Abstract
Bacterial microcompartments (BMCs) are organelles that encapsulate enzymes involved in CO2 fixation or carbon catabolism in a selectively permeable protein shell. Here, we highlight recent advances in the bioengineering of these protein-based nanoreactors in heterologous systems, including transfer and expression of BMC gene clusters, the production of template empty shells, and the encapsulation of non-native enzymes.
Introduction
A key goal of synthetic biology is to engineer metabolic pathways to produce bulk chemicals for medical, agricultural, and industrial purposes using microbial cell factories. Factors that reduce the efficiency of engineered pathways include crosstalk of metabolites, toxic intermediates, and inhibitory products. Eukaryotes have evolved compartmentalizing organelles to overcome these obstacles. Bacteria also have organelles, known as bacterial microcompartments (BMCs) [1–3]. BMCs contain enzymes that catalyze sequential reactions and a private pool of cofactors (e.g., NAD+/NADH, coenzyme A, and ATP) within a protein shell. The BMC shell serves as a selectively permeable interface between the encapsulated pathway and the cellular environment. Because they self-assemble entirely from proteins, BMCs are becoming a viable platform for engineering novel nanoreactors.
Functionally diverse BMCs are bioinformatically predicted to be present in at least 23 different bacterial phyla [2]. Cyanobacteria and some chemoautotrophs produce BMCs (α- or β-carboxysomes) that encapsulate carbonic anhydrase (CA) and ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) to enhance CO2 fixation. However, the majority of functionally diverse BMCs are catabolic (metabolosomes), utilized by heterotrophs to degrade a range of carbon compounds in niche environments. Most metabolosomes contain a signature enzyme, such as a propanediol dehydratase (PDH) [4], an ethanolamine-ammonia lyase (EAL) [5], or a glycyl-radical enzyme [6, 7], that defines the function of the BMC [e.g., propanediol utilization (PDU) or ethanolamine utilization (EUT) BMCs, or glycyl-radical enzyme microcompartment (GRM)]. Metabolosome cores also include four conserved [1, 2] enzymes: an aldehyde dehydrogenase (AldDH) [8], an alcohol dehydrogenase (AlcDH) [9], and a phosphotransacylase (PTAC) [10, 11]. In addition, many BMC loci encode ancillary proteins that support organelle function, such as the transport of substrates and recycling co-factor (e.g., ATP and vitamin B12) [1, 2].
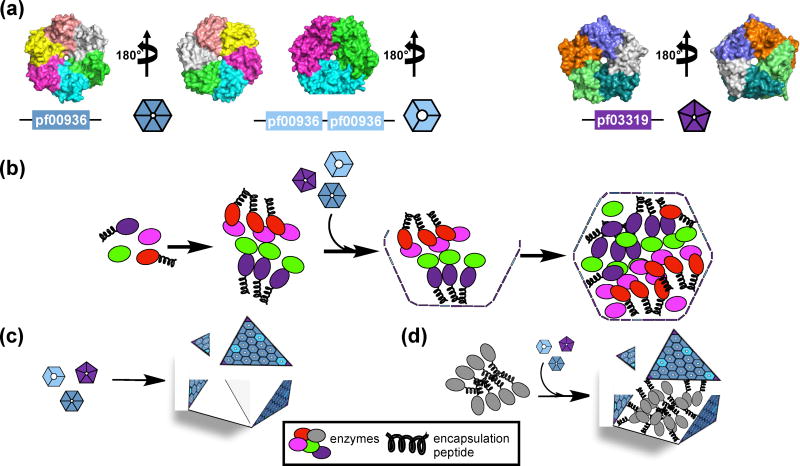
The core enzymes of carboxysomes and metabolosomes are encapsulated by a shell comprised of proteins that form hexamers (BMC-H) [12], pseudohexamers/trimers (BMC-T) [13, 14], and pentamers (BMC-P) [15, 16] (Figure 1a). The hexameric shell proteins typically contain a pore at the symmetry axis, with a diameter of 4 – 10 Å [17, 18••] and electrostatic properties [1, 6, 12, 14, 19] suited to the passage of small charged metabolites across the shell [1]. Confinement of sequential enzymatic reactions by the BMC shell facilitates substrate channeling, thereby enhancing catalytic efficiency. [1, 20–24]. The shell also acts as a barrier, preventing potentially toxic/volatile intermediates from diffusing into the cytoplasm [21–23•]. Most BMCs are predicted to form from the inside out, the core proteins coalesce into a bolus around which a shell assembles [1, 25] (Figure 1b). A short helical extension on a subset of core proteins, the encapsulation peptide (EP), facilitates the aggregation of the core enzymes [10, 26, 27] and their subsequent encapsulation by the shell [1, 25, 28–32]. The structure and native functions of BMCs have been reviewed elsewhere [1, 3, 20, 24, 33, 34]. The aim of this review is to highlight the recent efforts that adapt the BMC architectures for the development of novel nanoreactors in heterologous systems.
Figure 1.
