ABSTRACT
We followed the dynamics of capsid amino acid replacement among 403 Nigerian outbreak isolates of type 2 circulating vaccine-derived poliovirus (cVDPV2) from 2005 through 2011. Four different functional domains were analyzed: (i) neutralizing antigenic (NAg) sites, (ii) residues binding the poliovirus receptor (PVR), (iii) VP1 residues 1 to 32, and (iv) the capsid structural core. Amino acid replacements mapped to 37 of 43 positions across all 4 NAg sites; the most variable and polymorphic residues were in NAg sites 2 and 3b. The most divergent of the 120 NAg variants had no more than 5 replacements in all NAg sites and were still neutralized at titers similar to those of Sabin 2. PVR-binding residues were less variable (25 different variants; 0 to 2 replacements per isolate; 30/44 invariant positions), with the most variable residues also forming parts of NAg sites 2 and 3a. Residues 1 to 32 of VP1 were highly variable (133 different variants; 0 to 6 replacements per isolate; 5/32 invariant positions), with residues 1 to 18 predicted to form a well-conserved amphipathic helix. Replacement events were dated by mapping them onto the branches of time-scaled phylogenies. Rates of amino acid replacement varied widely across positions and followed no simple substitution model. Replacements in the structural core were the most conservative and were fixed at an overall rate ∼20-fold lower than the rates for the NAg sites and VP1 1 to 32 and ∼5-fold lower than the rate for the PVR-binding sites. Only VP1 143-Ile, a non-NAg site surface residue and known attenuation site, appeared to be under strong negative selection.
IMPORTANCE The high rate of poliovirus evolution is offset by strong selection against amino acid replacement at most positions of the capsid. Consequently, poliovirus vaccines developed from strains isolated decades ago have been used worldwide to bring wild polioviruses almost to extinction. The apparent antigenic stability of poliovirus obscures a dynamic of continuous change within the neutralizing antigenic (NAg) sites. During 7 years of a large outbreak in Nigeria, the circulating type 2 vaccine-derived polioviruses generated 120 different NAg site variants via multiple independent pathways. Nonetheless, overall antigenic evolution was constrained, as no isolate had fixed more than 5 amino acid differences from the Sabin 2 NAg sites, and the most divergent isolates were efficiently neutralized by human immune sera. Evolution elsewhere in the capsid was also constrained. Amino acids binding the poliovirus receptor were strongly conserved, and extensive variation in the VP1 amino terminus still conserved a predicted amphipathic helix.
KEYWORDS: poliovirus, vaccine-derived poliovirus, antigenic evolution, vaccines
INTRODUCTION
The World Health Organization Global Polio Eradication Initiative has brought wild polioviruses to the threshold of eradication (1). Wild poliovirus type 2 (WPV2) was last detected in 1999 (2) and WPV3 in 2012 (3), and since August 2014, WPV1 has been detected only in parts of Pakistan, Afghanistan, and Nigeria (4–7) (for updates, see http://www.polioeradication.org/polio-today/polio-now/this-week). Polioviruses (PVs) are defined by two distinct properties residing with the capsid protein: (i) the neutralizing antigenic (NAg) sites distinguishing the three PV serotypes (8, 9) and (ii) the capacity to enter human and simian cells via the poliovirus receptor (PVR), CD155 (10, 11). Genomic regions outside the capsid sequences, subject to frequent exchange during transmission by recombination with other human enteroviruses (12–14), are not PV specific and will persist in nature posteradication.
Immune protection against paralytic poliomyelitis is type specific (8). Although antigenic variation within PV serotypes has long been recognized (15–19), it is normally limited in extent, possibly because the steric requirements for efficient docking to the PVR impose structural constraints on the virion surface (10, 11). An important public health consequence is that antigenic variation is not an impediment to polio eradication (4), and immunization with vaccines (either inactivated poliovirus vaccine [IPV] or oral poliovirus vaccine [OPV]) developed from PV strains dating back 60 to 75 years (20, 21) confers protective immunity to all WPVs (4). Indeed, until the advent of molecular methods, comparison of antigenic properties was the mainstay for intratypic differentiation between OPV-related isolates and WPVs (22–24).
The NAg sites of each PV serotype have been defined by mapping amino acid replacements conferring resistance to neutralization by murine monoclonal antibodies (MAbs) onto the X-ray crystallographic structure of the PV capsid (9, 25–29). Four NAg sites (NAg1, NAg2, NAg3a, and NAg3b [alternatively designated NAg4]) have been described (26, 30), most comprehensively for types 1 (9, 26, 31, 32) and 3 (9, 27). The antigenic structure of PV2 has not been as extensively described but is similar overall to those of the other types (28). X-ray crystallographic studies have shown that the greatest structural differences in the capsids across PV serotypes reside in the exposed loops that form the NAg sites (26–29). Amino acid variability within PV serotypes is highest in the NAg sites and in the N terminus of VP1 (33–37).
Most of the sequence comparisons with PV2s have been with genetically divergent vaccine-derived polioviruses (VDPVs) (38–40), defined as having >1% nucleotide sequence divergence (types 1 and 3) or >0.6% (type 2) from their parental Sabin strains in the ∼900-nucleotide (nt) region encoding the major capsid protein, VP1 (41). This definition follows from the high rate of nucleotide sequence evolution in PV (∼1% per year) (35) and the normal period of PV excretion of less than 3 months (38, 42). VDPVs are categorized as (i) circulating VDPVs (cVDPVs) from outbreaks; (ii) immunodeficiency-associated VDPVs (iVDPVs), excreted over prolonged periods by persons with primary immunodeficiencies; and (iii) ambiguous VDPVs (aVDPVs), which cannot be definitively assigned to the other two distinct and well-defined categories (39, 41).
Starting in 2005, a large poliomyelitis outbreak associated with type 2 cVDPV (cVDPV2) occurred in northern Nigeria, where immunization coverage with trivalent OPV (tOPV) was low (41, 43, 44). Phylogenetic analysis of the capsid region sequences of 403 case isolates from 2005 to 2011 resolved the outbreak into 23 independent VDPV2 emergences (named 2004-1 to 2010-2 according to the year and estimated order of emergence); at least 7 of the emergences established circulating lineage groups (representing multiple chains of transmission) (13). The largest lineage group (2005-8; 361 isolates through 2011) is estimated to have circulated for nearly a decade (13, 41).
Although PVs have been the subject of intensive study for over a century (45, 46), quantitative information on the dynamics of PV capsid protein evolution is limited, and previous investigations have focused primarily on WPV1 (35), WPV3 (47), and iVDPVs (34, 40, 48–50) rather than cVDPVs (14, 37). Sensitive PV surveillance conducted during the cVDPV2 outbreak in Nigeria provided an opportunity for high-resolution studies of the evolution of the outbreak virus. Similar detailed evolutionary studies had not been possible for WPV2 because large WPV2 outbreaks were rare (51), and WPV2 circulation had stopped in many settings prior to the launch of regional (52) and global (1) polio eradication activities and the accompanying implementation of sensitive global PV surveillance. However, cVDPVs closely resemble WPVs in their biological properties (13, 39) and offer the additional advantage that the evolutionary pathways for each emergence can be independently traced back to the well-defined parental Sabin strains (53–55). In this report, we describe the multiple pathways of antigenic divergence among Nigerian cVDPV2 lineages, compare the rates and patterns of amino acid replacement in the NAg sites with those of other capsid domains, and show that all cVDPV2 antigenic variants were efficiently neutralized by human immune sera.
RESULTS
Distribution of nucleotide substitutions and amino acid replacements in the capsid region.
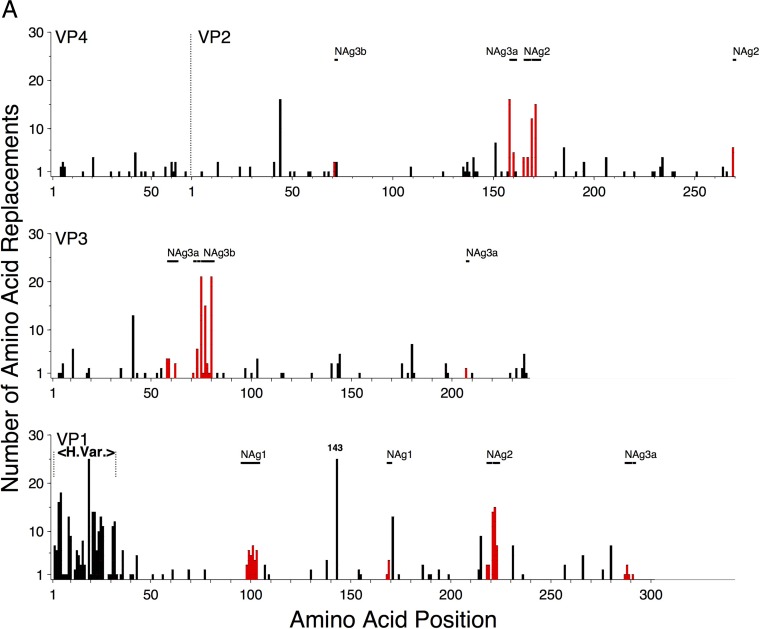
The estimated 8,431 total nucleotide substitutions (total substitutions [KT] = synonymous transitions [AS] + synonymous transversions [BS] + nonsynonymous transitions [AA] + nonsynonymous transversions [BA] [see Fig. S1 in the supplemental material]) across all 23 2005-to-2011 Nigerian VDPV2 emergences and lineage groups were nearly uniformly distributed along the capsid region. In contrast, the estimated 807 amino acid replacements (AA + BA) in the capsid protein mapped to 188 sites distributed in a more irregular pattern as short blocks of high variability (Fig. 1; see Fig. S1 and S2 in the supplemental material).
