ABSTRACT
Maraviroc is a CCR5 antagonist used in the treatment of HIV-1 infection. We and others have suggested that maraviroc could reactivate latent HIV-1. To test the latency-reversing potential of maraviroc and the mechanisms involved, we performed a phase II, single-center, open-label study in which maraviroc was administered for 10 days to 20 HIV-1-infected individuals on suppressive antiretroviral therapy (EudraCT registration no. 2012-003215-66). All patients completed full maraviroc dosing and follow-up. The primary endpoint was to study whether maraviroc may reactivate HIV-1 latency, eliciting signaling pathways involved in the viral reactivation. An increase in HIV-1 transcription in resting CD4+ T cells, estimated by levels of HIV-1 unspliced RNA, was observed. Moreover, activation of the NF-κB transcription factor was observed in these cells. To elucidate the mechanism of NF-κB activation by maraviroc, we have evaluated in HeLa P4 C5 cells, which stably express CCR5, whether maraviroc could be acting as a partial CCR5 agonist, with no other mechanisms or pathways involved. Our results show that maraviroc can induce NF-κB activity and that NF-κB targets gene expression by CCR5 binding, since the use of TAK779, a CCR5 inhibitor, blocked NF-κB activation and functionality. Taking the results together, we show that maraviroc may have a role in the activation of latent virus transcription through the activation of NF-κB as a result of binding CCR5. Our results strongly support a novel use of maraviroc as a potential latency reversal agent in HIV-1-infected patients.
IMPORTANCE HIV-1 persistence in a small pool of long-lived latently infected resting CD4+ T cells is a major barrier to viral eradication in HIV-1-infected patients on antiretroviral therapy. A potential strategy to cure HIV-1-infection is the use of latency-reversing agents to eliminate the reservoirs established in resting CD4+ T cells. As no drug has been shown to be completely effective so far, the search for new drugs and combinations remains a priority for HIV cure. We examined the ability of maraviroc, a CCR5 antagonist used as an antiretroviral drug, to activate latent HIV-1 in infected individuals on antiretroviral therapy. The study showed that maraviroc can activate NF-κB and, subsequently, induce latent HIV-1-transcription in resting CD4+ T cells from HIV-1-infected individuals on suppressive antiretroviral therapy. Additional interventions will be needed to eliminate latent HIV-1 infection. Our results suggest that maraviroc may be a new latency-reversing agent to interfere with HIV-1 persistence during antiretroviral therapy.
KEYWORDS: HIV-1, maraviroc, NF-κB, latency, LRA, reservoir, persistence, HIV infected
INTRODUCTION
The persistence of quiescent HIV-1 infection within a small population of long-lived CD4+ T cells is currently the major barrier for the eradication or the functional cure of HIV-1. Reversal of proviral latency without global T-cell activation is being pursued as a curative strategy for HIV-1 infection, with the hope that infected cells in which the virus is reactivated will subsequently be eliminated by viral cytopathic effects or by virus-specific cytolytic T lymphocytes (CTLs). Several drug families with different mechanisms of action have been proposed as being potentially useful for this purpose, and in particular, histone deacetylase (HDAC) inhibitors have been the subject of intensive research (1–3). Carefully designed clinical trials have shown the capacity of some of these latency-reversing agents (LRAs) to reactivate latent HIV-1 in vivo (4–7), but no LRA is likely to drive the elimination of the latent reservoir in vivo when administered individually (8). It has been argued that the potency of individual LRAs may be too low and that the combination of several drugs may be needed to achieve clinically meaningful results (9). However, potential toxicities and drug-drug interactions may limit the chances of combining these agents.
Maraviroc (MVC) is a potent antiretroviral agent approved for the treatment of HIV-1 infection that blocks interaction between the virus and the CCR5 coreceptor, a crucial step in the HIV-1 life cycle (10). Previous clinical trials have demonstrated the safety, tolerability, and efficacy of maraviroc in both treatment-naive and treatment-experienced patients (11, 12).
Given the safety and tolerability of the drug, we performed an open-label phase II clinical trial to evaluate the effect of 48 weeks of administration of maraviroc on the cellular HIV-1 reservoir in patients receiving antiretroviral therapy (ART) (ClinicalTrials.gov registration no. NCT01365065) (13). The rationale of the trial was that ART intensification with an entry inhibitor would help in reducing the HIV-1 latent reservoir in resting CD4+ T cells by suppressing the residual replication of HIV-1. Maraviroc was added to the suppressive ART administered to the patients. We found that intensification with maraviroc was associated with a trend to a decrease in the size of the latent HIV-1 reservoir in resting CD4+ T cells, with a transient increase in the residual viremia and in the episomal two-long-terminal-repeat (2LTR) DNA circles. The effect on the cell reservoir persisted for 24 weeks after discontinuation of maraviroc (14). These observations raised the hypothesis that maraviroc could increase transcriptional activation of the latent virus. To our knowledge, a residual agonistic effect of maraviroc on CCR5 in resting CD4+ T cells latently infected with HIV-1 had not been described (10).
We hypothesize that maraviroc could promote HIV-1 transcription in resting CD4+ T cells by downstream activation of CCR5-mediated intracellular signaling pathways. To test this hypothesis, we have conducted a clinical trial to explore whether maraviroc could trigger this effect in suppressed HIV-1-infected patients, thus potentially helping to accelerate the decay of the HIV-1 cell reservoir. Then, maraviroc could be used, in addition to as an antiretroviral drug, as part of a combination regimen of LRAs.
RESULTS
Study design and participants.
This was a phase II clinical trial to determine whether treatment with maraviroc for a short period of time (10 days) in long-term-treated HIV-1-infected patients with previously suppressed viral load leads to an increase in the transcription of latent HIV-1 and to study the intracellular signaling pathways by which the activation of the transcription of latent virus may occur. The study was conducted at the University Hospital Ramón y Cajal in Madrid, Spain, between October 2012 and January 2015.
The 20 HIV-1-infected patients who were included met all the following criteria: at least 2 years of receiving ART with three drugs, HIV-1 RNA levels of <37 copies/ml for at least 2 years, and a CD4+ T-cell count of >350 cells/μl. We excluded HIV-1-infected patients with recent use of immunomodulatory agents or valproic acid. The patients (19 male and 1 female) had a median age of 49 years (Table 1). The median baseline CD4+ T-cell count was 664 cells/μl, and the median duration of ART before study entry was 127 months. Most patients (16 out of 20) were receiving a nonnucleoside-based regimen and the remaining a protease inhibitor-based one. All patients had plasma HIV-1 RNA levels of <37 copies/ml. Viral tropism determination based on proviral DNA was R5 in 12 patients, non-R5 in 4 patients, and indeterminate in 4 patients. All enrolled subjects completed the study as planned. Maraviroc was administered at dosages usually used in clinical practice, adjusted for concomitant antiretroviral drugs. Overall, maraviroc was well tolerated by all the patients, with no discontinuations during the study.
TABLE 1.