Shell proteins (a) and assembly of BMCs (b–d). (a) Representatives of a BMC-H protein (PDB 5DJB) (left), a BMC-T protein (PDB 5DIH) (middle), and a BMC-P protein (PDB 2QW7) (right). Pf indicates Pfam identification. Individual polypeptide chains are colored differently. (b) Cartoon representation of BMC assembly from the inside out, where the primary role of the EP is in shell recruitment [25, 35••] (c) of empty BMC shell assembly, and (d) of targeting enzymes to the lumen of BMC shells using EPs [36••].
Overview of BMC engineering
Efforts to engineer BMCs have involved both the transfer and expression of BMC operons in heterologous systems, the production of empty shells (Figure 1c), and the encapsulation of heterologous cargo using encapsulation peptides (Figure 1d). Subsequent efforts have focused on building a core based on protein domain interactions and tuning shell permeability to support encapsulated metabolism.
Heterologous expression of BMC gene clusters in E. coli
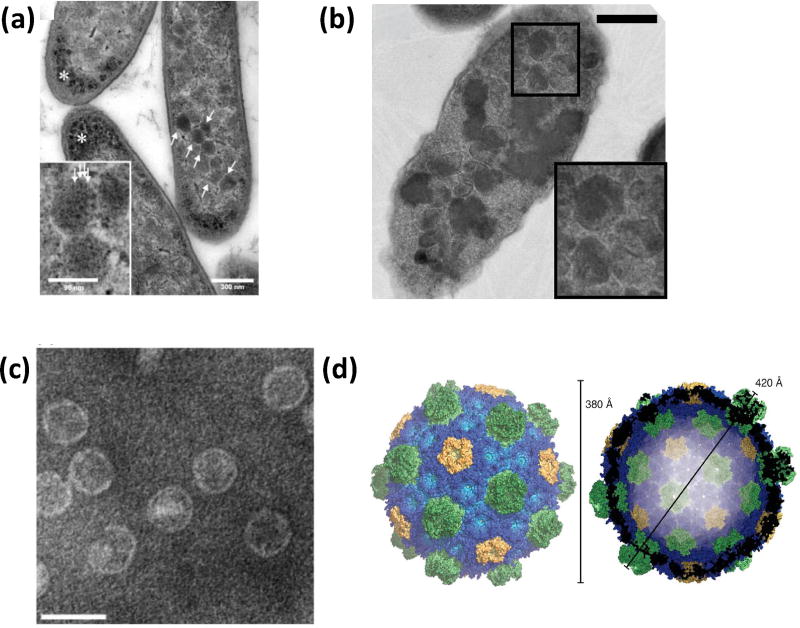
BMCs are encoded by gene clusters, providing a ready genetic module for heterologous expression. The 21-gene PDU operon of Citrobacter freundii was the first demonstration of the potential for “transplanting” a metabolic module in E. coli [37]. Electron microscopy of thin sections of strains expressing the operon revealed polyhedral bodies (Figure 2a) and recombinant metabolosome demonstrated diol dehydratase activity. Follow-up work showed that the recombinant PDU BMCs have similar morphology and mechanical properties as wildtype (WT) PDU BMCs [38]. Similarly, the α-carboxysome operon of a chemoautotroph was expressed in E. coli (Figure 2b), generating carboxysome-like particles and active RuBisCO [39].
Figure 2.
Physical characterization of engineered BMCs. (a) Thin section electron micrograph of E. coli expressing the complete PDU BMC operon (arrows) (scale bar 300 nm). Asterisks mark unknown granular dense matter. Inset is an enlarged view of the polyhedral bodies (scale bar 96 nm) that show regular substructures (arrows) [37]. (b) Thin sections electron micrograph of E. coli expressing the carboxysome genes of Halothiobacillus neapolitanus viewed by TEM (scale bar 500 nm) showing polyhedral bodies (magnified in inset) [39]. (c) Negatively stained electron micrograph of purified HO BMC shells (scale bar 50 nm). Reproduced with permission from [40]. Copyright Elsevier 2014. (d) Surface representation of the HO shell crystal structure. Shell proteins are colored blue (BMC-H), green (BMC-T), and yellow (BMC-P). Reproduced with permission from [41••]. Copyright The Association for the Advancement of Science 2017.
Heterologous expression of BMC shells
A variety of BMC shells from both carboxysomes [42] and metabolosomes have been shown to assemble in E. coli [37, 40, 43–45]. Compared to fully packaged native counterparts, recombinant empty shells tend to be smaller; only recombinant EUT shells, formed from all of the or a single EUT shell protein, were observed to be similar to WT EUT BMCs in size [45]. The number of these recombinant EUT shells were shown to increase when co-expressed with a putative cupin domain [46]. When the shell protein genes of a metabolosome of unknown function (from Haliangium ochraceum ) were expressed in E. coli, homogeneous, robust shells were formed and readily purified (Figure 2c) [40], enabling crystallization [41••]. The atomic resolutions structure of the 6.5 MDa empty shell is estimated to be able to accommodate threehundred 30 kDa proteins (Figure 2d). Interestingly, the structure revealed that the interactions between the shell proteins are largely governed by shape complementarity rather than salt bridges and hydrogen bonds between conserved residues. The structure provides scalable shell assembly principles that likely apply to all BMCs, and provides a blueprint for shell engineering.