FIG 1.
(A) Numbers of independent amino acid replacements into each position of the capsid regions of all 403 Nigerian VDPV2 variants isolated from 2005 to 2011. VP4, shown joined with VP2 (as in its uncleaved precursor, VP0), is structurally homologous to the N termini of VP3 and VP1. Amino acid replacements in NAg sites are shown in red. H.Var, the hypervariable N-terminal residues at VP1 positions 1 to 32 (1001 to 1032). (B) Detail of Fig. S3B in the supplemental material mapping amino acid replacements in NAg sites onto the branch structure of the MCC tree (13) for lineage group 2005-8. Amino acid residues are indicated by four-digit numbers; the first digit is the name of the viral protein (4, VP4; 2, VP2; 3, VP3; 1, VP1), followed consecutively from residue 1 of each protein.
Mapping amino acid replacements onto the branch structures of MCC trees.
NAg site replacements were mapped onto previously published maximum clade credibility (MCC) trees (13) to visualize the number, temporal order, and approximate dates of replacements along lineages (see Fig. S3 to S5 in the supplemental material). Three sets of trees were constructed showing replacements in different capsid functional domains (see below). Our approach rests on two basic assumptions: (i) the branch structures of MCC trees accurately reflect the temporal patterns of divergence of cVDPV transmission chains and (ii) the number of unobserved, superimposed, nonsynonymous substitutions was negligible. The first assumption is supported by the concordance of our Bayesian MCC assignments with those obtained by codon analysis based on maximum likelihood (CODEML) (56) and, most critically, by the finding that amino acid replacements fixed in isolates representing progenitor viruses were present in the progeny until any subsequent superimposed replacement(s). In a small number of instances (<20, representing <3% of replacements) the first assumption may not hold, because nodes were closely spaced and the highest posterior density (HPD) intervals overlapped (for example, H3077R in KTS09-13, KTS09-26, and NAS09-02; T2270A in KTS09-08 and KNS09-36) (see Fig. S3B in the supplemental material), and single replacements were probably counted more than once. In such instances, we accepted the trees as close approximations of the “true” trees and made no attempt to correct for the small potential overcount.
The second assumption could not be tested rigorously. However, of the estimated 807 total replacements, 47 (5.8%) were observed to be superimposed, 26 (3.2%) of which restored the original Sabin 2 residue (see Fig. S3 to S5 in the supplemental material). The relatively low frequencies of the observed superimposed replacements and the long time intervals between replacement events at the same site suggested that few superimposed replacements were missing from the annotated trees. Therefore, we consider our assumptions to be generally valid and the effects of the infrequent violations to be small.
Differential evolution of VDPV2 capsid functional domains.
The X-ray crystallographic structure of the native poliovirion was solved 3 decades ago (25), and capsid functional domains have been defined in three dimensions. The capsid proteins VP1, VP2, and VP3 each contain a structurally conserved core (∼329 residues) of eight-stranded β-barrels and two α-helices (25, 27, 29). These regular secondary-structural domains of the cores are joined by loops and flanked by terminal extensions (∼273 residues, excluding the N-terminal extensions) (see Fig. S2 in the supplemental material) (25, 29). VP4 and the N-terminal extensions of VP1, VP2, and VP3 (∼277 residues in total) intertwine in a complex network, forming the inner surface of the capsid (25). To evaluate the selective forces shaping the observed patterns of variability, we grouped capsid protein residues into four functional domains: (i) the NAg sites, (ii) the residues binding the PVR, (iii) the 32 hypervariable residues (1001 to 1032) at the N terminus of VP1, and (iv) the structural core. The more conserved N-terminal residues of VP2 and VP3 and all of VP4 were excluded from the analyses below because their structures are not well defined (25).
(i) NAg sites.
The NAg sites for PV2 have been previously defined by mapping MAb escape mutants (28, 57, 58) to the PV X-ray crystallographic structure (25, 27, 29), complemented by analysis of natural type 2 vaccine variants (34, 36, 40, 59) and by analogy with types 1 and 3 (9, 26, 60). Considering these various observations, we assigned PV2 NAg sites to the following amino acid positions: NAg1, 1095 to 1103, 1168, and 1169; NAg2, 1218, 1219, 1221 to 1223, 2161, 2166 to 2168, 2170, 2172, and 2270; NAg3a, 1287 to 1289, 1291, 2159, 3058 to 3062, 3071, 3073, and 3207; NAg3b, 2072 and 3075 to 3080. Sites NAg3a and NAg3b (which span adjacent pentamers), in structural proximity, have been antigenically linked by cross-neutralizing MAbs in types 1 (60) and 2 (28). Similarly, structurally adjacent parts of NAg2 and NAg3a have been antigenically linked in type 2 (28, 40, 61).
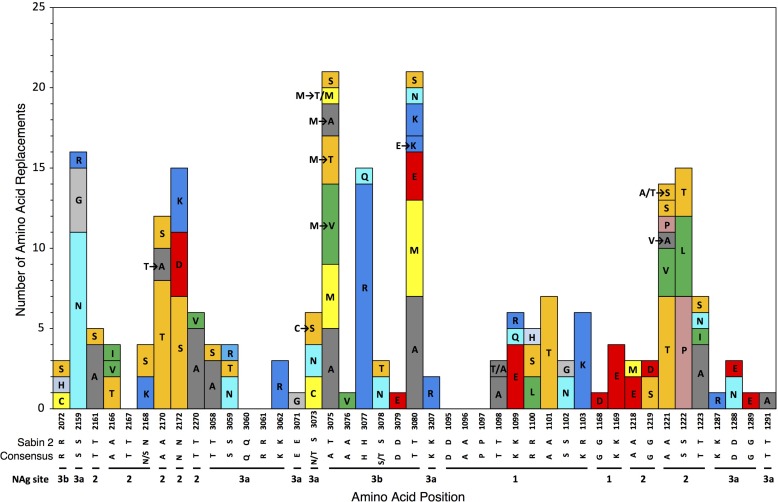
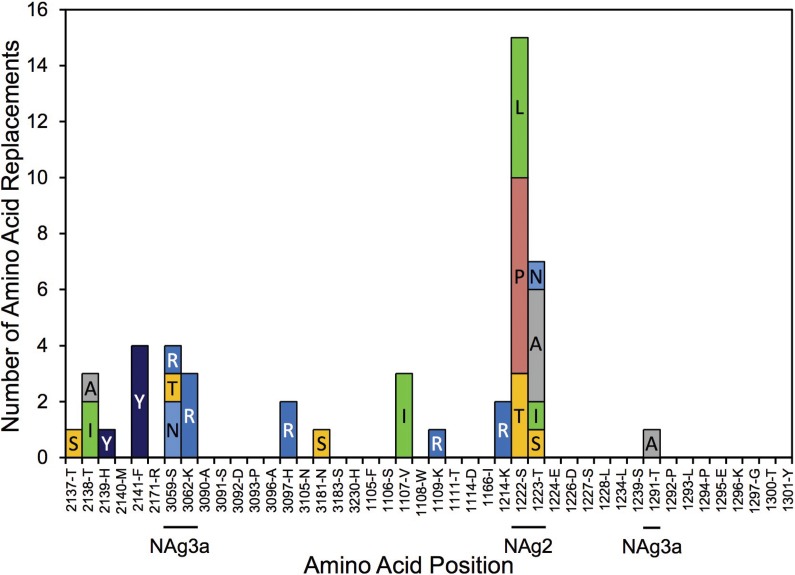
Although the NAg sites as defined here constitute only 4.9% (43/879) of the total capsid residues, 30% (231/778) of the observed amino acid replacements (excluding VP1 143) mapped to them (Fig. 1; see Fig. S2 in the supplemental material). Most (37 of 43) positions within NAg sites were variable (Fig. 2), with 6.24 replacements per variable residue (Table 1). The NAg sites accommodated a wider range (mean, 2.19) of amino acid replacements per variable position than the more conserved structural core (see below). Moreover, the NAg sites accommodated relatively high structural diversity among amino acid residues. In our comparisons, we used two alternative indices of amino acid exchangeability: (i) the Miyata distance (62), where lower values indicate higher rates of exchangeability (implying greater structural and functional similarity), and (ii) the LG amino acid replacement matrix (63), where higher values indicate higher rates of exchangeability. The weighted average Miyata distance per variable NAg site was 1.33 compared with 1.10 for the structural core, and the corresponding weighted LG values were 2.75 (NAg sites) and 5.63 (structural core) (Table 1). The overall rate of amino acid exchange in the NAg sites was 1.86 × 10−2 replacements/site/lineage/year, 17-fold higher than in the structural core and exceeded only by the rates in the hypervariable N terminus of VP1 (Table 1). Despite the potential for variability within the NAg sites, most isolates (98%; range, 0% to 90%) retained the parental Sabin 2 residue at each NAg site position (apart from position 3075), because 89% of the replacements mapped to peripheral branches, indicating that the viruses mapping to the branch tips had no observed progeny. The notable exception was T3075M, which was fixed early in lineage groups 2005-8, 2005-10, and 2007-10 and remained fixed in most progeny virus despite the occurrence of 11 subsequent replacements, 10 of which mapped to peripheral branches (Fig. 2; see Fig. S3B to D in the supplemental material).
