Abstract
Antisense oligonucleotides (AONs) are versatile molecules that can be used to modulate gene expression by binding to RNA. The therapeutic potential of AONs appears particularly high in the central nervous system, due to excellent distribution and uptake in brain cells, as well as good tolerability in clinical trials thus far. Nonetheless, immune stimulation in response to AON treatment in the brain remains a concern. For this reason we performed RNA sequencing analysis of brain tissue from mice treated intracerebroventricularly with phosphorothioate, 2′-O-methyl modified AONs. A significant upregulation of immune system associated genes was observed in brains of AON treated mice, with the striatum showing largest transcriptional changes. Strongest upregulation was seen for the antiviral enzyme 2′-5′-oligoadenylate synthase-like protein 2 (Oasl2) and Bone marrow stromal antigen 2 (Bst2). Histological analysis confirmed activation of microglia and astrocytes in striatum. The upregulation of immune system associated genes was detectable for at least 2 months after the last AON administration, consistent with a continuous immune response to the AON.
Keywords: : immune stimulation, antisense oligonucleotide, 2′-O-methyl, mouse brain
Introduction
Antisense oligonucleotides (AONs) are versatile molecules consisting of single strands of nucleic acids capable of binding to RNA through standard Watson–Crick interactions. AONs have been used since the 1960s [1], and a wide range of chemical modifications have been developed to tailor AONs to specific functions, as well as improve their drug-like properties such as: binding energy, stability, and tolerability [2]. Single-stranded AONs are mainly used for the downregulation of target transcripts through an RNase-H dependent mechanism or to induce steric blocking for modulation of transcript splicing or translational inhibition [3]. Making use of these mechanisms, AONs are investigated as therapeutic agents for a wide range of diseases and show particular promise for neurodegenerative disorders [3]. This is mainly due to the favorable distribution and excellent cellular uptake of AONs following administration in the cerebrospinal fluid [4–6]. An AON approach based on redirecting splicing of SMN2 to induce protein expression as a treatment strategy for spinal muscular atrophy (SMA) [7,8] has arguably been most successful thus far, with recent approval from the Food and Drug Administration (FDA) and European Medicines Agency (EMA) being achieved for the drug SPINRAZA [7]. AON-based therapies for other central nervous system (CNS) disorders have also shown very promising preclinical results in animal models, where, for instance, target downregulation for huntingtin [9] and SOD1 [10] protein was achieved very efficiently in mice. SOD1 downregulation was the first AON based therapy for the CNS to be successfully tested in phase 1 trial [11], and AON based downregulation of huntingtin, as well as SOD1, is both in phase 2 clinical trial at the moment.
Most AONs currently in clinical development for CNS disorders contain a phosphorothioate (PS) backbone modification to provide protection from nuclease degradation, improve cellular uptake, and to enhance AON stability [12]. Despite its many advantages, the main drawback of the PS backbone is its increased cellular toxicity compared to standard phosphodiester linkage [13]. These side effects may arise due to nonspecific binding to proteins [14,15] or complement activation [16]. Indeed, PS-AON is known to induce production of IL-6, TNF-α, IL-12, and CCL5 when added to splenocyte cultures [17]. At least some of these side effects also appear to occur in the CNS, where AON administration has been shown to induce an immune response in the rat brain [18]. This immune stimulation has a clear AON sequence dependent effect, which can be separate from RNA target engagement, as in particular the CpG motifs are known stimulants of the Toll-like receptor (TLR) 9 [19]. These immunostimulatory effects can fortunately be diminished by 2′-sugar and 5-methyl cytosine modifications [20–22], which also was shown to improve AON tolerability in the CNS [23].
Given the clinical advancement of AONs for disorders of the CNS, a comprehensive assessment of potential side effects is required. For this reason, we assessed the effect of a PS 2′-O-methyl modified AON when administered intracerebroventricularly (ICV) in the mouse brain. To assess changes induced at the transcript level, RNA sequencing analysis was performed for striatum, cortex, and cerebellum. Furthermore, immunohistological examination was performed on striatal tissue to determine microglia and astrocyte activation after AON administration.