Baseline characteristics of study participants
| Patient ID | Age (yr) | Gendera | HIV-1 RNA (copies/ml) | Baseline CD4+ T cells (cells/mm3) | Viral tropism | ART duration (mo) | Regimenb |
|---|---|---|---|---|---|---|---|
| MTS 1 | 58 | M | <37 | 390 | R5 | 118 | TDF/FTC/EFV |
| MTS 2 | 52 | M | <37 | 908 | R5 | 76 | ABC/3TC/EFV |
| MTS 3 | 50 | M | <37 | 792 | Indeterminate | 53 | TDF/FTC/EFV |
| MTS 4 | 35 | F | <37 | 641 | R5 | 36 | TDF/FTC/EFV |
| MTS 5 | 67 | M | <37 | 380 | Non-R5 | 138 | TDF/FTC/EFV |
| MTS 6 | 51 | M | <37 | 394 | R5 | 156 | TDF/FTC/EFV |
| MTS 7 | 58 | M | <37 | 1,147 | Indeterminate | 169 | ABC/3TC/ATV |
| MTS 8 | 72 | M | <37 | 794 | Non-R5 | 181 | ABC/3TC/ATVr |
| MTS 9 | 29 | M | <37 | 526 | Indeterminate | 51 | TDF/FTC/EFV |
| MTS 10 | 52 | M | <37 | 907 | R5 | 106 | TDF/FTC/EFV |
| MTS 11 | 44 | M | <37 | 788 | R5 | 95 | ABC/3TC/ATV |
| MTS 12 | 41 | M | <37 | 686 | R5 | 206 | ABC/3TC/NVP |
| MTS 13 | 54 | M | <37 | 517 | R5 | 137 | TDF/FTC/EFV |
| MTS 14 | 52 | M | <37 | 461 | R5 | 92 | TDF/FTC/RPV |
| MTS 15 | 42 | M | <37 | 400 | R5 | 23 | TDF/FTC/RPV |
| MTS 16 | 34 | M | <37 | 888 | R5 | 23 | TDF/FTC/RPV |
| MTS 17 | 43 | M | <37 | 399 | Indeterminate | 210 | TDF/3TC/ATVr |
| MTS 18 | 49 | M | <37 | 741 | R5 | 221 | ABC/3TC/EFV |
| MTS 19 | 44 | M | <37 | 740 | Non-R5 | 194 | TDF/FTC/RPV |
| MTS 20 | 50 | M | <37 | 456 | Non-R5 | 170 | TDF/FTC/EFV |
| Median (IQR) | 49 (42.5–51) | <37 | 664 (414.6–794.2) | 127.5 (58.7–181) |
M, male; F, female.
3TC, lamivudine; ABC, abacavir; ATV, atazanavir; ATVr, ritonavir-boosted atazanavir; EFV, efavirenz; FTC, emtricitabine; NVP, nevirapine; RPV, rilpivirine; TDF, tenofovir disoproxil fumarate.
Maraviroc induces an increase in viral transcription in resting CD4+ T cells.
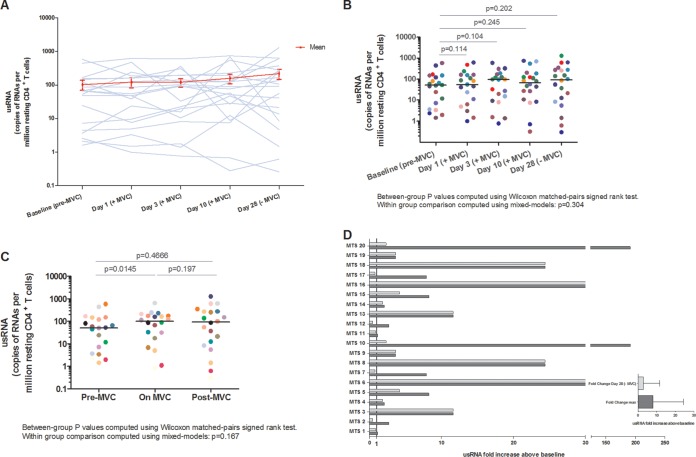
To assess the effects of maraviroc in the transcription of latent HIV-1, we evaluated the changes in unspliced RNA (usRNA) expression in the 20 participants at baseline, on days 1, 3, and 10 of maraviroc intensification, and after the withdrawal of maraviroc at day 28 of the study. A nonsignificant increase in the median number of copies of usRNA per resting CD4+ T cell during the study was observed (Fig. 1A and B). The increase in the expression of usRNA was detected as soon as at 24 h of maraviroc treatment (day 1) and persisted throughout the duration of treatment (P = 0.0145 for the comparison between baseline and during maraviroc treatment) and 18 days after the discontinuation of the drug, although differences were not significant at this time point (P = 0.197) (Fig. 1C). The median fold change in usRNA from baseline to peak in resting CD4+ T cells was 8.1 (interquartile range [IQR], 2.7 to 24.4) (Fig. 1D).
FIG 1.
Maraviroc promotes HIV-1 latency reversal in resting CD4+ T cells. HIV-1 latency reversal was estimated by the number of copies per million of HIV-1 usRNA in resting CD4+ T cells from all patients as determined by nested qRT-PCR. (A) Individual changes in usRNA levels are shown for each study participant. (B) Expression of the number of copies of usRNA per million resting CD4+ T cells per day of the study. Significant changes between time points were determined using the Wilcoxon signed-rank test. Central lines indicate the median. (C) Expression of usRNA before, during, and after 10-day intensification of maraviroc in resting CD4+ T cells of the patients. Samples were stratified into the following groups: baseline (Pre-MVC); day 1, day 3, day 10 (On MVC); and day 28 (Post-MVC). Comparisons of expression of usRNA among the premaraviroc, on-maraviroc, and off-maraviroc time periods were also performed using the Wilcoxon signed-rank test. (D) Fold change in usRNA following maraviroc treatment in resting CD4+ T cells compared to baseline. The maximum fold change in usRNA during the study (dark gray bar) and the change at day 28 (light gray bar) are shown for resting CD4+ T cells, and the median (IQR) change is shown for all participants. The solid line indicates 1-fold change. MVC, maraviroc.
Maraviroc activates NF-κB, but not AP-1 or NFAT, in resting CD4+ T cells.
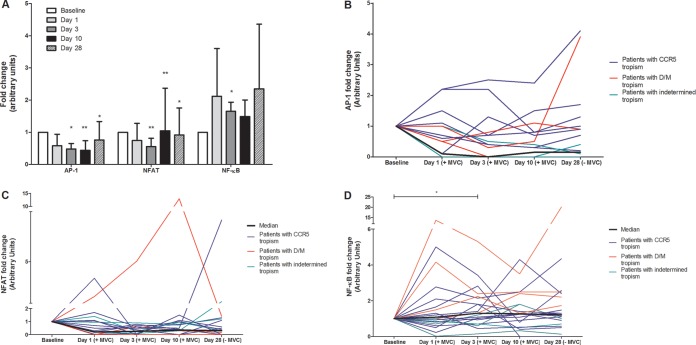
HIV-1 transcription is dependent on the activation of transcription factors, mainly AP-1, nuclear factor of activated T cells (NFAT), and NF-κB. We have determined the activities of these three factors in resting CD4+ T cells, using a DNA binding enzyme-linked immunosorbent assay (ELISA)-based assay (TransAM kits; Active Motif). The activities of the transcription factors were calculated as the fold change at each time point in comparison to baseline, before the administration of maraviroc (Fig. 2A). We did not establish a threshold at which the expression level of a transcription factor was considered to be changed. For NF-κB, for example, we considered that there was NF-κB induction when changes were associated with NF-κB target gene induction. These changes were in all cases higher than 1.3.