Encapsulation of non-native proteins using EPs
EPs on BMC core enzymes have been shown to interact with shell proteins [28, 29, 47•, 48•], facilitating their encapsulation. EPs are typically short amphipathic helical extensions connected by a poorly conserved linker to a subset of core proteins [1, 30, 31]. They have been identified on the essential β-carboxysome protein CcmN [28], and on the signature (e.g., PDH [31, 32], EAL [30, 31, 45], and aldolase [2, 31, 49]) and conserved core (e.g., AldDH [29, 30, 40, 50], AlcDH [30, 31], and PTAC [10, 11, 31]) enzymes of metabolosomes. Recently, designed EPs were developed for the PDU shell using both rational and library-based strategies [47•].
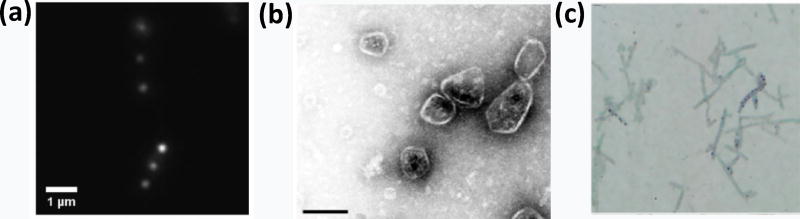
EPs have been employed to target non-native proteins to the interior of BMC shells. For example, fluorescent proteins (FPs) can be encapsulated inside native and recombinant carboxysome and metabolosome shells by full length core enzymes or by EPs alone [40, 42–45, 48•, 51, 52]. Green fluorescent protein fused to the EP of AldDH was used to develop an assay for the rapid quantification of encapsulated protein (Figure 3a) [51]. Likewise, various enzymes have been targeted to the interior of recombinant PDU shells, producing prototypes for the engineering of nanoreactors based on BMC architectures. For example, the EPs from the native enzymes of the PDU BMC were used to encapsulate exogenous pyruvate decarboxylase and AlcDH (Figure 3b) into synthetic shells made from PDU shell proteins to generate ethanolproducing BMCs in E. coli [50]. Recently, the EP of AldDH was used to separately encapsulate three different enzymes, one of which remained active in varying pH conditions in vitro [53•]. The EP of AldDH was also used to generate a polyphosphate-synthesizing BMC in E. coli (Figure 3c) [54••]. Because ATP is the substrate of the encapsulated enzyme, this study also shows that the shell of the engineered BMC is permeable to ATP (as is native PDU BMCs [1]), but the specific shell protein that allows ATP transit is unknown. Beyond the encapsulation of enzymes for biosynthesis, the EP of the AldDH was used to sequester a cytotoxic enzyme inside recombinant PDU shells, to restrict its toxic effects [55••].
Figure 3.
Heterologous expression of BMC shell proteins and cargo. (a) Fluorescent microscopy image of S. enterica expressing PDU BMC shell proteins and EPAldH-GFP. Reproduced with permission from [51]. Copyright John Wiley and Sons 2017. (b) Negatively stained electron micrograph of purified PDU BMC ethanol bioreactors from E. coli [50] (scale bar 100 nm). (c) Light microscopy of Neisser stained fixed E. coli cells expressing a polyphosphate kinase and PDU shell proteins, polyphosphate appears purple-black. Reproduced with permission from [54••]. Copyright John Wiley and Sons 2017.
Developing new methods for core assembly
In addition to targeting proteins for encapsulation, EPs cause their cognate enzyme to oligomerize [8, 10, 26, 27]. Taking advantage of this property, four enzymes tagged with the EP of PDH or AldDH resulted in a shell-free enzyme aggregate that was able to convert glycerol to 1,2-propanediol [36••]. Disadvantages of the EP-based approach include the loss of precise control of internal organization and stoichiometry. Control over core assembly may be obtained by visualizing the core components as an array of interacting protein domains. For example, utilizing the knowledge of core protein domain structures and their interactions in β-carboxysome formation [25], a chimeric protein (CcmC) was designed that could replace four gene products required for carboxysome assembly [35••]. CcmC is a synthetic protein consisting of domains to aggregate RuBisCO, a CA, and an EP for adherance to the carboxysome shell.
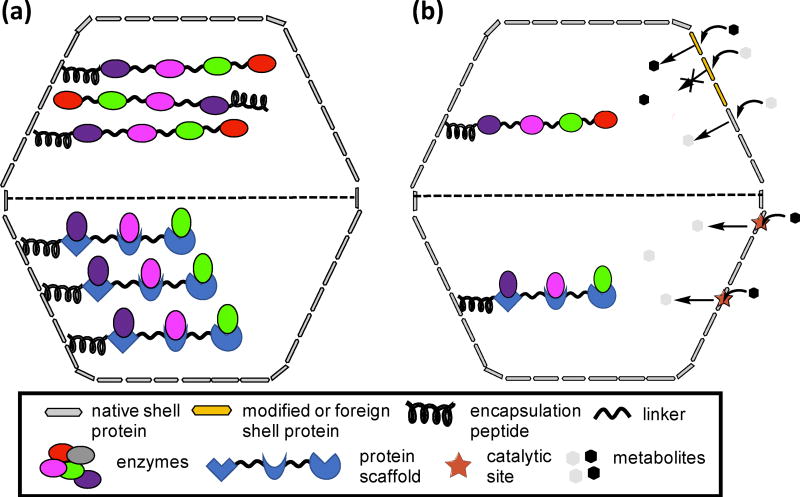
Domain-based engineering can be expanded to build chimeric cores that may improve the organization of the encapsulated enzymes. In addition, it has recently been observed that some core protein domains interact with the shell [42]. For instance, a chimeric core could be formed by the fusion of enzymatic domains from a specific metabolic reaction sequence; association with the shell could be mediated by one of these domains, or via an EP extension (Figure 4a, upper). The EP would facilitate the oligomerization of the fused domains and their subsequent encapsulation within the BMC shells. Alternatively, the stoichiometry of the enzymes may be controlled by utilizing a linear protein scaffold [56] fused to an EP (Figure 4a, lower). Enzymes are recruited to the core via specific protein-protein interactions with the scaffold domains.
