ABSTRACT
Type I interferons (IFNs) induce expression of more than 300 cellular genes that provide protection against viruses and other pathogens. For survival, viruses evolved defenses to prevent the IFN response or counteract the IFN-induced antiviral state. However, because viruses and cells coevolved, the dynamic relationship between virus and host is difficult to discern. In the present study, we demonstrated that vaccinia virus with a large deletion near the left end of the genome had a diminished ability to replicate in cells that had been pretreated with beta interferon (IFN-β), suggesting that one or more of the missing 17 open reading frames (ORFs) encode an antagonist of the IFN-induced antiviral state. By systematically deleting groups of ORFs and then individual ORFs, the C9L gene was shown to be required for IFN resistance. Replication of the C9L deletion mutant (vΔC9) was impaired in human cells that had been pretreated with IFN-β. Expression of viral early genes occurred, but subsequent events, including genome uncoating, genome replication, and postreplicative gene expression, were inhibited. Expression of the C9 protein occurred prior to genome replication, consistent with an early role in counteracting the IFN-induced antiviral state. C9 contains six ankyrin repeat motifs and a near C-terminal F-box. Mass spectrometry and immunoblotting identified host proteins that copurified with a functional epitope-tagged C9. The most abundant proteins were components of the SCF (CUL1, SKP1, F-box) and signalosome/deneddylation complexes, which interact with each other, suggesting a possible role in proteolysis of one or more interferon-induced proteins.
IMPORTANCE Poxviruses comprise a family of large DNA viruses that replicate in the cytoplasm of vertebrate and insect hosts and cause human and zoonotic diseases. In most cases the primary infection is moderated by innate immune defenses. Vertebrates, including fish, amphibians, reptiles, birds, and mammals, all produce type I interferon homologs. In humans, interferon stimulates the synthesis of more than 300 proteins thought to have roles in host defense. Conversely, viruses have evolved means to thwart the host defenses. We are attempting to deconstruct the established virus-host relationship in order to better understand the molecular mechanisms involved. In the present study, we identified a vaccinia virus gene that prevents interferon-mediated inhibition of very early stages of viral replication and is conserved in orthopoxviruses. The viral protein was shown to interact with host proteins involved in proteolysis, suggesting that vaccinia virus may subvert the cellular apparatus for its own defense.
KEYWORDS: cullin 1, interferon, orthopoxvirus, poxvirus, SCF complex, SKP1, deneddylation, signalosome
INTRODUCTION
Interferons (IFNs) and their induced products provide important innate cellular defenses against viruses (1). The IFN response is initiated by cell pattern recognition receptors upon sensing pathogen-associated molecular patterns. These interactions result in activation of signaling cascades, leading to transcription of genes encoding IFNs. The secreted IFNs engage specific receptors on the same or neighboring cells, which activate the JAK-STAT pathway to transcribe interferon-stimulated genes (ISGs). More than 300 type I ISGs have been identified, though the antiviral roles of only a minority have been characterized (2–5). For survival, viruses acquired defenses that neutralize the IFN response (6). Indeed, vaccinia virus (VACV), the prototype member of the poxvirus family, is highly resistant to type I IFNs in mouse and human cells (7). VACV encodes proteins that suppress the induction of IFN, down modulate signaling pathways, and counteract the antiviral state by inhibiting the action of ISGs (8). However, the majority of poxvirus IFN-defense proteins that have been identified thus far are involved in suppressing and down modulating signal pathways. The present study pertains to the ability of VACV to neutralize the IFN-induced antiviral state.
The basal expression of ISGs varies in cultured cells, providing a means to isolate host range mutants that are also IFN sensitive (9). Examples are VACV mutants that lack a functional E3 double-stranded RNA (dsRNA) binding protein, which sequesters dsRNA (10–12), or the D9/D10 decapping enzymes (13, 14), which reduce the amount of dsRNA by enhancing mRNA degradation (15). The dsRNA is a by-product of VACV transcription (16–18), and when E3 or the decapping enzymes are disabled, dsRNA activates oligoadenylate synthetase (OAS), which catalyzes the formation of oligonucleotides that stimulate RNase L, leading to the degradation of mRNA and rRNA (19). In addition, excess dsRNA activates protein kinase R (PKR), which phosphorylates the α subunit of eukaryotic initiation factor 2 (eIF2α), resulting in inhibition of translation (20). The multifunctional E3 protein may also directly inhibit PKR (21–24) and ISG15, a ubiquitin-like protein that has multiple antiviral functions (25, 26). Another VACV protein, K3, competes with activated PKR for association with eIF2α (27, 28), providing another layer of protection. The susceptibility of the VACV C7/K1 host range mutant to IFN in Huh7 cells can be reproduced by expression of IFN regulatory factor 1 (IRF1), which induces a subset of ISGs (29). Subsequent studies showed that C7 and K1 bind the ISG SAMD9 and that knockout of this protein allows replication of the C7/K1 mutant (30, 31). Recently, FAM111A was shown to be IFN inducible and to inhibit replication of the VACV SPI-1 (C12) host range mutant (32). Thus, six antiviral ISGs and opposing viral defense genes have been identified through the study of VACV host range mutants.
In the present study, we screened replication-competent VACV mutants with gene deletions for their sensitivity to beta interferon (IFN-β), as an alternative to investigating host range mutants. By treating the cells with IFN prior to infection, we focused on ISGs and the antiviral state. This approach led to the discovery that C9, a previously uncharacterized 75-kDa ankyrin repeat/F-box protein that is conserved in orthopoxviruses, as determined by a BLAST search, counteracts the action of IFN on early events in the replication cycle of VACV and interacts with components of the SCF (SKP, cullin 1 [CUL1], F-box) and SCN (COP9 signalosome/deneddylation) complexes in human cells.
RESULTS
IFN-sensitive VACV strains.
IFN sensitivity varies with the VACV strain and cell line employed. To initiate this project, we analyzed the replication of VACV strain Western Reserve (WR) in IFN-β-treated human lung carcinoma A549 cells as well as A549 double-knockout (DKO) cells in which the ISGs PKR and RNase L have been inactivated by clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 (12). The cells were treated for 24 h with 2 to 2,000 international units of IFN-β per ml and then infected with WR. After 3 h and 24 h, the virus yields were determined by plaque assay. The yields were higher in the A549 DKO cells than in the parent A549 cells, but even the highest concentrations of IFN-β caused relatively modest reductions in either cell line (Fig. 1A). The induction of the ISG OAS1 confirmed that the IFN was active in both A549 and A549 DKO cells (Fig. 1B). The IFN resistance of WR was likely due to the activity of virus-encoded immunomodulatory proteins that counteracted the antiviral state by neutralizing ISGs. In all succeeding experiments, the high dose of 2,000 units of IFN-β was used.
FIG 1.
Sensitivity of VACV strains to IFN-β. (A) Resistance of VACV WR to IFN-β. A549 and A549 RNase L and PKR double-knockout (DKO) cells were pretreated with 0, 2, 20, 200, or 2,000 IU/ml of IFN-β for 24 h and then infected with WR at 1 PFU/cell. After 3 h with no IFN or 24 h with no or various amounts of IFN, the virus yields were determined by plaque assay. (B) Mock-infected A549 and A549 DKO cells were treated with 0 to 2,000 IU/ml of IFN-β for 24 h, and lysates were analyzed by Western blotting using antibody to OAS1 and actin. (C) IFN-β sensitivity of attenuated VACV strains. A549 and A549 DKO cells were untreated or treated with 2,000 IU/ml of IFN-β for 24 h and infected with 0.01 PFU/cell of VACV WR, 6/2, ACAM2000, or Dryvax. After 48 h, the lysates were analyzed by Western blotting using antibody to VACV, the L1 late protein, and actin. Short and long exposures for L1 are shown. The positions of protein markers (in kilodaltons) are shown on the right of panels B and C.
Because of the resistance of VACV WR to IFN, our strategy was to identify highly sensitive mutant viruses as a way to investigate the IFN evasion proteins of the parent. We compared the effects of IFN on viral protein synthesis in cells infected with the well-characterized, pathogenic WR strain to the effects in cells infected with an attenuated WR mutant, referred to as 6/2, with a large spontaneous deletion near the left end of the genome (33) and to the effects in cells infected with Dryvax and the related ACAM2000 attenuated vaccine strains (34, 35). The infections were carried out by infecting A549 and A549 DKO cells with 0.01 PFU/cell of each virus for 48 h to allow virus spread. Viral protein synthesis, determined by Western blotting with broadly reactive antiserum to abundant VACV proteins, was used to assess replication. The numerous viral protein bands from lysates of A549 and A549 DKO cells that had been infected with WR in the absence or presence of IFN were similar in intensity, confirming the resistance to IFN (Fig. 1C, top). In contrast, the intensities of the viral bands from both A549 cells and A549 DKO cells infected with 6/2, ACAM2000, and Dryvax were diminished by IFN treatment (Fig. 1C, top). The effect of IFN on viral protein synthesis was also measured by probing the Western blots with a monoclonal antibody (MAb) specific to the late L1 protein. More L1 was detected in WR-infected A549 DKO cells that lacked RNase L and PKR than in A549 cells, but there was little effect of IFN in either case (Fig. 1C, bottom). The IFN sensitivity of 6/2 was greater than that of ACAM2000 or Dryvax and was not alleviated by the knockout of PKR and RNase L. The result obtained with the DKO cells suggested that novel ISGs might be involved. Subsequent experiments were designed to determine the basis for the IFN sensitivity of 6/2 compared to parental WR.