FIG 2.
Maraviroc induces NF-κB, but not AP-1 or NFAT, in resting CD4+ T cells. AP-1, NFAT, and NF-κB activities in resting CD4+ T cells were estimated by a DNA binding assay coupled with ELISA (Active Motif). (A) Fold change in transcription factor activity in resting CD4+ T cells following maraviroc treatment compared to baseline. Results are shown as geometric means, with error bars denoting 95% confidence intervals. (B to D) Fold changes in AP-1 (B), NFAT (C), and NF-κB (D) are shown for each participant (*, P < 0.05; **, P < 0.01). Blue lines represent patients infected with R5-tropic virus, red lines those infected with non-R5-tropic virus, and green lines those with indeterminate tropism; the solid black line indicates the median value. MVC, maraviroc.
AP-1 transcription factor activity was measured with the TransAM AP-1 kit (c-Jun detection) in resting CD4+ T cells from maraviroc-treated patients. Our results indicated AP-1 activation at one of the study visits in four of the 12 patients with CCR5 tropism and in one of four patients with indeterminate tropism (Fig. 2B; Table 2). However, no maintained and consistent activation of AP-1 in resting CD4+ T cells from all maraviroc-treated patients was observed. In fact, the median AP-1 activation was significantly lower at some time points than at baseline (Fig. 2A and B).
TABLE 2.
Numbers of patients with activation of transcription factors according to viral tropism
| Transcription factor | No. of patients with activation and with tropism: |
|||
|---|---|---|---|---|
| R5 (n = 12) | Non-R5 (n = 4) | Indeterminate (n = 4) | Total (n = 20) | |
| AP-1 | 4 | 1 | 5 | |
| NFAT | 3 | 1 | 2 | 6 |
| NF-κB | 10 | 4 | 2 | 16 |
NFAT transcription factor activity was also measured in resting CD4+ T cells from maraviroc-treated patients by using the TransAM NFATc1 kit. In this case, activation of NFAT was detected at at least one of the study visits in three of the patients with CCR5-tropic virus, in one of the patients with non-R5-tropic virus, and in two of the four patients with indeterminate tropism (Fig. 2A and C; Table 2). Again, no maintained and consistent activation of NFAT in resting CD4+ T cells from all-maraviroc treated patients was observed, and a significant decrease in the median NFAT activation was observed at most time points.
Importantly a clear trend of increasing activation of the NF-κB transcription factor (TransAM NF-κB p65 kit) in resting CD4+ T cells from maraviroc-treated patients was observed. Differences from baseline were determined at day 3 (Wilcoxon matched-pairs signed-rank test; P = 0.0363), with borderline statistical significance determined at day 10 (P = 0.0885). Using linear mixed models, we found no significant differences in overall changes during treatment. Remarkably, this activation was observed during the administration of maraviroc and after the drug was discontinued. NF-κB activation was detected in resting CD4+ T cells from treated patients at any of the study visits in 16 of the total population of 20 patients evaluated, in 10 of the 12 patients with CCR5-tropic virus, in all patients with non-R5-tropic virus, and in two of the four patients with indeterminate tropism (Fig. 2A and D; Table 2).
Altogether, these results indicate that maraviroc treatment induces NF-κB activity in maraviroc-treated patients but does not promote activation of the AP-1 or NFAT pathway. To assess the contribution of NF-κB activity to usRNA expression during MVC treatment, we evaluated the effects of the interaction between NF-κB and time on usRNA levels using mixed-effects modelling, and this association was not significant (P = 0.499).
Maraviroc increases expression of NF-κB target genes.
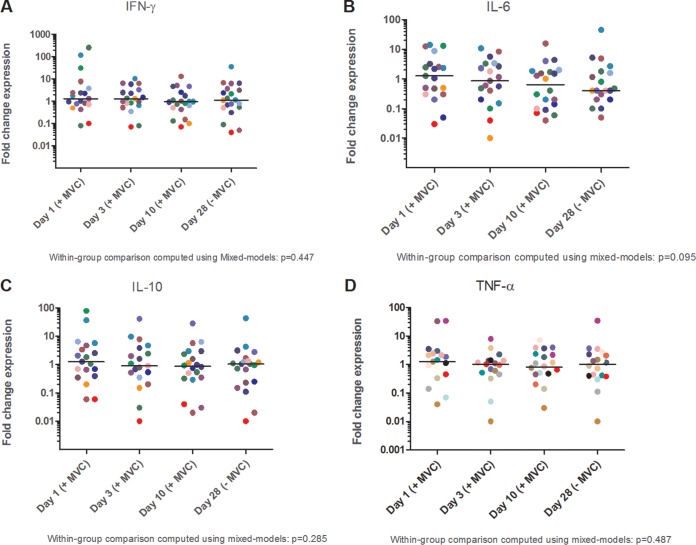
In order to confirm that the above-reported NF-κB activation is functional and leads to transcription of NF-κB target genes in resting CD4+ T cells from patients, we performed quantitative real-time PCR (qRT-PCR) of four NF-κB-dependent genes (encoding gamma interferon [IFN-γ], interleukin-6 [IL-6], IL-10, and tumor necrosis factor alpha [TNF-α]) (Fig. 3A to D). In the four studied genes, we detected an induction of expression at 24 h after initiating maraviroc treatment.
FIG 3.
Maraviroc induces expression of the NF-κB target genes for IFN-γ (A), IL-6 (B), IL-10 (C), and TNF-α (D) in resting CD4+ T cells. Significant changes between time points were determined using the Wilcoxon signed-rank test. Central lines indicate the median at each time point, and results are expressed as the fold change in mRNA expression compared to the baseline (without maraviroc). MVC, maraviroc.
Activation of NF-κB and subsequent transcription of latent HIV-1 is mediated by maraviroc binding to CCR5.