Figure 4.
Cartoon representation of domain-based cores (a) and of shells with selective permeability (b). (a) Top panel: a chimeric protein tagged with an EP comprised of protein domains for a metabolic reaction. Lower panel: a linear scaffold tagged with an EP that is able to form protein-protein interaction with enzymes of a metabolic pathway. (b). Top panel: introduction of a foreign or a modified shell protein to tune permeability for specific metabolites. Lower panel: engineering of catalytic sites in the pore of shell proteins that will convert metabolites to the form required by the encapsulated pathway.
Modifying the BMC shell to tune permeability
Fundamental to the engineering of pathways within BMCs is the ability to tailor the permeability of the shell as the interface with metabolism to conduct the requisite substrates and products. The pores in BMC shells selectively control the passage of metabolites; accordingly, shell permeability can be tuned by modifying residues that surround the pores. Mutation of residues surrounding the pores have been shown to cause permeability defects [57], or enhance BMC function, due to alteration of small molecule diffusion rates [58•]. Shell permeability has also been modified by creating shell chimeras (Figure 4b). Chimeric β-carboxysome shells were generated by incorporating an α-carboxysome BMC-H protein, which structurally rescued a carboxysome-minus strain [59]. PDU BMCs incorporating shell proteins from the EUT BMC or β-carboxysome also have been produced, albeit with impaired function [60]. Recently, chimeric PDU shells were formed by substituting a EUT shell protein that has a smaller and more charged pore than its PDU counterpart [58•]. Strains expressing the chimeric BMCs showed improved growth over WT, demonstrating enhanced permeability to the substrate.
The prevailing model of the BMC shell as merely a passive semipermeable barrier is being reconsidered through engineering. PduT, a BMC-T protein from the PDU BMC, is the sole natural example of shell protein that binds a redox-active [4Fe-4S] cluster [61, 62]. The pore of a BMC-T protein, naturally devoid of a cofactor, was engineered to conduct electrons by incorporating a stable and redox-active [4Fe-4S] cluster, providing an example of generating a shell protein with a non-native function [18••]. This work provides a major step forward in the development of a synthetic shell permeable to electrons. This approach could be extended to the engineering of catalytic metal centers in the pores of shells, generating metabolites consumed by the encapsulated enzymes (Figure 4b).
Summary and Outlook
BMCs are proteinaceous organelles that encapsulate enzymes involved in anabolic or catabolic reactions. BMCs are genetic, structural, and functional modules. Future efforts in bioengineering of BMCs could lead to nanoreactors applicable for use in agriculture and energy sectors. For instance, the first steps toward installing carboxysomes into chloroplasts has been taken, including the heterologous expression of cyanobacterial RuBisCO and shell proteins [63, 64]. Likewise, the development of microbial cell factories that harbor engineered BMCs encapsulating a pathway that produces renewable alternatives to petroleum-based commodities is now in reach. The capacity to engineer BMCs as nanoreactors will be of significant value for synthetic biology and beyond.
BMCs are natural metabolic modules found in diverse bacterial phyla.
BMCs are proteinaceous organelles that encapsulate enzymes for CO2 fixation or carbon catabolism.
Bioengineering of BMCs has led to the production of empty synthetic shells.
Encapsulation of enzymes led to BMC-based nanoreactors.
BMC-based nanoreactors will be of significant value for synthetic biology.
