Abstract
Candida albicans secretes aspartyl proteases (Saps) during infection. Although Saps are secretory proteins, little is known about the intracellular trafficking and secretion of these proteins. We previously cloned and analyzed the C. albicans pre-vacuolar protein sorting gene VPS4, and demonstrated that extracellular Sap2p is absent in the culture supernatants of the vps4Δ null mutant. We therefore investigated the role of the C. albicans pre-vacuolar secretion pathway in the trafficking of Sap4–6p and in vivo virulence. The C. albicans vps4Δ mutant failed to produce extracellular Sap4–6p. Next, when tested in a mouse model of disseminated candidiasis, the vps4Δ mutant was greatly attenuated in virulence. Histopathological analysis indicated that infection with the vps4Δ mutant did not cause renal microabscess formation, in contrast to the wild-type strain. Our results imply that VPS4 is required for extracellular secretion of Sap4–6p, and that C. albicans requires an intact pre-vacuolar secretory pathway for wild-type virulence in vivo.
Keywords: Candida albicans, Protein secretion, Secreted aspartyl proteases, Vacuole, Virulence, VPS4
Introduction
Candida albicans secretes several cell surface and extracellular proteins that are implicated in pathogenesis, such as secreted aspartyl proteases (Saps) [1–3] extracellular phospholipases [4] and secreted lipases [5]. Saps are encoded by a multi-gene family of 10 SAP genes, which are differentially secreted depending on strain and environmental conditions [6]. SAP2 is transcriptionally induced by protein nitrogen sources, such as hemoglobin or albumin. SAP1, SAP2, and SAP3 are expressed predominantly in yeast cells, whereas SAP4–6 are expressed in hyphal forms. Saps 1, 2, and 3p have optimal activity in a pH range of 2.5–5.0, in contrast to Saps 4–6p which have greater activity at a higher pH range [7]. It has been postulated that the Saps are initially synthesized intracellularly as prepropeptides, which are then liberated into the lumen of the endoplasmic reticulum, transferred to the Golgi apparatus, and then to secretory vesicles via the general secretory pathway [8–10]. Pulse-chase experiments of one undefined Sap (probably Sap2p) have shown that it originates as an intracellular, membrane-associated 45 kDa precursor protein which is processed to a 43 kDa protein, which is then secreted extracellularly [10]. Under many growth conditions, Sap2p is the dominant isoenzyme that is produced and secreted in vitro [6]. In vivo, C. albicans sap1Δ, sap2Δ, and sapΔ null mutants are modestly attenuated in virulence, and a triple sap4Δ, sap5Δ, and sap6Δ null mutant is greatly attenuated in virulence in a mouse model of disseminated candidiasis [3, 7].
In previous studies, we have demonstrated that Sap2p secretion in C. albicans depends on YPT1 and SEC4, which are components of the pre- and post-Golgi general secretory pathway, respectively [11, 12]. In addition, we have shown that trafficking of the multiple-transmembrane domain proteins Cdr1p and Ftr1p requires an intact general secretory pathway [13]. However, eukaryotic cells can transport cargo proteins to the cell surface via multiple exocytic pathways. For example, in Saccharomyces cerevisiae, it has been shown that sec1, sec4, and sec6 mutants accumulate two distinct populations of post-Golgi vesicles which contain different exocytic cargo proteins [14]. Light density post-Golgi vesicles contain the cell-wall enzyme endoglucanase and the plasma membrane-associated ATPase Pma1p. High density post-Golgi vesicles contain secreted periplasmic enzymes, such as invertase and acid phosphatase. Thus, exocytic cargo is sorted and transported by at least two different routes [14]. Furthermore, Harsay and Schekman [15] have shown that pre-vacuolar protein sorting genes play an important role in cargo transport in the pre-vacuolar branch of the exocytic pathway in S. cerevisiae. By isolating dense and light vesicle populations in S. cerevisiae vps1 sec6-4, vps4 sec6-4, and pep12 sec6-4 mutants, it was observed that mutants blocked in this pre-vacuolar pathway missorted marker proteins that are normally found in high-density post-Golgi vesicles into low-density vesicles. Gurunathan et al. [16] also demonstrated these findings in vps1 and pep12 mutants in a similar late secretory mutant background (snc1). These results indicate that some exocytic cargo, including the conditionally-regulated soluble secretory proteins invertase and acid phosphatase, is differentially sorted through a pre-vacuolar compartment (PVC) prior to exocytosis.
Saccharomyces cerevisiae VPS4 encodes a 48 kDa member of the AAA-type ATPase family. S. cerevisiae vps4 mutants accumulate vacuolar, endocytic, and late-Golgi marker proteins in an aberrant multilamellar compartment termed the class E compartment, which is thought to represent an abnormal PVC. Vps4p function is required for efficient transport out of the PVC whether this traffic is retrograde to the Golgi or antegrade to the vacuole. Studies of a C. albicans vps4Δ null mutant by Kullas et al. [17] have revealed minor defects in filamentation on M199 solid media but not in serum, and no overt growth defects in rich media. In assays intended for Sap secretion, we have previously demonstrated that the C. albicans vps4Δ null mutant degraded more extracellular BSA than did wild-type or reintegrant strains, which implied that this mutant secreted more extracellular protease activity [18]. However, using a series of protease inhibitors, the origin of this aberrant extracellular protease activity was identified as a serine protease, and genetic analyses using a C. albicans vps4Δ prc1Δ mutant identified this missorted vacuolar protease as carboxypeptidase Y. Unexpectedly, C. albicans Sap2p was not detected in the culture supernatants of the vps4Δ mutants compared to wild-type and reintegrant strains. We therefore hypothesized that C. albicans may also require VPS4 for trafficking of Sap4–6p.
