Abstract
Circulating microRNAs (ci-miRNAs) in blood have emerged as promising diagnostic, prognostic and predictive biomarkers in cancer. Many clinical studies currently incorporate studies that assess ci-miRNAs. Validation of the clinical significance of candidate biomarker miRNAs has proven to be difficult, potentially resulting from vast discrepancies in the detection methodology as well as biological variability. In the current study, the influence of several methodological factors on ci-miRNA detection was evaluated as well as short-term biological variability in patients with head and neck cancer. RNA was isolated from 124 serum and plasma samples originating from patients with head and neck cancer and healthy volunteers. The miRNA levels were measured using RT-qPCR and the influence of pre-analytical factors, different normalization strategies and temporal reproducibility was assessed. RNA carriers improved ci-miRNA detection in serum and plasma specimens. A prolonged pre-processing time correlated with an increased hemolytic index in serum samples only. Hemolysis differentially affected the detection of individual miRNAs. Optimal normalization was achieved using the averaged detection values of spike-in cel-miR-39-3p and endogenous miR-16-5p. Comparing biological replicates from patients with head and neck cancer, the intra-individual miRNA levels were relatively stable (average interval 7 days). Differences in the ci-miRNA detection methodology and limitations of currently used technologies can greatly affect the results and may explain inconsistent outcomes between studies. Prior to the implementation of ci-miRNAs as useful clinical biomarkers, further advances in the standardization of the detection methodology and reduction of technical variability are needed.
Introduction
MicroRNAs (miRNAs) are small non-coding RNAs of 17–25 nucleotides in length that regulate post-transcriptional expression of target genes. MiRNA-mediated epigenetic regulation is involved in essentially all physiological and pathological processes, including carcinogenesis.1, 2
The miRNA expression profiles of tumor tissue are distinct from those of adjacent normal tissue. As a consequence, cancer-specific miRNA signatures can serve as diagnostic biomarkers.3, 4 Furthermore, tumor miRNA expression profiles have demonstrated prognostic and predictive value in cancer.5, 6, 7
The disadvantage of a tissue-based biomarker is the need for (repeated) invasive biopsy procedures. The advancement of minimally invasive liquid biopsies for disease characterization and follow up has recently gained considerable momentum. Notably, a study including over 15 000 patients revealed that somatic DNA alterations in tumor tissues could be detected in matched blood samples. Several clinical outcome benefits were reported, such as the early capture of actionable resistance mutations at the time of progression.8 Similar to circulating cell-free DNA (cfDNA) or circulating tumor cells (CTCs), circulating miRNAs (ci-miRNAs) can be detected in blood samples.9, 10, 11 Ci-miRNAs are contained in microvesicles or are bound to circulating proteins.12 The limited size of these molecules prevents degradation by RNAses. These characteristics promote the stability of ci-miRNAs in blood and have fueled research efforts using liquid biopsies for ci-miRNA profiling.13 It has been hypothesized that ci-miRNA-based diagnostic tests can be developed to aid in clinical decision-making. These blood-based tests may replace or supplement tissue-based diagnostics, diminishing the need for invasive tissue procurement.
Several specific ci-miRNAs have been described as biomarkers for solid tumors, and studies have shown that the tumor burden correlates with the level of ci-miRNAs in blood.14, 15, 16 Interpretation of reported results is however challenging, as contradictory findings are common when comparing different studies.14, 17
Extensive variation of pre-analytical conditions and detection methods may account for the conflicting results concerning specific candidate biomarker ci-miRNAs.14, 17, 18 As an example, the pre-processing time may be an influence, as miRNAs can be released from blood cells into a sample during in vitro hemolysis, thus affecting the amount of ci-miRNA detected in serum or plasma.18, 19, 20 No standard endogenous control for the normalization of ci-miRNA levels in blood has been established. Several ci-miRNAs have been proposed and a synthetic non-primate miRNA is frequently spiked in during RNA isolation to correct for technical variations in RNA recovery.10, 21 Finally, the choice of sample type, that is, serum or plasma, is likely important. Although the results from previous studies indicate a high concordance between serum and plasma ci-miRNA levels, this idea is disputed by other studies.10, 12, 22, 23
Potential clinical applications of ci-miRNAs depend on the reproducibility and stability of the profile. Previous studies have demonstrated the limited short-term variation of ci-miRNAs in healthy individuals.18, 22 The intra-individual biological variation of ci-miRNAs in untreated patients with head and neck squamous cell carcinoma (HNSCC) has not yet been reported to our knowledge.
We performed a methodological study assessing ci-miRNAs in blood samples from patients with HNSCC. On the basis of the findings, we provide several methodological recommendations to reduce the technical variability of ci-miRNA detection in future studies.
Materials and methods
Blood samples from patients and healthy subjects
All samples were procured by using a local Institutional Review Board-approved biobank protocol. Informed consent was obtained from all participants. For the present study, a combined total of 124 serum and plasma RNA isolations were performed from 100 unique samples originating from 15 subjects (10 patients with HNSCC and 5 healthy subjects). From 10 patients with HNSCC, 2 serum and 2 plasma samples were collected at two different time points (minimum time interval of 24 h). Serum or plasma from 10 patients with HNSCC was pooled and used to assess the effect of RNA carriers on miRNA detection. From 5 healthy subjects, 2 serum and 2 plasma samples were collected at two different time points (interval 7 days). In addition, at another time point, 4 serum and 4 plasma tubes were drawn from 5 healthy subjects for a pre-processing interval time course study. For this experiment, the samples were processed at 1, 2, 4 and 24 h post collection (see also Supplementary Figure 1).