Identification of C9 as a VACV IFN evasion protein.
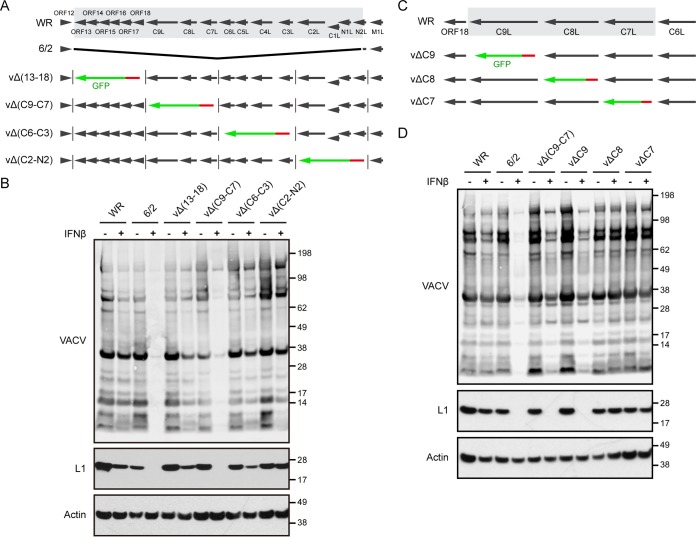
Previous studies (33) suggested that 6/2 suffered a deletion of 17 contiguous open reading frames (ORFs) near the left end of the genome and the addition of 2 ORFs at the right end during a spontaneous transposition event. No additional changes were detected by restriction endonuclease analysis. Although 6/2 replicates well in cell culture, the virus is attenuated in mice, consistent with a defect in immune defense. We considered that one or more ORFs that had been deleted from 6/2 were responsible for IFN sensitivity. A comparison of the ORFs present in WR but missing from 6/2 is shown in Fig. 2A. To narrow the number of possible genes responsible for IFN resistance, we deleted blocks of 3 to 5 ORFs from WR by homologous recombination with DNA encoding the enhanced green fluorescent protein (GFP) regulated by a late VACV promoter. Recombinant virus plaques were identified by their fluorescence and clonally purified. The absence of the deleted genes was confirmed by PCR and DNA sequencing. The IFN resistance of the four deletion mutants was compared to that of the parental WR strain and mutant 6/2 by Western blotting of extracts from infected A549 cells (Fig. 2B). The greatest IFN sensitivity occurred when the deletion encompassed ORFs C7, C8, and C9.
FIG 2.
Identification of C9 as an IFN antagonist. (A) The diagram at the top shows the left portion of the WR genome, with the 17 ORFs deleted from the 6/2 mutant shaded gray. Black arrows indicate the direction of transcription. Lower diagrams depict four WR mutants, vΔ(13–18), vΔ(C9-C7), vΔ(C6-C3), and vΔ(C2-N2), with deletions of multiple ORFs. The green arrows and vertical lines indicate the replacement of WR ORFs by GFP regulated by the P11 promoter, in red. (B) IFN sensitivity of the viruses with deletions of multiple ORFs. A549 cells were untreated (lanes −) or treated with 2,000 IU/ml of IFN-β for 24 h (lanes +) and infected with 0.01 PFU/cell of the indicated virus. After 48 h, the lysates were analyzed by Western blotting using antibody to VACV, the L1 protein, and actin as a loading control. The positions of protein markers (in kilodaltons) are shown on the right. (C) The diagram at the top shows a portion of the WR genome containing ORFs 18, C9L, C8L, C7L, and C6L, with black arrows indicating the direction of transcription. The shaded ORFs were individually deleted to construct three deletion mutants, vΔC9, vΔC8, and vΔC7. The green arrows depict the replacement of an individual WR ORF by GFP regulated by the P11 promoter, in red. (D) The IFN sensitivity of the deletion mutants was determined as described in the legend to panel B.
Using a similar protocol, the C7, C8, and C9 genes were individually replaced with GFP, and the recombinant viruses were isolated (Fig. 2C). Western blotting revealed that only the C9 deletion mutant (vΔC9) exhibited IFN sensitivity comparable to that of 6/2 (Fig. 2D). The IFN sensitivity of vΔC9 was confirmed by demonstrating a >6-fold decrease in the 24-h virus yield (P < 0.0001) following infection of A549 cells at a multiplicity of 3 PFU/cell in triplicate, whereas a 1.5-fold decrease in yield of WR was insignificant (P = 0.165).
Synthesis of intermediate and late proteins is reduced in IFN-treated cells infected by 6/2 and vΔC9.
Expression of VACV proteins is transcriptionally regulated, and the program can be divided into early, intermediate, and late stages. The early proteins include enzymes and factors for DNA replication and transcription of intermediate genes; intermediate proteins include factors for transcription of late genes; late proteins include factors that are packaged in virus particles for transcription of early genes. Since the early genes are transcribed within the viral core, neither de novo protein synthesis nor DNA synthesis is required. Genes that are transcribed only from replicated DNA are referred to as postreplicative genes, and many have both intermediate and late promoters (36). In our initial screening for IFN sensitivity, virus replication was assessed using a low-multiplicity spread assay in which VACV late protein synthesis was measured. Further experiments were designed to analyze early, intermediate, and late protein synthesis under synchronous one-step infection conditions.
Untreated or IFN-β-treated A549 cells were mock infected or infected with 3 PFU per cell of WR, 6/2, or vΔC9 and lysed after 8 h. The lysates were analyzed by Western blotting with antibodies to the E3, D13, A3, and L1 proteins. E3 is an early protein expressed as major and minor products from the first and second AUG of the ORF, respectively (37); D13 and A3 are both regulated by promoters with dual intermediate and late functionality, but A3 has a relatively stronger late component; and L1 is a late protein (36, 38). In the cells infected with WR, IFN treatment slightly enhanced synthesis of the early protein and slightly diminished synthesis of the intermediate and late proteins (Fig. 3A). Although IFN treatment also slightly increased early protein synthesis by 6/2 and vΔC9, the synthesis of intermediate and late proteins was almost entirely abrogated (Fig. 3A).
FIG 3.
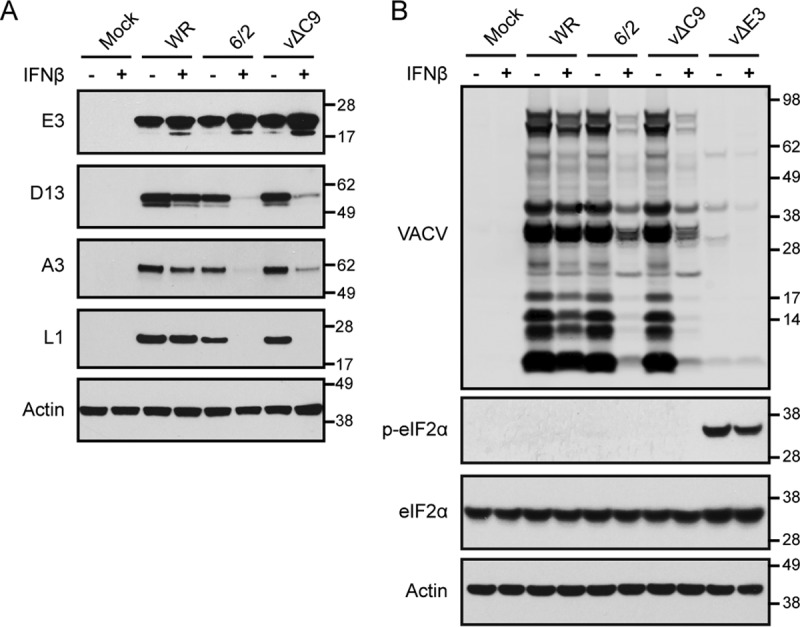
Effect of IFN-β on viral early, intermediate, and late protein synthesis by 6/2 and vΔC9. (A) Effect of IFN-β on the synthesis of representative early, intermediate, and late proteins. A549 cells were untreated (lanes −) or treated with 2,000 IU/ml of IFN-β for 24 h (lanes +) and then infected with 3 PFU/cell of the indicated viruses for 8 h. The lysates were analyzed by Western blotting using antibody to the E3 early protein, the D13 intermediate protein, the A3 late protein, the L1 late protein, and cellular actin. (B) Analysis of eIF2α phosphorylation. A549 cells were treated and infected as described in the legend to panel A, and the Western blots were probed with antibody to VACV, phosphorylated eIF2α (p-eIF2α), total eIF2α, and actin. The positions of protein markers (in kilodaltons) are shown on the right of panels A and B.
The inhibition of intermediate and late protein synthesis in cells infected with some other VACV mutants has a translational basis and correlates with phosphorylation of the translation initiation factor eIF2α. We therefore compared the phosphorylation of eIF2α in cells infected with 6/2 and vΔC9 with that in cells infected with an E3 deletion mutant in the absence and presence of IFN. Unlike 6/2 and vΔC9, viral protein synthesis was reduced in untreated or IFN-treated cells infected with the E3 mutant, and this correlated with phosphorylation of eIF2α (Fig. 3B). Ιn contrast, phosphorylated eIF2α was undetectable under either condition in cells infected with 6/2 or vΔC9 (Fig. 3B). However, the amount of total eIF2α remained constant (Fig. 3B).
IFN reduces transcription of intermediate and late genes by vΔC9.