The experiments described below were therefore designed to elucidate whether: (i) the pre-vacuolar protein sorting gene VPS4 is required for Sap4–6p secretion, and (ii) an intact pre-vacuolar secretory pathway is required for in vivo virulence.
Methods
Strains and Media
Candida albicans strains CAF-2 (URA3/ura3Δ:: imm434) and CAI4 (ura3Δ::imm434/ura3Δ::imm434) (both from W. Fonzi, Georgetown University) were grown at 30°C in YPD (1% yeast extract, 2% peptone, and 2% glucose) supplemented with uridine (80 μg/ml), or in minimal glucose (0.67% yeast nitrogen base without amino acids [YNB], 2% glucose). Uracil auxotrophs were selected on 5-fluoro-orotic acid (FOA) medium (minimal glucose, 0.1 mg of uridine ml−1, and 0.7 mg of FOA ml−1). Plasmids were expanded in Escherichia coli DH5α or TOP10F′ cells grown in LB medium + ampicillin (100 μg ml−1) at 37°C. Solid media were prepared by adding 2% agar (Table 1).
Table 1.
Candida albicans strains used in this study
| Strain | Parent | Relevant genotype | Source/Reference |
|---|---|---|---|
| SC5314 | Wild-type | [19] | |
| CAF-2 | SC5314 | URA3/ura3Δ::imm434 | [20] |
| CAI-4 | CAF-2 | ura3Δ::imm434/ura3Δ::imm434 | [20] |
| CAL1–4 | CAI-4 | ura3Δ/ura3Δ arg4Δ/arg4Δ his1Δ/his1Δ VPS4/vps4Δ::dpl200-URA3-dpl200 | This study |
| CAL1–4F | CAL1–1 | ura3Δ/ura3Δ arg4Δ/arg4Δ his1Δ/his1Δ VPS4/vps4Δ::dpl200 | This study |
| CAL2–4 | CAL1–2 | ura3Δ/ura3Δ arg4Δ/arg4Δ his1Δ/his1Δ vps4Δ::dpl200/vps4Δ::dpl200-URA3-dpl200 | This study |
| CAL2–4F | CAL1–3 | ura3Δ/ura3Δ arg4Δ/arg4Δ his1Δ/his1Δ vps4Δ::dpl200/vps4Δ::dpl200 | This study |
| CAL2–4R | CAL1–4 | ura3Δ/ura3Δ arg4Δ/arg4Δ his1Δ/his1Δ vps4Δ::dpl200/vps4Δ::dpl200 + VPS4::URA3 | This study |
| CAL1–4U | CAL1–4F | ura3Δ/URA3 arg4Δ/arg4Δ his1Δ/his1Δ VPS4/vps4Δ::dpl200 | This study |
| CAL2–4U | CAL2–4F | ura3Δ/URA3 arg4Δ/arg4Δ his1Δ/his1Δ vps4Δ::dpl200/vps4Δ::dpl200 | This study |
Targeted Disruption of C. albicans VPS4
The C. albicans vps4Δ null mutant in background strain CAI4 was generated by disrupting both chromosomal alleles of C. albicans VPS4 using a PCR-based gene disruption strategy which employs cis-recombination to recycle the URA3 marker [21]. PCR-generated amplicons were generated using the synthetic oligonucleotides shown in Table 2 and plasmid pDDB57 (from A. Mitchell, Columbia University) as the template. C. albicans CAI4 was transformed directly with the PCR reaction mixtures using the lithium acetate method [22]. Uridine prototrophs were selected and purified on synthetic media lacking uracil and uridine, genomic DNA was extracted by vortexing the transformants with glass beads in phenol–chloroform, and homologous integration of each gene targeting cassette was verified by allele-specific PCR, using one primer upstream and one primer downstream of the open reading frame and outside of the targeting region of the disruption cassette (Table 2). Next, C. albicans genomic DNA was digested with HhaI and transferred to nylon membranes. The membranes were hybridized in 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 65°C with a 417 bp digoxigenin-labeled PCR product generated with primers VPS4-5SOU and VPS4-3SOU using SC5314 genomic DNA as a template, after which the membranes were washed in 0.2 × SSC-0.1% sodium dodecyl sulfate (SDS) at 65°C and analyzed using the DIG DNA detection kit (Boehringer Mannheim) according to manufacturer’s instructions.
Table 2.