Acknowledgments
This work was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (NIAID) grant 1R01AI114975-01 and the U.S. Department of Energy, Basic Energy Sciences, Contract DE-FG02-91ER20021.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest : None
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Kerfeld CA, Erbilgin O. Bacterial microcompartments and the modular construction of microbial metabolism. Trends Microbiol. 2015;23:22–34. doi: 10.1016/j.tim.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Axen SD, Erbilgin O, Kerfeld CA. A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Comput Biol. 2014;10:e1003898. doi: 10.1371/journal.pcbi.1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chowdhury C, Sinha S, Chun S, Yeates TO, Bobik TA. Diverse bacterial microcompartment organelles. Microbiol Mol Biol Rev. 2014;78:438–468. doi: 10.1128/MMBR.00009-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. The Propanediol Utilization (pdu) Operon ofSalmonella enterica Serovar Typhimurium LT2 Includes Genes Necessary for Formation of Polyhedral Organelles Involved in Coenzyme B12- Dependent 1,2-Propanediol Degradation. J Bateriol. 1999;181:5967–5975. doi: 10.1128/jb.181.19.5967-5975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brinsmade SR, Paldon T, Escalante-Semerena JC. Minimal functions and physiological conditions required for growth of salmonella enterica on ethanolamine in the absence of the metabolosome. J Bacteriol. 2005;187:8039–8046. doi: 10.1128/JB.187.23.8039-8046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zarzycki J, Erbilgin O, Kerfeld CA. Bioinformatic Characterization of Glycyl Radical Enzyme- Associated Bacterial Microcompartments. Appl Environ Microbiol. 2015;81:8315–8329. doi: 10.1128/AEM.02587-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craciun S, Balskus EP. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci U S A. 2012;109:21307–21312. doi: 10.1073/pnas.1215689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leal NA, Havemann GD, Bobik TA. PduP is a coenzyme-a-acylating propionaldehyde dehydrogenase associated with the polyhedral bodies involved in B12-dependent 1,2-propanediol degradation by Salmonella enterica serovar Typhimurium LT2. Arch Microbiol. 2003;180:353–361. doi: 10.1007/s00203-003-0601-0. [DOI] [PubMed] [Google Scholar]
- 9.Cheng S, Fan C, Sinha S, Bobik TA. The PduQ enzyme is an alcohol dehydrogenase used to recycle NAD+ internally within the Pdu microcompartment of Salmonella enterica. PLoS One. 2012;7:e47144. doi: 10.1371/journal.pone.0047144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erbilgin O, Sutter M, Kerfeld CA. The Structural Basis of Coenzyme A Recycling in a Bacterial Organelle. PLoS Biol. 2016;14:e1002399. doi: 10.1371/journal.pbio.1002399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Jorda J, Yeates TO, Bobik TA. The PduL Phosphotransacylase Is Used To Recycle Coenzyme A within the Pdu Microcompartment. J Bacteriol. 2015;197:2392–2399. doi: 10.1128/JB.00056-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. Protein structures forming the shell of primitive bacterial organelles. Science. 2005;309:936–938. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- 13.Klein MG, Zwart P, Bagby SC, Cai F, Chisholm SW, Heinhorst S, Cannon GC, Kerfeld CA. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J Mol Biol. 2009;392:319–333. doi: 10.1016/j.jmb.2009.03.056. [DOI] [PubMed] [Google Scholar]
- 14.Cai F, Sutter M, Cameron JC, Stanley DN, Kinney JN, Kerfeld CA. The structure of CcmP, a tandem bacterial microcompartment domain protein from the beta-carboxysome, forms a subcompartment within a microcompartment. J Biol Chem. 2013;288:16055–16063. doi: 10.1074/jbc.M113.456897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO. Atomic-Level Models of the Bacterial Carboxysome Shell. Science. 2008;319:1083–1086. doi: 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- 16.Sutter M, Wilson SC, Deutsch S, Kerfeld CA. Two new high-resolution crystal structures of carboxysome pentamer proteins reveal high structural conservation of CcmL orthologs among distantly related cyanobacterial species. Photosynthesis Research. 2013;118:9–16. doi: 10.1007/s11120-013-9909-z. [DOI] [PubMed] [Google Scholar]
- 17.Sutter M, Faulkner M, Aussignargues C, Paasch BC, Barrett S, Kerfeld CA, Liu LN. Visualization of Bacterial Microcompartment Facet Assembly Using High-Speed Atomic Force Microscopy. Nano Lett. 2015 doi: 10.1021/acs.nanolett.5b04259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18••.Aussignargues C, Pandelia ME, Sutter M, Plegaria JS, Zarzycki J, Turmo A, Huang J, Ducat DC, Hegg EL, Gibney BR, et al. Structure and Function of a Bacterial Microcompartment Shell Protein Engineered to Bind a [4Fe-4S] Cluster. J Am Chem Soc. 2015 doi: 10.1021/jacs.5b11734. The pore of a BMC-T protein, which lacks a cofactor, was engineered to bind a [4Fe-4S] cluster. The redox properties of the cluster mimics low-potential ferredoxins, while its coordination environment resembles radical S-adenosymethionine enzymes. This is the first example of imparting a shell protein with a non-native function. [DOI] [PubMed] [Google Scholar]