Materials and Methods
Animals
Wild-type FVB mice and C57BL/6 mice were obtained from Jackson Laboratories (Bar Harbor, ME). The test cohort of FVB mice used for RNA sequencing consisted of a total of 18 mice from 3 to 4 months of age, 3 of which received a total of 215 μg AON, 6 received 665 μg AON, and 9 received phosphate buffered saline (PBS). The independent validation cohort consisted of 12 C57BL/6 mice, 8 of which received a total dose of 500 μg of the AON, and 4 animals received PBS. Animal experiments were carried out in accordance with European Communities Council Directive 2010/63/EU and were approved by the Leiden University and Maastricht University Animal Ethics Committees. Mice were housed individually during experimental procedures, with food and drinking water available ad libitum and with a reverse 12-h light/12-h dark cycle. Only male mice were used for experiments.
AON administration
The AON consisted of a 19-mer (5′-CUGAACUGGUCUACAGCUC-3′) [24,25] and followed a steric blocking design with a full PS backbone and uniform 2′-O-methyl ribose modifications. Nucleotides were not 5-methyl modified. The AON was designed to be noncomplementary to the mouse genome, with a maximum of 17/19 nucleotide complementarity. AONs were dissolved in sterile PBS without calcium or magnesium and diluted to a maximum concentration of 40 μg/μL for injection. Injections of AONs was performed ICV, following previously described procedures [24,26]. In brief, mice were fixed in a stereotactic frame under isoflurane anesthesia, and a burr hole was drilled in the skull. A 26 gauge cannula (Plastics, Anaheim, CA) was implanted at coordinates (relative to bregma) AP: −0.4, ML: 1.0, DV: −1.7 mm and fixed with dental composite (OptiBond® All-In-One; Kerr Dental, Bioggio, Switzerland). AONs or PBS were administered under general anesthesia at an infusion rate of ∼1 μL/min for a total dose of 215 or 665 μg. The simplified administration scheme is depicted (Fig. 1A). The 215 μg dose was achieved through a total of six injections (15–50 μg per injection) during a 2-month period. Animals dosed to 665 μg received two extra injections of 250 and 200 μg during a 1.5 month period. Animals from the C57Bl/6 validation cohort received repeated injections of 5 μL of the same nontargeting AON at 20 μg/μL. Five injections for a total dose of 500 μg was performed in these mice during a 10-week period.
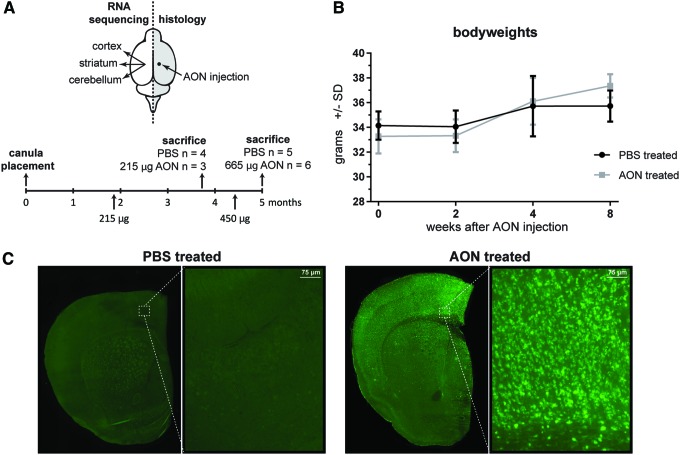
FIG. 1.
Study design and AON cellular uptake. (A) During a 5-month period, FVB mice of the test cohort were injected ICV with a total of 215 μg AON (achieved with a total of six injections) or 625 μg AON (achieved with two additional injections). Two and 8 weeks after the last injection, mice were sacrificed, and the right hemisphere was fixed for histology. Cortex, striatum, and cerebellum were isolated from the left hemisphere for molecular analyses. (B) ICV AON injection did not result in altered body weight compared to PBS injected mice in the FVB test cohort. Data depicted from mice treated with 215 μg AON (n = 4) or PBS (n = 5). (C) Histological analysis showing AON distribution throughout the brain 2 weeks after the last injection. AON, antisense oligonucleotide; ICV, intracerebroventricularly; PBS, phosphate buffered saline. Color images available online at www.liebertpub.com/nat
Tissue collection
Mice were sacrificed either 2 weeks (n = 11) or 8 weeks (n = 7) postinjection (Fig. 1A). Mice from the C57Bl/6 validation cohort were sacrificed 4 months after the last AON injection (n = 12). Brains were removed, and striatum, cortex, and cerebellum of the left hemisphere were snap frozen in liquid nitrogen and stored at −80°C until RNA isolation. The entire right hemisphere was fixed in 4% paraformaldehyde overnight at 4°C. Tissue was then placed in 30% sucrose for 1 or 2 days and stored in PBS containing 0.02% sodium azide at 4°C until sectioning.