Primer sequences used in this study
| Primer | Primer sequence |
|---|---|
| VPS4-5DRB | TCAAACAACAATATGGAGGAAATT TTTGCCCTTAGAAAGTTTCCAAC TAAAAT TAATAACGCCACCTCC ACCACCACCACGTTTTCCCAGTC ACGACGTT |
| VPS4-3DRB | GAGAATTGATAGCGAAGACGTAT CATATAGACAAAGATGAAACAA TAGTACAATGGTTTGTTTCAAT TGTAATAGTACTTGTGTGGAATT GTGAGCGGATA |
| VPS4-5DETB | AGTCCAAACTTACCCCAAACTG |
| VPS4-3DETB | TGATCAAGAAGATTTGTTTGCTG |
| VPS4-5SOU | TCTCATTGGTGATCTCGATC |
| VPS4-3SOU | ACTGAAGAACTGGATAGTGGTG |
To disrupt the second VPS4 allele, selected C. albicans VPS4/vps4Δ::dpl200-URA3-dpl200 mutants (strain CAL1-4) were expanded in YPD with uridine to permit loss of URA3 by cis-recombination between the flanking dpl200 repeats, uracil auxotrophs were selected on FOA medium, and these strains’ genotypes were confirmed by PCR and by Southern hybridization as described. The resulting C. albicans VPS4/vps4Δ::dpl200 strains (strain CAL1-4F) were transformed again with the PCR-generated gene disruption cassette, uracil prototrophs were selected, and their genotypes were analyzed by PCR and by Southern hybridization to identify strains with a vps4Δ::dpl200-URA3-dpl200/>vps4Δ::dpl200 genotype (strain CAL2-4).
To confirm that the phenotypic results observed were a direct result of loss of VPS4 function, one copy of wild-type VPS4 was sub-cloned into pGEM-URA3 (from A. Mitchell, Columbia University), digested with NotI, and transformed into a vps4Δ:: dpl200/vps4Δ::dpl200 strain (CAL2-4F). Correct integration of the wild-type VPS4 gene was confirmed by allele-specific PCR in multiple independent transformants, to generate the reintegrant strain CAL2–4R. Finally, to restore URA3 prototrophy in its original locus, plasmid pLUBP bearing URA3 (from W. Fonzi, Georgetown University) was linearized and reintegrated into the heterozygous VPS4/vps4Δ mutant strain CAL1-4F, and the vps4Δ/vps4Δ null mutant CAL2-4F, to generate prototrophic, isogenic strains CAL1-4U, and CAL2-4U, respectively. Strains CAF-2, CAL1-4U, CAL2-4U, and CAL2-4R constituted the final set of prototrophic strains all bearing a single copy of URA3 used for phenotypic studies and in vivo virulence studies.
Enzyme Assays
Sap2p expression by C. albicans was assayed using BSA plate assays, as previously described [23]. Phospholipase activity was assessed on egg-yolk agar medium, and lipase activity was visualized on YNB agar containing 2.5% (v/v) Tween 80. In addition, Sap2p secretion was examined in 0.34% YNB without ammonium sulfate and with 0.2% bovine serum albumin (BSA), 0.2% yeast extract, and 2% glucose (glucose–BSA–YE). Sap2p expression was induced by growing the C. albicans transformants to stationary phase in minimal glucose, after which the cells were washed and resuspended at OD600 > 20, as previously described [18]. Sap4-6p expression was induced by shifting overnight cultures in YPD to RPMI-1640 at 37°C, at a starting concentration of 1.0 × 106 cells/ml. The cell suspensions were shaken at 30°C (for Sap2p) or 37°C (for Sap4-6p), and cell-free supernatants obtained after 2 to 48 h were tested for residual BSA by SDS-polyacrylamide gel electrophoresis with Coomassie blue staining and for immunoreactive Sap by Western blotting, using anti-Sap2p or anti-Sap4-6p polyclonal antibody (from M. Monod, University of Lausanne), after transferring to polyvinylidene fluoride (PVDF) membranes (Biorad), and detection using chemiluminescence (ECL Detection Kit, Amersham Biosciences) and autoradiography.
Murine Model of Disseminated Candidiasis
To assess virulence in a standard mouse tail vein model of invasive candidiasis, the C. albicans mutant strains in the CAI4 background (CAL1-4U, CAL2-4U, and CAL2-4R) and control strain CAF-2, all bearing a single copy of URA3, were grown to midlog phase in YPD at 30°C, and yeast-phase cells were harvested, washed, counted, and re-suspended in sterile 0.9% (w/v) NaCl. Next, groups of 5 BALB/c female mice (from Charles River Laboratories) were injected intravenously with 0.2 ml of each cell suspension at 1.0 × 106 cells per animal, and survival over 30 days was assessed. At time of death or sacrifice, the kidney organs were removed aseptically and weighed, and colony burden was determined by plating serial dilutions on Sabouraud’s dextrose agar. Finally, histologic sections of selected organs were stained (H & E and GMS) and examined to assess the extent and severity of infection.
