE Consortium, supported by: U01MH103339, U01MH103365, U01MH103392, U01MH103340, U01MH103346, R01MH105472, R01MH094714, R01MH105898, R21MH102791, R21MH105881, R21MH103877, and P50MH106934 awarded to: Schahram Akbarian (Icahn School of Medicine at Mount Sinai), Gregory Crawford (Duke), Stella Dracheva (Icahn School of Medicine at Mount Sinai), Peggy Farnham (USC), Mark Gerstein (Yale), Daniel Geschwind (UCLA), Thomas M. Hyde (LIBD), Andrew Jaffe (LIBD), James A. Knowles (USC), Chunyu Liu (UIC), Dalila Pinto (Icahn School of Medicine at Mount Sinai), Nenad Sestan (Yale), Pamela Sklar (Icahn School of Medicine at Mount Sinai), Matthew State (UCSF), Patrick Sullivan (UNC), Flora Vaccarino (Yale), Sherman Weissman (Yale), Kevin White (UChicago) and Peter Zandi (JHU). RNAseq data from the CommonMind Consortium used in this study (Synapse accession syn2759792) was supported by funding from Takeda Pharmaceuticals Company Limited, F. Hoffman-La Roche Ltd and NIH grants R01MH085542, R01MH093725, P50MH066392, P50MH080405, R01MH097276, RO1-MH-075916, P50M096891, P50MH084053S1, R37MH057881 and R37MH057881S1, HHSN271201300031C, AG02219, AG05138 and MH06692. Brain tissue for the study was obtained from the following brain bank collections: the Mount Sinai NIH Brain and Tissue Repository, the University of Pennsylvania Alzheimer’s Disease Core Center, the University of Pittsburgh NeuroBioBank and Brain and Tissue Repositories, the Harvard Brain Bank as part of the Autism Tissue Project (ATP), the Stanley Medical Research Institute, and the NIMH Human Brain Collection Core. Summary statistics for the ASD GWAS performed on data from the iPSYCH consortium are available in Data Table S5. Data and analysis code are available at https://github.com/mgandal/Shared-molecular-neuropathology-across-major-psychiatric-disorders-parallels-polygenic-overlap. The authors thank Sepideh Parhami, Hyejung Won, Jason Stein, Damon Poliodakis, Jonathan Flint, Roel Ophoff and members of the Geschwind laboratory for critical reading of earlier versions of this manuscript. The authors also thank Bogdan Pasaniuc for his helpful comments and for assistance with module heritability analyses.
Footnotes
References and Notes
- 1.Geschwind DH, Flint J. Genetics and genomics of psychiatric disease. Science. 2015;349:1489–1494. doi: 10.1126/science.aaa8954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gandal MJ, Leppä V, Won H, Parikshak NN, Geschwind DH. The road to precision psychiatry: translating genetics into disease mechanisms. Nat Neurosci. 2016;19:1397–1407. doi: 10.1038/nn.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garbett K, et al. Immune transcriptome alterations in the temporal cortex of subjects with autism. Neurobiol Dis. 2008;30:303–311. doi: 10.1016/j.nbd.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow ML, et al. Age-Dependent Brain Gene Expression and Copy Number Anomalies in Autism Suggest Distinct Pathological Processes at Young Versus Mature Ages. PLoS Genet. 2012;8:e1002592. doi: 10.1371/journal.pgen.1002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C, et al. Two gene co-expression modules differentiate psychotics and controls. Mol Psychiatry. 2012;18:1308–1314. doi: 10.1038/mp.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maycox PR, et al. Analysis of gene expression in two large schizophrenia cohorts identifies multiple changes associated with nerve terminal function. Mol Psychiatry. 2009;14:1083–1094. doi: 10.1038/mp.2009.18. [DOI] [PubMed] [Google Scholar]
- 8.Iwamoto K, Bundo M, Kato T. Altered expression of mitochondria-related genes in postmortem brains of patients with bipolar disorder or schizophrenia, as revealed by large-scale DNA microarray analysis. Hum Mol Genet. 2005;14:241–253. doi: 10.1093/hmg/ddi022. [DOI] [PubMed] [Google Scholar]
- 9.Narayan S, et al. Molecular profiles of schizophrenia in the CNS at different stages of illness. Brain Res. 2008;1239:235–248. doi: 10.1016/j.brainres.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang LC, et al. A Conserved BDNF, Glutamate- and GABA-Enriched Gene Module Related to Human Depression Identified by Coexpression Meta-Analysis and DNA Variant Genome-Wide Association Studies. PLoS ONE. 2014;9:e90980. doi: 10.1371/journal.pone.0090980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene Coexpression Networks in Human Brain Identify Epigenetic Modifications in Alcohol Dependence. J Neurosci. 2012;32:1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.See supplemental materials.