FIG 2.
Numbers of independent amino acid replacements in NAg sites of all 403 Nigerian VDPV2 variants isolated from 2005 to 2011. The positions are ordered starting from the N terminus of the capsid, aligned with the corresponding residues of Sabin 2 and the consensus among 12 isolates representing distinct WPV2 genotypes. Amino acids are colored by physicochemical properties according to the Amino/Shapely scheme (A [nonpolar, small aliphatic], dark gray; C and M [nonpolar, sulfur-containing], yellow; D and E [charged, acidic], bright red; F and Y [polar and nonpolar aromatic], mid-blue; G [nonpolar, no side chain], light gray; H [charged, imidazole], pale blue; I, L, and V [nonpolar, aliphatic], green; K and R [charged, basic], blue; N and Q [polar, amide], cyan; P [nonpolar, cyclic imino], flesh; S and T [polar, hydroxyl], orange; W [nonpolar, aromatic, indole], pink) (http://acces.ens-lyon.fr/biotic/rastop/help/colour.htm#shapelycolours).
TABLE 1.
Amino acid replacements by capsid functional domain
| Capsid region | Proportion of variable residue positions | No. of replacements/variable residue positiona | No. of alternative replacement residues/variable residue position | Weighted avg Miyata distance/variable residueb | Weighted avg LG distance/ variable residuec |
Replacement rate (no. of replacements/ site/lineage/yr) (10−3) |
Relative replacement rate |
|---|---|---|---|---|---|---|---|
| Total capsid | 0.217 | 4.07 | 1.60 | 1.32 | 3.86 | 3.06 | 2.88 |
| Capsid without N termini | 0.200 | 3.82 | 1.51 | 1.34 | 3.86 | 2.64 | 2.48 |
| Structural core | 0.125 | 2.46 | 1.10 | 1.06 | 5.63 | 1.06 | 1.00 |
| β-Strands | 0.127 | 1.97 | 1.13 | 1.07 | 6.15 | 0.95 | 0.89 |
| α-Helices | 0.115 | 4.11 | 1.00 | 1.05 | 4.63 | 1.64 | 1.54 |
| Loops and C termini | 0.271 | 4.58 | 1.74 | 1.42 | 3.33 | 4.29 | 4.04 |
| NAg sites | 0.860 | 6.24 | 2.19 | 1.33 | 2.75 | 18.57 | 17.47 |
| PVR-binding sites | 0.318 | 3.21 | 1.57 | 1.35 | 3.56 | 4.76 | 4.47 |
| PVR-binding sites without NAg sites | 0.231 | 1.67 | 1.11 | 1.36 | 5.07 | 1.81 | 1.70 |
| VP1 1–32 | 0.844 | 8.11 | 2.59 | 1.36 | 3.67 | 23.66 | 22.25 |
Mixed-base positions were counted as a single replacement.
Theoretical range, 0.47 to 5.38 (mean, 2.256); observed range, 0.47 to 2.69; a low value indicates frequent replacement.
Theoretical range, 0.008705 to 10.64911 (mean, 1.022213); observed range, 0.037967 to 10.64911; a high value indicates frequent replacement.
(ii) PVR-binding sites.
Capsid residues interacting directly with the PVR have been mapped by X-ray crystallography and cryoelectron microscopic reconstruction of virus-receptor complexes (10, 11, 64). A total of 25 different PVR-binding site sequence variants (1 to 2 differences from Sabin 2) were observed (see Fig. S6 and S7 in the supplemental material). Most (30 of 44) PVR-binding site positions were invariant, and the most variable residues also formed parts of NAg sites 2 and 3a (Fig. 3). The K1109R replacement mapped to the same position as a Sabin 2 mutant selected for resistance to neutralization by a human MAb that complexes with the PVR-binding site (58). It has been suggested that the steric requirements for efficient binding to the PVR may limit the evolution of the PV surface and restrict PV to three serotypes (11). When the NAg site residues are excluded, the PVR-binding site residues were nearly as well conserved as those of the structural core, with infrequent exchanges among a limited set of similar amino acids (Table 1).
FIG 3.
Frequencies and identities of amino acid replacements in residues binding the PVR aligned to the Sabin 2 residues.
(iii) Hypervariable residues 1 to 32 of VP1.
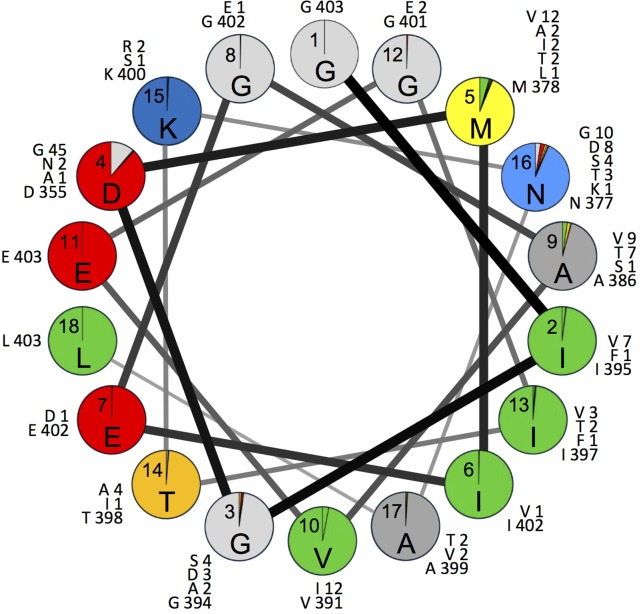
As previously observed with wild PV1 (35), 28% (219/778) of all capsid amino acid replacements mapped to the hypervariable (53) N-terminal residues 1001 to 1032, which constitute only 3.6% of the total capsid residues. A total of 133 different sequence variants (1 to 6 differences from Sabin 2) were observed for residues 1001 to 1032 (see Fig. S8 and S9 in the supplemental material). Included among the 5 invariant residues was G1001 of the absolutely conserved Q/G dipeptide forming the VP3/VP1 cleavage site (65) (see Fig. S2 in the supplemental material); the remainder had an average of 8.11 replacement events, with 2.59 alternative replacement residues (Table 1). The amino acid replacement rate was ∼22-fold higher than in the structural core. During the early stages of viral infection, the N-terminal VP1 residues are externalized to form a predicted amphipathic helix (66). Despite the high variability and rapid evolution of this domain (Table 1), the amphipathic properties of the first 5 turns (residues 1001 to 1018) of the predicted helical wheel were conserved (Fig. 4). Like the NAg sites, most replacements mapped to peripheral branches. The main exception was a P1021L replacement that occurred early in the evolution of lineage group 2005-8 and remained fixed in most progeny, but with 7 secondary replacements (including back-replacements) mapping primarily to peripheral branches (see Fig. S4B in the supplemental material).
FIG 4.
Proportions of isolates with Sabin 2 amino acid residues (shown inside each circle) or replacement residues (colored sectors) in the helix wheel of hypervariable positions 1001 to 1018. The numbers correspond to VP1 residue positions; the letters indicate the most frequently detected residue at each position. The numbers outside the circles are the numbers by position of each residue among the 403 cVDPV2 isolates analyzed.
(iv) Structural core.
The structural core constitutes 37% (329/879) of capsid residues, but only 12% of its amino acid positions were variable, with a mean of 2.46 replacements per variable position (Table 1). Most variable positions had only 1 alternative residue (see Fig. S2 in the supplemental material), and the replacements tended to be structurally conservative (for example, 28% of replacements were I↔V exchanges). The overall rate of amino acid exchange in the structural core was 1.06 × 10−3 replacements/site/lineage/year, with a <2-fold difference between the evolution rates for β-strands (76% of the structural core) and α-helices (24% of the structural core) (Table 1).
(v) VP1 143.
Excluded from the above-mentioned analyses was an important capsid determinant of the attenuated and temperature-sensitive phenotypes of Sabin 2 (55, 67), I1143. This residue in the D-E loop (see Fig. S2 in the supplemental material) is exposed on the virion surface but is not known to contribute to the NAg sites. Residue I1143, which is unstable to replication in the human intestine (67), had reverted in all Nigerian VDPV2 isolates, and the reversions remained fixed in all but 2 of the progeny (13) (see Fig. S5 in the supplemental material).
Differential evolution of amino acid positions within functional capsid domains.