Results
Effects of the vps4Δ Null Mutation on Extracellular Proteases, Phospholipases, and Sap2p Secretion
We first compared the abilities of wild-type C. albicans, vps4Δ null mutant, and reintegrant strains to secrete catalytically-active proteases out of the cell. When these strains were spotted on BSA agar [23], the vps4Δ null mutant produced a larger zone of proteolysis than did CAF-2 or the reintegrant strain (Fig. 1a). Similarly, when these strains were incubated in liquid YNB-glucose with BSA, SDS-PAGE analyses of culture supernatants showed that the vps4Δ mutant degraded more extracellular BSA than did the wild-type or isogenic reintegrant strains (Fig. 1b). These results were identical to our previous observations in a similar genetic background [18]. Prior genetic and biochemical studies have indicated that this increased extracellular proteolysis is caused by extracellular missorting of the vacuolar enzyme carboxypeptidase (CPY) [18]. Next, when extracellular supernatants of the strains of interest were analyzed by Western blotting with polyclonal antibodies to Sap2p (from M. Monod, University of Lausanne), we found that both the wild-type and reintegrant strains secreted abundant amounts of immunoreactive Sap2p out of the cell, whereas the vps4Δ null mutant did not (Fig. 1c), confirming our results seen previously in the BWP17 genetic background [18].
Fig. 1.
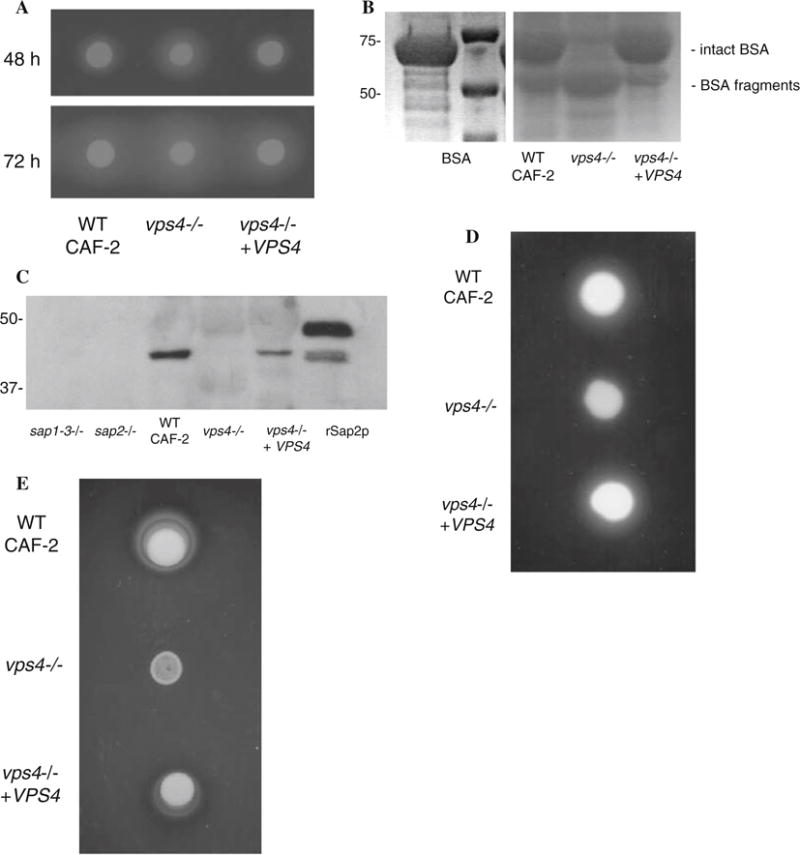
C. albicans vps4Δ mutants secrete increased extracellular protease and phospholipase activity. (a) Sap secretion by C. albicans was assayed using a plate assay for BSA degradation. Overnight cultures were spotted on BSA plates. Plates were visualized at 24 h (not shown), 48 h, and 72 h intervals. The zone of proteolysis indicates the relative amount of extracellular protease secretion. (b) Cell-free supernatants were obtained from stationary phase cells incubated in liquid BSA media, and 20 ll of each sample were analyzed by reducing SDS-polyacrylamide gel electrophoresis and Coo-massie blue staining. C. albicans vps4Δ null mutants incubated in BSA at 30°C for 14 h degraded much more extracellular BSA than wild-type strains. The smaller sized bands in this protein gel represent partially degraded BSA. This experiment indicates that the vps4Δ mutant degrades BSA to a greater extent than the control or reintegrant strains at the same time point. (c) Native Sap2p is not detected in culture supernatants of Ca vps4Δ mutants. Culture supernatants from the indicated time points were analyzed by reducing SDS-PAGE and Western blotting using polyclonal antibodies to Sap2p. rSap2p indicates purified recombinant Sap2p (from M. Monod) used as a positive control. A sap1–3Δ and a sap2Δ null mutant are used as negative controls in this experiment. (d) The vps4Δ mutant secretes reduced extracellular phospholipase activity. Overnight cultures were spotted onto egg-yolk agar plates and incubated at 37°C. The relative amount of extracellular phospholipase activity is indicated by the halo surrounding the fungal colony. (e) The vps4Δ mutant grows poorly on Tween 80 agar. Overnight cultures were spotted onto YNB–Tween 80 plates incubated at 37°C in order to assay for lipolytic activity. Unexpectedly, the vps4Δ mutant grew poorly, in comparison to the control and reintegrant strains
We next examined phospholipase secretion on egg yolk agar plates (Fig. 1d). The vps4Δ null mutant secreted a reduced zone of precipitation, compared to the wild-type control or reintegrant strains, suggesting a partial defect in phospholipase secretion. We next assayed for lipase secretion on Tween 80 plates. Although there was a reduced zone of lipolysis compared the wild-type and reintegrant strains, interestingly, the vps4Δ mutant grew poorly on Tween 80 plates (Fig. 1e). Thus, although we were unable to assay for lipase activity using this approach, the vps4Δ mutant showed a clear growth defect on Tween 80 agar.