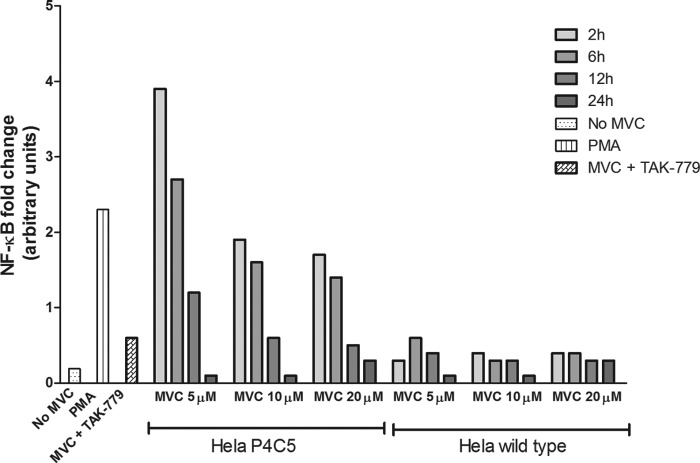
In order to elucidate whether the NF-κB activation is dependent on maraviroc binding to CCR5 or an alternative pathway, we used an in vitro model of stable CCR5 overexpression in HeLa cells. HeLa P4 C5 cells were exposed to maraviroc treatment for 2, 6, 12, and 24 h, at different concentrations (5, 10, and 20 μM). Nontreated HeLa P4 C5 cells and treated wild-type HeLa cells (null for CCR5 expression) were used as controls.
NF-κB activity was observed after 2 and 6 h of maraviroc treatment at all the concentrations tested in HeLa P4 C5 cells (Fig. 4). We did not detect activation of NF-κB at 12 and 24 h at any of the concentrations used in this study. The greatest increase in NF-κB activity was observed in HeLa P4 C5 cells at 2 h of maraviroc treatment, with the maximum activity at 5 μM maraviroc. This NF-κB activation was CCR5 dependent, since activation was not observed in wild-type HeLa cells at any of the concentrations and times tested or in HeLa P4 C5 cells when TAK-779, a CCR5 inhibitor, was also present in the tissue culture medium (Fig. 4). As expected, phorbol 12-myristate 13-acetate (PMA) treatment of HeLa cells led to NF-κB activation.
FIG 4.
Maraviroc induces NF-κB activity in CCR5-overexpressing HeLa P4 C5 cells. The cells were treated with 5 µM, 10 µM, and 20 µM maraviroc for 2, 6, 12, and 24 h. NF-κB p65 activity was significantly higher in HeLa P4 C5 cells at 2 h with 5 µM maraviroc. PMA was used as a positive control. NF-κB activity was not observed in HeLa wild-type cells at any of the concentrations and times or in HeLa P4 C5 cells treated with TAK-779, a CCR5 inhibitor, for 2 h with maraviroc (5 µM). Mean values were calculated from triplicates of a representative experiment. MVC, maraviroc; PMA, phorbol 12-myristate 13-acetate.
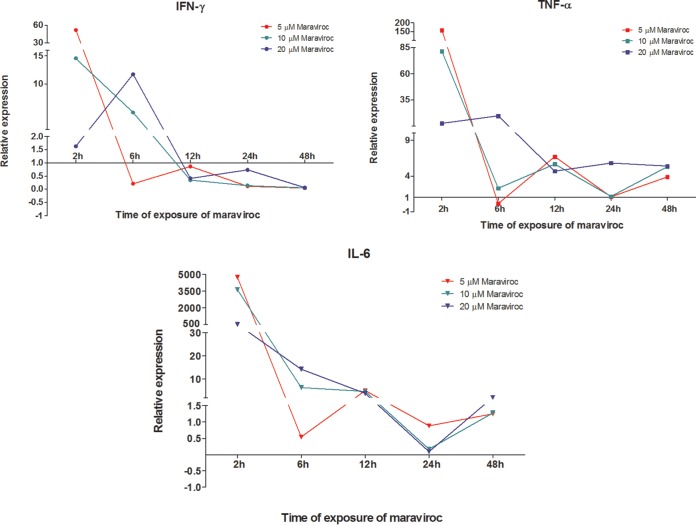
In order to demonstrate that this NF-κB activation drives transcription, we determined the effect of maraviroc on NF-κB target gene expression in HeLa P4 C5 cells (Fig. 5). At low concentrations of maraviroc (5 μM), we observed a remarkable upregulation of target genes. This upregulation was detected after 2 h of maraviroc treatment and reached levels similar to those found under PMA treatment. However, when TAK-779 was present in the medium, the upregulation was abolished.
FIG 5.
Maraviroc induces NF-κB target genes for IFN-γ, IL-6 and TNF-α in HeLa P4 C5 cells. Representative results of qRT-PCR experiments for relative expression of NF-κB target genes in HeLa P4 C5 cells upon maraviroc treatment are shown. MVC, maraviroc.
Altogether, these results strongly suggest that the NF-κB activation and activity reported here are dependent on maraviroc binding to CCR5. Moreover, the robust results observed at a low concentration and short time strongly suggest that the maraviroc effect on NF-κB activity reported here is highly specific and very efficient.
DISCUSSION
We show here that the administration of maraviroc for 10 days induces significant viral reactivation in HIV-1-infected patients receiving suppressive antiretroviral therapy. The observed increase in usRNA appears to be mediated by increased activity of NF-κB after the binding of maraviroc to CCR5. Thus, maraviroc not only inhibits HIV-1 by blocking the interaction between the virus and the coreceptor but also may act as an LRA through a residual agonistic effect on CCR5.
During the development of maraviroc, no association with intracellular signaling pathways was found, although the evaluation was limited to the activation state of the CCR5 signaling pathway mediated by calcium mobilization, and neither was there interference in the receptors of other chemokines (CCR1, -2, -2b, -3, -4, -7, and -8 and CXCR1 and -2) or of the IL-2, IL-8, and IL-4 cytokines. In addition, maraviroc did not induce the internalization of CCR5, thus behaving as an inhibitor (functional antagonist or inverse agonist) of the receptor. The affinity of the drug was not significantly affected by increases in chemokine concentrations, which is also consistent with allosteric inhibition (10). Combined, the data provided confirmed the binding specificity and antagonistic action of maraviroc but did not exclude the potential of the drug to trigger the activation of other CCR5 signaling pathways mediated by transcription factors. We present evidence that supports that maraviroc consistently induced HIV-1-replication in latently infected CD4+ T cells in most patients receiving the drug in a clinical trial. The effect was observed mainly during the administration of the drug but persisted after its cessation. The persistence of the transcription of latent HIV-1 after discontinuing the drug has been observed in clinical trials with other LRAs, including HDAC inhibitors and disulfiram (4–7, 15).
Interestingly, the effect of maraviroc on HIV-1 transcription was independent of viral tropism. It was observed in patients infected with viruses with either R5, non-R5, or indeterminate tropism. This was not unexpected. In ex vivo studies performed to measure the stimulation of the intracellular signaling pathways mediated by the gp120 viral surface glycoprotein, it was found that the majority of cellular signaling events occur independently of tropism but with different intensities (16). They also indicated a differential pattern in the triggered signaling, for example, on the induction of different transcription factors. In this sense, we have found that the increased transcription observed with the administration of maraviroc is mediated mainly by an increase of NF-κB activity but not of that of NFAT or AP-1. The activation of these transcription factors, particularly NF-κB, was observed to be independent of the virus tropism, indicating that even when maraviroc could not be used to treat HIV-1 infection due to non-R5 viral tropism, it could be given to all the patients to lead the activation of transcription factors.