The inhibition of intermediate and late protein synthesis could be explained by a translational block that did not involve phosphorylation of eIF2α or a transcriptional defect. Digital droplet PCR (ddPCR) was used to determine the amounts of representative early (E3) and intermediate/late (D13 and A3) mRNAs in untreated and IFN-treated cells at 0, 2, and 6 h after infection with WR or vΔC9. In the absence of IFN, the amounts of early mRNA in cells infected with WR or vΔC9 were similar at 2 and 6 h (Fig. 4A). For both viruses, the values for early E3 mRNA at 6 h actually increased severalfold in the presence of IFN, similar to the increase in early protein synthesis. In the untreated cells, the amounts of the D13 mRNA were 100- to 1,000-fold higher at 6 h than at 2 h for each virus (Fig. 4B). IFN reduced the amounts of D13 mRNA made by WR and vΔC9 by 2.3- and 10.4-fold, respectively. The difference was even more striking for the A3 mRNA, which has a relatively stronger late promoter component than D13, as IFN reduced the amounts for WR and vΔC9 by 2.4- and 20.6-fold, respectively. Similar reductions of intermediate and late mRNAs occurred in IFN-treated cells infected with 6/2 (data not shown). Thus, the IFN sensitivity of vΔC9 correlated with a severe reduction in transcription of postreplicative genes.
FIG 4.
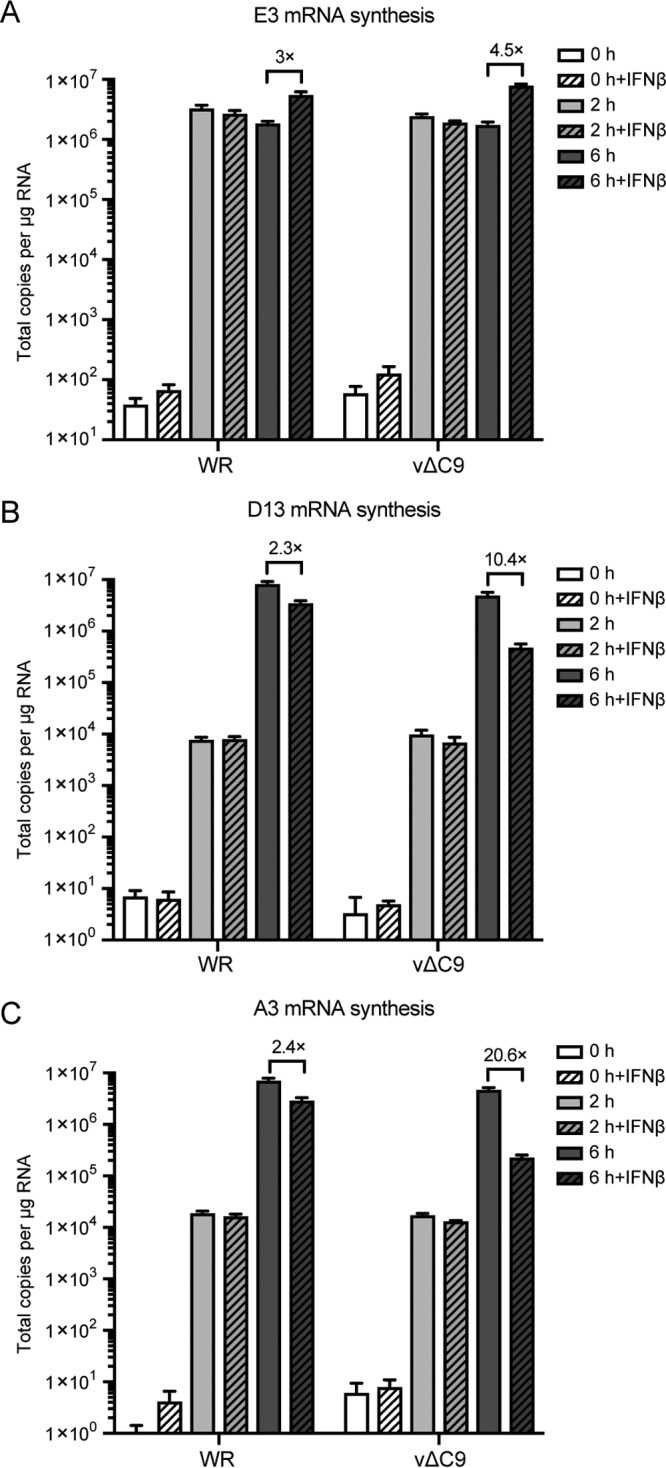
Effect of IFN-β on early, intermediate, and late mRNA synthesis. A549 cells were untreated or treated with 2,000 IU/ml of IFN-β for 24 h and infected with 3 PFU/cell of purified WR or vΔC9. At 0, 2, or 6 h after infection, the RNA was extracted and reverse transcribed. Primers specific for E3 mRNA (A), D13 mRNA (B), and A3 mRNA (C) were used for quantification by ddPCR. The experiments were repeated three times, and the data were combined to make the figure. The error bars represent SEMs.
IFN diminishes viral genome and plasmid synthesis in cells infected with vΔC9.
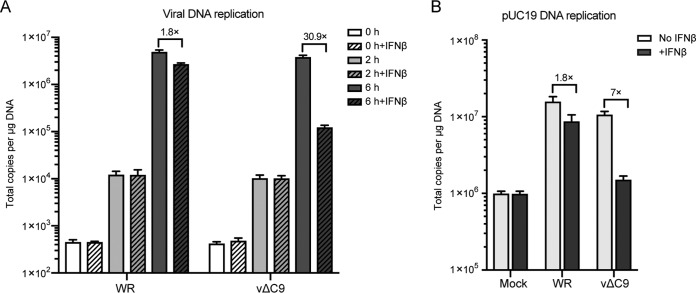
A defect in viral genome replication, which is mediated by early proteins, could account for the reduced transcription of intermediate and late genes. To test this hypothesis, untreated and IFN-treated A549 cells were infected with WR or vΔC9, and the numbers of genome copies were determined by ddPCR at 0, 2, and 6 h. Since the zero time sample was obtained by washing cells immediately after addition of virus, the DNA at the 2-h time point could represent DNA predominantly from attached virions or cytoplasmic cores rather than replicated DNA. In each case, DNA copies increased more than 100-fold between 2 and 6 h due to replication (Fig. 5A). IFN reduced the number of genome copies by less than 2-fold in cells infected with WR but by more than 30-fold in cells infected with vΔC9. Similar results were obtained with 6/2 (data not shown). Thus, the inhibition of DNA replication could account for the decreased postreplicative transcription and protein synthesis.
FIG 5.
Effect of IFN-β on DNA replication. (A) Viral DNA replication. A549 cells were untreated or treated with 2,000 IU/ml of IFN-β for 24 h and then infected with 3 PFU/cell of purified WR or vΔC9. At 0, 2, and 6 h after infection, DNA was extracted with a DNeasy blood and tissue kit (Qiagen). Viral DNA was quantified by ddPCR using primers for the E11L ORF. (B) Plasmid replication. A549 cells were transfected with pUC19 DNA. After 6 h, the cells were untreated or treated with IFN-β for 18 h and mock infected or infected with purified WR or vΔC9. At 6 h after infection, total DNA was isolated and digested with the BamHI and DpnI enzymes. The unmethylated pUC19 DNA was quantified by ddPCR. The experiments were repeated three times, and the data were combined to make the figure. The error bars represent SEMs.
The viral genome must be released from the virus core for its replication to occur. However, the poxvirus cytoplasmic DNA synthesis system has the ability to replicate any transfected circular DNA (39, 40). Although all of the viral proteins directly required for genome replication are also required for plasmid replication (41), there are important distinctions: no viral sequences are in the plasmid, and DNA uncoating is unnecessary. We used a recently described ddPCR assay (42) to quantify plasmid replication. The assay is based on the difference between transfected DNA, which is methylated, and replicated DNA, which is not. A549 cells were transfected with pUC19 and 6 h later were incubated with or without IFN, followed by mock infection or infection with WR or vΔC9. At 6 h after infection, DNA was extracted and treated with DpnI and BamHI to digest the methylated input plasmid and to linearize the DNA, respectively. In the absence of IFN, the amount of plasmid sequences in both WR- and vΔC9-infected cells increased more than 10-fold over that in mock-infected cells (Fig. 5B). IFN reduced the amount of plasmid sequences by 1.8- and 7-fold in the cells infected with WR and vΔC9, respectively. The 7-fold reduction value was only 2-fold higher than the value for mock-infected cells, indicating a profound inhibition of plasmid replication in the cells infected by vΔC9. Since the plasmid transfection assay bypasses the genome uncoating step, the data suggest a direct effect of IFN treatment on DNA synthesis. Nevertheless, this does not rule out the possibility of effects on both genome uncoating and DNA synthesis.
Inhibition of genome uncoating of vΔC9 by IFN.