Effects of the vps4Δ Mutation on Sap4-6p Secretion
We next tested secretion of Sap4-6p in vitro at 37°C in RPMI, which are in vitro conditions considered generally similar to bloodstream infection. When extracellular supernatants of the strains of interest were analyzed by Western blotting with polyclonal antibodies to Sap4-6p (from M. Monod, University of Lausanne), we found that the wild-type strain secreted abundant amounts of immunoreactive Sap4-6p out of the cell, whereas the vps4Δ null mutant did not (Fig. 2). As expected, no extracellular Sap2p was detected in these strains under the same conditions, by Western blotting using polyclonal anti-Sap2p antibodies (data not shown).
Fig. 2.
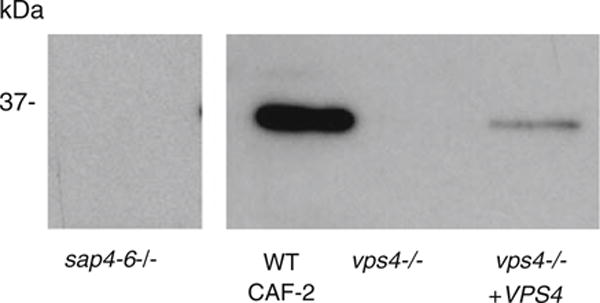
C. albicans vps4 mutants lack extracellular Sap4-6p. C. albicans cells were grown in RPMI at 37°C in vitro, conditions which are known to induce expression of Sap4p, Sap5p, and Sap6p. Western blotting of culture supernatants from control strain CAF-2, the vps4Δ mutant, and VPS4 reintegrant strain indicates that the vps4Δ mutant fails to produce extracellular Sap4-6p. Anti-Sap2p antibodies were used as a negative control in these growth conditions, and no extracellular Sap2p was detected in any strain (data not shown)
Effects of the vps4Δ Mutation on Virulence in a Mouse Model of Disseminated Candidiasis
These in vitro studies suggested that extracellular secretion of Sap4-6p is defective and extracellular phospholipase is reduced in the vps4Δ mutant. Thus, we next investigated the effects of the vps4Δ null mutation on virulence in a standard mouse model of hematogenously disseminated candidiasis (Fig. 3a). All mice inoculated with wild-type C. albicans died as expected. In contrast, all mice injected with the vps4Δ null mutant survived, which was statistically significant (P < 0.001). Heterozygous and reintegrant mutant strains bearing a single copy of VPS4 displayed an intermediate survival phenotype. In the kidney, colony burden was greatest for the wild-type strain, but markedly reduced in the vps4Δ mutant (P < 0.001; Fig. 3b). Both the heterozygous mutant and reintegrant strains produced an intermediate renal colony burden. These studies indicate that C. albicans VPS4 is required for wild-type virulence in a standard murine model of disseminated candidiasis.
Fig. 3.
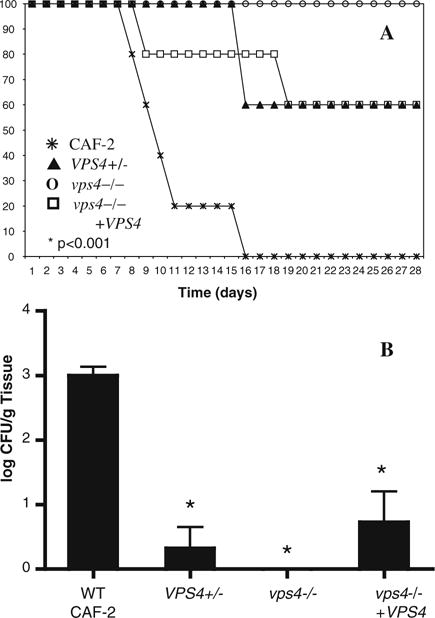
C. albicans vps4 mutants are hypovirulent in a mouse model of disseminated candidiasis. (a) C. albicans cells were injected via the tail vein into groups of 5 mice in a standard mouse model of hematogenously disseminated candidiasis. Survival is indicated over time. All mice injected with wild-type C. albicans CAF-2 died as expected. In contrast, all mice injected with the null mutant survived (P < 0.001 by Chi square analysis). Heterozygous and reintegrant mutant strains bearing a single copy of VPS4 displayed an intermediate phenotype. (b) Colony burden in the kidney is shown. There is a statistically significant reduction in colony burden in the kidney of the vps4Δ null mutant compared to the wild-type CAF-2 strain (* P < 0.001 by one-way ANOVA). The renal colony burdens of the heterozygous and reintegrant strains were reduced to a lesser extent compared to wild-type CAF-2 (* P < 0.001 by one-way ANOVA)
Histopathology of selected kidneys demonstrated that the wild-type strain formed multiple renal microabscesses predominantly in the papilla and calices, with minimal involvement of the cortical areas (Fig. 4a, b). In contrast, the vps4Δ mutant failed to form any renal microabscesses, in any part of the kidneys (Fig. 4c, d).