Immunofluorescent stainings and assessment of glial fibrillary acidic protein and Aif1 levels
Fixed mouse brain tissue was sectioned on a vibratome at 25 μm thickness, and coronal sections were collected in PBS containing 0.02% sodium azide. Sections were then washed with PBS containing 0.2% Triton X-100 for 10 min and incubated overnight with one of the following primary antibodies diluted in 1% normal donkey serum: rabbit anti-PS-backbone (kind gift of Ionis Pharmaceuticals, Carlsbad, CA; 1:1,000), rabbit anti-glial fibrillary acidic protein (GFAP) (Dako; 1:1,000), or rabbit anti-Aif1 (1:500; Wako Chemicals USA, Inc.). Sections were washed with PBS and incubated with secondary antibodies, donkey anti-rabbit-Alexa Fluor 488 1:500 (Life technologies, Paisley, UK), for fluorescent optical density determination. After washing, sections were mounted on Superfrost plus coated microscope slides (Fisher Emergo, Landsmeer, The Netherlands). The slides were then immediately coverslipped using EverBrite Hardset mounting medium with DAPI (Biotium, Hayward). For cell morphology assessment, the sections were incubated overnight with the primary antibody with the same dilution, followed by a secondary antibody 1:800 biotinylated donkey anti-rabbit (Jackson ImmunoResearch Laboratories, West Grove), and ABC-step (avidin–biotin peroxidase complex) diluted 1:800, Elite ABC-Kit, (VECTASTAIN, Burlingame, CA). To visualize the horseradish peroxide reaction product, the sections were incubated with DAB (3,3′-diaminobenzidine tetrahydrochloride) with nickel chloride intensification. Slides were washed, dehydrated, and coverslipped using PERTEX (Histolab Products AB, Goteborg, Sweden). Optical density determination through fluorescence was performed on three coronal sections per mouse (five AON treated vs. five PBS treated) at 10× magnification using ImageJ software. Coronal sections of the striatum at 60× magnification were scored for Aif1 levels to assess microglia activation and for GFAP levels to assess astrocyte number and state of activation. Morphology of cells was assessed for cells in five images from three different sections per mouse. The scoring of microglia activation state was done based on morphology assessment from previous studies [27,28]. Aif1 positive cells (microglia) were categorized as resting or active state. Resting state microglia was characterized by small and smooth-surfaced cell bodies with thin ramified processes. Active microglia had an irregularly shaped cell body with thicker ramified processes. Activation of astrocytes was assessed similar to previously described [29]. Resting state astrocytes had low levels of GFAP expression, with stellate morphology and numerous spatially distinct, thin branched processes. Active astrocytes had increased GFAP expression, with a hypertrophic appearance and thicker, highly branched overlapping processes. Images of AON distribution were obtained as described previously [26].
RNA isolation and sequencing
RNA was isolated from mouse striatum, cortex, and cerebellum using the TRIzol method and PureLink RNA Mini Kit (Ambion; Thermo Fisher Scientific, Waltham, MA). Genomic DNA was degraded using a DNase incubation according to manufacturer's protocol, and concentration and purity of RNA were determined by NanoDrop spectrophotometry. Library preparation and RNA sequencing were done at deCODE Genetics (Reykjavik, Iceland). Starting material was ∼1 μg of total RNA, with an average RIN value of 7.6 (SD ±0.3). Quality of RNA was determined using the LabChip GX (Perkin Elmer, Waltham). Sample preparation was performed nonstrand specifically, using the TruSeq Poly-A v2 Kit (Illumina, San Diego). Capture of mRNA was performed with magnetic poly-T beads, followed by fragmentation. Synthesis of cDNA was then performed with random hexamer primers using the SuperScript II Kit (Invitrogen, Carlsbad, CA). Subsequently, second strand cDNA synthesis was performed in conjunction with RNase-H treatment. Indexing adapters were ligated to the ds-cDNA, and amplification was performed using PCR. Insert size and sample diversity were assessed through pool sequencing on a MiSeq instrument (Illumina). Sequencing (read length 2 × 125 cycles) was then performed using the HiSeq 2500 with v4 SBS Sequencing Kits. Illumina scripts (bcl2fastq v1.8) were used for demultiplexing and FASTQ file generation. Counts were aligned to genes of the mouse reference genome build 10 (GRCm38/mm10).