An NF-κB DNA binding element is essential in the regulation of HIV-1 LTR induction, thus making NF-κB the major transcription factor involved in the initiation of HIV-1 replication (17–19). NFAT, another transcription factor able to bind the HIV-1 promoter, is crucial for the initiation of LTR transcription during the T-cell activation process (20), although in the absence of NF-κB. Moreover, AP-1 also modulates HIV-1 transcription, but it appears to be dependent on NF-κB and more related to the elongation process. Several authors have suggested that both NFAT and AP-1 play a role in HIV-1 transcription but always in cooperation with activated NF-κB (21, 22). NF-κB transcriptional activity in resting CD4+ T cells from patients treated with maraviroc has been demonstrated in our study. The functionality of activated NF-κB was confirmed by the increased expression of NF-κB target genes. It is worth noting the robustness of the NF-κB activity reported here, especially taking into account that it is observed after the oral administration, metabolism, and incorporation by CD4+ T cells of a drug. To our knowledge, no previous clinical studies showing such a potent molecular effect in NF-κB activity after the oral administration of any NF-κB agonist have been reported.
In contrast, significant AP-1 or NFAT transcriptional activity was not detected in resting CD4+ T cells. The activation of AP-1 was evaluated by measuring the levels of phosphorylated c-Jun, one of its major components. The levels of c-Jun are low or absent in resting CD4+ T cells (23–28). Therefore, it is conceivable that AP-1 activation by maraviroc cannot easily occur. However, we have not evaluated c-fos/JunD heterodimers, so a residual AP-1 activity cannot be discounted. In vivo studies of CCR5 activation by gp120 showed that NFATc1, an NFAT protein expressed in resting CD4+ T cells, leads to replication of HIV-1 (38). Our results demonstrate that maraviroc cannot consistently induce NFAT activation, although slight activation of NFAT was detected after withdrawal of maraviroc. Cicala et al. have previously shown that minimal or no NFAT activation is observed when CD4+ T cells are treated with MIP-1β or SDF-1, natural ligands of the HIV-1 receptors CCR5 and CXCR4, respectively (29). Failure to detect a pattern of activation of this transcription factor is in agreement with the absence of residual partial agonistic activity by mobilization of calcium, discarded when antiretroviral activity of maraviroc was evaluated, since NFAT is involved in the mobilization of calcium (10).
We have also evaluated the mechanisms underlying the maraviroc-associated activation of NF-κB with subsequent transcription of latent HIV-1. It could be hypothesized that maraviroc could promote an increase in the circulating levels of the natural CCR5 ligands (RANTES, MIP-1α, and MIP-1β), which would lead to activation of T cells, as well as monocytes and neutrophils, through binding to other coreceptors, such as CCR3, CCR4, and CCR1. Indeed, an increase in some of these cytokines has been shown in patients treated with maraviroc (30, 31). Alternatively, maraviroc could increase NF-κB activity just through the binding to CCR5 and the subsequent downstream signaling. Our results support this latter hypothesis. Maraviroc induced NF-κB activation in cells which expressed CCR5 but no other receptor and in the absence of other cytokines. In addition, maraviroc appears to be highly efficient for NF-κB activation in this cell model, since this activation is observed at low drug concentrations and short treatment times compared with those in previous reports (32). Moreover, this activation appears specific since it was abolished in the presence of the CCR5 antagonist TAK-779. These in vitro experimental approaches strongly support that maraviroc has an agonistic effect on CCR5, leading to NF-κB activation and HIV-1 transcription.
The small sample size is a significant limitation of the present study, as it does not allow for rigorous analysis of data and generalization of results. A statistical analysis was not feasible based on the small sample size for Fig. 4 and 5, thus limiting the strength of the conclusions. In summary, our study shows for the first time that a safe and already available antiretroviral agent can activate latent HIV-1 and could be used for cure efforts. Maraviroc increases the transcription of HIV-1 in resting CD4+ T cells through the activation of NF-κB by downstream signaling after binding to CCR5. Given its well-known safety and tolerability, maraviroc should be further evaluated in combination with other LRAs that could efficiently induce virus production and ultimately lead to the elimination of latent HIV-1 infection in resting CD4+ T cells.
MATERIALS AND METHODS
Patients.
Study patients were sampled from a clinical trial of 10 days of maraviroc intensification in 20 HIV-1-infected adults on suppressive ART (MARAVITRANS, EudraCT registration no. 2012-003215-66). The patient dosage of maraviroc was 150 mg, 300 mg, or 600 mg twice daily, according to the current ART. Maraviroc was provided by Pfizer Inc. This is a phase II, single-center, open-label clinical trial conducted at Ramón y Cajal University Hospital (Madrid, Spain) between October 2012 and January 2015. The main objective was to determine if treatment with maraviroc during a short period of time added to the current ART in patients with previously suppressed HIV-1 RNA leads to an increase in transcription of latent virus and to study the intracellular signaling pathways by which the activation of the transcription of latent virus may occur. The 20 patients met all of the following inclusion criteria: an undetectable plasma viral load (pVL) determined by ultrasensitive techniques (<37 copies HIV-1 RNA/ml) for at least 2 years, serologically documented HIV-1 infection, at least 2 years receiving triple ART, and a CD4+ T lymphocyte count above 350 cells/mm3. We excluded patients with recent use of immunomodulatory agents or with potential reversing activity of HIV-1 latency or valproic acid.
Ethics statement.
The clinical trial was performed according to the principles of the Declaration of Helsinki and the Good Clinical Practice Guidelines and was approved by the AEMPS (Spanish Agency for Medications and Health Products) and by the Ethics Committee of the University Hospital Ramón y Cajal (ceic.hrc@salud.madrid.org). All patients were adults and provided written informed consent for participation, sample collection, and laboratory determinations prior to initiation of study procedures.
Specimen collection.
Blood samples from patients were drawn at baseline (premaraviroc control data), after 1, 3, and 10 days on maraviroc, and 18 days after maraviroc withdrawal. A total of 100 ml of whole blood with EDTA (BD Vacutainer; Becton Dickinson, Franklin Lakes, NJ, USA) was drawn to obtain plasma and isolate peripheral blood mononuclear cells (PBMCs).
Proviral DNA coreceptor tropism testing.
Viral tropism in HIV-1 was estimated based on V3 sequences obtained from proviral DNA from PBMCs collected from individuals. The V3 region was amplified using a nested PCR method reported elsewhere (33). Tropism assignments (R5 or non-R5) were made with the geno2pheno algorithm (34) with a false-positive rate (FPR) of 10% (non-R5, ≤10%) the recommended cutoff when triplicate tropism determinations are employed (35).
Isolation of resting CD4+ T cells.
PBMCs from patient blood samples were isolated using Ficoll-Hypaque density gradient (Comercial Rafer S.L., Zaragoza, Spain). CD4+ T cells were isolated from PBMCs by negative selection using MACS MS columns (Miltenyi Biotech, Friedrich, Germany). To purify resting CD4+ T cells, cells were further enriched through negative depletion of cells expressing CD25 or HLA-DR (CD25 MicroBeads [Miltenyi Biotec] or Anti-HLA-DR MicroBeads [Miltenyi Biotec]). Resting CD4+ T cells were typically greater than 98% pure.