Fig. 4.
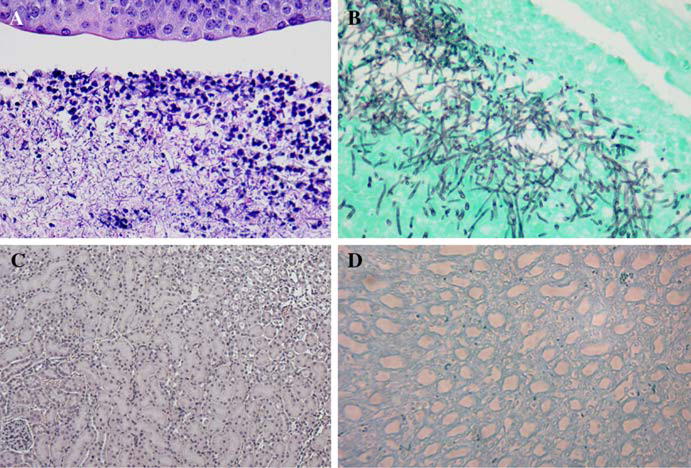
C. albicans vps4 mutants are less invasive than wild-type controls. Histopathologic examination was performed using hematoxylin and eosin (H & E) and Gomori-methanamine silver (GMS) staining on selected kidneys from the murine disseminated candidiasis experiments. In the wild-type strain, (a, b) multiple fungal microabscesses have formed in the renal papilla and calices, with minimal involvement of the cortical area. In contrast, there are no fungal microabscesses seen in the vps4Δ null mutant in any part of the kidneys. Representative sections are shown (c, d). Panels A and C are stained using H&E, panels B and D are strained with GMS
Discussion
The major goals of this study were to determine if the pre-vacuolar branch of the secretory pathway is required for Sap secretion, and if an intact pre-vacuolar secretory pathway is required for virulence in vivo. In S. cerevisiae, invertase and acid phosphatase are missorted from high-to low-density vesicles in vps1, vps4, and pep12 mutants via a pre-vacuolar branch of exocytosis [14–16]. While these missorted proteins still reach the cell surface in S. cerevisiae, we were unable to detect Sap2p or Sap4-6p in the culture supernatant of the C. albicans vps4Δ mutant. It has been previously reported that C. albicans class C vps11Δ mutants do not appear to secrete Saps or other proteases extracellularly when assayed by a BSA plate assay, as no zone of extracellular proteolysis is detected [24], perhaps because VPS11 may regulate late Golgi trafficking in addition to its well-described “class C” function at the stage of vesicle–vacuolar fusion. Additionally, C. albicans vps34Δ mutants have reduced Sap secretion, when measured by an ELISA assay [25]. Studies of a C. albicans vac1Δ mutant have also shown reduced Sap secretion [26]. Recently, we investigated the role of the C. albicans homolog of S. cerevisiae VPS1 in secretion of Saps and lipases [27]. S. cerevisiae VPS1 encodes a dynamin-like GTPase that mediates budding of vesicles from the late Golgi that are diverted from the general secretory pathway to the vacuole, or alternatively to a pre-vacuolar exocytic pathway [15, 28]. Interestingly, using a regulable tetracycline promoter, lack of VPS1 expression in C. albicans resulted in markedly reduced Sap2p and lipase secretion, and impaired filamentation and biofilm formation in vitro, suggesting a role of the pre-vacuolar pathway in several important virulence-related phenotypes [28].
Our results in this study imply that defects in the pre-vacuolar branch of the post-Golgi pathway can cause both (i) mistargeting of vacuolar proteins out of the cell, as would be predicted for vacuolar CPY [18] and vacuolar PrA (Lee et al. unpublished data), and (ii) inability to secrete other proteins out of the cell. Mutational analysis has shown that a 12-aa region in the propeptide of C. albicans Sap1p is necessary and sufficient for secretion of C. albicans Sap1p out of the cell [8]. Based on our previous studies, we have shown that ER-Golgi secretion (regulated by the rab-like GTPase Ypt1p), and post-Golgi secretion (regulated by the GTPase Sec4p) are required for Sap secretion [11, 12]. These studies suggest a model in which Saps are diverted to a pre-vacuolar branch of secretion prior to exocytosis. Further studies with additional pre-vacuolar mutants are in progress to determine where in the pre-vacuolar secretory pathway Saps are trafficked and sorted. However, our efforts to identify intracellular forms of native or epitope-tagged Saps have not yet been consistently successful.