Analysis of differential gene expression
Analysis of gene expression was performed only on genes exceeding an average of >4 counts per million (CPM). Gene counts were normalized using the trimmed mean of M-values (TMM) normalization method [30]. Differential gene expression was performed using the R package edgeR, version 3.14.0 [31]. A principal component analysis was performed to assess clustering of samples per brain region, and 6 out of 53 samples were excluded based on abnormal clustering (Supplementary Fig. S2A; Supplementary Data are available online at www.liebertpub.com/nat). One additional sample was excluded based on the fact that there was a large percentage of reads originating from a single gene (Supplementary Fig. S2B). The design model matrix for edgeR was described as model.matrix (∼treatment_group*brainregion) to allow for detection of AON treatment effects at the transcript level accounting for the different brain regions. Dispersion was estimated using the design matrix, after which a generalized linear model (GLM) was fitted using the glmFit function. Likelihood ratio test was performed using the glmLRT function on the coefficients of AON treatment and the interaction term treatment*brainregion. To assess the effect of the AON treatment within each brain region, a coefficient was assigned for each brain region and treatment group. The glmLRT function was then performed with contrast argument to allow pairwise treatment effect comparison for each brain region. Genes were considered significantly differentially expressed when the false discovery rate (FDR, Benjamini–Hochberg) was below 0.05. Data were plotted using ggplot2 or GraphPad Prism version 7.0.
Pathway analysis
Genes found to be significantly differentially expressed between PBS and AON treated mice at FDR <0.01 were included for pathway analysis using Ingenuity (Ingenuity Systems, Inc., Redwood City, CA; www.ingenuity.com). Functional annotation of genes was performed using DAVID bioinformatics resources 6.8 (https://david.ncifcrf.gov). Pathways and biological processes with corrected P value <0.05 (Benjamini–Hochberg) were considered significantly enriched in the dataset.
Validation with droplet digital PCR
To validate the RNA sequencing results, droplet digital PCR (ddPCR) was performed on striatum RNA from the same samples as used for RNA sequencing for the FVB test cohort and using RNA from a separate C57Bl/6 validation cohort. cDNA was generated as previously described [32] using Transcriptor First Strand cDNA Synthesis Kit (Roche, Mannheim, Germany) with random hexamer primers. The amount of cDNA that was used per reaction was the equivalent of 30 ng of RNA for ddPCR for genes of interest. Due to high expression, 5 ng of input was used for reference genes. PCR product sizes ranged between ∼100 and 200 bp (primer sequences are listed in Supplementary Table S1). PCRs were set up using QX200 ddPCR EvaGreen Supermix (Bio-Rad, Hercules, CA), with 100 nM of forward and reverse primer in an end volume of 22 μL. Between 15,000 and 20,000 droplets were generated using an AutoDG droplet generator and DG32 cartridges (Bio-Rad). PCR was then performed as follows: 10 min denaturation at 95°C followed by 40 cycles of 30 s at 94°C and 1 min at 58°C. The reactions were ended for 5 min at 4°C, 5 min at 90°C, and then cooled to 10°C. The ramp rate for all steps was 2°C/s. Droplets were quantified using the QX200 droplet reader (Bio-Rad) and analyzed with QuantaSoft analysis software (Bio-Rad). Expression was normalized to reference gene expression level, and copies of target transcript per μL are reported.