Sample collection
For serum, 6.0 ml of whole blood was collected in a BD vacutainer tube (Becton Dickinson, Breda, Netherlands) and incubated upright for 1 h at room temperature (RT). Serum samples processed after 2, 4 and 24 h were stored on ice 1 h post collection until centrifugation. The samples were centrifuged at 1500 g for 10 min at RT. For plasma, 6 ml of whole blood was collected in a BD vacutainer EDTA tube and stored upright at RT until a maximum of 4 h before centrifugation. Platelet-rich plasma was harvested by centrifugation at 120 g for 20 min at RT. Subsequently, the supernatant was centrifuged at 360 g for 20 min at RT. Finally, to remove any remaining platelets, the supernatant was centrifuged at 2700 g for 10 min at RT. To avoid contamination of fibroblasts, leukocytes and activation markers, after centrifugation of both serum and plasma samples, a layer of ~2–5 mm of the specimen was left inside the collection tube for disposal. All samples were aliquoted into 1 ml cryovials. (Thermo Scientific, Leusden, Netherlands) and immediately stored at −80 °C until further analysis.
Hemolytic index
The hemolytic index (HI) of serum and plasma samples was measured using the Roche Modular Cobas 8000 automated platform (Roche, Indianapolis, IN, USA). Reported HI values represent semi-quantitative estimates of the cell-free hemoglobin concentration, with higher values indicating a higher degree of hemolysis.24
RNA isolation
RNA fragments of less than 1000 nucleotides were isolated from 300 μl serum or plasma samples using the miRCURY RNA Isolation Kit—Biofluids (Exiqon, Vedbaek, Denmark). RNA isolation was performed according to the manufacturer’s instructions and included the use of recombinant DNAse. Twenty femtomolar of cel-miR-39-3p (Qiagen, Hilden, Germany) was spiked in during lysis of the sample. RNA was eluted in 30 μl of nuclease-free H2O and stored at −80 °C. To test the role of different RNA carriers, we isolated RNA under four different conditions: (1) no RNA carrier; (2) addition of 20 μg of glycogen (20 mg ml−1) (Roche); (3) addition of 1 μg of MS2 (0.8 μg μl−1) (Roche); and (4) joint addition of 20 μg of glycogen and 1 μg of MS2. Isolations were performed in triplicate.
RT-qPCR
cDNA synthesis was performed using the miRCURY LNA Universal RT microRNA PCR kit (Exiqon) according to the manufacturer’s instructions with the exception of withholding UniSp6. For cDNA synthesis, 2 μl of RNA was used. cDNA was diluted 1:40 for qPCR on a Bio-Rad CFX96 Real-Time PCR detection system (Bio-Rad Laboratories, Hercules, CA, USA) using the ExiLENT SYBR Green master mix and LNA PCR primers sets (Exiqon). Each sample was measured in duplicate. The Cq value was defined using Bio-Rad CFX Manager 3.1 software (the threshold was manually set at 300 relative fluorescent units (RFU) for each experiment). RNA isolated from the HT-29 colorectal cancer cell line (ATCC) was used as a positive control and nuclease-free H2O was used as a non-template negative control in each experiment. The maximum accepted variation between duplicates was set at a 0.6 standard deviation (s.d.). qPCR was repeated if the s.d. of the duplicate was >0.6. RNA isolation was repeated when the Cq value of spike-in cel-miR-39-3p was >30.
miRNAs
For this methodological study, empirical selection of ci-miRNAs was performed as those ci-miRNAs were previously reported to be abundantly present in the serum and plasma of patients with cancer, including HNSCC: miR-21-5p, miR-92a-3p and miR-142-3p.25, 26, 27, 28
Normalization
The miR-16-5p, cel-miR-39-3p and the average of miR-16-5p and cel-miR-39-3p detection values were tested as normalizers. miR-16-5p was selected, as it has been used as a normalizer in previous studies encompassing various tumor types.26, 27, 29 In addition, no differences in the circulating miR-16-5p levels were previously observed between patients with HNSCC and healthy volunteers.30
Two normalization algorithms, NormFinder and BestKeeper, were tested.31, 32 NormFinder identifies the optimal normalizer using a model-based variance estimation approach and can be applied to any experimental design. The lower the stability value (SV) calculated with this model, the higher the stability of the normalizer. BestKeeper calculates the s.d. and coefficient of variance (CV) of each candidate normalizer. The lower the s.d. and CV calculated, the higher the stability of the normalizer used.
As an input, 50 averaged duplicate qPCR reactions were used: 20 reactions from 10 patients with HNSCC obtained from at least two separate time points, 10 reactions from 5 healthy subjects obtained from at least two separate time points and 20 reactions from 5 healthy subjects obtained at the same time point (the latter 20 were used for the experiment, as presented in Figure 2 of the manuscript) (the breakdown shown in Supplementary Figure 1). The Cq values were log-transformed for analysis with NormFinder. The relative ci-miRNA levels were analyzed using the 2−ΔCq and 2−ΔΔCq method.33
Results
Addition of an RNA carrier to improve ci-miRNA detection
One of the challenges of extracting ci-miRNAs from blood samples is the low ci-miRNA concentration (femtomolar range) compared to tissue samples (nanomolar range).34 To improve the recovery of ci-miRNAs from blood samples, an RNA carrier can be added during the isolation procedure. Pooled serum and plasma samples from 10 patients with HNSCC were used to assess the effect of two distinct RNA carriers, MS2 RNA and glycogen, on ci-miRNA detection (see Table 1 for the patient and healthy subject characteristics). ci-miRNA detection in plasma and serum samples using a single carrier or both carriers was compared to that using no carrier (negative control).