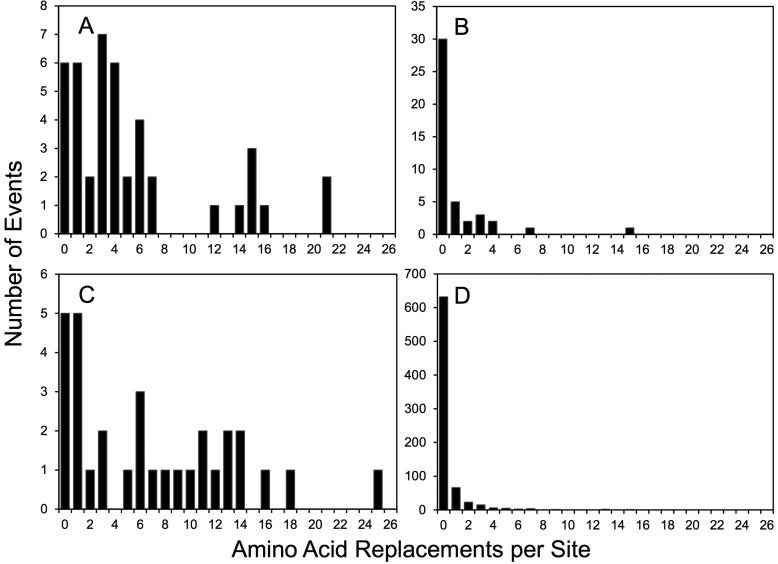
Although the mean amino acid replacement rates in Table 1 provide an approximate guide to the differential evolution of the four functional domains, they do not reflect the wide differences in replacement rates at specific positions. Unlike synonymous substitutions in the capsid region, which could be modeled as a Poisson process (35), nonsynonymous substitutions did not follow a simple substitution model (Fig. 5). For example, among the 43 amino acid positions forming the NAg sites, 6 were invariant (1095 to 1097, 2167, 3060, and 3061), while 2 (3075 and 3080) had 21 independent replacement events each (Fig. 2 and 5A). In the PVR-binding sites, 68% of positions had no replacements, but 2 positions (1222 and 1223; both part of NAg2) had 15 and 7 replacements, respectively (Fig. 3 and 5B). Similarly, among positions 1001 to 1032, 5 were invariant and 1 (1019) had 25 replacements (Fig. 5C; see Fig. S2 in the supplemental material). Some positions (e.g., 1004 and 1021) had frequent fluctuations between 2 alternative amino acids (see Fig. S2 and S5 in the supplemental material). Of the remaining 804 amino acid positions (excluding 1143), 671 (84%) were invariant, 67 (8.4%) had 1 replacement, and 22 (2.8%) had 2 replacements. However, 3 positions (2045 and 3041 [both internalized N-terminal extensions] and 1171 [a partially exposed E-F α-helix adjacent to part of NAg1]) were outliers, with 13 to 15 independent replacements (Fig. 5D; see Fig. S2 in the supplemental material).
FIG 5.
Numbers of independent amino acid replacements per site by functional domain for NAg sites (A); PVR-binding sites (B); residues 1001 to 1032 (C); and the remainder of the capsid, including the structural core (D).
Intensity of selection at individual capsid amino acid positions.
The high conservation of most capsid amino acid positions during widespread circulation is indicative of strong selection against replacements at those sites. One measure of the intensity of selection at a site is the ratio (ω) of nonsynonymous to synonymous substitutions (KA/KS or dN/dS) (35, 37, 68). When ω is >1, the site is inferred to be under positive selection; when ω is equal to 1, the site is inferred to be under no selection (neutral change); and when ω is <1, the site is inferred to be under negative (purifying) selection. Global ω values differed among functional domains estimated for the main emergences; ω estimates for the structural core had the lowest values (range, 0.01 to 0.02), whereas NAg sites had the highest values (range, 4.37 to 51.6). The PVR-binding sites and the hypervariable residues had similar global ω values, ranging from 0.67 to 3.84. Estimates of ω values were significantly influenced by site-to-site variation of both synonymous and nonsynonymous rates within the capsid region. Rate variation between the structural core domain and the NAg sites was slightly higher for increased nonsynonymous substitutions than for decreased synonymous substitutions. When the capsid sequences of isolates from the main lineage groups were analyzed under 6 different models of codon evolution (68), only 3 Sabin 2 positions showed evidence of positive selection, although it was not significant (3077 [NAg3b], ω = 1.47 to 1.66; 3080 [NAg3b], ω = 1.68 to 1.82; 1222 [NAg2], ω = 1.65 to 1.75), and 6 other sites (2159, 2170, 2172, 3075, 1100, and 1221; all within NAg sites) were inferred to be under no selection (ω = 0.91 to 1.13). All the other sites, including 1143 and 1001 to 1032, were initially found to be under negative (purifying) selection. However, ω values obtained over the entire period of observation could underestimate the intensity of selection if a positively selected residue was fixed early in the evolutionary pathway, as was the case for the genetically unstable Sabin 2 residue I1143 (67). All of the original Sabin 2 I1143 residues were exchanged for threonine, valine, or asparagine during the estimated time between the initiating tOPV dose and the first isolate (3 to 21 months) or earliest diverging node (2 to 6 months) of each VDPV2 emergence (13). The estimated branch-specific ω values for 1143 were initially very high (ωmean was equal to 43 across all emergences at the earliest Sabin 2 branch) but fell to <0.8 in subsequent lineages, as synonymous substitutions accumulated with no observed replacements back to I1143, and only 2 superimposed replacements (T1143A and T1143N) were fixed in 2 divergent isolates (13). Replacements at another position, 3075, were also fixed early (13) in 7 emergences (T3075M, within 2 to 21 months in 4 emergences; T3075A, within 3 to 10 months in 2 emergences; T3075S, within 21 months in 1 emergence), but the Sabin 2 T3075 residue was retained in the 16 other independent emergences. Unlike the early replacements at position 1143, which remained fixed in nearly all progeny, numerous superimposed replacements occurred at position 3075 (Fig. 2), consistent with no selection.
Multiplicity of antigenic variants among VDPV2 isolates.
The NAg sites changed continuously as the cVDPV2 spread through the Nigerian population. A total of 231 independent amino acid replacements, incorporating 86 alternative residues, mapped to the 37 variable positions in the NAg sites (Fig. 2). Only ∼8.7% (20/231) of the replacements restored the consensus residue of WPV2 isolates (Fig. 2) (34, 69), and none of the VDPV2 variants had overall NAg sequences matching the WPV2 consensus or MEF-1, the WPV2 component of conventional IPV (20). Although some recently emergent isolates from 5 independent emergences and 3 lineage groups (2004-1 [2 of 3 isolates], 2005-5 [1 of 2 isolates], and 2007-5 [2 isolates]) had apparently not accumulated NAg site replacements, no reversions back to the Sabin 2 NAg sequences were observed among the other isolates.
The 403 Nigerian VDPV2 isolates from 2005 to 2011 comprised 120 different variants (see Fig. S10 in the supplemental material) of replacement in the four NAg sites, each of which is represented on a maximum-parsimony tree (Fig. 6). Although 15 amino acid positions were highly polymorphic and variable, having 3 to 7 different alternative replacements (including back-replacements), the most divergent individual VDPV2 variants differed from Sabin 2 at no more than 5 NAg site positions (Fig. 6; see Fig. S10 in the supplemental material). The high diversity of NAg site variants and the star-like shape of the NAg site tree underscore the point that evolution of the NAg sites occurs primarily by genetic drift along multiple pathways rather than by positive selection for particular antigenic variants with increased fitness for transmission. The star-like topology of the cVDPV2 NAg site tree contrasts with the tree of the HA1 domain of influenza A virus, which has a strong central trunk with limited branching, indicative of antigenic selection (70). The trees for the PVR-binding sites (see Fig. S7 in the supplemental material) and for positions 1001 to 1032 (see Fig. S9 in the supplemental material) were also star-like.
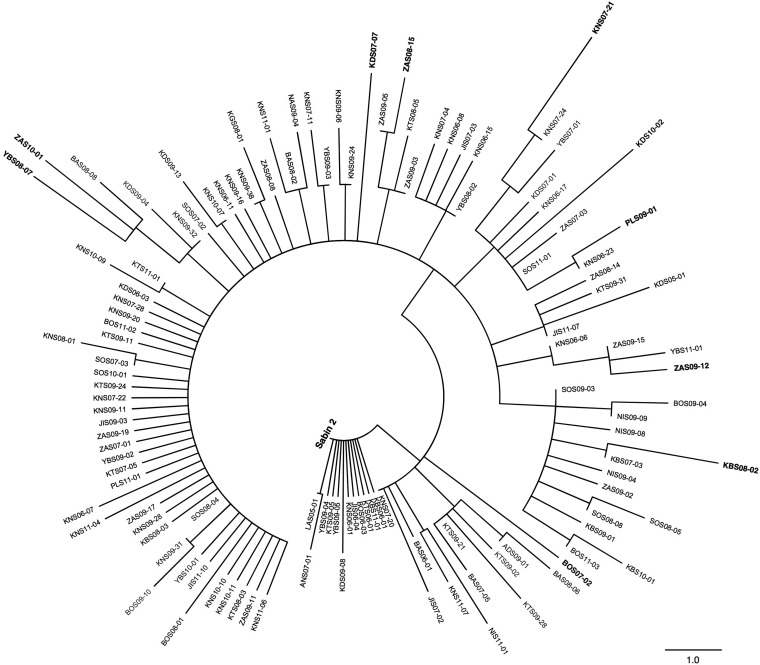
FIG 6.
Maximum-parsimony tree of amino acid sequences within all 4 NAg sites (see Fig. S1 in the supplemental material) of Sabin 2 and representatives of the 120 distinct NAg site variants among the 403 cVDPV2 isolates (see Fig. S10 in the supplemental material). Isolates used in serologic assays are shown in boldface.