- 19.Kinney JN, Axen SD, Kerfeld CA. Comparative analysis of carboxysome shell proteins. Photosynth Res. 2011;109:21–32. doi: 10.1007/s11120-011-9624-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng S, Liu Y, Crowley CS, Yeates TO, Bobik TA. Bacterial microcompartments: their properties and paradoxes. Bioessays. 2008;30:1084–1095. doi: 10.1002/bies.20830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penrod JT, Roth JR. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J Bacteriol. 2006;188:2865–2874. doi: 10.1128/JB.188.8.2865-2874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sampson EM, Bobik TA. Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J Bacteriol. 2008;190:2966–2971. doi: 10.1128/JB.01925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23•.Jakobson CM, Tullman-Ercek D, Slininger MF, Mangan NM. A systems-level model reveals that 1,2-Propanediol utilization microcompartments enhance pathway flux through intermediate sequestration. PLoS Comput Biol. 2017;13:e1005525. doi: 10.1371/journal.pcbi.1005525. A computational model was developed to study the flux of metabolites across the PDU BMC. The authors found that the PDU shell functions mainly to decouple the cytosolic concentration of the intermediate from the concentrations in the shell, thus enhancing metabolite channeling via the generation of large concentration gradients across the shell. From their model, the authors also suggested that the encapsulation of heterologous metabolic pathways in BMC shells may gain significant benefits for pathway flux and for toxicity mitigation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerfeld CA, Heinhorst S, Cannon GC. Bacterial microcompartments. Annu Rev Microbiol. 2010;64:391–408. doi: 10.1146/annurev.micro.112408.134211. [DOI] [PubMed] [Google Scholar]
- 25.Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA. Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell. 2013;155:1131–1140. doi: 10.1016/j.cell.2013.10.044. [DOI] [PubMed] [Google Scholar]
- 26.Tobimatsu T, Kawata M, Toraya T. The N-terminal regions of beta and gamma subunits lower the solubility of adenosylcobalamin-dependent diol dehydratase. Biosci Biotechnol Biochem. 2005;69:455–462. doi: 10.1271/bbb.69.455. [DOI] [PubMed] [Google Scholar]
- 27.Akita K, Hieda N, Baba N, Kawaguchi S, Sakamoto H, Nakanishi Y, Yamanishi M, Mori K, Toraya T. Purification and some properties of wild-type and N-terminal-truncated ethanolamine ammonia-lyase of Escherichia coli. J Biochem. 2010;147:83–93. doi: 10.1093/jb/mvp145. [DOI] [PubMed] [Google Scholar]
- 28.Kinney JN, Salmeen A, Cai F, Kerfeld CA. Elucidating essential role of conserved carboxysomal protein CcmN reveals common feature of bacterial microcompartment assembly. J Biol Chem. 2012;287:17729–17736. doi: 10.1074/jbc.M112.355305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan CG, Cheng SQ, Sinha S, Bobik TA. Interactions between the termini of lumen enzymes and shell proteins mediate enzyme encapsulation into bacterial microcompartments. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:14995–15000. doi: 10.1073/pnas.1207516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan C, Cheng S, Liu Y, Escobar CM, Crowley CS, Jefferson RE, Yeates TO, Bobik TA. Short N-terminal sequences package proteins into bacterial microcompartments. Proc Natl Acad Sci U S A. 2010;107:7509–7514. doi: 10.1073/pnas.0913199107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aussignargues C, Paasch BC, Gonzalez-Esquer R, Erbilgin O, Kerfeld CA. Bacterial microcompartment assembly: The key role of encapsulation peptides. Communicative & Integrative Biology. 2015;8:e1039755. doi: 10.1080/19420889.2015.1039755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan C, Bobik TA. The N-terminal region of the medium subunit (PduD) packages adenosylcobalamin-dependent diol dehydratase (PduCDE) into the Pdu microcompartment. J Bacteriol. 2011;193:5623–5628. doi: 10.1128/JB.05661-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bobik TA, Lehman BP, Yeates TO. Bacterial microcompartments: widespread prokaryotic organelles for isolation and optimization of metabolic pathways. Mol Microbiol. 2015;98:193–207. doi: 10.1111/mmi.13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerfeld CA, Melnicki MR. Assembly, function and evolution of cyanobacterial carboxysomes. Curr Opin Plant Biol. 2016;31:66–75. doi: 10.1016/j.pbi.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 35••.Gonzalez-Esquer CR, Shubitowski TB, Kerfeld CA. Streamlined Construction of the Cyanobacterial CO2-Fixing Organelle via Protein Domain Fusions for Use in Plant Synthetic Biology. Plant Cell. 2015;27:2637–2644. doi: 10.1105/tpc.15.00329. The core of β-carboxysomes was redesigned to contain a chimeric protein (CcmC), replacing four gene products required for carboxysome assembly. CcmC consists of scaffolding domains (small subunit-like domains that nucleates RuBisCO), an enzymatic carbonic anhydrase domain (CA), and an EP that interacts with the carboxysome shell. The chimeric carboxysomes are morphologically comparable to wild type carboxysomes and supported photosynthesis. This work provides an example of domain-based engineering of a BMC core. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36••.Lee MJ, Brown IR, Juodeikis R, Frank S, Warren MJ. Employing bacterial microcompartment technology to engineer a shell-free enzyme-aggregate for enhanced 1,2-propanediol production in Escherichia coli. Metab Eng. 2016;36:48–56. doi: 10.1016/j.ymben.2016.02.007. The N-terminal EPs of two conserved enzymes in the PDU BMC were tagged to glycerol dehydrogenase, dihydroxyacetone kinase, methylglyoxal synthase and 1,2-propanediol oxidoreductase, producing a shell-free enzyme aggregate. The aggregate was able to convert glycerol to 1,2-propanediol. This work demonstrated that the propensity of EPs to cause proteins to oligomerize can be used to construct a metabolically active aggregate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parsons JB, Dinesh SD, Deery E, Leech HK, Brindley AA, Heldt D, Frank S, Smales CM, Lunsdorf H, Rambach A, et al. Biochemical and structural insights into bacterial organelle form and biogenesis. J Biol Chem. 2008;283:14366–14375. doi: 10.1074/jbc.M709214200. [DOI] [PubMed] [Google Scholar]