Nuclear extract preparation and AP-1, NFAT, and NF-κB transcription factor activity assays.
Nuclear extracts were obtained from resting CD4+-T cells at each time point of the clinical trial using TransAM AP-1 Family, TransAM NFATc1, and TransAM NF-κB p65 Family kits (Active Motif, Carlsbad, CA, USA) according to the manufacturer's instructions. A similar protocol was used for NF-κB activity studies in HeLa P4 C5 and HeLa cells. Briefly, the transcription factor contained in nuclear extracts (2 μg) binds specifically to the immobilized oligonucleotides in a 96-well plate and is detected using an antibody against either p65 (NF-κB), c-Jun (AP-1), or NFATc1 (NFAT). The wild-type consensus oligonucleotides of each transcription factor were used as a competitor for NF-κB, AP-1, or NFAT binding in order to monitor the specificity of the assay. Conversely, the mutated consensus oligonucleotides should have no effect on transcription factor binding and served as negatives control. Each sample was assayed in triplicate. The coefficient of variation (CV) of these assays was <20%.
RNA preparation.
RNA from resting CD4+ T cells was extracted by using the RNeasy minikit (Qiagen, Hilden, Germany) and converted into cDNAs using the SuperScript III first-strand synthesis system (Invitrogen, Karlsruhe, Germany). In all assays, a control without reverse transcriptase (RT) was used. If there was any amplification from the no-RT control, i.e., evidence of DNA contamination, a second stored sample was reextracted. If contaminating DNA persisted, the reading was excluded. Repeat extraction was required for only 3 of the total samples analyzed for this study.
qRT-PCR for NF-κB target genes.
We measured the mRNA expression levels of four NF-κB-related genes by quantitative real-time PCR (qRT-PCR) using the LightCycler 480 real-time PCR system (Roche Applied Science, Penzberg, Germany) according to the manufacturer's instructions along with the Universal Probe Library (Roche) probes and specific primers for IFN-γ, IL-6, IL-10, and TNF-α. Gene expression was normalized using the β-actin gene as a housekeeping gene. All PCRs were performed in triplicates. Relative gene expression was calculated with the 2−ΔΔCT method (36). We considered that there was target gene induction when a 1.5-fold or greater change was shown.
Seminested real-time PCR for usRNA quantitation.
For seminested real-time PCR for usRNA, the eluted cellular RNA was directly subjected to two rounds of PCR amplification as previously described (37). The primer pair used in the first PCR, MH535 (5′-AACTAGGGAACCCACTGCTTAAG-3′) and SL20 (5′-TCTCCTTCTAGCCTCCGCTAGTC-3′), amplifies a region within the HIV-1 gag gene. The first round of the PCR was performed on a conventional thermal cycler as follows: 95°C for 10 min, followed by 15 cycles of 94°C for 20 s, 55°C for 40 s, and 72°C for 40 s. The product of the first PCR was subsequently used as a template in the second, seminested, real-time PCR amplification, performed on the LightCycler 480 using SYBR green detection. Real-time PCR settings were as follows: 95°C for 10 min, followed by 45 cycles of 94°C for 20 s and 55°C for 40 s. The seminested real-time PCR of the usRNA assay was performed with the primers SL19 (5′-TCTCTAGCAGTGGCGCCCGAACA-3′) and SL20 and was conducted in triplicate for each experimental condition tested. The intra-assay CV was 17.39%. As external standards, synthetic runoff RNA transcripts corresponding to the HIV-1 gag region were used. The RNA standards were kindly provided by Sharon R. Lewin (Department of Infectious Diseases, Alfred Hospital and Monash University, Melbourne, Australia). Serial dilutions of standards between 1 copy and 4.4 × 1011 input copies were made. The amplicon sizes were 286 bp for the first PCR and 160 bp for the second (real-time) PCR. PCR results are expressed as usRNA copies per 106 resting CD4+ T cells.
Viral load measurements.
Plasma HIV-1 RNA was measured using kPCR (Versant HIV-1 RNA v1.0 assay; Siemens Healthcare, Erlangen, Germany) in the Microbiology Service of the Ramón y Cajal University Hospital.
Cell cultures and treatments.
HeLa cells (ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco) with 10% (vol/vol) heat-inactivated fetal calf serum (FCS) (Gibco) and 100 units/ml penicillin-streptomycin (Sigma-Aldrich) and were incubated at 37°C with CO2. CD4- and CCR5-expressing HeLa P4 C5 cells were cultured at 37°C under CO2 in DMEM supplemented with 10% (vol/vol) FCS, 100 units/ml penicillin (Sigma-Aldrich), 100 μg/ml streptomycin (Sigma-Aldrich), 1 mg/ml G418 (Sigma-Aldrich), and 300 μg/ml hygromycin B (Sigma-Aldrich). For maraviroc treatment, all seeded cells were treated with different concentrations of maraviroc (5, 10, and 20 μM) (Sigma-Aldrich) added to the cultured cells, and cells were harvested at different time points (2, 6, 12 and 24 h). These concentrations were chosen considering the in vitro 90% inhibitory concentration (IC90) of MVC to inhibit CCR5-tropic viral entry and maraviroc plasma concentrations in treated patients. As controls, we used TAK-779 (1 μM), a CCR5 antagonist, and PMA (12.5 ng/ml and 4 h) (Sigma-Aldrich), also added to the culture medium. All experiments were performed in triplicate wells for each condition and repeated at least twice. For Fig. 4 and 5, we presented the mean value at each time point from triplicates of a representative experiment.
Statistical analysis.
Qualitative variables were reported as a frequency distribution, whereas quantitative variables were described as medians and interquartile ranges (IQRs) and represented as individual points in rainbow plots. Interaction terms (time-versus-treatment arm) were created to assess whether these changes over time differed significantly. We used linear mixed models with a random effect for each patient to allow for correlations caused by repeated observations to assess whether longitudinal changes in continuous outcome measures were overall significantly different from baseline. An interaction term was created to assess whether changes in intracellular HIV RNA over time depended significantly on NF-κB activation during maraviroc treatment. A robust variance estimator was used, given the limited sample size and the deviation from normal. We used the Wilcoxon signed-rank matched-pairs test to evaluate differences in numerical outcomes between specific time points. We used multilevel mixed-effects logistic regression to assess whether longitudinal changes in categorical outcome measures differed over time. Continuous variables were log transformed when necessary to satisfy model assumptions. GraphPad Prism 5.0 (GraphPad, San Diego, CA) and Stata 15 (StataCorp LP, College Station, TX) were used for statistical analyses and figure preparation. Data are expressed as mean ± standard error of the mean (SEM) unless mentioned otherwise. Comparisons of copies and fold change in usRNA and of fold change in NF-κB between baseline and subsequent time points across all patients and between groups used a nonparametric Wilcoxon signed-rank test. P values of <0.05 were considered statistically significant.