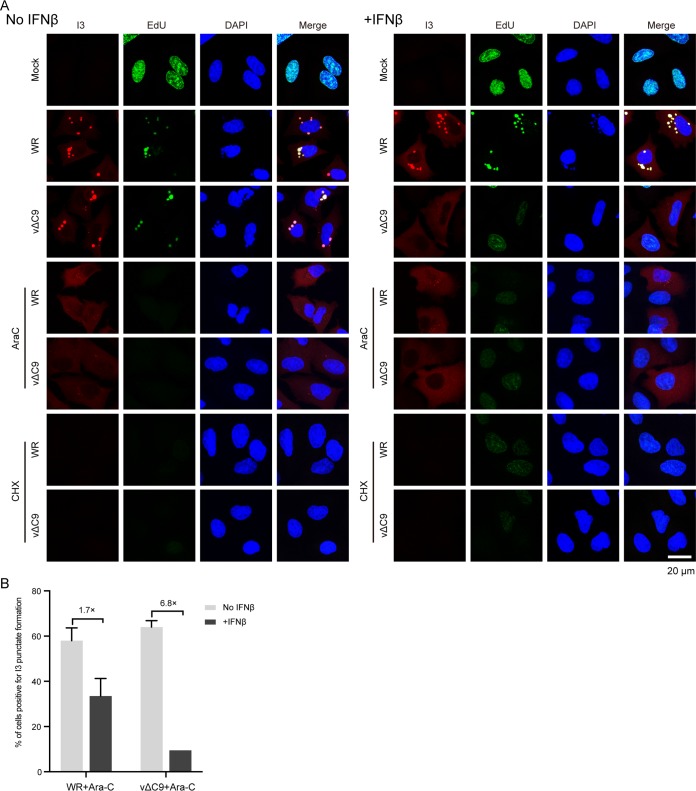
The first part of the experiment described in this section confirmed the IFN-mediated inhibition of viral DNA replication in cells infected with vΔC9. 5-Ethynyl-2′-deoxyuridine (EdU), a thymidine analog that can be linked to a fluor by click chemistry, has been used to label newly synthesized VACV DNA in cytoplasmic factories and demonstrate the shutdown of cellular DNA synthesis in the nucleus (43). As shown in Fig. 6A, EdU was incorporated into the nuclei of uninfected cells regardless of IFN treatment. However, nuclear incorporation was almost entirely eliminated following infection of untreated or IFN-treated cells with WR. The VACV I3 single-stranded DNA binding protein is expressed early in infection and prominently colocalized with the cytoplasmic replicated viral DNA in untreated and IFN-treated cells infected with WR (Fig. 6A). In the absence of IFN, cells infected with vΔC9 exhibited EdU and I3 staining patterns similar to those of cells infected with WR. In contrast, no viral DNA synthesis was detected in the cytoplasm of IFN-treated cells infected with vΔC9, and therefore, I3 was distributed diffusely throughout the cytoplasm. Thus, the effect of IFN on the synthesis of vΔC9 genomic DNA shown by ddPCR was confirmed by the inhibition of EdU incorporation.
FIG 6.
Effect of IFN-β on release of the vΔC9 genome from viral cores. (A) Confocal microscopy. A549 cells were pretreated with 2,000 IU/ml of IFN-β for 24 h and then mock infected or infected with 3 PFU/cell of purified WR or vΔC9 in the presence or absence of AraC or CHX. After 3 h, the cells were incubated with 10 μM EdU for 1 h, fixed, and permeabilized. EdU was reacted with Alexa Fluor 647 azide by click chemistry, and the cells were stained with antibody to the I3 protein followed by Alexa Fluor 568 secondary antibody and DAPI and examined by confocal microscopy. Bar, 20 μm. (B) Automatic tiling was used to select fields of cells, and the percentages of AraC-treated cells positive for I3 punctate spot formation relative to the total number of cells expressing I3 were determined manually. Approximately 300 I3-positive cells were counted. The error bars represent standard deviations.
As noted above, even though our data suggested that IFN treatment has a direct effect on DNA synthesis in cells infected with vΔC9, this did not preclude the possibility of an additional effect on genome uncoating. Indeed, several recent studies have suggested that genome uncoating and genome replication are intertwined (44–46). We modified a previously described uncoating assay that detects viral genomes released from viral cores by binding to the early I3 protein in the absence of viral DNA replication (46). The inhibition of viral DNA replication in A549 cells treated with cytosine-d-arabinofuranoside (AraC) was confirmed by the absence of EdU incorporation (Fig. 6A). However, AraC did not prevent uncoating, as shown by the bright punctate spots superimposed on diffuse I3 staining in both the WR- and vΔC9-infected cells that had not been treated with IFN. Importantly, the I3 punctae were still detected in the cells that had been treated with both AraC and IFN and infected with WR but were rare in the cells infected with vΔC9, indicating that the genome had not been released. We also treated cells with cycloheximide (CHX) to prevent protein synthesis and confirmed that there was insignificant background staining with the I3 antibody (Fig. 6A).
To quantify the data, the number of AraC-treated cells that had I3 punctae was determined (Fig. 6B). IFN reduced the percentage by 1.7-fold and 6.8-fold in the cells infected with WR and vΔC9, respectively. Taken together, our results indicate that uncoating and viral DNA replication were both reduced in IFN-treated cells that were infected with vΔC9.
C9 is an early protein, and ectopic expression overcomes IFN-mediated inhibition of vΔC9.
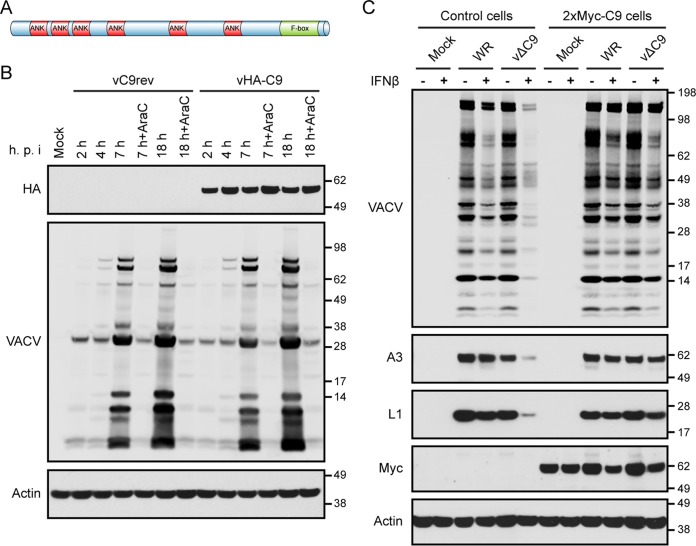
The VACV WR C9L ORF is predicted to encode a protein of 634 amino acids with 6 ankyrin (ANK) motifs and an F-box near the C terminus (Fig. 7A). In order to investigate the expression of the putative C9 protein, a recombinant virus encoding an epitope-tagged C9 protein was constructed. The P11 promoter-GFP ORF in vΔC9 (Fig. 2C) was replaced with C9 or C9 with an N-terminal hemagglutinin (HA) tag regulated by the original C9 promoter sequence. The recombinant viruses were identified by the loss of fluorescence and were plaque purified. The HA-C9 protein of vHA-C9, but not the C9 protein of the control virus vC9rev, was detected with a MAb to the epitope tag at 2 h and subsequent times even in the presence of AraC (Fig. 7B). In contrast, synthesis of the late proteins detected with broadly reactive VACV antiserum occurred later and was inhibited by AraC (Fig. 7B). Thus, C9 belongs to the early class of VACV proteins.
FIG 7.
Characterization of C9 protein and IFN resistance in C9-expressing cells. (A) Predicted domain structure of C9 protein showing ankyrin (ANK) repeats and the C-terminal F-box domain. (B) Expression of HA-tagged C9 protein. A549 cells were mock infected or infected with vC9rev or vHA-C9 in the absence or presence of AraC. At the indicated hours postinfection (h.p.i.), the cells were harvested, and lysates were analyzed by Western blotting with antibody to the HA epitope, VACV, or actin. (C) Ectopic expression of 2×Myc-C9 overcomes the IFN-β-sensitive phenotype of vΔC9. A549 control cells and A549 cells stably expressing 2×Myc-C9 were untreated (lanes −) or treated with 2,000 IU/ml of IFN-β for 24 h (lanes +) and then were mock infected or infected with 0.01 PFU/cell of purified WR or vΔC9. At 48 h after infection, the lysates were analyzed by Western blotting using antibody to VACV, A3, L1, the Myc epitope, and actin. The positions of protein markers (in kilodaltons) are shown on the right of panels B and C.
Next, we wanted to determine whether ectopically expressed C9 could overcome the IFN sensitivity of vΔC9. A549 cells expressing C9 with 2 tandem N-terminal Myc tags (2×Myc-C9 cells) were generated by retroviral transduction and identified by Western blotting with antibody specific to the Myc tag. The function of the epitope-tagged C9 was demonstrated by treating the cells with IFN and infecting them with WR and vΔC9. In contrast to the control cells, the IFN-treated 2×Myc-C9 cells supported late protein synthesis by vΔC9 (Fig. 7C). Thus, expression of the epitope-tagged C9 allowed vΔC9 to resist IFN.
Association of cellular proteins with C9.