- 38.Mayer MJ, Juodeikis R, Brown IR, Frank S, Palmer DJ, Deery E, Beal DM, Xue WF, Warren MJ. Effect of bio-engineering on size, shape, composition and rigidity of bacterial microcompartments. Sci Rep. 2016;6:36899. doi: 10.1038/srep36899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonacci W, Teng PK, Afonso B, Niederholtmeyer H, Grob P, Silver PA, Savage DF. Modularity of a carbon-fixing protein organelle. Proc Natl Acad Sci U S A. 2012;109:478–483. doi: 10.1073/pnas.1108557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lassila JK, Bernstein SL, Kinney JN, Axen SD, Kerfeld CA. Assembly of robust bacterial microcompartment shells using building blocks from an organelle of unknown function. J Mol Biol. 2014;426:2217–2228. doi: 10.1016/j.jmb.2014.02.025. [DOI] [PubMed] [Google Scholar]
- 41••.Sutter M, Greber B, Aussignargues C, Kerfeld CA. Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell. Science. 2017;356:1293–1297. doi: 10.1126/science.aan3289. The crystal structure of an intact metabolosome shell of Haliangium_ochraceum was solved at an atomic scale resolution. The 6.5 MDa HO shell self-assembled into a pseudo (T= 9) icosahedral shell. The structure revealed that the interactions between the shell proteins are largely governed by shape complementarity rather than salt bridges and hydrogen bonds. This structure provides shell assembly principles that likely apply to all BMCs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai F, Bernstein SL, Wilson SC, Kerfeld CA. Production and Characterization of Synthetic Carboxysome Shells with Incorporated Luminal Proteins. Plant Physiol. 2016 doi: 10.1104/pp.15.01822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parsons JB, Frank S, Bhella D, Liang M, Prentice MB, Mulvihill DP, Warren MJ. Synthesis of empty bacterial microcompartments, directed organelle protein incorporation, and evidence of filament-associated organelle movement. Mol Cell. 2010;38:305–315. doi: 10.1016/j.molcel.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 44.Sargent F, Davidson FA, Kelly CL, Binny R, Christodoulides N, Gibson D, Johansson E, Kozyrska K, Lado LL, Maccallum J, et al. A synthetic system for expression of components of a bacterial microcompartment. Microbiology. 2013;159:2427–2436. doi: 10.1099/mic.0.069922-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choudhary S, Quin MB, Sanders MA, Johnson ET, Schmidt-Dannert C. Engineered protein nano-compartments for targeted enzyme localization. PLoS One. 2012;7:e33342. doi: 10.1371/journal.pone.0033342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Held M, Kolb A, Perdue S, Hsu SY, Bloch SE, Quin MB, Schmidt-Dannert C. Engineering formation of multiple recombinant Eut protein nanocompartments in E. coli. Sci Rep. 2016;6:24359. doi: 10.1038/srep24359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47•.Jakobson CM, Slininger Lee MF, Tullman-Ercek D. De novo design of signal sequences to localize cargo to the 1,2-propanediol utilization microcompartment. Protein Sci. 2017;26:1086–1092. doi: 10.1002/pro.3144. The amphipathic motif of EPs was used to construct new PDU EPs through both rational and library-based strategies. This work showed that a canonical leucine-zipper motif can also function as an EP, Fluorescence microscopy analysis on strains expressing EP-GFP fusion showed that the designed EPs are able to localize GFP to PDU shells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48•.Quin MB, Perdue SA, Hsu SY, Schmidt-Dannert C. Encapsulation of multiple cargo proteins within recombinant Eut nanocompartments. Appl Microbiol Biotechnol. 2016;100:9187–9200. doi: 10.1007/s00253-016-7737-8. The number of recombinant EUT shells expressed in E. coli increased when EUT shell proteins were co-expressed with a putative cupin domain protein (EutQ). The authors proposed that EutQ serves as nucleation point for EUT BMC assembly, which is supported by its interaction with a shell protein EutM. [DOI] [PubMed] [Google Scholar]
- 49.Erbilgin O, McDonald KL, Kerfeld CA. Characterization of a planctomycetal organelle: a novel bacterial microcompartment for the aerobic degradation of plant saccharides. Appl Environ Microbiol. 2014;80:2193–2205. doi: 10.1128/AEM.03887-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lawrence AD, Frank S, Newnham S, Lee MJ, Brown IR, Xue WF, Rowe ML, Mulvihill DP, Prentice MB, Howard MJ, et al. Solution structure of a bacterial microcompartment targeting peptide and its application in the construction of an ethanol bioreactor. ACS Synth Biol. 2014;3:454–465. doi: 10.1021/sb4001118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim EY, Tullman-Ercek D. A rapid flow cytometry assay for the relative quantification of protein encapsulation into bacterial microcompartments. Biotechnol J. 2014;9:348–354. doi: 10.1002/biot.201300391. [DOI] [PubMed] [Google Scholar]