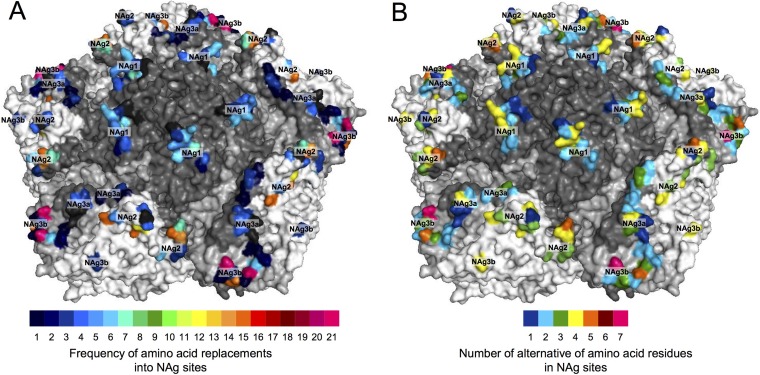
We mapped the NAg sites onto the crystal structure of the poliovirion (see Fig. S11 in the supplemental material) and color-coded individual positions in structural heat maps according to the frequency of amino acid replacement (Fig. 7A) or the number of alternative replacements (Fig. 7B). Replacements were most frequent (≥10 independent events/position) in NAg sites 2 (positions 2159, 2170, 2172, 1221, and 1222), 3a (3075), and 3b (3080) (Fig. 2 and 7A). The most polymorphic positions (≥4 alternative replacements) were in NAg sites 2 (1221 and 1223), 3a (3075), and 3b (3080) (Fig. 2 and 7B).
FIG 7.
Heat maps illustrating the frequency of NAg amino acid replacements (A) and the number of alternative amino acid residues (B) in the NAg sites of all 403 cVDPV2 isolates mapped to the X-ray crystallographic structure of the poliovirus type 2 capsid pentamer. Capsid proteins (see Fig. S7 in the supplemental material) are shaded as follows: VP1, dark gray; VP2, white; VP3, light gray.
Neutralization of VDPV2 antigenic variants by immune human sera.
We selected 10 isolates with the most divergent NAg site sequences, in addition to Sabin 2 and MEF-1, for serologic testing (Fig. 6; see Fig. S10 in the supplemental material). Because the maximum-parsimony algorithm may occasionally count the same replacement twice and some sites are noninformative (71), branch distances between some sequence pairs on the NAg site tree were not strictly proportional to the pairwise differences indicated in the difference table (see Table S1 in the supplemental material). Nonetheless, the most divergent isolates mapped to the periphery of the tree (Fig. 6) and differed from Sabin 2 at 2 to 5 positions and from each other at 4 to 8 positions (see Table S1 in the supplemental material). Median neutralizing titers against cVDPV2s in 95 serum samples collected from infants in Kano state, northern Nigeria, in 2011 were comparable to those against Sabin 2 (7.00 log2; P < 0.05) and MEF-1 (6.83 log2; P < 0.05). Seropositivity rates against all cVDPV2s ranged from 71.9% to 77.1% compared to 70.8% for both Sabin 2 and MEF-1 (see Table S2 in the supplemental material).
DISCUSSION
Despite many decades of PV research, information remains limited on the rates, patterns, and dynamics of PV capsid protein evolution during person-to-person transmission (34, 35). Intensive PV surveillance conducted during a large, prolonged, and widespread outbreak, coupled with application of a robust molecular clock (13), set the stage for our high-resolution evolutionary studies. The mapping of individual amino acid replacement events to time-scaled MCC trees illustrated the evolutionary trajectories of different capsid functional domains. In many instances, the dates of fixation of new amino acid residues could be estimated with an accuracy of a few months by inspection of the MCC trees. Furthermore, the likely geographic settings for replacement events could often be localized to particular Nigerian states. However, the timing and locations of the earliest replacement events (72, 73), such as the I1143T reversion, could not be estimated with precision because they occurred during the initial stages of emergence, before circulating lineages could be clearly distinguished from vaccine virus progeny with more limited spread. The resolving power of our analyses was also limited by the inherently low paralytic attack rate of PV2 infections (74), compounded by some gaps in case-based surveillance. Nonetheless, the genetic data were sufficiently robust to permit critical evaluation of key assumptions underpinning our replacement rate data. While our analyses covered the peak periods of the outbreak, less intense transmission of the major lineage group, 2005-8, continued at least until March 2015, when an isolate of that lineage group was last detected, during which time an additional 138 cVDPV2s from 2005-8 were detected (19 from poliomyelitis patients and their contacts; 119 from environmental samples) (13, 41), including new NAg site variants. Low-level transmission of more recently emergent cVDPV2 lineages continued in northern Nigeria at least until November 2016 (41, 75).
A remarkable finding from this study was the large number of distinct NAg site variants that emerged during the cVDPV2 outbreak in Nigeria. Extensive NAg site evolution for WPV was first described for WPV3 variants that circulated during the 1984-1985 outbreak in Finland (47). However, the Finnish outbreak took place within a population that had high rates of poliovirus vaccine coverage but had been vaccinated with a weakly immunogenic IPV preparation, conditions that might have accelerated the rate of antigenic evolution (76). In contrast, in Nigeria, the cVDPV2 circulated in settings of deficient OPV coverage and low population immunity to PV2 (43, 44), similar to prevaccine era WPV2 outbreaks, and selection by preexisting anti-PV2 neutralizing antibodies probably played at most a limited role in antigenic evolution.
Our studies have confirmed and extended those of previous investigations into the variability of PV NAg sites (34, 37, 40, 47, 48, 50). The 6 invariant NAg positions in our set have been found to be variable in other Sabin 2-derived viruses (28, 33, 34, 36). On the other hand, 37 NAg site replacements were first described here, while ∼50 previously described replacements were not found in our set (33, 34, 36, 40, 58, 59). The observed replacements in NAg1 (K1099E/Q and R1100L/S) and NAg2 (G1219S/D and T1223A/I) were identical to those described for PV2 MAb escape variants (28). As first reported by Macadam et al. (67), the non-NAg site surface residue I1143 is subject to the strongest selective pressure for change in Sabin 2 (13, 72).
Antigenic evolution of cVDPVs appears to be slower and less extensive than that of iVDPVs (and environmental aVDPVs that are probably iVDPVs) (37, 39, 77). Apart from single codon deletions found in some iVDPVs (36, 40), the NAg sites of both cVDPV and iVDPV isolates accommodate similar alternative amino acid residues. However, the NAg sites of individual cVDPV2 isolates described here incorporated no more than 5 replacements, unlike the more highly divergent NAg sites of iVDPV isolates with similar degrees of nucleotide divergence (36, 38, 40). The differences in the extent of NAg site divergence between cVDPVs and iVDPVs may provide further insights into the epidemiological significance of environmental aVDPVs in settings where the source patient(s) remains unidentified (33, 39, 40, 77–80).
As observed with the capsids of WPV1 and aVDPV2 (35, 37), most cVDPV2 NAg sites were under strong purifying selection, with no clear directionality to their evolution. Although the rate of PV nucleotide evolution is similar to that of influenza A virus (35), structural protein evolution during infections of humans, including that of the NAg sites, is much slower. Many of the replacements in the cVDPV2 NAg sites were back-mutations to the preceding residue, suggesting that the antigenic repertoire within a PV serotype, while wide, is nonetheless constrained. In agreement with previous reports (34, 40), both cVDPVs and WPVs are efficiently neutralized by human immune sera, and their transmission can be effectively controlled by high rates of OPV coverage (41). Accordingly, the geographic range of individual WPV genotypes and cVDPV outbreaks is circumscribed (39, 81), and no pandemic antigenic variants have ever been observed. Although long-range importations of WPV have occasionally occurred (81), further spread can be prevented by high rates of population immunity, especially mucosal immunity (82), even for antigenically divergent WPVs (21, 47).
Detailed understanding of the structural and functional rules governing capsid protein evolution, especially of the NAg sites, may offer critical insights into why some viral vaccines, such as OPV and IPV, are broadly protective whereas others, such as those for another picornavirus, foot-and-mouth disease virus, require frequent updating (83). It may also be possible to apply these rules to construct more accurate PV protein molecular clocks (35, 84), using more slowly evolving variable positions to probe deeper into the relationships among WPV genotypes and thus obtain a wider overview of the evolutionary history of three serotypes of human viruses now approaching extinction (2–4, 21, 46).
MATERIALS AND METHODS
Virus isolation and characterization.
The 2005–2011 Nigerian VDPV2 isolates were isolated from the stools of patients with acute flaccid paralysis and characterized by capsid region nucleotide sequencing, as previously described (13).
Phylogenetic analysis.
Sequence relationships (all nucleotide substitutions [KT]) in the capsid region (nt 748 to 3384) among all VDPV2 isolates (GenBank accession numbers JX274980 to JX275382) were summarized in Bayesian MCC trees constructed by Bayesian Markov chain Monte Carlo (MCMC) analysis implemented in BEAST v. 1.7, as described previously (13, 85). Site- and branch-specific selection analyses were performed in likelihood calculations under the random-sites models (86) and the fixed-effects models (87) implemented in PAML (56) and HyPhy (88), respectively.
Pattern and distribution of substitutions.
Capsid nucleotide substitutions by category (total substitutions [KT], synonymous transitions [AS], synonymous transversions [BS], nonsynonymous transitions [AA], and nonsynonymous transversions [BA]) along all branches of the phylogenetic trees were calculated as described previously (35). Inferred amino acid replacements were mapped and labeled onto corresponding branches of the MCC trees. The first digit of a residue position identifies the viral protein (4, VP4; 2, VP2; 3, VP3; 1, VP1), followed consecutively from residue 1 of each protein. Original and replacement amino acids are indicated at the beginning and at the end of the 4-digit number, respectively.
Estimation of the frequency of amino acid replacements in NAg sites.
Rate estimates were performed for the well-defined cVDPV2 lineage groups described previously (13). The overall rate of fixation of replacements in NAg sites was calculated by dividing the total number of NAg replacements by the total branch lengths of all the trees (estimated using the program TreeStat v. 1.2 [http://tree.bio.ed.ac.uk/software/treestat/]) based on the estimated dates of the initiating tOPV dose (13).