Table 1. Patient and healthy subject characteristics.
| Tumor site | Gender | Age (years) | TNM | Collection interval (days) | |
|---|---|---|---|---|---|
| pt1 | Hypopharynx | Male | 66 | cT4aN1M0 | 6 |
| pt2 | Base of tongue | Male | 56 | cT2N1M0 | 30 |
| pt3 | Base of tongue | Male | 62 | cT1N2aM0 | 6 |
| pt4 | Soft palate | Male | 56 | cT4aN0M0 | 4 |
| pt5 | Posterior pharyngeal wall | Male | 58 | cT2N2cM0 | 5 |
| pt6 | Tonsil | Female | 66 | cT4bN1M0 | 6 |
| pt7 | Supraglottic larynx | Male | 62 | cT3N2cM0 | 1 |
| pt8 | Tonsil | Male | 68 | cT2N2bM0 | 1 |
| pt9 | Floor of mouth | Male | 67 | cT3N0M0 | 6 |
| pt10 | Base of tongue | Male | 69 | cT4aN2cM0 | 7 |
| H01 | N/A | Female | 30 | N/A | 7 |
| H02 | N/A | Male | 53 | N/A | 7 |
| H03 | N/A | Female | 35 | N/A | 7 |
| H04 | N/A | Female | 28 | N/A | 7 |
| H05 | N/A | Male | 51 | N/A | 7 |
Abbreviations: H, healthy subject; pt, patient; N/A, not applicable.
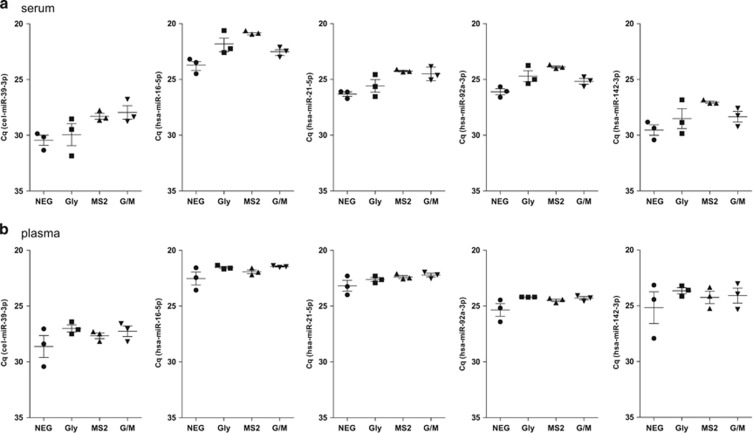
The Cq values are inversely proportional to the amount of target nucleic acid in the sample. As shown in Figure 1, addition of a single carrier or both carriers resulted in increased ci-miRNA recovery as reflected by a decrease in the average Cq value of all of the tested ci-miRNAs as well as spike-in cel-miR-39-3p.
Figure 1.
Effect of different RNA carriers on ci-miRNA detection. The results of triplicate RNA isolations from serum (a) and plasma (b) pools. Each value represents the raw Cq value of duplicate qPCR measurements from three independent RNA isolations. The standard error of the mean (s.e.m.) is indicated as error bars. NEG (negative), no RNA carrier; Gly (glycogen), addition of glycogen; MS2, addition of MS2 RNA; G/M, addition of glycogen and MS2 RNA.
In serum samples, considering all of the tested miRNAs, the average decrease in the Cq value compared to the negative control was 1.11 (±0.56 (standard error of mean; s.e.m.)), 2.36 (±0.34) and 1.53 (±0.63) for glycogen, MS2 and the combination of glycogen and MS2, respectively (Figure 1a).
In plasma samples, the effect of adding a carrier was less pronounced, with an average decrease in Cq value compared to the negative control of 1.17 (±0.43), 0.83 (±0.14) and 1.11 (±0.15) for glycogen, MS2 and the combination, respectively (Figure 1b).
The addition of MS2 resulted in a decrease of the s.e.m. of the average Cq value for all of the tested ci-miRNAs in serum and for 2 out of 5 miRNAs in plasma (cel-miR-39-3p and miR-21-5p). Consequently, MS2 was included in all subsequent RNA isolations from serum and plasma, as it was shown to improve ci-miRNA detection and/or the reproducibility of ci-miRNA detection, outperforming glycogen or the MS2/glycogen combination.
Normalization of the ci-miRNA levels
Endogenous small nucleolar RNAs, such as RNU44, RNU48 or other stably expressed miRNAs, are typically used for the normalization of target miRNA expression in tissue samples. For ci-miRNAs, there are no established normalizers, thereby hindering a comparison of the results within a single study as well as between different studies. A synthetic spike-in, such as cel-miR-39-3p, can be added during the RNA extraction procedure for technical normalization of RT-qPCR. Hereby, the amount of cel-miR-39-3p recovered after RNA extraction is directly correlated to the RNA extraction efficiency. To normalize for the total amount of RNA in a specific sample, an endogenous miRNA should be used. Considering biological variability, the endogenous control is likely to be different depending on the disease studied. Endogenous miR-16-5p has been suggested by several other studies to be a suitable candidate for normalization of the ci-miRNA levels.26, 27, 29 As miR-16-5p is enriched in red blood cells, it is important to prevent hemolysis during sample collection.19, 20
We established a pragmatic normalization method using the spike-in cel-miR-39-3p and/or miR-16-5p Cq values for normalization. We first assessed the variability of the measured cel-miR-39-3p and miR-16-5p Cq values, comparing the s.d. and CV using BestKeeper. The lowest variability was observed for the combined average of the miR-16-5p and cel-miR-39-3p Cq values. Similar results were obtained for both serum (s.d.=0.700; CV=2900) and plasma samples (s.d.=0.530; CV=2.180) (Supplementary Table 1). Application of the NormFinder algorithm demonstrated that the average of miR-16-5p and cel-miR-39-3p is the optimal normalizer for serum (SV=0.288) and plasma (SV=0.247) (Supplementary Table 1). In addition, the variability between measurements, as indicated by the s.e.m., was also lowest using this method for serum (s.e.m.=0.126) and plasma samples (s.e.m.=0.099) (Supplementary Table 1). On the basis of these results, the average Cq value of miR-16-5p and cel-miR-39-3p was subsequently used as the normalizer for the determination of ci-miRNA levels in subsequent experiments.