- 13.Prilutsky D, et al. iPSC-derived neurons as a higher-throughput readout for autism: promises and pitfalls. Trends Mol Med. 2014;20:91–104. doi: 10.1016/j.molmed.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 15.Sekar A, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fromer M, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang B, et al. Integrated Systems Approach Identifies Genetic Nodes and Networks in Late-Onset Alzheimer’s Disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salter MW, Stevens B. Microglia emerge as central players in brain disease. Nat Med. 2017;23:1018–1027. doi: 10.1038/nm.4397. [DOI] [PubMed] [Google Scholar]
- 20.Gold PW. The organization of the stress system and its dysregulation in depressive illness. Mol Psychiatry. 2014;20:32–47. doi: 10.1038/mp.2014.163. [DOI] [PubMed] [Google Scholar]
- 21.Winden KD, et al. The organization of the transcriptional network in specific neuronal classes. Mol Syst Biol. 2009;5:291. doi: 10.1038/msb.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cross-Disorder Group of the Psychiatric Genomics Consortium. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genetics. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pedersen CB, et al. The iPSYCH2012 case-cohort sample: New directions for unravelling genetic and environmental architectures of severe mental disorders. bioRxiv. 2017:146670. doi: 10.1038/mp.2017.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finucane HK, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 27.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 28.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 29.Oldham MC, Langfelder P, Horvath S. Network methods for describing sample relationships in genomic datasets: application to Huntington’s disease. BMC Syst Biol. 2012;6:63. doi: 10.1186/1752-0509-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2006;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 31.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller JA, et al. Strategies for aggregating gene expression data: The collapseRows R function. BMC Bioinformatics. 2011;12:322. doi: 10.1186/1471-2105-12-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 34.Parikshak NN, et al. Integrative Functional Genomic Analyses Implicate Specific Molecular Pathways and Circuits in Autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mason MJ, Fan G, Plath K, Zhou Q, Horvath S. Signed weighted gene co-expression network analysis of transcriptional regulation in murine embryonic stem cells. BMC Genomics. 2009;10:327. doi: 10.1186/1471-2164-10-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zambon AC, et al. GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics. 2012;28:2209–2210. doi: 10.1093/bioinformatics/bts366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reimand J, Arak T, Vilo J. g:Profiler–a web server for functional interpretation of gene lists (2011 update) Nucleic Acids Res. 2011;39:W307–W315. doi: 10.1093/nar/gkr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dougherty JD, Schmidt EF, Nakajima M, Heintz N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res. 2010;38:4218–4230. doi: 10.1093/nar/gkq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu X, Wells AB, O’Brien DR, Nehorai A, Dougherty JD. Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J Neurosci. 2014;34:1420–1431. doi: 10.1523/JNEUROSCI.4488-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ripke S, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2012;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium. Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol Autism. 2017;8:21. doi: 10.1186/s13229-017-0137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schumann G, et al. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci USA. 2011;108:7119–7124. doi: 10.1073/pnas.1017288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okbay A, et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet. 2016;48:624–633. doi: 10.1038/ng.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okbay A, et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533:539–542. doi: 10.1038/nature17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLoS Comput Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munk-Jørgensen P, Mortensen PB. The Danish Psychiatric Central Register. Dan Med Bull. 1997;44:82–84. [PubMed] [Google Scholar]
- 50.Pedersen CB, et al. A comprehensive nationwide study of the incidence rate and lifetime risk for treated mental disorders. JAMA Psychiatry. 2014;71:573–581. doi: 10.1001/jamapsychiatry.2014.16. [DOI] [PubMed] [Google Scholar]
- 51.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manichaikul A, et al. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang CC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Ligt J, et al. Diagnostic Exome Sequencing in Persons with Severe Intellectual Disability. N Engl J Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 57.De Rubeis S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fromer M, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Girard SL, et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- 60.Gulsuner S, et al. Spatial and Temporal Mapping of De Novo Mutations in Schizophrenia to a Fetal Prefrontal Cortical Network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iossifov I, et al. De Novo Gene Disruptions in Children on the Autistic Spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rauch A, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 66.