- 52.Jakobson CM, Kim EY, Slininger MF, Chien A, Tullman-Ercek D. Localization of proteins to the 1,2-propanediol utilization microcompartment by non-native signal sequences is mediated by a common hydrophobic motif. J Biol Chem. 2015;290:24519–24533. doi: 10.1074/jbc.M115.651919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53•.Wagner HJ, Capitain CC, Richter K, Nessling M, Mampel J. Engineering bacterial microcompartments with heterologous enzyme cargos. Engineering in Life Sciences. 2017;17:36–46. doi: 10.1002/elsc.201600107. β-galactosidase, glycerol dehydrogenase, and esterase were each tagged with the EP of AldDH and were individually targeted to the lumen of recombinant PDU shells. An in vitro assay showed the encapsulation of β-galactosidase with the PDU shell protected its catalytic activity against pH stress. This work also found that hydrophilic compounds can easily diffuse across the recombinant PDU shell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54••.Liang M, Frank S, Lunsdorf H, Warren MJ, Prentice MB. Bacterial microcompartment-directed polyphosphate kinase promotes stable polyphosphate accumulation in E. coli. Biotechnol J. 2017;12 doi: 10.1002/biot.201600415. With the aim of recovering phosphorus from wastewater, a polyphosphate-synthesizing BMC was developed in E. coli. The EP of AldDH was used to target a polyphosphate kinase (PPK1) to the lumen of recombinant PDU shells In vivo assays showed that encapsulation of PPK1 resulted in continuous phosphate uptake and an increase in cellular polyphosphate levels throughout cell growth. [DOI] [PubMed] [Google Scholar]
- 55••.Yung MC, Bourguet FA, Carpenter TS, Coleman MA. Re-directing bacterial microcompartment systems to enhance recombinant expression of lysis protein E from bacteriophage varphiX174 in Escherichia coli. Microb Cell Fact. 2017;16:71. doi: 10.1186/s12934-017-0685-x. The ö X174 toxic lysis protein E tagged with the EP of AldDH was targeted X174 toxic lysis protein E tagged with the EP of AldDH was targeted to the iinteriior of recombinant PDU shells. Even though co-expression of E with PDU shell proteins produced irregularly shaped BMCs, the expression of protein E increased by 20–50%. The encapsulation protein E, in part, reduced its toxic effects. The authors suggested that BMCs can be used to enhance the expression of other toxic proteins in heterologous systems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KL, Keasling JD. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 57.Sinha S, Cheng S, Sung YW, McNamara DE, Sawaya MR, Yeates TO, Bobik TA. Alanine scanning mutagenesis identifies an asparagine-arginine-lysine triad essential to assembly of the shell of the Pdu microcompartment. J Mol Biol. 2014;426:2328–2345. doi: 10.1016/j.jmb.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58•.Slininger Lee MF, Jakobson CM, Tullman-Ercek D. Evidence for Improved Encapsulated Pathway Behavior in a Bacterial Microcompartment through Shell Protein Engineering. ACS Synth Biol. 2017 doi: 10.1021/acssynbio.7b00042. Chimeric and modified PDU shells were produced in S. enterica by substituting EutM for PduA and by mutating residues lining the pore of PduA to resemble the pore of EutM, respectively. EutM and the PduA mutant were each incorporated in the PDU BMCs. Strains expressing the chimeric or the modified PDU BMCs showed improved growth over wild type. This work showed that an encapsulated pathway was enhanced by altering the permeability of the shell. [DOI] [PubMed] [Google Scholar]
- 59.Cai F, Sutter M, Bernstein SL, Kinney JN, Kerfeld CA. Engineering bacterial microcompartment shells: chimeric shell proteins and chimeric carboxysome shells. ACS Synth Biol. 2015;4:444–453. doi: 10.1021/sb500226j. [DOI] [PubMed] [Google Scholar]
- 60.Sturms R, Streauslin NA, Cheng S, Bobik TA. In Salmonella enterica, Ethanolamine Utilization Is Repressed by 1,2-Propanediol To Prevent Detrimental Mixing of Components of Two Different Bacterial Microcompartments. J Bacteriol. 2015;197:2412–2421. doi: 10.1128/JB.00215-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parsons JB, Lawrence AD, McLean KJ, Munro AW, Rigby SE, Warren MJ. Characterisation of PduS, the pdu metabolosome corrin reductase, and evidence of substructural organisation within the bacterial microcompartment. PLoS One. 2010;5:e14009. doi: 10.1371/journal.pone.0014009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pang A, Warren MJ, Pickersgill RW. Structure of PduT, a trimeric bacterial microcompartment protein with a 4Fe-4S cluster-binding site. Acta Crystallogr D Biol Crystallogr. 2011;67:91–96. doi: 10.1107/S0907444910050201. [DOI] [PubMed] [Google Scholar]
- 63.Lin MT, Occhialini A, Andralojc PJ, Devonshire J, Hines KM, Parry MA, Hanson MR. beta-Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J. 2014;79:1–12. doi: 10.1111/tpj.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin MT, Occhialini A, Andralojc PJ, Parry MA, Hanson MR. A faster Rubisco with potential to increase photosynthesis in crops. Nature. 2014;513:547–550. doi: 10.1038/nature13776. [DOI] [PMC free article] [PubMed] [Google Scholar]