Influence of pre-processing time on ci-miRNA detection in serum
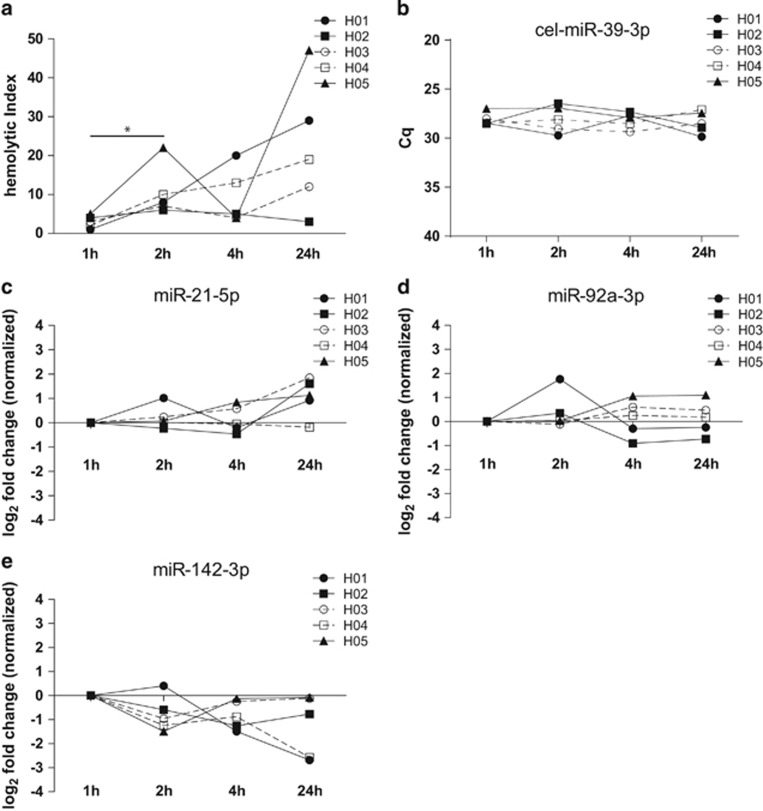
A prolonged interval between serum collection and processing (pre-processing time) is generally associated with a higher degree of hemolysis. As shown in the present study, all samples had an increase in the HI when the pre-processing time was prolonged from 1 to 2 h. A paired-grouped comparison revealed that this difference was significant (P=0.040) (Figure 2a). At later time points, serum samples from two healthy subjects (H01, H04) demonstrated an increase in the HI over time, whereas other samples demonstrated a stable HI (H02) or fluctuation of the HI (H03, H05) over time.
Figure 2.
Hemolysis and ci-miRNA levels according to the serum pre-processing time. Hemolytic index (HI) measured with Roche Modular clinical chemistry platform (a). Cq levels of cel-miR-39-3p (b). miR-21-5p, miR-92a-3p and miR-142-3p levels normalized to the average of miR-16-5p and cel-miR-39-3p and presented as fold change (FCs) compared to 1 h samples (c–e).
To assess the influence of the serum pre-processing time on the ci-miRNA levels, the ci-miRNA levels after 1, 2, 4 and 24 h of pre-processing time were measured. A <1.5-fold change (FC) between samples from the same subject was arbitrarily set as the threshold for no difference based on a previous study that used a <2-fold replicate ci-miRNA detection variability.23 A serum pre-processing time of 1 h was considered to be the standard protocol.
The Cq values of cel-miR-39-3p were comparable in all healthy volunteers at all time points, and all of the Cq values were below 30 (average Cq value 28.17±0.95 (s.d.)), indicating technically robust and reproducible RNA recovery (Figure 2b). A >1.5 increase in the miR-21-5p level was observed after 24 h in two subjects (H02, FC: 1.61 and H03 FC: 1.85). The FCs for all of the miR-21-5p measurements ranged from −0.23 to 1.85 (Figure 2c). No difference over time was observed for the miR-92a-3p levels, except for an increase at the 2- h time point for one out of five samples (H02, FC: 1.76) (FCs ranged from −0.91 to 1.76) (Figure 2d). A decrease in the miR-142-3p level was observed after 24 h in two samples (H01, FC: −2.68 and H04, FC: −2.57). The FCs ranged from −2.68 to 0.40 (Figure 2e).
miR-16-5p, miR-21-5p and miR-92a-3p are enriched in red blood cells, and consequently, their detected levels can be affected by hemolysis.20
Therefore, we correlated the measured Cq values with the hemolytic index values. The expression levels of miR-16-5p (P=0.02) and miR-21-5p (P=0.02) were significantly correlated with the HI values, whereas the miR-92a-3p values showed a non-significant trend (P=0.07). As expected, the cel-mir-39-3p and miR-142-3p values did not correlate with the HI (Supplementary Figure 2). These results show a hemolysis-associated increase of the miR-16-5p and miR-21-5p levels in serum with a prolonged coagulation time, suggesting the release of these miRNAs from in vitro rupture of blood cells after collection.