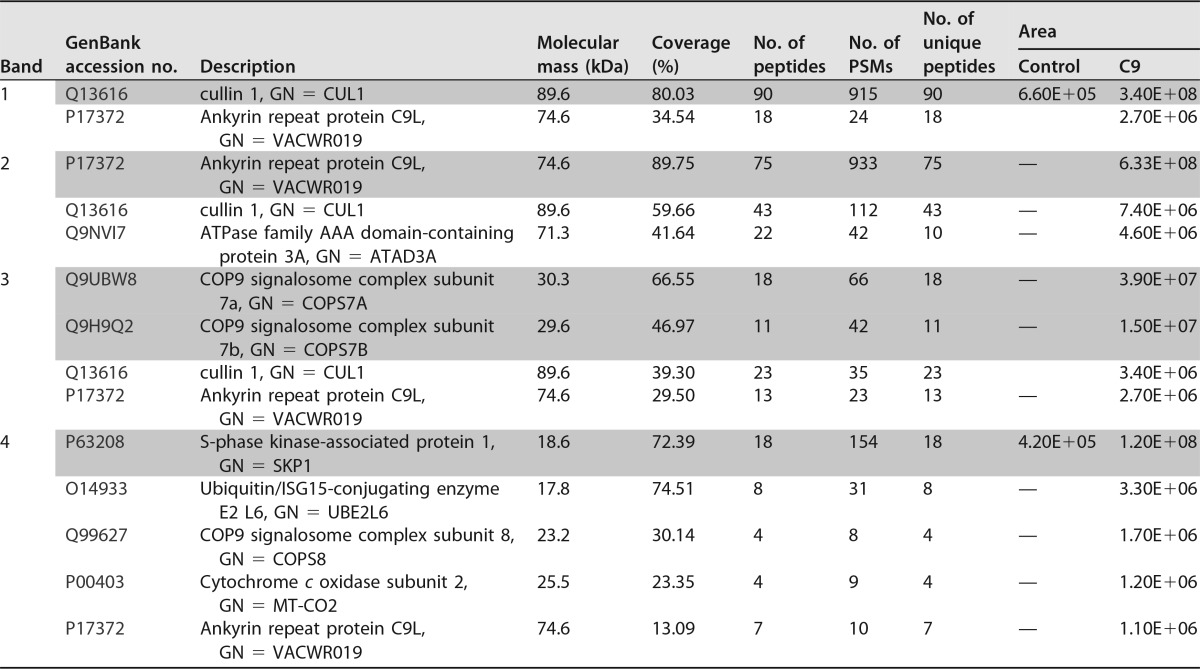
The main purpose of making a cell line expressing Myc-tagged C9 was for the isolation of C9-associated cellular proteins. Having established that the Myc-tagged C9 was functional, lysates of IFN-treated control cells and 2×Myc-C9 cells were incubated with Myc-Trap agarose beads to capture C9 and associated proteins. After extensive washing, the bound proteins were eluted, subjected to SDS-PAGE, and stained with Coomassie blue. Inspection of the gel revealed four bands that were more intense in the sample from the 2×Myc-C9 cells than in the sample from the control cells (Fig. 8A). The bands were excised and trypsinized, and the peptides were analyzed by label-free nano-liquid chromatography-tandem mass spectrometry. For the initial analysis, we set the following criteria: at least three unique peptides were required for protein identification, and the extracting ion current (XIC) peak areas of the three most abundant peptides had to meet a threshold of 105 for significance. In addition, the peptides had to be undetected in the samples from the control cells, or the average areas of the ion peaks from the 2×Myc-C9 cells had to be at least 10-fold greater than those from the control cells. Proteins that exceeded the above-described criteria with areas of 106 or more are shown in Table 1. CUL1, C9, COP9 signalosome subunits 7A and 7B (COPS7A and COPS7B), and S-phase kinase-associated protein 1 (SKP1) were the most abundant proteins in bands 1, 2, 3, and 4, respectively. The presence of CUL1 and C9 in smaller amounts in additional bands probably reflects their abundance and some degradation.
FIG 8.
Identification of proteins associated with C9. (A) Coomassie blue-stained protein bands. Control and A549 2×Myc-C9 cells were pretreated with 2,000 IU/ml of IFN-β for 24 h. The cells were lysed, and proteins were affinity purified using Myc-Trap agarose beads. The bound proteins were eluted and resolved by SDS-PAGE, and the gel was stained with Coomassie blue. Gel bands 1 to 4 were excised individually from the 2×Myc-C9 cells and the corresponding positions of control cells and analyzed by mass spectrometry. A ladder of protein markers (in kilodaltons) is on the left. (B) Protein interaction network. Proteins identified by mass spectrometry and listed in Table 1 were analyzed using the String Protein-Protein Interaction Network. Connecting lines indicate known and predicted protein interactions. Node colors (Gene Ontology terms) are as follows: red, proteolysis (GO: 0006508); blue, cullin deneddylation (GO: 0010388) and COP9 signalosome (GO: 0008180); green, SCF ubiquitin ligase complex (GO: 0019005); white, unassociated proteins. (C) Verification of the mass spectrometry results. Proteins from lysates of control or A549 2×Myc-C9 cells with (lanes +) or without (lanes −) IFN-β treatment were captured with Myc-Trap beads and analyzed by Western blotting using antibodies to the Myc epitope, SKP1, CUL1, and COPS7A.
TABLE 1.
Proteins associated with 2×Myc-C9a
Abbreviations and symbols: GN, gene name; PSM, peptide spectral mass; —, undetected; Area, XIC peak area in arbitrary units. The information for the most abundant proteins from each band is shaded.
It seemed unlikely that C9 could interact individually with each of the proteins in Table 1. To assess whether these proteins interacted with each other to form a complex, all the gene names were entered into the String Protein-Protein Interaction Network. Six of the eight proteins were in the proteolysis (GO: 0006508) Gene Ontology (GO) biological process subset, and of these, three were also assigned to the cullin deneddylation/COP9 signalosome (GO: 0010388 and 0008180) and two were assigned to the SCF ubiquitin ligase complex (GO: 0019005). The interaction network of the six proteins is shown in Fig. 8B. Another subunit of the signalosome complex, GPS1 (SCN1), which interacts with COPS7A, COPS7B, and CUL1, was also detected by mass spectrometry but not included in Table 1 because its abundance was slightly below the imposed cutoff. The two proteins that were not associated with the complex, ATAD3A and MT-CO2 (Fig. 8B), were present in smaller amounts than the ones in the complex (Table 1), and their significance, if any, remains to be determined.
The experiment was repeated with untreated and IFN-treated control and 2×Myc-C9 cells. Antibodies to two representatives of the SCF complex (SKP1, CUL1) and one representative of the cullin deneddylation/COP9 signalosome complex (COPS7A) confirmed the mass spectrometry data by Western blotting. These proteins were detected only in the affinity-purified samples from the 2×Myc-C9 cells (Fig. 8C). The interaction occurred, however, regardless of whether the cells were treated with IFN. UBE2L6 was detected by Western blotting only in lysates of IFN-treated cells; however, the signal was less intense than that for the other proteins and was not detected after affinity purification (data not shown).
DISCUSSION
Through coevolution, viruses and their hosts have arrived at a balanced state which obscures the underlying dynamics. For example, despite the existence of more than 300 ISGs, VACV WR can fully replicate in IFN-treated human cells. However, the number of viral genes needed to achieve IFN resistance is unknown. In the first part of this study, we screened attenuated vaccine strains (ACAM2000 and Dryvax) and a VACV WR mutant (6/2) with a large DNA deletion and determined that they exhibited decreased replication in IFN-treated human cells compared to WR. The previously characterized 6/2 mutant has a spontaneous deletion encompassing 17 ORFs near the left end of the genome (33). Interestingly, Paez and Esteban (47) had found that the presence of IFN in the culture medium prevented the appearance of spontaneous deletions at the left end of the genome during persistent infection. By systematically deleting groups of genes and individual genes from the parent WR virus, we demonstrated that the IFN sensitivity of 6/2 could be mimicked by loss of the C9L gene alone. Furthermore, the VACV C9L deletion mutant, vΔC9, could replicate in the presence of IFN in a transformed cell line expressing Myc-tagged C9, indicating that C9 was sufficient to restore resistance. Homologs of the C9L ORF are conserved in orthopoxviruses, including modified VACV Ankara (MVA), which has numerous deletions, suggesting an important role in their replication. Since C9 is encoded by ACAM2000, a clonal isolate of Dryvax (35), there is likely to be another basis for its IFN sensitivity, although we did not actually measure C9 expression.
In the second part of this study, we showed that vΔC9 synthesized early proteins but not intermediate or late proteins in IFN-treated cells. The defect in protein synthesis was not associated with phosphorylation of PKR or the translation initiation factor eIF2α, nor was it relieved in cells in which both RNase L and PKR were inactivated. By working backwards, we found that the block in protein synthesis resulted from a profound inhibition of intermediate and late mRNA synthesis and that the latter corresponded to a failure of genome replication. For viral genome replication, the DNA must be released from the core and then acted upon by a large number of viral early proteins (43, 48, 49). We found that the release of viral DNA from cytoplasmic cores was reduced in IFN-treated cells that were infected with vΔC9. However, under these conditions, we also found a strong reduction in the replication of plasmid DNA, for which uncoating is not required. Taken together, the IFN sensitivity of the C9L deletion mutant appears to be due to defects in uncoating and DNA synthesis, unlike other IFN-sensitive mutants that exhibit more direct defects in protein synthesis. This dual effect was not too surprising, as other studies have provided evidence for the intertwining of uncoating and DNA replication, both of which require ubiquitin/proteasome functions (44–46, 50). In the absence of IFN, the essential D5 protein (51) and three proteins (B18, C5, and M2) with mutually redundant roles (42) are involved in both uncoating and DNA replication, although the mechanisms are not yet understood. The present study adds a requirement for an additional protein, C9, in IFN-treated cells.