Results
AONs lead to upregulation of the antiviral enzyme Oasl2
A total dose of 200 μg AON administered ICV did not result in any negative effect on the body weights of the mice in the FVB test cohort (Fig. 1B). However, in the C57Bl/6 validation cohort a significantly reduced body weight was observed already after 100 μg of AON, which persisted until the end of the study (Supplementary Fig. S1). Despite this, epileptic seizures were sporadically observed directly after AON administration in our FVB test cohort, but not the C57Bl/6 validation cohort. This observation is consistent with previous reports of increased sensitivity to epileptic seizures in mice of FVB background [33]. Immunohistological staining for the PS backbone revealed widespread AON distribution throughout the isolated brain regions (Fig. 1C). Striatum, cortex, and cerebellum tissue from mice treated ICV with either AON or PBS was collected for RNA sequencing (Fig. 1A). When analyzing the 8-week (215 μg) and 2-week (665 μg) AON treatment groups from the FVB test cohort, no major differences in gene expression were found between these dosage groups. For this reason it was decided to combine the 2- and 8-week time point groups for analysis of differential gene expression to obtain increased power for the comparison of AON versus PBS treated. After exclusion of samples with low number of reads, poor alignment, or outliers in principal component analysis (Supplementary Fig. S2A), a total of 44 samples were included for analysis of differential gene expression (Table 1). A median of 7.9 million reads per sample was aligned to the mouse reference genome (GRCm38/mm10). Genes with low expression (CPM <4) were excluded, leading to inclusion of 12,160 genes for differential gene expression analysis. None of the mouse genes containing >16 nucleotide homology in BLAST [34] analysis were observed to be downregulated in the RNA sequencing analysis.
Table 1.
Overview of RNA Sequencing Samples from FVB Test Cohort
| PBS treated | AON treated | |||
|---|---|---|---|---|
| Cerebellum | n = 7 | week 2: n = 4 | n = 8 | week 2: n = 6 |
| week 8: n = 3 | week 8: n = 2 | |||
| Cortex | n = 8 | week 2: n = 5 | n = 7 | week 2: n = 5 |
| week 8: n = 3 | week 8: n = 2 | |||
| Striatum | n = 6 | week 2: n = 2 | n = 8 | week 2: n = 5 |
| week 8: n = 4 | week 8: n = 3 | |||
AON, antisense oligonucleotide; PBS, phosphate buffered saline.
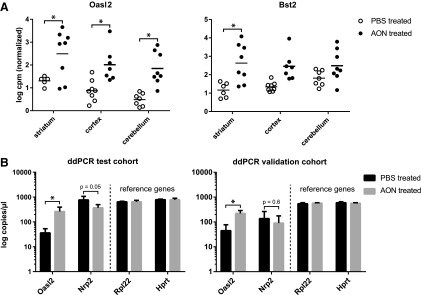
Gene expression analysis revealed a total of 925 genes differentially expressed (FDR <0.05) in at least one brain region of animals treated with AON. The top 10 differentially expressed genes and corresponding fold changes per brain region are depicted in Table 2. The largest and most consistent change observed for each brain region was an upregulation of 2′-5′-oligoadenylate synthase-like protein 2 (Oasl2) (Fig. 2A), which is a dsRNA-activated antiviral enzyme involved in the innate antiviral response. Oasl2 requires double-stranded RNA as a cofactor [35], although the enzyme does not have a characteristic dsRNA binding motif [36].
Table 2.
Top 10 Significantly Altered Gene Expression in Brain After Antisense Oligonucleotide Treatment
| Gene | Name | FDR | Cerebellum log2-fold change | Cortex log2-fold change | Striatum log2-fold change | Protein function |
|---|---|---|---|---|---|---|
| Oasl2 | 2′-5′-oligoadenylate synthase-like protein 2 | 1.3E-07 | 2.2a | 1.9 | 2.5a | IFN-induced, dsRNA-activated antiviral enzyme |
| Sphkap | A-kinase anchor protein SPHKAP | 1.3E-07 | 0.1 | 0.3 | −0.9a | Regulation of protein kinase A signaling |
| Lgals3bp | Galectin-3-binding protein | 1.9E-07 | 1.4 | 1.5a | 1.3a | Scavenger receptor activity |
| Bst2 | Bone marrow stromal antigen 2 | 1.2E-06 | 1.3 | 2.0 | 2.7a | Defense response to virus |
| H2-Q4 | Histocompatibility 2, Q region locus 4 | 3.9E-06 | 0.9 | 0.9 | 1.8a | Antigen processing and presentation of peptide antigen through MHC class I |
| Ifit3 | Interferon-induced protein with tetratricopeptide repeats 3 | 4.3E-06 | 1.4 | 1.4 | 1.8a | Cellular response to interferon-alpha |
| Kcns1 | Potassium voltage-gated channel subfamily S member 1 | 5.6E-06 | 0.0 | −0.4 | 1.9a | Potassium ion transport |
| Dgat2 | Diacylglycerol O-acyltransferase 2 | 5.8E-06 | 0.1 | 0.2 | 0.9a | Triacylglycerol biosynthesis |
| Kctd9 | BTB/POZ domain-containing protein KCTD9 | 6.8E-06 | −0.3 | −0.1 | −0.8a | Adapter of E3 ubiquitin-protein ligase complex |
| Vipr1 | Vasoactive intestinal polypeptide receptor 1 | 7.3E-06 | −0.1 | 0.2 | 1.5a | Cell surface receptor signaling pathway |
Gene is differentially expressed within individual brain region.