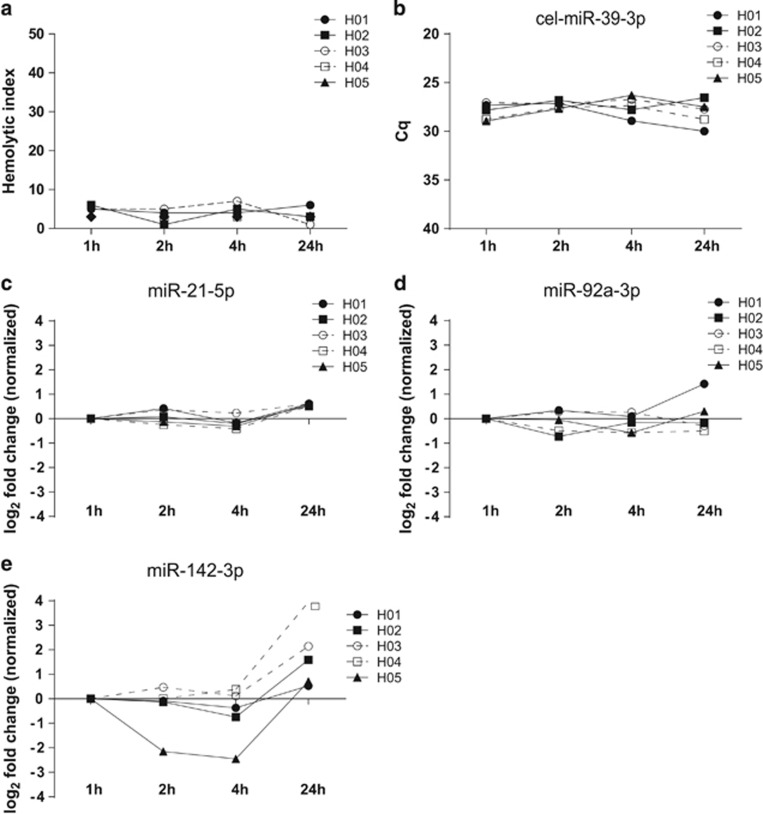
Influence of pre-processing time on ci-miRNA detection in plasma
The HI did not increase with prolonged plasma pre-processing time, indicating that no significant in vitro hemolysis occurred (Figure 3a). Platelets are an abundant source of miRNAs and could potentially influence ci-miRNA expression in plasma. Thus, we used a processing protocol that resulted in platelet poor plasma. As a quality control, we measured the number of platelets in five random plasma samples after processing.35, 36 No platelets were detected in any of these samples (Supplementary Figure 3).
Figure 3.
Hemolysis and ci-miRNA levels according to the plasma pre-processing time. Hemolytic index (HI) measured with Roche Modular clinical chemistry platform (a). Cq levels of cel-miR-39-3p (b). miR-21-5p, miR-92a-3p and miR-142-3p levels normalized to the average of miR-16-5p and cel-miR-39-3p and presented as FCs compared to 1 h samples (c–e).
The influence of a prolonged plasma pre-processing time on the ci-miRNA levels was assessed in plasma samples processed at 1, 2, 4 and 24 h after collection. The Cq values of cel-miR-39-3p were comparable and below the defined threshold of a Cq value of 30 for all subjects at all time points (mean Cq 27.69±0.95 (s.d.)) (Figure 3b). No >1.5-FCs of circulating miR-21-5p and miR-92a-3p levels were observed for any subjects at all four time points (Figure 3c and d). The level of miR-142-3p decreased in 1 out of 5 samples after 2 h (H05, FC: −2.51) and 1 out of 5 samples after 4 h (H05, FC: −2.45) and increased in 3 out of 5 samples after 24 h (H02, FC: 1.59, H03, FC: 2.14, H04, FC: 3.95) (FCs ranged from −2.45 to 3.95 Cq) (Figure 3e). These results suggest that the observed differences of the ci-miRNA concentrations due to a prolonged pre-processing time are miRNA-dependent.
Intra-individual short-term temporal variation of the ci-miRNA levels in patients with cancer
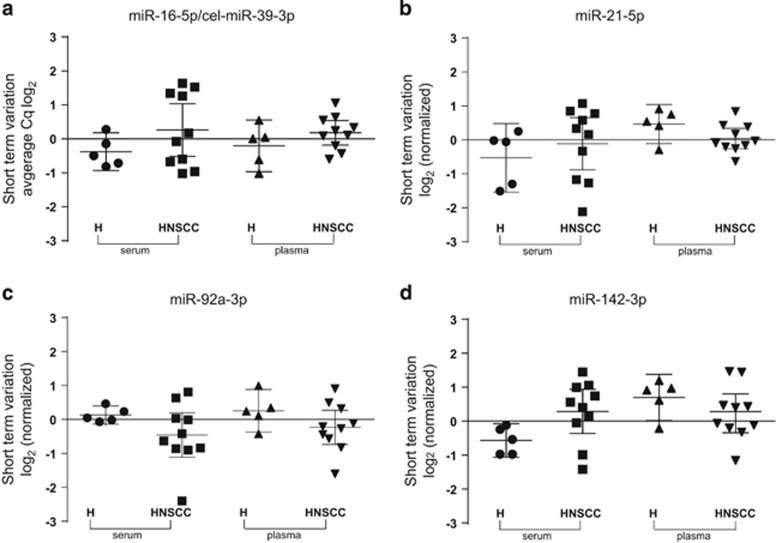
To examine the short-term temporal variation of ci-miRNAs, two blood samples were collected at separate time points from clinically stable patients with cancer and healthy subjects. Patients were not treated between blood collections. The average time between the two sample draws was 8 days (range: 1–30); for healthy subjects, the average time was 7 days (see Table 1).