As anticipated from the early requirement for C9, the protein was expressed prior to viral DNA replication. Although we do not yet know how C9 contributes to genome uncoating and DNA replication, there are some clues. Using a cell line that expresses 2×Myc-tagged C9, we affinity purified C9 with associated proteins. The abundant proteins identified by mass spectrometry included the CUL1 and SKP1 components of the SCF complex and the COPS7A, COPS7B, and COPS8 components of the cullin deneddylation/COP9 signalosome complex. String Protein-Protein Interaction Network and Gene Ontology analyses confirmed that these proteins are all associated with each other and have related functions. SCF is an E3 ubiquitin ligase complex that catalyzes the ubiquitination of proteins, usually for proteosomal degradation (52). The COP9 signalosome is homologous to the 26S proteasome, has isopeptidase activity, and is responsible for the removal of a ubiquitin-like protein, Nedd8 (hence the term deneddylation), from the CUL1 component of SCF, which regulates its activity (53). The specificity of the SCF is determined by F-box proteins, which bind to CUL1 and a target protein. The human genome encodes at least 69 F-box proteins, although not all have been confirmed functionally and the roles of most are unknown (54). Intriguingly, C9 is predicted to contain an F-box that is likely responsible for the interaction with SKP1, leading to the copurification of the additional associated proteins. An important remaining question is whether C9 directs proteosomal degradation of a specific cell protein, perhaps an ISG. This specificity could be provided by the six ANK repeats in C9. The combination of ANK repeats and F-box-like domains (called Ank/PRANC) is found in proteins encoded by nearly all poxvirus genera (55). However, in very few cases have the interactions of the poxvirus ANK repeats been determined (56). The rapid equilibrium between existing SCF complexes that can occur following cell lysis (52, 57) may contribute to the difficulty in analysis.
In summary, we demonstrated the utility of using a virus with a large deletion to probe the resistance of VACV to IFN, identified a previously uncharacterized VACV gene that is conserved in orthopoxviruses to be an antagonist of the antiviral state, showed that inhibition occurs at the stages of genome uncoating and replication, and implicated the SCF and signalosome complexes as effectors.
MATERIALS AND METHODS
Cells.
Cells of the A549 and A549 RNase L and PKR CRISPR-Cas9 double-knockout (DKO) cell lines (12) and BS-C-1 cells were grown in Dulbecco's modified Eagle's medium–F-12 medium (Life Technologies) or Eagle's minimum essential medium (EMEM) containing 10% fetal bovine serum (FBS; Sigma-Aldrich), and 2 mM l-glutamine, 100 units of penicillin, and 100 μg of streptomycin per ml (Quality Biologicals, Inc.).
Antibodies and chemicals.
The antibodies used included the following: mouse monoclonal antibodies to E3 (58), I3 (59), and L1 (60); rabbit antibodies to VACV strain WR (61), A3 (unpublished), and D13 (62); phospho-(Ser51)-eIF2α (catalog number 3398), eIF2α (catalog number 5324), and OAS1 (catalog number 14498) from Cell Signaling Technology; horseradish peroxidase (HRP)-conjugated c-Myc antibody (9E10; catalog number sc-40 HRP), CUL1 (D-5; catalog number sc-17775), SKP1 p19 (H-6; catalog number sc-5281), CSN7a (D-6; catalog number sc-398882), and actin (catalog number sc-47778) from Santa Cruz Biotechnology; and HA (catalog number 901501; BioLegend). CHX (catalog number C4859) and cytosine-d-arabinofuranoside (AraC; catalog number C1768) were from Sigma-Aldrich. Human IFN-β1a (catalog number HC99921B) was from Antigenix America.
Viruses.
The VACV mutants 6/2 (33) and vΔE3 (63) were described previously. The following recombinant VACVs were also derived from the WR strain of VACV (ATCC VR-1354) by deleting the ORFs in parentheses: vΔ(13–18), vΔ(C9-C7), vΔ(C6-C3), vΔ(C2-N2), vΔC9, vΔC8, and vΔC7. In each case, the ORFs were deleted by homologous recombination with DNA constructed by overlap PCR with a mixture containing the GFP ORF regulated by the P11 late promoter flanked by portions of the adjacent genes. The revertant C9 virus (vC9rev) and HA-tagged C9 virus (vHA-C9) were generated by replacing the P11-GFP ORF in vΔC9 with a wild-type C9L or the C9 ORF with an HA tag at the N terminus regulated by the natural promoter. Basically, homologous recombination was carried out by infecting BS-C-1 cells with 1 PFU/cell of WR or vΔC9, followed by transfection with assembled PCR products using the Lipofectamine 2000 reagent (Thermo Fisher). After 24 h, cells were harvested and lysed by three freeze-thaw cycles. The lysates were diluted 10-fold and used to infect BS-C-1 cell monolayers. Green or colorless recombinant plaques were distinguished from the parental plaques and clonally purified five times. The purities of the recombinant viruses were confirmed by PCR amplification and sequencing of the modified region. The WR and recombinant viruses were propagated in BS-C-1 cells.
Purification of virus particles.
Recombinant viruses grown in BS-C-1 cells were purified by centrifugation through a 36% sucrose cushion, followed by centrifugation through a 24% to 40% sucrose gradient, as described previously (64), and used for infections after determination of infectivity by plaque assay in BS-C-1 cells.
Plaque assay and virus yield determination.
Virus samples were disrupted in a chilled water bath sonicator with two 30-s periods of vibration, followed by 10-fold serial dilutions in EMEM supplemented with 2.5% FBS. Diluted viruses were distributed onto BS-C-1 cell monolayers. After adsorption for 1.5 h, the medium was aspirated and replaced with medium containing 0.5% methylcellulose. After 48 h, the cells were stained with crystal violet at room temperature for 20 min and dried overnight and the plaques were counted.
Construction of the A549 2×Myc-C9 cell line.
A549 cells expressing the 2×Myc tagged C9 protein were created by retroviral transduction. A eukaryotic codon-optimized C9 ORF with an N-terminal 2×Myc tag (2×Myc-C9) was cloned into pQCXIP (Clontech) to generate pQCXIP-2×Myc-C9. Retrovirus particles were produced by cotransfecting pQCXIP or pQCXIP-2×Myc-C9 (transfer plasmid), pMLV-Gag-Pol (packaging plasmid), and pVSV-G (vesicular stomatitis virus G protein [VSV-G] envelope plasmid) into 293T cells using Lipofectamine 2000. A549 cells were infected with the retroviruses in the presence of 5 μg/ml Polybrene (Sigma-Aldrich) by spinoculation at 450 × g for 30 min at room temperature. The cells were subcultured and passaged several times in selection medium containing 1 μg/ml of puromycin (Sigma-Aldrich). The expression of C9 protein was determined by Western blotting using HRP-conjugated anti-Myc antibody.
Affinity purification and mass spectrometry.
A549 and A549 2×Myc-C9 cells from 4 10-cm dishes were pretreated with 2,000 IU/ml IFN-β for 24 h. The cells were washed twice with cold phosphate-buffered saline (PBS) on ice, harvested by scraping, and lysed in immunoprecipitation (IP) buffer (20 mM Tris [pH 7.4], 150 mM NaCl, 2 mM EDTA, 1% Triton X-100) containing protease inhibitor and phosphatase inhibitor (Roche) on wet ice for 30 min with frequent agitation. Lysates were centrifuged for 10 min at 20,000 × g at 4°C, and the supernatant was incubated with 120 μl of control agarose beads (ChromoTek) for 1 h at 4°C on a rotating wheel. The precleared lysates were incubated with 80 μl of Myc-Trap agarose beads (ChromoTek). The lysate and beads were rotated at 4°C for 3 h, followed by washing six times with lysis buffer. The bound proteins were eluted with 1× reducing sample buffer and resolved by SDS-PAGE. The gel was stained with Coomassie blue or silver reagent, the bands of interest were excised from the gel, digested with trypsin, and analyzed by mass spectrometry, and the proteins were identified essentially as described previously (43).
Western blotting.
Cells were harvested, washed, and lysed in lysis buffer (20 mM Tris [pH 7.4], 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, protease inhibitor) on wet ice for 30 min with frequent agitation. Cell lysates were cleared by centrifugation at 13,000 × g for 10 min at 4°C; the proteins were resolved on 4 to 12% NuPAGE bis-Tris gels (Thermo Fisher) and transferred to a nitrocellulose membrane with an iBlot2 system (Thermo Fisher). The membrane was blocked with 5% nonfat milk in Tris-buffered saline (TBS) for 1 h, washed with TBS with 0.1% Tween 20 (TBST), and then incubated with the primary antibody in 5% nonfat milk or 5% bovine serum albumin (BSA) in TBST overnight at 4°C. The membrane was washed with TBST and incubated with the secondary antibody conjugated with horseradish peroxidase (Jackson ImmunoResearch) in TBST with 5% nonfat milk for 1 h. After the membrane was washed, the bound proteins were detected with SuperSignal West Dura substrates (Thermo Scientific). For detection of phosphorylated eIF2α, cells were lysed with buffer containing phosphatase inhibitors, as follows: 2 mM sodium orthovanadate, 2 mM sodium pyrophosphate, 50 mM glycerol-2-phosphate disodium, 50 mM sodium fluoride, and phosphatase inhibitor.
Genome uncoating.