Our results also suggest that an intact pre-vacuolar secretory pathway is required for wild-type virulence in vivo. We show direct evidence that Sap4-6p is absent extracellularly, which can contribute to a loss of virulence [3, 7]. Moreover, the vps4Δ mutant appears to secrete reduced amounts of extracellular phospholipase. However, the C. albicans vps4Δ mutant is greatly reduced in virulence, and we are now investigating the proteomics of secreted proteins in the vps4Δ mutant, to determine if secretion of additional virulence-associated proteins requires an intact pre-vacuolar pathway. In preliminary studies, for example, we have demonstrated that extracellular mp65 is markedly reduced in the vps4Δ mutant strain (Thomas et al. unpublished data). Further investigation of secreted proteins in the vps4Δ mutant using proteomic approaches will allow us to identify specific changes in pre-vacuolar secretory proteins and understand the overall importance of pre-vacuolar secretion in virulence.
Acknowledgments
We thank William Fonzi (Georgetown University) for providing strains CAF-2 and CAI4, and plasmid pLUBP, Aaron Mitchell (Columbia University) for providing plasmids pDDB57 and pGEM-URA3; and Michel Monod for providing anti-Sap antibodies and recombinant Sap2p. We also thank Brian Wong (Oregon University of the Health Sciences) for helpful advice. Sequence data for Candida albicans was obtained from the Stanford DNA Sequencing and Technology Center website at http://www-sequence.stanford.edu/group/candida. Sequencing of Candida albicans was accomplished with the support of the NIDR and the Burroughs Wellcome Fund. This work was supported by grants from the Department of Veterans’ Affairs (MERIT award to Samuel A. Lee) and the Biomedical Research Institute of New Mexico (to Samuel A. Lee).
Contributor Information
Samuel A. Lee, Division of Infectious Diseases, University of New Mexico Health Science Center, Albuquerque, NM, USA Division of Infectious Diseases, New Mexico Veterans Healthcare System, 1501 San Pedro SE, Mail Code: 111-J Albuquerque, NM 87108, USA.
Jason Jones, Division of Infectious Diseases, University of Texas Health Science Center at San Antonio, San Antonio, TX, USA.
Sarah Hardison, Division of Infectious Diseases, University of New Mexico Health Science Center, Albuquerque, NM, USA; Division of Infectious Diseases, New Mexico Veterans Healthcare System, 1501 San Pedro SE, Mail Code: 111-J Albuquerque, NM 87108, USA.
John Kot, Infectious Diseases Section, Yale University School of Medicine, New Haven, CT, USA.
Zachary Khalique, Infectious Diseases Section, Yale University School of Medicine, New Haven, CT, USA.
Stella M. Bernardo, Division of Infectious Diseases, University of New Mexico Health Science Center, Albuquerque, NM, USA Division of Infectious Diseases, New Mexico Veterans Healthcare System, 1501 San Pedro SE, Mail Code: 111-J Albuquerque, NM 87108, USA.
Anna Lazzell, South Texas Center for Emerging Infectious Diseases, University of Texas at San Antonio, San Antonio, TX, USA.
Carlos Monteagudo, Department of Pathology, University of Valencia, Valencia, Spain.
Jose Lopez-Ribot, South Texas Center for Emerging Infectious Diseases, University of Texas at San Antonio, San Antonio, TX, USA.
References
- 1.Hoegl L, Ollert M, Korting HC. The role of Candida albicans secreted aspartic proteinase in the development of candidoses. Mol Med. 1996;74:135–42. doi: 10.1007/BF01575445. [DOI] [PubMed] [Google Scholar]
- 2.Hube B, Sanglard D, Odds FC, Hes D, Monod M, Schafer W, et al. Disruption of each of the secreted aspartyl proteinase genes SAP1, SAP2, and SAP3 of Candida albicans attenuates virulence. Infect Immun. 1997;65:3529–38. doi: 10.1128/iai.65.9.3529-3538.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanglard D, Hube B, Monod M, Odds F, Gow N. A triple deletion of the secreted aspartyl proteinase genes SAP4, SAP5, and SAP6 of Candida albicans causes attenuated virulence. Infect Immun. 1997;65:3539–46. doi: 10.1128/iai.65.9.3539-3546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghannoum MA. Potential role of phospholipases in virulence, and fungal pathogenesis. Clin Microbiol Rev. 2000;13:122–43. doi: 10.1128/cmr.13.1.122-143.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hube B, Stehr F, Bossenz M, Mazur A, Kretschmar M, Schafer W. Secreted lipases of Candida albicans: cloning, characterization and expression analysis of a new gene family with at least ten members. Arch Microbiol. 2000;174:362–74. doi: 10.1007/s002030000218. [DOI] [PubMed] [Google Scholar]
- 6.White TC, Agabian N. Candida albicans secreted aspartyl proteinases: isoenzyme pattern is determined by cell type, and levels are determined by environmental factors. J Bacteriol. 1995;177:5215–21. doi: 10.1128/jb.177.18.5215-5221.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hube B, Monod M, Schofield DA, Brown AJ, Gow NA. Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol Microbiol. 1994;14:87–99. doi: 10.1111/j.1365-2958.1994.tb01269.x. [DOI] [PubMed] [Google Scholar]