The short-term variation was determined by subtracting the measured concentration at day 1 from the measured concentration at day 2. First, the variation in the average Cq values of miR-16-5p and cel-miR-39-3p was analyzed. The mean difference in the Cq value using serum from healthy volunteers and patients with cancer was −0.38 (95% CI: −0.93 to 0.18) and 0.26 (95% CI: −0.51 to 1.04) for miR-16-5p and cel-miR-39-3p, respectively. The mean difference in the Cq value using plasma from healthy subjects and patients with cancer was −0.20 (95% CI: −0.97 to 0.56) and 0.18 (95% CI: −0.18 to 0.54) for miR-16-5p and cel-miR-39-3p, respectively (Figure 4a). The mean short-term variation in the Cq value observed for miR-21-5p detection in serum was −0.53 (95% CI: −1.54 to 0.48) in healthy volunteers vs −0.11 (95% CI: −0.88 to 0.65) in patients with cancer. In plasma samples, the observed variation in the Cq value was 0.47 (95% CI: −0.11 to 1.04) in healthy volunteers vs 0.04 (95% CI: −0.26 to 0.34) in patients with cancer. In general, the miR-21-5p short-term variation was less pronounced in plasma samples compared to serum samples. For two subjects (H05, −1.51 Cq and patient (pt)7, −2.12 Cq), the short-term serum miR-21-5p variation was >1.50 Cq, while this result was not observed in plasma (Figure 4b). The mean serum miR-92a-3p short-term variation was slightly less for healthy subjects compared to patients with cancer: 0.13 Cq value (95% CI: −0.14 to 0.40) vs −0.46 Cq value (95% CI: −1.11 to 0.20). The mean plasma miR-92a-3p short-term variation was similar between healthy subjects and patients with cancer: 0.25 Cq (95% CI: −0.37 to 0.88) vs −0.23 Cq value (95% CI: −0.74 to 0.27). Among all paired serum and plasma samples, a short-term variation of more than 1.50 Cq value was observed in two subjects: patient 2 (plasma) (−1.60 Cq) and patient 7 (serum) (−2.40 Cq), (Figure 4c). Notably, hemolysis was visually observed in the second plasma sample of patient 2 and second serum sample of patient 7.
Figure 4.
Short-term variation of serum and plasma miRNAs in patients with head and neck squamous cell carcinoma (HNSCC) and healthy subjects. Average Cq values of miR-16-5p and cel-miR-39-3p (a). Short-term variation of selected miRNAs relative to the average variation of miR-16-5p and cel-miR-39-3p (normalizer) (b–d). H, healthy subjects (n=5), patients with HNSCC (n=10).
The miR-142-3p levels did not show short-term variations in all subjects. The mean short-term difference in the serum ci-miRNA Cq value was −0.57 (95% CI: −1.06 to −0.07) for healthy subjects and 0.29 (95% CI: −0.36 to 0.94) for patients with cancer. The plasma ci-miRNA levels from healthy subjects demonstrated a mean difference of 0.70 Cq value (95% CI: 0.02 to 1.38) and 0.23 Cq value (95% CI: −0.34 to 0.80) (Figure 4d). These findings indicate that the ci-miRNA levels are stable in healthy subjects and clinically stable patients with HNSCC who are not undergoing treatment.
Discussion
Along with circulating tumor cells, peptides and extracellular vesicles, tumor-derived cell-free nucleic acids in blood are emerging as promising biomarkers in cancer.14 Ci-miRNAs form a subset of circulating nucleic acids. miRNAs are particularly attractive as biomarkers as they are relatively stable and can be easily detected in serum or plasma from a single tube of blood, allowing repeated sampling for minimally invasive disease monitoring. Discrepancies in detection methodologies are numerous when reviewing published ci-miRNA studies.14, 17 For example, the centrifugation steps and serum or plasma pre-processing time are often not mentioned, although these pre-analytical factors can greatly influence the measured ci-miRNA levels.36, 37 As analytical consistency is crucial for independent validation of results as well as comparisons across different studies, the present study aimed to develop a pragmatic and reproducible method to measure ci-miRNAs in patients with cancer.
The column-based Exiqon miRCURY biofluids kit was selected for RNA extraction based on its superior performance in previous comparative studies of various commercial isolation kits.22, 38 RNA carriers, such as MS2 and glycogen, have been reported to improve miRNA recovery.38, 39 A direct comparison between these two RNA carriers has not been performed, and the combined use of these two carriers has not been reported. The present results indicate that the RNA carriers glycogen and MS2 both improved ci-miRNA detection in serum and plasma. Limited by the small sample size, this effect may be more pronounced in serum samples compared to plasma samples. A potential explanation for this sample-type-dependent effect might be the RNA concentration differences between serum and plasma. The RNA concentrations tend to be lower in serum compared to plasma, and the relative effect of MS2 can be greater at lower RNA concentrations.22, 23, 40 Differences in the RNA isolation conditions resulted in comparable detection differences of spike-in cel-miR-39-3p and endogenous miRNAs. Therefore, the addition of a spike-in non-primate small RNA specimen as a technical control during the RNA isolation procedure is recommended.
The most widely used normalization approaches measuring ci-miRNAs employ either an endogenous (biological) control, exogenous spike-in (non-biological control) or geometric mean of all measured miRNAs.21, 41, 42 Because of the limited number of measured target miRNAs, we could only compare the normalization approach using the endogenous and/or exogenous control. The data showed that taking the average of both endogenous miR-16-5p and exogenous cel-miR-39-3p was a better normalization approach for the present study compared to using only an endogenous or exogenous normalizer. Using miR-16-5p as an endogenous control is also supported by the results of other studies analyzing ci-miRNAs in patients with different tumor types.26, 27, 29 However, when selecting a miRNA for normalization, it is crucial to realize that there can be disease-specific differences in expression patterns because of the biological variability between different tumor types.20 For example Hu et al.43 observed variable concentrations of miR-16-5p in patients with breast cancer, while this variation has not been observed in patients with HNSCC.30
As certain miRNAs are enriched in blood cells, hemolysis has previously been reported to be an important confounding factor in ci-miRNA detection.19, 20, 23 We observed an increase in hemolysis, coinciding with a change in ci-miRNA levels, when serum samples were processed at later time points. Therefore, a strict standard pre-processing time of 1 h for serum samples is proposed. In contrast to serum, negligible hemolysis was observed at all time points in plasma samples. The plasma ci-miRNA concentrations were stable when samples were processed up to 4 h post collection. The increase of miR-142-3p after 24 h might be related to the release of this miRNA from platelets.36 On the basis of these results, we recommend a plasma pre-processing time of 4 h maximum.