ACKNOWLEDGMENTS
This work was supported by an Investigator Initiated Research Grant from ViiV Healthcare and the RD12/0017/0017 project as part of Plan Nacional R + D + I and cofunded by Instituto de Salud Carlos III (ISCIII)-Subdirección General de Evaluación and Fondo Europeo de Desarrollo Regional (FEDER).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We acknowledge all the study participants who contributed to this work as well as the clinical research staff who made this study possible. We thank Sharon R. Lewin and Ajantha Solomon at the Department of Infectious Diseases, Alfred Hospital and Monash University, Melbourne, Australia, for providing RNA standards for seminested real-time PCR of the usRNA assay. HeLa P4 C5 cells and TAK-779 were kindly provided by Jose Alcamí, Javier García-Pérez, and Nuria González (AIDS Immunopathogenesis Unit, National Centre of Microbiology, Instituto de Salud Carlos III, Madrid, Spain). Javier García-Pérez provided valuable advice for maraviroc and TAK-779. We thank Virginia Sacristán and Laura Jiménez-Tormo for excellent technical assistance.
REFERENCES
- 1.Wei DG, Chiang V, Fyne E, Balakrishnan M, Barnes T, Graupe M, Hesselgesser J, Irrinki A, Murry JP, Stepan G, Stray KM, Tsai A, Yu H, Spindler J, Kearney M, Spina CA, McMahon D, Lalezari J, Sloan D, Mellors J, Geleziunas R, Cihlar T. 2014. Histone deacetylase inhibitor romidepsin induces HIV expression in CD4 T cells from patients on suppressive antiretroviral therapy at concentrations achieved by clinical dosing. PLoS Pathog 10:e1004071. doi: 10.1371/journal.ppat.1004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rasmussen TA, Søgaard OS, Brinkmann C, Wightman F, Lewin SR, Melchjorsen J, Dinarello C, Østergaard L, Tolstrup M. 2013. Comparison of HDAC inhibitors in clinical development. Hum Vaccin Immunother 9:993–1001. doi: 10.4161/hv.23800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wightman F, Lu HK, Solomon AE, Saleh S, Harman AN, Cunningham AL, Gray L, Churchill M, Cameron PU, Dear AE, Lewin SR. 2013. Entinostat is a histone deacetylase inhibitor selective for class 1 histone deacetylases and activates HIV production from latently infected primary T cells. AIDS 27:2853–2862. doi: 10.1097/QAD.0000000000000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rasmussen TA, Tolstrup M, Brinkmann CR, Olesen R, Erikstrup C, Solomon A, Winckelmann A, Palmer S, Dinarello C, Buzon M, Lichterfeld M, Lewin SR, Østergaard L, Søgaard OS. 2014. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV 1:e13–e21. doi: 10.1016/S2352-3018(14)70014-1. [DOI] [PubMed] [Google Scholar]
- 5.Elliott JH, Wightman F, Solomon A, Ghneim K, Ahlers J, Cameron MJ, Smith MZ, Spelman T, McMahon J, Velayudham P, Brown G, Roney J, Watson J, Prince MH, Hoy JF, Chomont N, Fromentin R, Procopio FA, Zeidan J, Palmer S, Odevall L, Johnstone RW, Martin BP, Sinclair E, Deeks SG, Hazuda DJ, Cameron PU, Sékaly R-P, Lewin SR. 2014. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog 10:e1004473. doi: 10.1371/journal.ppat.1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Archin NM, Bateson R, Tripathy MK, Crooks AM, Yang K-H, Dahl NP, Kearney MF, Anderson EM, Coffin JM, Strain MC, Richman DD, Robertson KR, Kashuba AD, Bosch RJ, Hazuda DJ, Kuruc JD, Eron JJ, Margolis DM. 2014. HIV-1 expression within resting CD4+ T cells after multiple doses of vorinostat. J Infect Dis 210:728–735. doi: 10.1093/infdis/jiu155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM. 2012. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF. 2014. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat Med 20:425–429. doi: 10.1038/nm.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laird GM, Bullen CK, Rosenbloom DIS, Martin AR, Hill AL, Durand CM, Siliciano JD, Siliciano RF. 2015. Ex vivo analysis identifies effective HIV-1 latency-reversing drug combinations. J Clin Invest 125:1901–1912. doi: 10.1172/JCI80142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, Mori J, Rickett G, Smith-Burchnell C, Napier C, Webster R, Armour D, Price D, Stammen B, Wood A, Perros M. 2005. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob Agents Chemother 49:4721–4732. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper DA, Heera J, Goodrich J, Tawadrous M, Saag M, Dejesus E, Clumeck N, Walmsley S, Ting N, Coakley E, Reeves JD, Reyes-Teran G, Westby M, Van Der Ryst E, Ive P, Mohapi L, Mingrone H, Horban A, Hackman F, Sullivan J, Mayer H. 2010. Maraviroc versus efavirenz, both in combination with zidovudine-lamivudine, for the treatment of antiretroviral-naive subjects with CCR5-tropic HIV-1 infection. J Infect Dis 201:803–813. doi: 10.1086/650697. [DOI] [PubMed] [Google Scholar]
- 12.Gulick RM, Lalezari J, Goodrich J, Clumeck N, DeJesus E, Horban A, Nadler J, Clotet B, Karlsson A, Wohlfeiler M, Montana JB, McHale M, Sullivan J, Ridgway C, Felstead S, Dunne MW, van der Ryst E, Mayer H, MOTIVATE Study Teams . 2008. Maraviroc for previously treated patients with R5 HIV-1 infection. N Engl J Med 359:1429–1441. doi: 10.1056/NEJMoa0803152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutiérrez C, Díaz L, Vallejo A, Hernández-Novoa B, Abad M, Madrid N, Dahl V, Rubio R, Moreno AM, Dronda F, Casado JL, Navas E, Pérez-Elías MJ, Zamora J, Palmer S, Muñoz E, Muñoz-Fernández MÁ, Moreno S. 2011. Intensification of antiretroviral therapy with a CCR5 antagonist in patients with chronic HIV-1 infection: effect on T cells latently infected. PLoS One 6:e27864. doi: 10.1371/journal.pone.0027864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutiérrez C, Hernández-Novoa B, Vallejo A, Serrano-Villar S, Abad-Fernández M, Madrid N, Díaz L, Moreno A, Dronda F, Zamora J, Muñoz-Fernández MA, Moreno S. 2013. Dynamics of the HIV-1 latent reservoir after discontinuation of the intensification of antiretroviral treatment: results of two clinical trials. AIDS 27:2081–2088. doi: 10.1097/QAD.0b013e328361d0e1. [DOI] [PubMed] [Google Scholar]