Mapping replacement frequencies and diversity in the NAg sites of the crystal structure of PV2.
To map the NAg sites, homology modeling was performed by using Modeller (v. 9), since the Sabin 2 structure is not deposited in the Protein Data Bank (PDB). Modeller implements comparative protein structure modeling by satisfaction of spatial restraints (89, 90), and it requires a structural template and a target protein sequence. A pentamer from the X-ray crystallographic structure of poliovirus type 2 strain MEF-1 (frequently described as Lansing; PDB no. 1EAH) with a resolution of 2.9 Å (29) was used as the template. In-house Perl scripts were developed for data processing (e.g., preparing the input PDB file and sequence alignment data) required by Modeller. Although Modeller embeds with the structure refinements and loop optimizations (91), the calculated structure was further checked by ProCheck (92) for the stereochemical (e.g., Ramachandran plot) quality of the structure and was visually inspected, as well. The all-atom root mean square deviation between the calculated Sabin 2 structure and the MEF-1 template was 0.257, which implies that there are no significant conformational differences between the two. PyMol (version 1.1) was used for visualizing the structure and mapping the NAg sites onto the structure. The frequency of NAg amino acid replacements and the number of alternative amino acid residues in the NAg sites were illustrated in heat maps projected onto the Sabin 2 capsid pentamer.
Neutralization assays with human immune sera.
Human sera were collected in 2011 from children aged 6 to 9 months or 36 to 47 months in Kano, Nigeria (n = 95) (93). Deidentified (coded) specimens were tested for neutralizing antibodies against a panel of 10 Nigerian cVDPV2 isolates and two control antigens (MEF-1 and Sabin 2) by microneutralization assay (94, 95). Briefly, 50 to 150 50% cell culture infectious doses (CCID50) of each poliovirus and 2-fold serial dilutions of serum were combined and preincubated at 35°C for 3 h before the addition of HEp-2(C) cells. After incubation for 5 days at 35°C and 5% CO2, the plates were stained with crystal violet and cell viability was measured by optical density in a plate spectrophotometer. Each specimen was run in triplicate, and neutralization titers were estimated by the Spearman-Kärber method (96). Seropositivity was defined as a log2 neutralization titer of ≥3 log2 (1:8 serum dilution). Statistical comparisons of neutralization titers against cVDPV2s to the control antigens were performed using a nonparametric one-way analysis of variance (ANOVA) test with Dunn's posttest (GraphPad [La Jolla, CA] Prism v. 6). The project was reviewed and did not constitute human subject research for CDC.
Supplementary Material
ACKNOWLEDGMENTS
We thank the field surveillance teams who investigated acute flaccid paralysis cases, laboratory staff in the WHO National Polio Laboratories in Maiduguri and Ibadan, Ray Campagnoli, and Karen Ching (CDC) for excellent technical assistance and Javier Martín for helpful discussions.
The findings and conclusions in this report are ours and do not necessarily represent the views or the official position of the Centers for Disease Control and Prevention.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01949-17.
REFERENCES
- 1.Wassilak SGF, Oberste MS, Tangermann RH, Diop OM, Jafari HS, Armstrong GL. 2014. Progress toward global interruption of wild poliovirus transmission, 2010–2013, and tackling the challenges to complete eradication. J Infect Dis 210(Suppl 1):S5–S15. doi: 10.1093/infdis/jiu456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2001. Apparent global interruption of wild poliovirus type 2 transmission. MMWR Morb Mortal Wkly Rep 50:222–224. [PubMed] [Google Scholar]
- 3.Kew OM, Cochi SL, Jafari HS, Wassilak SGF, Mast EE, Diop OM, Tangermann RH, Armstrong GL. 2014. Possible eradication of wild poliovirus type 3—worldwide, 2012. MMWR Morb Mortal Wkly Rep 63:1031–1033. [PMC free article] [PubMed] [Google Scholar]
- 4.Morales M, Tangermann RH, Wassilak SG. 2016. Progress toward polio eradication—worldwide, 2015–2016. MMWR Morb Mortal Wkly Rep 65:470–473. doi: 10.15585/mmwr.mm6518a4. [DOI] [PubMed] [Google Scholar]
- 5.Elhamidi Y, Mahamud A, Safdar M, Al Tamimi W, Jorba J, Mbaeyi C, Hsu CH, Wadood Z, Sharif S, Ehrhardt D. 2017. Progress toward poliomyelitis eradication—Pakistan, January 2016–September 2017. MMWR Morb Mortal Wkly Rep 66:1276–1280. doi: 10.15585/mmwr.mm6646a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez M, Shukla H, Nikulin J, Wadood MZ, Hadler S, Mbaeyi C, Tangermann R, Jorba J, Ehrhardt D. 2017. Progress toward poliomyelitis eradication—Afghanistan, January 2016–June 2017. MMWR Morb Mortal Wkly Rep 66:854–858. doi: 10.15585/mmwr.mm6632a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nnadi C, Damisa E, Esapa L, Braka F, Waziri N, Siddique A, Jorba J, Nganda GW, Ohuabunwo C, Bolu O, Wiesen E, Adamu U. 2017. Continued endemic wild poliovirus transmission in security-compromised areas—Nigeria, 2016. Morbid Mortal Wkly Rep 66:190–193. doi: 10.15585/mmwr.mm6607a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Committee on Typing of the National Foundation for Infantile Paralysis. 1951. Immunologic classification of poliomyelitis viruses: a cooperative program for the typing of one hundred strains. Am J Hyg 54:191–204. [PubMed] [Google Scholar]
- 9.Minor PD, Ferguson M, Evans DMA, Almond JW, Icenogle JP. 1986. Antigenic structure of polioviruses of serotypes 1, 2, and 3. J Gen Virol 67:1283–1291. doi: 10.1099/0022-1317-67-7-1283. [DOI] [PubMed] [Google Scholar]
- 10.Belnap DM, McDermott BM, Filman DJ, Cheng N-Q, Trus BL, Zuccola HJ, Racaniello VR, Hogle JM, Steven AC. 2000. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc Natl Acad Sci U S A 97:73–78. doi: 10.1073/pnas.97.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He Y, Mueller S, Chipman PR, Bator CM, Peng X, Bowman VD, Mukhopadhyay S, Wimmer E, Kuhn RJ, Rossmann MG. 2003. Complexes of poliovirus serotypes with their common cellular receptor, CD155. J Virol 77:4827–4835. doi: 10.1128/JVI.77.8.4827-4835.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georgescu MM, Delpeyroux F, Crainic R. 1995. Tripartite genome organization of a natural type 2 vaccine/nonvaccine recombinant poliovirus. J Gen Virol 76:2343–2348. doi: 10.1099/0022-1317-76-9-2343. [DOI] [PubMed] [Google Scholar]
- 13.Burns C, Shaw J, Jorba J, Bukbuk D, Adu F, Gumede N, Iber J, Chen Q, Vincent A, Chenoweth P, Henderson E, Wannemeuhler K, Campagnoli R, Pate MA, Abanida E, Gasasira A, Shimizu H, Williams AJ, Kilpatrick D, Wassilak S, Tomori O, Pallansch M, Kew O. 2013. Multiple independent emergences of type 2 vaccine-derived polioviruses during a large outbreak in northern Nigeria. J Virol 87:4907–4922. doi: 10.1128/JVI.02954-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joffret ML, Jégouic S, Bessaud M, Balanant J, Tran C, Caro V, Holmblat B, Razafindratsimandresy R, Reynes J-M, Rakoto-Andrianarivelo M, Delpeyroux F. 2012. Common and diverse features of co-circulating type 2 and 3 recombinant vaccine-derived polioviruses isolated from patients with poliomyelitis and healthy children. J Infect Dis 205:1363–1373. doi: 10.1093/infdis/jis204. [DOI] [PubMed] [Google Scholar]
- 15.Minor PD, John A, Ferguson M, Icenogle JP. 1986. Antigenic and molecular evolution of the vaccine strain of type 3 poliovirus during the period of excretion by a primary vaccinee. J Gen Virol 67:693–706. doi: 10.1099/0022-1317-67-4-693. [DOI] [PubMed] [Google Scholar]
- 16.Crainic R, Couillin P, Blondel B, Cabau N, Boué A, Horodniceanu F. 1983. Natural variation of poliovirus neutralization epitopes. Infect Immun 41:1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakano JH, Gelfand HM, Cole JT. 1963. The use of a modified Wecker technique for the serodifferentiation of type 1 polioviruses related and unrelated to Sabin's vaccine strain. II. Antigenic segregation of isolates from specimens collected in field studies. Am J Hyg 78:214–226. [DOI] [PubMed] [Google Scholar]
- 18.Nakano JH, Cole JT. 1970. Antigenic segregation of type 2 poliovirus isolants related and unrelated to Sabin's vaccine strain with modified Wecker technique. Am J Epidemiol 91:317–327. doi: 10.1093/oxfordjournals.aje.a121142. [DOI] [PubMed] [Google Scholar]
- 19.Wenner HA, Kamitsuka P, Lenahan M, Melnick JL. 1956. A comparative study of type 2 poliomyelitis viruses. II. Antigenic differences relating to 18 type 2 strains J Immunol 77:220–231. [PubMed] [Google Scholar]
- 20.Vidor E. 2018. Poliovirus vaccine—inactivated, p 841–865. In Plotkin SA, Orenstein WA, Offit PA, Edwards KM (ed), Vaccines, 7th ed Elsevier, Philadelphia, PA. [Google Scholar]