To our knowledge, the short-term temporal variation and stability of ci-miRNAs levels in patients with head and neck cancer not undergoing treatment has not been previously investigated. Studies have previously demonstrated that ci-miRNAs can be detected at similar levels over time in healthy donors and that specific ci-miRNAs are more abundant in patients with cancer.14, 18, 22 In addition, an association between the ci-miRNA levels and tumor burden has been demonstrated for different tumor types, suggesting the secretion of miRNAs by tumor cells.44, 45 The precise mechanism of the release of tumor-derived miRNAs into circulation is unknown. Two mechanisms are proposed: active secretion (exosomes) and passive secretion (cell death).46 Potentially, a rapid increase of tumor-derived miRNAs in circulation could function as a marker for early response to treatments, such as chemotherapy or radiotherapy. To detect such treatment-induced dynamic changes in ci-miRNA levels, the variability of the ci-miRNA levels in clinically stable, untreated patients with cancer should be limited. To assess the pre-treatment variability of ci-miRNAs from patients with cancer, we collected two samples at two different time points from patients not undergoing treatment. As this patient group only consisted of patients with HNSCC, we selected miRNAs that were previously reported to be overexpressed in tissue from patients with HNSCC.47, 48 Using this method, we reduced the technical variability and demonstrated that the ci-miRNA levels are relatively stable and reproducibly measurable in patients with head and neck cancer when comparing replicates obtained on separate days prior to initiating treatment. Incidental differences are likely caused by sample hemolysis. Although the age of the healthy subject group was lower than that of the patient group, the intra-individual variation in both groups was comparable, suggesting that with all of the limitations of a small study, age-related factors do not affect the intra-individual variability of ci-miRNAs.
In conclusion, differences in detection methodologies can greatly affect the outcome of ci-miRNA measurements. Similar technical challenges are also observed for the analysis of other promising biofluid-derived biomarkers, such as circulating endothelial cells and circulating tumor cells.49, 50 Here, we list several methodological recommendations to reduce the technical variability that is typically observed in ci-miRNA profiling studies (Table 2). This study is the first to show a minimal intra-individual short-term variation of ci-miRNA levels in untreated, clinically stable patients with head and neck cancer. Temporal changes in ci-miRNA levels are therefore hypothesized to reflect treatment effects and/or disease behavior. Further studies using a standardized approach for miRNA profiling are warranted to advance its promising use as a prognostic and predictive biomarker in patients with head and neck cancer.
Table 2. Recommendations for ci-miRNA detection studies.
| Serum | Plasma | |
|---|---|---|
| Sample handling | ||
| Pre-processing time | 1 h upright at RT | Max. 4 h upright at RT |
| Centrifugation | 1500 g for 10 min | Platelet poor protocola |
| Quantify platelets | No | Yes |
| RNA isolation | ||
| Volume | Fixed (e.g., 300 μl) | |
| RNA carrier | Yes (MS2 or glycogen) | |
| Exogenous spike-in | 20 fmoles cel-miR-39-3p in current | |
| Use DNAse | Yes | |
| Normalization | ||
| Endogenous control | Stably expressed miRNAb | |
| Exogenous control | Spike-in, non-primate miRNA | |
See methods section for details.
Can be tumor specific; test selected normalization strategy or normalizer per study.
Acknowledgments
This work was financially supported by unrestricted donations from Aegon N.V. and the Blokker-Verwer foundation, The Netherlands.
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors declare no conflict of interest.
Supplementary Material
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 2006; 6: 259–269. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435: 834–838. [DOI] [PubMed] [Google Scholar]
- Neerincx M, Sie DL, van de Wiel MA, van Grieken NC, Burggraaf JD, Dekker H et al. MiR expression profiles of paired primary colorectal cancer and metastases by next-generation sequencing. Oncogenesis 2015; 4: e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006; 9: 189–198. [DOI] [PubMed] [Google Scholar]
- Zhang JX, Song W, Chen ZH, Wei JH, Liao YJ, Lei J et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol 2013; 14: 1295–1306. [DOI] [PubMed] [Google Scholar]
- Hwang JH, Voortman J, Giovannetti E, Steinberg SM, Leon LG, Kim YT et al. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS ONE 2010; 5: e10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zill OA, Mortimer S, Banks KC, Nagy RJ, Chudova D, Jackson C et al. Somatic genomic landscape of over 15,000 patients with advanced-stage cancer from clinical next-generation sequencing analysis of circulating tumor DNA. J Clin Oncol 2016; 34:15_suppl, LBA11501-LBA11501. [Google Scholar]
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008; 18: 997–1006. [DOI] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 2008; 105: 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol 2008; 141: 672–675. [DOI] [PubMed] [Google Scholar]
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA 2011; 108: 5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M, Schaefer A, Steiner I, Kempkensteffen C, Stephan C, Erbersdobler A et al. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin Chem 2010; 56: 998–1006. [DOI] [PubMed] [Google Scholar]
- He Y, Lin J, Kong D, Huang M, Xu C, Kim TK et al. Current State of Circulating MicroRNAs as Cancer Biomarkers. Clin Chem 2015; 61: 1138–1155. [DOI] [PubMed] [Google Scholar]
- Hsu CM, Lin PM, Wang YM, Chen ZJ, Lin SF, Yang MY. Circulating miRNA is a novel marker for head and neck squamous cell carcinoma. Tumour Biol 2012; 33: 1933–1942. [DOI] [PubMed] [Google Scholar]