A549 cells were pretreated with IFN-β for 24 h and then mock infected or infected with 3 PFU/cell of purified virus in the presence of AraC (44 μg/ml) or CHX (100 μg/ml). At 3 h after infection, the cells were incubated with 10 μM 5-ethynyl-2′-deoxyuridine (EdU; Thermo Fisher) for 1 h, after which the cells were fixed with 4% formaldehyde for 15 min and permeabilized with 0.5% Triton X-100 for 20 min. EdU was detected by a click reaction with Alexa Fluor 647 azide according to the manufacturer's protocol (Thermo Fisher). Samples were then stained with antibody to the I3 protein followed by Alexa Fluor 568 secondary antibody (Life Technologies). Nuclei and virus factories were stained with 4′,6-diamidino-2-phenylindole (DAPI). Coverslips were mounted on slides using ProLong Gold reagent (Life Technologies). Images were collected on a Leica DMI6000 confocal microscope with a 63× oil immersion objective and processed using Imaris software, version 9.0.2 (Bitplane AG), to adjust the brightness. For quantification, images were collected using an automated tiling method to obtain an unbiased data pool from 50 tiles. Acquired images were further analyzed using Imaris image-processing software to count the number of cells with I3 punctae and the number of cells expressing the I3 protein.
ddPCR for quantification of the copies of the viral genome and specific mRNAs.
A549 cells were untreated or treated with IFN-β for 24 h and infected with 3 PFU/cell of purified viruses. After 0, 2, and 6 h, infected cells were harvested for extraction of DNA and RNA. DNA was extracted using a DNeasy blood and tissue kit (Qiagen). Total RNA was extracted using the TRIzol reagent (Invitrogen), treated with DNase I (Invitrogen), and then reverse transcribed with a SuperScript III first-strand synthesis system for reverse transcription-PCR (Invitrogen). The DNA was serially diluted and used as a template for ddPCR (Bio-Rad). The PCR was carried out with primers binding to individual ORFs following the protocol described previously (65). For analysis of genome replication, the primer pair was (5′ to 3′) E11 Forward (GGTTCGTCAAAGACATAAAACTCATT) and E11 Reverse (GAATACATTCACATTGACCAATCAGAA). For analysis of E3 mRNA the following primer pair was used: E3 Forward (CGTCGATGTCTACACAGGCA) and E3 Reverse (GTGCTAACCCTGTCACCGTT). The primer sequences for D13 and A3 were described previously (66). After 40 reaction cycles, the droplets were digitally analyzed with a droplet reader (Bio-Rad), and absolute DNA or mRNA copy numbers were determined.
Analysis of plasmid replication.
The plasmid replication assay was performed as previously described (42). A549 cells were transfected with pUC19 DNA, treated with or without IFN for 18 h, and mock infected or infected with purified viruses for 6 h. Total DNA was isolated using a DNeasy blood and tissue kit (Qiagen). DNA (2 μg) was digested with the BamHI and DpnI enzymes. The digested DNA was purified using a QIAquick DNA purification kit (Qiagen). The replication of pUC19 was determined by ddPCR using the following primer pair: pUC19 Forward (TCGTTTGGTATGGCTTCATTC) and pUC19 Reverse (GCGGCCAACTTACTTCTGAC).
ACKNOWLEDGMENTS
We thank Catherine Cotter and Jeffrey Americo for maintaining cell lines and advice on ddPCR, respectively. Sundar Ganesan assisted with the confocal microscopy, and Ming Zhao and Lisa R. Olano carried out the mass spectroscopy analysis. Stuart Isaacs and David Evans kindly provided antibodies, and John Connor supplied the E3-deletion virus.
The research was supported by the Division of Intramural Research, NIAID.
REFERENCES
- 1.Isaacs A, Lindenmann J. 1957. Virus interference. I. The interferon. Proc R Soc B Biol Sci 147:258–267. doi: 10.1098/rspb.1957.0048. [DOI] [PubMed] [Google Scholar]
- 2.de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, Williams BR. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol 69:912–920. [PubMed] [Google Scholar]
- 3.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu SY, Sanchez DJ, Aliyari R, Lu S, Cheng G. 2012. Systematic identification of type I and type II interferon-induced antiviral factors. Proc Natl Acad Sci U S A 109:4239–4244. doi: 10.1073/pnas.1114981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haller O, Kochs G, Weber F. 2006. The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344:119–130. doi: 10.1016/j.virol.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paez E, Esteban M. 1984. Resistance of vaccinia virus to interferon is related to an interference phenomenon between the virus and the interferon system. Virology 134:12–28. doi: 10.1016/0042-6822(84)90268-X. [DOI] [PubMed] [Google Scholar]
- 8.Smith GL, Benfield CTO, de Motes CM, Mazzon M, Ember SWJ, Ferguson BJ, Sumner RP. 2013. Vaccinia virus immune evasion: mechanisms, virulence and immunogenicity. J Gen Virol 94:2367–2392. doi: 10.1099/vir.0.055921-0. [DOI] [PubMed] [Google Scholar]
- 9.Bratke KA, McLysaght A, Rothenburg S. 2013. A survey of host range genes in poxvirus genomes. Infect Genet Evol 14:406–425. doi: 10.1016/j.meegid.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang HW, Watson JC, Jacobs BL. 1992. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc Natl Acad Sci U S A 89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang H-W, Jacobs BL. 1993. Identification of a conserved motif that is necessary for binding of the vaccinia virus E3L gene products to double-stranded RNA. Virology 194:537–547. doi: 10.1006/viro.1993.1292. [DOI] [PubMed] [Google Scholar]
- 12.Liu R, Moss B. 2016. Opposing roles of double-stranded RNA effector pathways and viral defense proteins revealed with CRISPR/Cas9 knock-out cell lines and vaccinia virus mutants. J Virol 90:7864–7879. doi: 10.1128/JVI.00869-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parrish S, Resch W, Moss B. 2007. Vaccinia virus D10 protein has mRNA decapping activity, providing a mechanism for control of host and viral gene expression. Proc Natl Acad Sci U S A 104:2139–2144. doi: 10.1073/pnas.0611685104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parrish S, Moss B. 2007. Characterization of a second vaccinia virus mRNA-decapping enzyme conserved in poxviruses. J Virol 81:12973–12978. doi: 10.1128/JVI.01668-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu SW, Katsafanas GC, Liu R, Wyatt LS, Moss B. 2015. Poxvirus decapping enzymes enhance virulence by preventing the accumulation of dsRNA and the induction of innate antiviral responses. Cell Host Microbe 17:320–331. doi: 10.1016/j.chom.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colby C, Jurale C, Kates JR. 1971. Mechanism of synthesis of vaccinia virus double-stranded ribonucleic acid in vivo and in vitro. J Virol 7:71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boone RF, Parr RP, Moss B. 1979. Intermolecular duplexes formed from polyadenylated vaccinia virus RNA. J Virol 30:365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z, Bruno DP, Martens CA, Porcella SF, Moss B. 2010. Simultaneous high-resolution analysis of vaccinia virus and host cell transcriptomes by deep RNA sequencing. Proc Natl Acad Sci U S A 107:11513–11518. doi: 10.1073/pnas.1006594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silverman RH. 2007. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol 81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfaller CK, Li Z, George CX, Samuel CE. 2011. Protein kinase PKR and RNA adenosine deaminase ADAR1: new roles for old players as modulators of the interferon response. Curr Opin Immunol 23:573–582. doi: 10.1016/j.coi.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharp TV, Moonan F, Romashko A, Joshi B, Barber GN, Jagus R. 1998. The vaccinia virus E3L gene product interacts with both the regulatory and the substrate binding regions of PKR: implications for PKR autoregulation. Virology 250:302–315. doi: 10.1006/viro.1998.9365. [DOI] [PubMed] [Google Scholar]
- 22.Romano PR, Zhang F, Tan SL, Garcia-Barrio MT, Katze MG, Dever TE, Hinnebusch AG. 1998. Inhibition of double-stranded RNA-dependent protein kinase PKR by vaccinia virus E3: role of complex formation and the E3 N-terminal domain. Mol Cell Biol 18:7304–7316. doi: 10.1128/MCB.18.12.7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White SD, Jacobs BL. 2012. The amino terminus of vaccinia virus E3 protein is necessary to inhibit the interferon response. J Virol 86:5895–5914. doi: 10.1128/JVI.06889-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thakur M, Seo EJ, Dever TE. 2014. Variola virus E3L Z alpha domain, but not its Z-DNA binding activity, is required for PKR inhibition. RNA 20:214–227. doi: 10.1261/rna.042341.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerra S, Caceres A, Knobeloch KP, Horak I, Esteban M. 2008. Vaccinia virus E3 protein prevents the antiviral action of ISG15. PLoS Pathog 4:e1000096. doi: 10.1371/journal.ppat.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eduardo-Correia B, Martinez-Romero C, Garcia-Sastre A, Guerra S. 2014. ISG15 is counteracted by vaccinia virus E3 protein and controls the proinflammatory response against viral infection. J Virol 88:2312–2318. doi: 10.1128/JVI.03293-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beattie E, Tartaglia J, Paoletti E. 1991. Vaccinia virus encoded eIF-2 alpha homolog abrogates the antiviral effect of interferon. Virology 183:419–422. doi: 10.1016/0042-6822(91)90158-8. [DOI] [PubMed] [Google Scholar]
- 28.Carroll K, Elroy-Stein O, Moss B, Jagus R. 1993. Recombinant vaccinia virus K3L gene product prevents activation of double-stranded RNA-dependent, initiation factor 2 alpha-specific protein kinase. J Biol Chem 268:12837–12842. [PubMed] [Google Scholar]