- 8.Beggah S, Lechenne B, Reichard U, Foundling S, Monod M. Intra- and intermolecular events direct the propeptide-mediated maturation of the Candida albicans secreted aspartic proteinase Sap1p. Microbiology. 2000;146:2765–73. doi: 10.1099/00221287-146-11-2765. [DOI] [PubMed] [Google Scholar]
- 9.Homma M, Kanbe T, Hiroji C, Tanaka K. Detection of intracellular forms of secretory aspartic proteinase in Candida albicans. J Gen Microbiol. 1991;138:627–33. doi: 10.1099/00221287-138-3-627. [DOI] [PubMed] [Google Scholar]
- 10.Homma M, Chiban H, Tanaka K. Induction of extracellular proteinase in Candida albicans. J Gen Microbiol. 1993;139:1187–93. doi: 10.1099/00221287-139-6-1187. [DOI] [PubMed] [Google Scholar]
- 11.Lee SA, Mao YM, Zhang Z, Wong B. Overexpression of a dominant-negative allele of YPT1 inhibits growth and aspartyl protease secretion in Candida albicans. Microbiology. 2001;147:1961–70. doi: 10.1099/00221287-147-7-1961. [DOI] [PubMed] [Google Scholar]
- 12.Mao Y, Kalb VF, Wong B. Overexpression of a dominant-negative allele of SEC4 inhibits growth, and protein secretion in Candida albicans. J Bacteriol. 1999;181:7235–42. doi: 10.1128/jb.181.23.7235-7242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SA, Khalique Z, Gale CA, Wong B. Intracellular trafficking of fluorescently tagged proteins associated with pathogenesis in Candida albicans. Med Mycol. 2005;43:423–30. doi: 10.1080/13693780400013340. [DOI] [PubMed] [Google Scholar]
- 14.Harsay E, Bretscher A. Parallel secretory pathways to the cell surface in yeast. J Cell Biol. 1995;131:297–310. doi: 10.1083/jcb.131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harsay E, Shekman R. A subset of yeast vacuolar protein sorting mutants is blocked in one branch of the exocytic pathway. J Cell Biol. 2002;156:271–85. doi: 10.1083/jcb.200109077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurunathan S, David D, Gerst JE. Dynamin, and clathrin are required for the biogenesis of a distinct class of secretory vesicles in yeast. EMBO J. 2002;21:602–14. doi: 10.1093/emboj/21.4.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kullas AL, Li M, Davis DA. Snf7p, a component of the ESCRT-III protein complex, is an upstream member of the RIM101 pathway in Candida albicans. Eukaryot Cell. 2004;3:1609–18. doi: 10.1128/EC.3.6.1609-1618.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SA, Jones J, Khalique Z, Kot J, Alba M, Bernardo S, et al. A functional analysis of the Candida albicans homolog of Saccharomyces cerevisiae VPS4. FEMS Yeast Res. 2007;7:973–85. doi: 10.1111/j.1567-1364.2007.00253.x. [DOI] [PubMed] [Google Scholar]
- 19.Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-50-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–82. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 20.Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–28. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson RB, Davis D, Enloe BM, Mitchell AP. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast. 2000;16:65–70. doi: 10.1002/(SICI)1097-0061(20000115)16:1<65::AID-YEA508>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 22.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, et al. Current protocols in molecular biology. New York: Wiley; 1987. [Google Scholar]
- 23.Crandall M, Edward JE. Segregation of proteinase-negative mutants from heterozygous Candida albicans. J Gen Microbiol. 1987;133:2817–24. doi: 10.1099/00221287-133-10-2817. [DOI] [PubMed] [Google Scholar]
- 24.Palmer GE, Cashmore A, Sturtevant J. Candida albicans VPS11 is required for vacuole biogenesis, and germ tube formation. Eukaryot Cell. 2003;2:411–21. doi: 10.1128/EC.2.3.411-421.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitanovic A, Nguyen M, Vogl G, Hartmann A, Günther J, Würzner R, et al. Phosphatidylinositol 3-kinase VPS34 of Candida albicans is involved in filamentous growth, secretion of aspartic proteases, and intracellular detoxification. FEMS Yeast Res. 2005;5:431–39. doi: 10.1016/j.femsyr.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Franke K, Nguyen M, Härtl A, Dahse HM, Vogl G, Würzner R, et al. The vesicle transport protein Vac1p is required for virulence of Candida albicans. Microbiology. 2006;152:3111–21. doi: 10.1099/mic.0.29115-0. [DOI] [PubMed] [Google Scholar]
- 27.Bernardo SM, Jones JK, Kot J, Khalique ZK, Hardison S, Lee SA. Candida albicans VPS1 contributes to protease secretion, filamentation, and biofilm formation. Fungal Genet Biol. 2008;45:861–77. doi: 10.1016/j.fgb.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bryant NJ, Stevens TH. Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol Mol Biol Rev. 1998;62:230–47. doi: 10.1128/mmbr.62.1.230-247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]