- 15.Elliott JH, McMahon JH, Chang CC, Lee SA, Hartogensis W, Bumpus N, Savic R, Roney J, Hoh R, Solomon A, Piatak M, Gorelick RJ, Lifson J, Bacchetti P, Deeks SG, Lewin SR. 2015. Short-term administration of disulfiram for reversal of latent HIV infection: a phase 2 dose-escalation study. Lancet HIV 2:e520-9. doi: 10.1016/S2352-3018(15)00226-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cicala C, Arthos J, Martinelli E, Censoplano N, Cruz CC, Chung E, Selig SM, Van Ryk D, Yang J, Jagannatha S, Chun TW, Ren P, Lempicki RA, Fauci AS. 2006. R5 and X4 HIV envelopes induce distinct gene expression profiles in primary peripheral blood mononuclear cells. Proc Natl Acad Sci U S A 103:3746–3751. doi: 10.1073/pnas.0511237103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alcamí J, Laín de Lera T, Folgueira L, Pedraza MA, Jacqué JM, Bachelerie F, Noriega AR, Hay RT, Harrich D, Gaynor RB, et al. 1995. Absolute dependence on kappa B responsive elements for initiation and Tat-mediated amplification of HIV transcription in blood CD4 T lymphocytes. EMBO J 14:1552–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan JK, Greene WC. 2012. Dynamic roles for NF-κB in HTLV-I and HIV-1 retroviral pathogenesis. Immunol Rev 246:286–310. doi: 10.1111/j.1600-065X.2012.01094.x. [DOI] [PubMed] [Google Scholar]
- 19.Wolschendorf F, Bosque A, Shishido T, Duverger A, Jones J, Planelles V, Kutsch O. 2012. Kinase control prevents HIV-1 reactivation in spite of high levels of induced NF-κB activity. J Virol 86:4548–4558. doi: 10.1128/JVI.06726-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosque A, Planelles V. 2009. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood 113:58–65. doi: 10.1182/blood-2008-07-168393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinoshita S, Su L, Amano M, Timmerman LA, Kaneshima H, Nolan GP. 1997. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity 6:235–244. doi: 10.1016/S1074-7613(00)80326-X. [DOI] [PubMed] [Google Scholar]
- 22.Yang X, Chen Y, Gabuzda D. 1999. ERK MAP kinase links cytokine signals to activation of latent HIV-1 infection by stimulating a cooperative interaction of AP-1 and NF-kappaB. J Biol Chem 274:27981–27988. doi: 10.1074/jbc.274.39.27981. [DOI] [PubMed] [Google Scholar]
- 23.Foletta VC, Segal DH, Cohen DR. 1998. Transcriptional regulation in the immune system: all roads lead to AP-1. J Leukoc Biol 63:139–152. doi: 10.1002/jlb.63.2.139. [DOI] [PubMed] [Google Scholar]
- 24.Hughes CC, Pober JS. 1993. Costimulation of peripheral blood T cell activation by human endothelial cells. Enhanced IL-2 transcription correlates with increased c-fos synthesis and increased Fos content of AP-1. J Immunol 150:3148–3160. [PubMed] [Google Scholar]
- 25.Lupino E, Ramondetti C, Piccinini M. 2012. IκB kinase β is required for activation of NF-κB and AP-1 in CD3/CD28-stimulated primary CD4(+) T cells. J Immunol 188:2545–2555. doi: 10.4049/jimmunol.1102938. [DOI] [PubMed] [Google Scholar]
- 26.Padhan K, Varma R. 2010. Immunological synapse: a multi-protein signalling cellular apparatus for controlling gene expression. Immunology 129:322–328. doi: 10.1111/j.1365-2567.2009.03241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rozek D, Pfeifer GP. 1993. In vivo protein-DNA interactions at the c-jun promoter: preformed complexes mediate the UV response. Mol Cell Biol 13:5490–5499. http://mcb.asm.org/content/13/9/5490.short. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Dam H, Duyndam M, Rottier R, Bosch A, de Vries-Smits L, Herrlich P, Zantema A, Angel P, van der Eb AJ. 1993. Heterodimer formation of cJun and ATF-2 is responsible for induction of c-jun by the 243 amino acid adenovirus E1A protein. EMBO J 12:479–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cicala C, Arthos J, Censoplano N, Cruz C, Chung E, Martinelli E, Lempicki RA, Natarajan V, VanRyk D, Daucher M, Fauci AS. 2006. HIV-1 gp120 induces NFAT nuclear translocation in resting CD4+ T-cells. Virology 345:105–114. doi: 10.1016/j.virol.2005.09.052. [DOI] [PubMed] [Google Scholar]
- 30.Passaes CP, Sáez-Cirión A. 2014. HIV cure research: advances and prospects. Virology 454–455:340–352. [DOI] [PubMed] [Google Scholar]
- 31.Hunt PW, Shulman NS, Hayes TL, Dahl V, Somsouk M, Funderburg NT, McLaughlin B, Landay AL, Adeyemi O, Gilman LE, Clagett B, Rodriguez B, Martin JN, Schacker TW, Shacklett BL, Palmer S, Lederman MM, Deeks SG. 2013. The immunologic effects of maraviroc intensification in treated HIV-infected individuals with incomplete CD4+ T-cell recovery: a randomized trial. Blood 121:4635–4646. doi: 10.1182/blood-2012-06-436345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minami R, Takahama S, Kaku Y, Yamamoto M. 2017. Addition of maraviroc to antiretroviral therapy decreased interferon-γ mRNA in the CD4+ T cells of patients with suboptimal CD4+ T-cell recovery. J Infect Chemother 23:29–34. doi: 10.1016/j.jiac.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Barroso H, Taveira N. 2005. Evidence for negative selective pressure in HIV-2 evolution in vivo. Infect Genet Evol 5:239–246. doi: 10.1016/j.meegid.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 34.Lengauer T, Sander O, Sierra S, Thielen A, Kaiser R. 2007. Bioinformatics prediction of HIV coreceptor usage. Nat Biotechnol 25:1407–1410. doi: 10.1038/nbt1371. [DOI] [PubMed] [Google Scholar]
- 35.Vandekerckhove LPR, Wensing AMJ, Kaiser R, Brun-Vézinet F, Clotet B, De Luca A, Dressler S, Garcia F, Geretti AM, Klimkait T, Korn K, Masquelier B, Perno CF, Schapiro JM, Soriano V, Sönnerborg A, Vandamme A-M, Verhofstede C, Walter H, Zazzi M, Boucher CAB, European Consensus Group on Clinical Management of Tropism Testing . 2011. European guidelines on the clinical management of HIV-1 tropism testing. Lancet Infect Dis 11:394–407. doi: 10.1016/S1473-3099(10)70319-4. [DOI] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Lewin SR, Vesanen M, Kostrikis L, Hurley A, Duran M, Zhang L, Ho DD, Markowitz M. 1999. Use of real-time PCR and molecular beacons to detect virus replication in human immunodeficiency virus type 1-infected individuals on prolonged effective antiretroviral therapy. J Virol 73:6099–6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cicala C, Arthos J, Selig SM, Dennis G Jr, Hosack DA, Van Ryk D, Spangler ML, Steenbeke TD, Khazanie P, Gupta N, Yang J, Daucher M, Lempicki RA, Fauci AS. 2002. HIV envelope induces a cascade of cell signals in non-proliferating target cells that favor virus replication. Proc Natl Acad Sci U S A 99:9380–9385. doi: 10.1073/pnas.142287999. [DOI] [PMC free article] [PubMed] [Google Scholar]