- Komatsu S, Ichikawa D, Hirajima S, Kawaguchi T, Miyamae M, Okajima W et al. Plasma microRNA profiles: identification of miR-25 as a novel diagnostic and monitoring biomarker in oesophageal squamous cell carcinoma. Br J Cancer 2014; 111: 1614–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer KW. Circulating microRNA biomarker studies: pitfalls and potential solutions. Clin Chem 2015; 61: 56–63. [DOI] [PubMed] [Google Scholar]
- MacLellan SA, MacAulay C, Lam S, Garnis C. Pre-profiling factors influencing serum microRNA levels. BMC Clin Pathol 2014; 14: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner MB, Kao SC, Edelman JJ, Armstrong NJ, Vallely MP, van Zandwijk N et al. Haemolysis during sample preparation alters microRNA content of plasma. PLoS ONE 2011; 6: e24145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res 2012; 5: 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 2010; 50: 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan GW, Khoo AS, Tan LP. Evaluation of extraction kits and RT-qPCR systems adapted to high-throughput platform for circulating miRNAs. Sci Rep 2015; 5: 9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem 2011; 57: 833–840. [DOI] [PubMed] [Google Scholar]
- Darby D, Broomhead C. Interference with serum indices measurement, but not chemical analysis, on the Roche Modular by Patent Blue V. Ann Clin Biochem 2008; 45: 289–292. [DOI] [PubMed] [Google Scholar]
- Freedman JE, Gerstein M, Mick E, Rozowsky J, Levy D, Kitchen R et al. Diverse human extracellular RNAs are widely detected in human plasma. Nat Commun 2016; 7: 11106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zhang Q. The expression and clinical significance of circulating microRNA-21 in serum of five solid tumors. J Cancer Res Clin Oncol 2012; 138: 1659–1666. [DOI] [PubMed] [Google Scholar]
- Liu GH, Zhou ZG, Chen R, Wang MJ, Zhou B, Li Y et al. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumour Biol 2013; 34: 2175–2181. [DOI] [PubMed] [Google Scholar]
- Summerer I, Unger K, Braselmann H, Schuettrumpf L, Maihoefer C, Baumeister P et al. Circulating microRNAs as prognostic therapy biomarkers in head and neck cancer patients. Br J Cancer 2015; 113: 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Yu J, Kim H, Wolfgang CL, Canto MI, Hruban RH et al. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin Cancer Res 2013; 19: 3600–3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victoria MB, Dhahbi JM, Nunez Lopez YO, Lamperska K, Golusinski P, Luczewski L et al. Circulating small non-coding RNA signature in head and neck squamous cell carcinoma. Oncotarget 2015; 6: 19246–19263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 2004; 64: 5245–5250. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol Lett 2004; 26: 509–515. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Williams Z, Ben-Dov IZ, Elias R, Mihailovic A, Brown M, Rosenwaks Z et al. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc Natl Acad Sci USA 2013; 110: 4255–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalla S, Shaw C, Kong X, Kondkar AA, Edelstein LC, Ma L et al. Platelet microRNA-mRNA coexpression profiles correlate with platelet reactivity. Blood 2011; 117: 5189–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HH, Yi HS, Kim Y, Kroh EM, Chien JW, Eaton KD et al. Plasma processing conditions substantially influence circulating microRNA biomarker levels. PLoS ONE 2013; 8: e64795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koberle V, Kakoschky B, Ibrahim AA, Schmithals C, Peveling-Oberhag J, Zeuzem S et al. Vesicle-associated microRNAs are released from blood cells on incubation of blood samples. Transl Res 2016; 169: 40–46. [DOI] [PubMed] [Google Scholar]
- McAlexander MA, Phillips MJ, Witwer KW. Comparison of methods for miRNA extraction from plasma and quantitative recovery of RNA from cerebrospinal fluid. Front Genet 2013; 4: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen D, Fog JU, Biggs W, Salomon J, Dahslveen IK, Baker A et al. Improved microRNA quantification in total RNA from clinical samples. Methods 2010; 50: S6–S9. [DOI] [PubMed] [Google Scholar]
- El-Khoury V, Pierson S, Kaoma T, Bernardin F, Berchem G. Assessing cellular and circulating miRNA recovery: the impact of the RNA isolation method and the quantity of input material. Sci Rep 2016; 6: 19529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De PK, Pattyn F, Poppe B, Van RN, De PA et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002; 3:research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA 2008; 14: 844–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Dong J, Wang LE, Ma H, Liu J, Zhao Y et al. Serum microRNA profiling and breast cancer risk: the use of miR-484/191 as endogenous controls. Carcinogenesis 2012; 33: 828–834. [DOI] [PubMed] [Google Scholar]
- Okajima W, Komatsu S, Ichikawa D, Miyamae M, Kawaguchi T, Hirajima S et al. Circulating microRNA profiles in plasma: identification of miR-224 as a novel diagnostic biomarker in hepatocellular carcinoma independent of hepatic function. Oncotarget 2016; 7: 53820–53836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TF, Carlsen AL, Heegaard NH, Sorensen FB, Jakobsen A. Changes in circulating microRNA-126 during treatment with chemotherapy and bevacizumab predicts treatment response in patients with metastatic colorectal cancer. Br J Cancer 2015; 112: 624–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol 2014; 11: 145–156. [DOI] [PubMed] [Google Scholar]
- Chang SS, Jiang WW, Smith I, Poeta LM, Begum S, Glazer C et al. MicroRNA alterations in head and neck squamous cell carcinoma. Int J Cancer 2008; 123: 2791–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui AB, Lenarduzzi M, Krushel T, Waldron L, Pintilie M, Shi W et al. Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res 2010; 16: 1129–1139. [DOI] [PubMed] [Google Scholar]
- Alix-Panabieres C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer 2014; 14: 623–631. [DOI] [PubMed] [Google Scholar]
- Manzoni M, Comolli G, Torchio M, Mazzini G, Danova M. Circulating endothelial cells and their subpopulations: role as predictive biomarkers in antiangiogenic therapy for colorectal cancer. Clin Colorectal Cancer 2015; 14: 11–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.