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu B, et al. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet. 2012;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mi G, Di Y, Emerson S, Cumbie JS, Chang JH. Length Bias Correction in Gene Ontology Enrichment Analysis Using Logistic Regression. PLoS ONE. 2012;7:e46128. doi: 10.1371/journal.pone.0046128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moreno-De-Luca D, et al. Using large clinical data sets to infer pathogenicity for rare copy number variants in autism cohorts. Mol Psychiatry. 2012;18:1090–1095. doi: 10.1038/mp.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanders SJ, et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Akbarian S, et al. The PsychENCODE project. Nat Neurosci. 2015;18:1707–1712. doi: 10.1038/nn.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hansen KD, Irizarry RA, WU Z. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics. 2012;13:204–216. doi: 10.1093/biostatistics/kxr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jaffe AE, et al. qSVA framework for RNA quality correction in differential expression analysis. Proc Natl Acad Sci USA. 2017;114:7130–7135. doi: 10.1073/pnas.1617384114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reinhart V, et al. Evaluation of TrkB and BDNF transcripts in prefrontal cortex, hippocampus, and striatum from subjects with schizophrenia, bipolar disorder, and major depressive disorder. Neurobiol Dis. 2015;77:220–227. doi: 10.1016/j.nbd.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 76.Cope L, Zhong X, Garrett E, Parmigiani G. MergeMaid: R tools for merging and cross-study validation of gene expression data. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1046. Article29–13. [DOI] [PubMed] [Google Scholar]
- 77.Parmigiani G, Garrett-Mayer ES, Anbazhagan R, Gabrielson E. A cross-study comparison of gene expression studies for the molecular classification of lung cancer. Clin Cancer Res. 2004;10:2922–2927. doi: 10.1158/1078-0432.ccr-03-0490. [DOI] [PubMed] [Google Scholar]
- 78.Martin MV, Mirnics K, Nisenbaum LK, Vawter MP. Olanzapine Reversed Brain Gene Expression Changes Induced by Phencyclidine Treatment in Non-Human Primates. Mol Neuropsychiatry. 2015;1:82–93. doi: 10.1159/000430786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yao P, et al. Coexpression networks identify brain region-specific enhancer RNAs in the human brain. Nat Neurosci. 2015;18:1168–1174. doi: 10.1038/nn.4063. [DOI] [PubMed] [Google Scholar]
- 80.The GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van A, Granlund B, et al. Whole genome gene expression meta-analysis of inflammatory bowel disease colon mucosa demonstrates lack of major differences between Crohn’s disease and ulcerative colitis. PLoS One. 2013;8:e56818. doi: 10.1371/journal.pone.0056818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Noble CL, et al. Regional variation in gene expression in the healthy colon is dysregulated in ulcerative colitis. Gut. 2008;57:1398–1405. doi: 10.1136/gut.2008.148395. [DOI] [PubMed] [Google Scholar]
- 83.Girirajan S, et al. Refinement and Discovery of New Hotspots of Copy-Number Variation Associated with Autism Spectrum Disorder. Am J Hum Genet. 2013;92:221–237. doi: 10.1016/j.ajhg.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Malhotra D, Sebat J. CNVs: Harbingers of a Rare Variant Revolution in Psychiatric Genetics. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sanders SJ, et al. Multiple Recurrent De Novo CNVs, Including Duplications of the 7q11.23 Williams Syndrome Region, Are Strongly Associated with Autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weiss LA, et al. Association between Microdeletion and Microduplication at 16p11.2 and Autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 87.Marshall CR, et al. Structural Variation of Chromosomes in Autism Spectrum Disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moreno-De-Luca D, et al. Deletion 17q12 Is a Recurrent Copy Number Variant that Confers High Risk of Autism and Schizophrenia. Am J Hum Genet. 2010;87:618–630. doi: 10.1016/j.ajhg.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Glessner JT, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Green EK, et al. Copy number variation in bipolar disorder. Mol Psychiatry. 2015;21:89–93. doi: 10.1038/mp.2014.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Glessner JT, et al. Duplication of the SLIT3 Locus on 5q35.1 Predisposes to Major Depressive Disorder. PLoS One. 2010;5:e15463. doi: 10.1371/journal.pone.0015463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stefansson H, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Corvin A, et al. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Levinson DF, et al. Copy Number Variants in Schizophrenia: Confirmation of Five Previous Findings and New Evidence for 3q29 Microdeletions and VIPR2 Duplications. Am J Psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vacic V, et al. Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature. 2011;471:499–503. doi: 10.1038/nature09884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rujescu D, et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18:988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McCarthy SE, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mulle JG, et al. Microdeletions of 3q29 Confer High Risk for Schizophrenia. Am J Hum Genet. 2010;87:229–236. doi: 10.1016/j.ajhg.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