- 21.Sutter RW, Kew OM, Cochi SL, Aylward RB. 2018. Poliovirus vaccine—live, p 866–917. In Plotkin SA, Orenstein WA, Offit PA, Edwards KM (ed), Vaccines, 7th ed Elsevier, Philadelphia, PA. [Google Scholar]
- 22.Nakano JH, Hatch MH, Thieme ML, Nottay B. 1978. Parameters for differentiating vaccine-derived and wild poliovirus strains. Prog Med Virol 24:78–206. [PubMed] [Google Scholar]
- 23.van der Avoort HGAM, Hull BP, Hovi T, Pallansch MA, Kew OM, Crainic R, Wood DJ, Mulders MN, van Loon AM. 1995. A comparative study of five methods of intratypic differentiation of polioviruses. J Clin Microbiol 33:2562–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Wezel AL, Hazendonk AG. 1979. Intratypic serodifferentiation of poliomyelitis virus by strain-specific antisera. Intervirology 11:2–8. doi: 10.1159/000149005. [DOI] [PubMed] [Google Scholar]
- 25.Hogle JM, Chow M, Filman DJ. 1985. The three-dimensional structure of poliovirus at 2.9 Å resolution. Science 229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 26.Page GS, Mosser AG, Hogle JM, Filman DJ, Rueckert RR, Chow M. 1988. Three-dimensional structure of poliovirus serotype 1 neutralizing determinants. J Virol 62:1781–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Filman DJ, Syed R, Chow M, Macadam AJ, Minor PD, Hogle JM. 1989. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J 8:1567–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel V, Ferguson M, Minor PD. 1993. Antigenic sites on type 2 poliovirus. Virology 192:361–364. doi: 10.1006/viro.1993.1044. [DOI] [PubMed] [Google Scholar]
- 29.Lentz KN, Smith AD, Geisler SC, Cox S, Buontempo P, Skelton A, DeMartino J, Rozhon E, Schwartz J, Girijavallabhan V, O'Connell J, Arnold E. 1997. Structure of poliovirus type 2 Lansing complexed with antiviral agent SCH48973: comparison of the structural and biological properties of three poliovirus serotypes. Structure 5:961–978. doi: 10.1016/S0969-2126(97)00249-9. [DOI] [PubMed] [Google Scholar]
- 30.Minor PD. 1990. Antigenic structure of picornaviruses. Curr Top Microbiol Immunol 161:121–154. [DOI] [PubMed] [Google Scholar]
- 31.Blondel B, Crainic R, Fichot O, Dufraisse G, Candréa A, Diamond D, Girard M, Horaud F. 1986. Mutations conferring resistance to neutralization with monoclonal antibodies in type 1 poliovirus can be located outside or inside the antibody-binding site. J Virol 57:81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiegers K, Uhlig H, Dernick R. 1989. N-AgIB of poliovirus type 1: a discontinuous epitope formed by two loops of VP1 comprising residues 96–104 and 141–152. Virology 170:583–586. doi: 10.1016/0042-6822(89)90452-2. [DOI] [PubMed] [Google Scholar]
- 33.Shulman L, Manor J, Handsher R, Delpeyroux F, McDonough M, Halmut T, Silberstein I, Alfandari J, Quay J, Fisher T, Robinov J, Kew O, Crainic R, Mendelson E. 2000. Molecular and antigenic characterization of a highly evolved derivative of the type 2 oral poliovaccine strain isolated from sewage in Israel. J Clin Microbiol 38:3729–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yakovenko ML, Cherkasova EA, Rezapkin GV, Ivanova OE, Ivanov AP, Eremeeva TP, Baykova OY, Chumakov KM, Agol VI. 2006. Antigenic evolution of vaccine-derived polioviruses: changes in individual epitopes and relative stability of the overall immunological properties. J Virol 80:2641–2653. doi: 10.1128/JVI.80.6.2641-2653.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jorba J, Campagnoli R, De L, Kew O. 2008. Calibration of multiple poliovirus molecular clocks covering an extended evolutionary range. J Virol 82:4429–4440. doi: 10.1128/JVI.02354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeVries AS, Harper J, Murray A, Lexau C, Bahta L, Christensen J, Cebelinski E, Fuller S, Kline S, Wallace GS, Shaw JH, Burns CC, Lynfield R. 2011. Vaccine-derived poliomyelitis 12 years after infection in Minnesota. N Engl J Med 364:2316–2323. doi: 10.1056/NEJMoa1008677. [DOI] [PubMed] [Google Scholar]
- 37.Hovi T, Savolainen-Kopra C, Smura T, Blomqvist S, Al-Hello H, Roivainen M. 2013. Evolution of type 2 vaccine derived poliovirus lineages. Evidence for codon-specific positive selection at three distinct locations on capsid wall. PLoS One 8:e66836. doi: 10.1371/journal.pone.0066836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kew OM, Sutter RW, de Gourville EM, Dowdle WR, Pallansch MA. 2005. Vaccine-derived polioviruses and the endgame strategy for global polio eradication. Annu Rev Microbiol 59:587–635. doi: 10.1146/annurev.micro.58.030603.123625. [DOI] [PubMed] [Google Scholar]
- 39.Burns CC, Diop OM, Sutter RW, Kew OM. 2014. Vaccine-derived polioviruses. J Infect Dis 210(Suppl 1):S283–S293. doi: 10.1093/infdis/jiu295. [DOI] [PubMed] [Google Scholar]
- 40.Dunn G, Klapsa D, Wilton T, Stone L, Minor PD, Martín J. 2015. Twenty-eight years of poliovirus replication in an immunodeficient individual: impact on the global polio eradication initiative. PLoS Pathog 11:e1005114. doi: 10.1371/journal.ppat.1005114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jorba J, Diop OM, Iber J, Sutter RW, Wassilak SG, Burns CC. 2016. Update on vaccine-derived polioviruses—worldwide, January 2015–May 2016. MMWR Morb Mortal Wkly Rep 65:763–769. doi: 10.15585/mmwr.mm6530a3. [DOI] [PubMed] [Google Scholar]
- 42.Alexander JP Jr, Gary HE Jr, Pallansch MA. 1997. Duration of poliovirus excretion and its implications for acute flaccid paralysis surveillance: a review of the literature. J Infect Dis 175(Suppl 1):S176–S182. doi: 10.1093/infdis/175.Supplement_1.S176. [DOI] [PubMed] [Google Scholar]
- 43.Wassilak S, Pate MA, Wannemuehler K, Jenks J, Burns C, Chenoweth P, Abanida EA, Adu F, Baba M, Gasasira A, Iber J, Mkanda P, Williams AJ, Shaw J, Pallansch M, Kew O. 2011. Outbreak of type 2 vaccine-derived poliovirus in Nigeria, 2005–2009: emergence and widespread circulation in an underimmunized population. J Infect Dis 203:898–909. doi: 10.1093/infdis/jiq140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jenkins HE, Aylward RB, Gasasira A, Donnelly CA, Mwanza M, Garnier S, Chauvin C, Abanida E, Pate MA, Adu F, Baba M, Grassly NC. 2010. Implications of a circulating vaccine-derived poliovirus in Nigeria. N Engl J Med 362:2360–2369. doi: 10.1056/NEJMoa0910074. [DOI] [PubMed] [Google Scholar]
- 45.Racaniello VR. 2013. Picornaviridae: the viruses and their replication, p 453–489. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 1 Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 46.Kew OM. 2014. Enteroviruses: polio, p 277–336. In Kaslow RA, Stanberry LR, Leduc JW (ed), Viral infections of humans, 5th ed Springer, New York, NY. [Google Scholar]
- 47.Huovilainen A, Kinnunen L, Ferguson M, Hovi T. 1988. Antigenic variation among 173 strains of type 3 poliovirus isolated in Finland during the 1984 to 1985 outbreak. J Gen Virol 69:1941–1948. doi: 10.1099/0022-1317-69-8-1941. [DOI] [PubMed] [Google Scholar]
- 48.Martín J, Dunn G, Hull R, Patel V, Minor PD. 2000. Evolution of the Sabin strain of type 3 poliovirus in an immunodeficient patient during the entire 637-day period of virus excretion. J Virol 74:3001–3010. doi: 10.1128/JVI.74.7.3001-3010.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang C-F, Chen H-Y, Jorba J, Sun H-C, Yang S-J, Lee H-C, Huang Y-C, Lin T-Y, Chen P-J, Shimizu H, Nishimura Y, Utama A, Pallansch M, Miyamura T, Kew O, Yang J-Y. 2005. Intratypic recombination among lineages of type 1 vaccine-derived poliovirus emerging during chronic infection of an immunodeficient patient. J Virol 79:12623–12634. doi: 10.1128/JVI.79.20.12623-12634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Odoom JK, Yunus Z, Dunn G, Minor PD, Martín J. 2008. Changes in population dynamics during long-term evolution of Sabin type 1 poliovirus in an immunodeficient patient. J Virol 82:9179–9190. doi: 10.1128/JVI.00468-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patriarca PA, Wright PF, John TJ. 1991. Factors affecting the immunogenicity of oral poliovirus vaccine in developing countries: review. Rev Infect Dis 13:926–939. doi: 10.1093/clinids/13.5.926. [DOI] [PubMed] [Google Scholar]
- 52.de Quadros CA, Andrus JK, Olivé JM, Guerra de Macedo C, Henderson DA. 1992. Polio eradication from the Western Hemisphere. Annu Rev Public Hlth 13:239–252. doi: 10.1146/annurev.pu.13.050192.001323. [DOI] [PubMed] [Google Scholar]
- 53.Toyoda H, Kohara MM, Kataoka Y, Suganuma T, Omata T, Imura N, Nomoto A. 1984. Complete nucleotide sequences of all three poliovirus serotype genomes: implication for genetic relationship, gene function and antigenic determinants. J Mol Biol 174:561–585. doi: 10.1016/0022-2836(84)90084-6. [DOI] [PubMed] [Google Scholar]