- 29.Meng XZ, Schoggins J, Rose L, Cao JX, Ploss A, Rice CM, Xiang Y. 2012. C7L family of poxvirus host range genes inhibits antiviral activities induced by type I interferons and interferon regulatory factor 1. J Virol 86:4538–4547. doi: 10.1128/JVI.06140-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, McFadden G. 2015. SAMD9 is an innate antiviral host factor with stress response properties that can be antagonized by poxviruses. J Virol 89:1925–1931. doi: 10.1128/JVI.02262-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sivan G, Ormanoglu P, Buehler EC, Martin SE, Moss B. 2015. Identification of restriction factors by human genome-wide RNA interference screening of viral host range mutants exemplified by discovery of SAMD9 and WDR6 as inhibitors of the vaccinia virus K1L−C7L− mutant. mBio 6:e01122-15. doi: 10.1128/mBio.01122-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panda D, Fernandez DJ, Lal M, Buehler E, Moss B. 2017. Triad of human cellular proteins, IRF2, FAM111A, and RFC3, restrict replication of orthopoxvirus SPI-1 host-range mutants. Proc Natl Acad Sci U S A 114:3720–3725. doi: 10.1073/pnas.1700678114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kotwal GJ, Moss B. 1988. Analysis of a large cluster of nonessential genes deleted from a vaccinia virus terminal transposition mutant. Virology 167:524–537. [PubMed] [Google Scholar]
- 34.Hayasaka D, Ennis FA, Terajima M. 2007. Pathogeneses of respiratory infections with virulent and attenuated vaccinia viruses. Virol J 4:22. doi: 10.1186/1743-422X-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monath TP, Caldwell JR, Mundt W, Fusco J, Johnson CS, Buller M, Jian L, Gardner B, Downing G, Blum PS, Kemp T, Nichols R, Weltzin R. 2004. ACAM2000 clonal Vero cell culture vaccinia virus (New York City Board of Health strain)—a second-generation smallpox vaccine for biological defense. Int J Infect Dis 8:S31–S44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z, Reynolds SE, Martens CA, Bruno DP, Porcella SF, Moss B. 2011. Expression profiling of the intermediate and late stages of poxvirus replication. J Virol 85:9899–9908. doi: 10.1128/JVI.05446-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuwen H, Cox JH, Yewdell JW, Bennink JR, Moss B. 1993. Nuclear localization of a double-stranded RNA-binding protein encoded by the vaccinia virus E3l gene. Virology 195:732–744. doi: 10.1006/viro.1993.1424. [DOI] [PubMed] [Google Scholar]
- 38.Yang Z, Maruri-Avidal L, Sisler J, Stuart C, Moss B. 2013. Cascade regulation of vaccinia virus gene expression is modulated by multistage promoters. Virology 447:213–220. doi: 10.1016/j.virol.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeLange AM, McFadden G. 1986. Sequence-nonspecific replication of transfected plasmid DNA in poxvirus-infected cells. Proc Natl Acad Sci U S A 83:614–618. doi: 10.1073/pnas.83.3.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merchlinsky M, Moss B. 1988. Sequence-independent replication and sequence-specific resolution of plasmids containing the vaccinia virus concatemer junction: requirements for early and late trans-acting factors, p 87–93. In Kelly T, Stillman B (ed), Cancer cells 6/eukaryotic DNA replication. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 41.De Silva FS, Moss B. 2005. Origin-independent plasmid replication occurs in vaccinia virus cytoplasmic factories and requires all five known poxvirus replication factors. Virol J 2:23. doi: 10.1186/1743-422X-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu B, Panda D, Mendez-Rios J, Ganesan S, Wyatt LS, Moss B. 17 January 2018. Identification of poxvirus genome uncoating and DNA replication factors with mutually redundant roles. J Virol. doi: 10.1128/JVI.02152-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Senkevich TG, Katsafanas G, Weisberg A, Olano LR, Moss B. 2017. Identification of vaccinia virus replisome and transcriptome proteins by isolation of proteins in nascent DNA coupled with mass spectrometry. J Virol 91:e01015-17. doi: 10.1128/JVI.01015-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Satheshkumar PS, Anton LC, Sanz P, Moss B. 2009. Inhibition of the ubiquitin-proteasome system prevents vaccinia virus DNA replication and expression of intermediate and late genes. J Virol 83:2469–2479. doi: 10.1128/JVI.01986-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teale A, Campbell S, Van Buuren N, Magee WC, Watmough K, Couturier B, Shipclark R, Barry M. 2009. Orthopoxviruses require a functional ubiquitin-proteasome system for productive replication. J Virol 83:2099–2108. doi: 10.1128/JVI.01753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mercer J, Snijder B, Sacher R, Burkard C, Bleck CK, Stahlberg H, Pelkmans L, Helenius A. 2012. RNAi screening reveals proteasome- and cullin3-dependent stages in vaccinia virus infection. Cell Rep 2:1036–1047. doi: 10.1016/j.celrep.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 47.Paez E, Esteban M. 1985. Interferon prevents the generation of spontaneous deletions at the left terminus of vaccinia virus DNA. J Virol 56:75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moss B. 2013. Poxvirus DNA replication. Cold Spring Harb Perspect Biol 5:a010199. doi: 10.1101/cshperspect.a010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Czarnecki MW, Traktman P. 2017. The vaccinia virus DNA polymerase and its processivity factor. Virus Res 234:193–206. doi: 10.1016/j.virusres.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sivan G, Martin SE, Myers TG, Buehler E, Szymczyk KH, Ormanoglu P, Moss B. 2013. Human genome-wide RNAi screen reveals a role for nuclear pore proteins in poxvirus morphogenesis. Proc Natl Acad Sci U S A 110:3519–3524. doi: 10.1073/pnas.1300708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kilcher S, Schmidt FI, Schneider C, Kopf M, Helenius A, Mercer J. 2014. siRNA screen of early poxvirus genes identifies the AAA+ ATPase D5 as the virus genome-uncoating factor. Cell Host Microbe 15:103–112. doi: 10.1016/j.chom.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 52.Reitsma JM, Liu X, Reichermeier KM, Moradian A, Sweredoski MJ, Hess S, Deshaies RJ. 2017. Composition and regulation of the cellular repertoire of SCF ubiquitin ligases. Cell 171:1326–1339.e14. doi: 10.1016/j.cell.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deshaies RJ, Emberley ED, Saha A. 2010. Control of cullin-ring ubiquitin ligase activity by nedd8. Subcell Biochem 54:41–56. doi: 10.1007/978-1-4419-6676-6_4. [DOI] [PubMed] [Google Scholar]
- 54.Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. 2004. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev 18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mercer AA, Fleming SB, Ueda N. 2005. F-box-like domains are present in most poxvirus ankyrin repeat proteins. Virus Genes 31:127–133. doi: 10.1007/s11262-005-1784-z. [DOI] [PubMed] [Google Scholar]
- 56.Herbert MH, Squire CJ, Mercer AA. 2015. Poxviral ankyrin proteins. Viruses 7:709–738. doi: 10.3390/v7020709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pierce NW, Lee JE, Liu X, Sweredoski MJ, Graham RL, Larimore EA, Rome M, Zheng N, Clurman BE, Hess S, Shan SO, Deshaies RJ. 2013. Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell 153:206–215. doi: 10.1016/j.cell.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weaver JR, Shamim M, Alexander E, Davies DH, Felgner PL, Isaacs SN. 2007. The identification and characterization of a monoclonal antibody to the vaccinia virus E3 protein. Virus Res 130:269–274. doi: 10.1016/j.virusres.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin YC, Li J, Irwin CR, Jenkins H, DeLange L, Evans DH. 2008. Vaccinia virus DNA ligase recruits cellular topoisomerase II to sites of viral replication and assembly. J Virol 82:5922–5932. doi: 10.1128/JVI.02723-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolffe EJ, Vijaya S, Moss B. 1995. A myristylated membrane protein encoded by the vaccinia virus L1R open reading frame is the target of potent neutralizing monoclonal antibodies. Virology 211:53–63. doi: 10.1006/viro.1995.1378. [DOI] [PubMed] [Google Scholar]
- 61.Davies DH, Wyatt LS, Newman FK, Earl PL, Chun S, Hernandez JE, Molina DM, Hirst S, Moss B, Frey SE, Felgner PL. 2008. Antibody profiling by proteome microarray reveals the immunogenicity of the attenuated smallpox vaccine modified vaccinia virus Ankara is comparable to that of Dryvax. J Virol 82:652–663. doi: 10.1128/JVI.01706-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sodeik B, Griffiths G, Ericsson M, Moss B, Doms RW. 1994. Assembly of vaccinia virus: effects of rifampin on the intracellular distribution of viral protein p65. J Virol 68:1103–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Simpson-Holley M, Kedersha N, Dower K, Rubins KH, Anderson P, Hensley LE, Connor JH. 2011. Formation of antiviral cytoplasmic granules during orthopoxvirus infection. J Virol 85:1581–1593. doi: 10.1128/JVI.02247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cotter CA, Earl PL, Wyatt LS, Moss B. 2017. Preparation of cell cultures and vaccinia virus stocks. Curr Protoc Protein Sci 89:5.12.1–15.12.18. doi: 10.1002/cpps.34. [DOI] [PubMed] [Google Scholar]
- 65.Americo JL, Earl PL, Moss B. 2017. Droplet digital PCR for rapid enumeration of viral genomes and particles from cells and animals infected with orthopoxviruses. Virology 511:19–22. doi: 10.1016/j.virol.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hyun SI, Weisberg A, Moss B. 2017. Deletion of the vaccinia virus I2 protein interrupts virion morphogenesis, leading to retention of the scaffold protein and mislocalization of membrane-associated entry proteins. J Virol 91:e00558-17. doi: 10.1128/JVI.00558-17. [DOI] [PMC free article] [PubMed] [Google Scholar]