- 54.Pollard SR, Dunn G, Cammack N, Minor PD, Almond JW. 1989. Nucleotide sequence of a neurovirulent variant of the type 2 oral poliovirus vaccine. J Virol 63:4949–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ren R, Moss EG, Racaniello VR. 1991. Identification of two determinants that attenuate vaccine-related type 2 poliovirus. J Virol 65:1377–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Z. 2007. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol 24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 57.La Monica N, Kupsky WJ, Racaniello VR. 1987. Reduced mouse neurovirulence of poliovirus type 2 Lansing antigenic variants selected with monoclonal antibodies. Virology 161:429–437. doi: 10.1016/0042-6822(87)90136-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Z, Fischer ER, Kouiavskaia D, Hansen BT, Ludtke SJ, Bidzhieva B, Makiya M, Agulto L, Purcell RH, Chumakov K. 2013. Cross-neutralizing human anti-poliovirus antibodies bind the recognition site for cellular receptor. Proc Natl Acad Sci U S A 110:20242–20247. doi: 10.1073/pnas.1320041110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shulman LM, Manor Y, Sofer D, Handsher R, Swartz T, Delpeyroux F, Mendelson E. 2006. Neurovirulent vaccine-derived polioviruses in sewage from highly immune populations. PLoS One 1:e69. doi: 10.1371/journal.pone.0000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiegers K, Dernick R. 1992. Molecular basis of antigenic structures of poliovirus: implications for their evolution during morphogenesis. J Virol 66:4597–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bannwarth L, Girerd-Chambaz Y, Arteni A, Guigner JM, Ronzon F, Manin C, Vénien-Bryan C. 2015. Mapping of the epitopes of poliovirus type 2 in complex with antibodies. Mol Immunol 67:233–239. doi: 10.1016/j.molimm.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 62.Xia X, Xie Z. 2002. Protein structure, neighbor effect, and a new index of amino acid dissimilarities. Mol Biol Evol 19:58–67. doi: 10.1093/oxfordjournals.molbev.a003982. [DOI] [PubMed] [Google Scholar]
- 63.Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 64.Zhang P, Mueller S, Morais MC, Bator CM, Bowman VD, Hafenstein S, Wimmer E, Rossmann MG. 2008. Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses. Proc Natl Acad Sci U S A 105:18284–18289. doi: 10.1073/pnas.0807848105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Larsen GR, Anderson CW, Dorner AJ, Semler BL, Wimmer E. 1982. Cleavage sites within the poliovirus capsid protein precursors. J Virol 41:340–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strauss M, Levy HC, Bostina M, Filman DJ, Hogle JM. 2013. RNA transfer from poliovirus 135S particles across membranes is mediated by long umbilical connectors. J Virol 87:3903–3914. doi: 10.1128/JVI.03209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Macadam AJ, Pollard SR, Ferguson G, Skuce R, Wood D, Almond JW, Minor PD. 1993. Genetic basis of attenuation of the Sabin type 2 vaccine strain of poliovirus in primates. Virology 192:18–26. doi: 10.1006/viro.1993.1003. [DOI] [PubMed] [Google Scholar]
- 68.Yang Z, Nielsen R. 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol 17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- 69.La Monica N, Meriam C, Racaniello VR. 1986. Mapping of sequences required for mouse neurovirulence of poliovirus type 2 Lansing. J Virol 57:515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bush RM, Bender CA, Subbarao K, Cox NJ, Fitch WM. 1999. Predicting the evolution of human influenza A. Science 286:1921–1925. doi: 10.1126/science.286.5446.1921. [DOI] [PubMed] [Google Scholar]
- 71.Felsenstein J. 2004. Inferring phylogenies. Sinauer Associates, Sunderland, MA. [Google Scholar]
- 72.Famulare M, Chang S, Iber J, Zhao K, Adeniji JA, Bukbuk D, Baba M, Behrend M, Burns CC, Oberste MS. 2015. Sabin vaccine reversion in the field: a comprehensive analysis of Sabin-like poliovirus isolates in Nigeria. J Virol 90:317–331. doi: 10.1128/JVI.01532-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stern A, Yeh MT, Zinger T, Smith M, Wright C, Ling G, Nielsen R, Macadam A, Andino R. 2017. The evolutionary pathway to virulence of an RNA virus. Cell 169:35–46.e19. doi: 10.1016/j.cell.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nathanson N, Kew OM. 2010. From emergence to eradication: the epidemiology of poliomyelitis deconstructed. Am J Epidemiol 172:1213–1229. doi: 10.1093/aje/kwq320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jorba J, Diop OM, Iber J, Henderson E, Sutter RW, Wassilak SGF, Burns CC. 2017. Update on vaccine-derived polioviruses—worldwide, January 2016–June 2017. MMWR Morb Mortal Wkly Rep 66:1185–1191. doi: 10.15585/mmwr.mm6643a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hovi T, Cantell K, Huovalainen A, Kinnunen E. 1986. Outbreak of paralytic poliomyelitis in Finland: widespread circulation of antigenically altered poliovirus type 3 in a vaccinated population. Lancet i:1427–1432. [DOI] [PubMed] [Google Scholar]
- 77.Al-Hello H, Jorba J, Blomqvist S, Raud R, Kew O, Roivainen M. 2013. Highly divergent types 2 and 3 vaccine-derived polioviruses isolated from sewage in Tallinn, Estonia. J Virol 87:13076–13080. doi: 10.1128/JVI.01174-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blomqvist S, Savolainen C, Laine P, Hirttio P, Lamminsalo E, Penttila E, Joks S, Roivainen M, Hovi T. 2004. Characterization of a highly evolved vaccine-derived poliovirus type 3 isolated from sewage in Estonia. J Virol 78:4876–4883. doi: 10.1128/JVI.78.9.4876-4883.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roivainen M, Blomqvist S, Al-Hello H, Paananen A, Delpeyroux F, Kuusi M, Hovi T. 2010. Highly divergent neurovirulent vaccine-derived polioviruses of all three serotypes are recurrently detected in Finnish sewage. Euro Surveil 15:pii/19566. [PubMed] [Google Scholar]
- 80.Hovi T, Paananen A, Blomqvist S, Savolainen-Kopra C, Al-Hello H, Smura T, Shimizu H, Nadova K, Sobotova Z, Gavrilin E, Roivainen M. 2013. Characteristics of an environmentally monitored prolonged type 2 vaccine derived poliovirus shedding episode that stopped without intervention. PLoS One 8:e66849. doi: 10.1371/journal.pone.0066849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kew O. 2012. Reaching the last one percent: progress and challenges in global polio eradication. Curr Opin Virol 2:188–198. doi: 10.1016/j.coviro.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 82.Shulman LM, Mendelson E, Anis E, Bassal R, Gdalevich M, Hindiyeh M, Kaliner E, Kopel E, Manor Y, Moran-Gilad J, Ram D, Sofer D, Somekh E, Tasher D, Weil M, Gamzu R, Grotto I. 2014. Laboratory challenges in response to silent introduction and sustained transmission of wild poliovirus type 1 in Israel during 2013. J Infect Dis 210(Suppl 1):S304–S314. doi: 10.1093/infdis/jiu294. [DOI] [PubMed] [Google Scholar]
- 83.Rowlands DJ, Brown F. 2002. Antigenic variation in foot-and-mouth disease virus, p 51–58. In Semler BL, Wimmer E (ed), Molecular biology of picornaviruses. ASM Press, Washington, DC. [Google Scholar]
- 84.Zuckerkandl E, Pauling L. 1965. Evolutionary divergence and convergence in proteins, p 97–166. In Bryson V, Vogel HJ (ed), Evolving genes and proteins. Academic Press, New York, NY. [Google Scholar]
- 85.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang Z. 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol 15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- 87.Kosakovsky Pond SL, Frost SDW. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol 22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 88.Pond SL, Frost SD, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 89.Webb B, Sali A. 2014. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics 47:5.6.1–5.6.32. doi: 10.1002/0471250953.bi0506s47. [DOI] [PubMed] [Google Scholar]
- 90.Sali A, Blundell TL. 1993. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 91.Fiser A, Sali A. 2003. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol 374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- 92.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 93.Iliyasu Z, Nwaze E, Verma H, Mustapha AO, Weldegebriel G, Gasasira A, Wannemuehler KA, Pallansch MA, Gajida AU, Pate M, Sutter RW. 2014. Survey of poliovirus antibodies in Kano, Northern Nigeria. Vaccine 32:1414–1420. doi: 10.1016/j.vaccine.2013.08.060. [DOI] [PubMed] [Google Scholar]
- 94.World Health Organization. 2004. Polio laboratory manual, 4th ed World Health Organization, Geneva, Switzerland. [Google Scholar]
- 95.World Health Organization. 1991. Report of a WHO informal consultation on polio neutralization antibody assays, Nashville, 5–6 December 1991 WHO, Geneva, Switzerland. [Google Scholar]
- 96.Kärber G. 1931. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Arch Exp Pathol Pharmakol 162:480–483. doi: 10.1007/BF01863914. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.