IFN, interferon; FDR, false discovery rate; MHC, major histocompatibility complex.
FIG. 2.
Increased expression of Oasl2 in brain of AON treated mice. (A) RNA sequencing shows expression of Oasl2 and Bst2 most strongly upregulated in response to AON, in particular in the striatum. *FDR <0.05, based on eight versus eight mice, analysis on samples of 2 and 8 weeks after AON injection. (B) ddPCR validation of top upregulated (Oasl2) and top downregulated (Nrp2) genes in striatum at 2 weeks after AON injection (FVB test cohort) and 4 months after AON injection (C57Bl/6 independent validation cohort). Based on ddPCR of 4 versus 4 mice. *P < 0.05 using Student's t-test. ddPCR, droplet digital PCR; FDR, false discovery rate.
Other top differentially upregulated genes were also involved in the immune and antiviral response, such as Bone marrow stromal antigen 2 (Bst2), Histocompatibility 2, Q region locus 4 (H2-Q4), and interferon (IFN)-induced protein with tetratricopeptide repeats 3 (Ifit3). In general, the upregulation of these genes was observed to a larger extent in the striatum than in the cortex and cerebellum and was altered at both the 2- and 8-week time points after AON injection. Since AON uptake is comparable between striatum and cortex, it is unlikely that this difference is due to uptake efficiency of the AON. The top upregulated (Oasl2) and a downregulated (Nrp2) gene was validated using ddPCR on the same striatum RNA as was used for RNA sequencing. Both transcripts showed similar expression differences as detected in the RNA sequencing analysis, with Oasl2 showing strong significant upregulation in AON treated mice (log2FC = 2.87) and Nrp2 showing significant downregulation (log2FC = −1.05). A similar upregulation of Oasl2 (Fig. 2B) and several other immune system associated genes (Supplementary Table S2) was also confirmed in the C57Bl/6 validation cohort through ddPCR. The immunostimulatory effect of the AON appears to be long lasting since mice of the validation cohort were sacrificed 4 months after the last AON administration (Supplementary Fig. S1) and seems to be independent of the genetic background of the mice.
Immune system pathways are activated in AON treated brain
To determine which biological processes are altered in the mouse brain in response to AON administration, we performed pathway analysis using Ingenuity [37] and DAVID [38] on the significantly altered genes with FDR <0.01 observed in the RNA sequencing analysis. This approach led to inclusion of 379 genes with relatively large changes that showed a most consistent change across the three brain regions. The significant canonical pathways and biological processes (KEGG) that are altered in response to the AON are listed in Table 3. Pathways involving the immune system, such as IFN and B cell receptor signaling, were activated in the AON treated mice.
Table 3.
Pathways and Biological Processes Altered After Antisense Oligonucleotide Treatment
| No. of genes | FDR | Activation z-score | |
|---|---|---|---|
| Pathway (ingenuity) | |||
| IFN signaling | 5 | 3.2E-04 | 2.0 |
| G-protein coupled receptor signaling | 13 | 7.9E-04 | NA |
| GABA receptor signaling | 6 | 1.1E-03 | NA |
| B cell receptor signaling | 10 | 1.5E-03 | 2.3 |
| IGF-1 signaling | 7 | 2.1E-03 | NA |
| Leukocyte extravasation signaling | 10 | 3.2E-03 | 2.3 |
| Neuropathic pain signaling in dorsal horn neurons | 7 | 3.4E-03 | 1.9 |
| Complement system | 4 | 3.4E-03 | 2.0 |
| No. of genes | FDR | Identifier | |
|---|---|---|---|
| Biological process (KEGG) | |||
| Immune system process | 11 | 2.8E-05 | GO:0002376 |
| Cellular response to IFN-beta | 5 | 5.2E-04 | GO:0035456 |
| Antigen processing and presentation of peptide antigen through MHC class I | 4 | 2.6E-03 | GO:0002474 |
| Defense response to virus | 6 | 0.01 | GO:0051607 |
| Innate immune response | 7 | 0.03 | GO:0045087 |
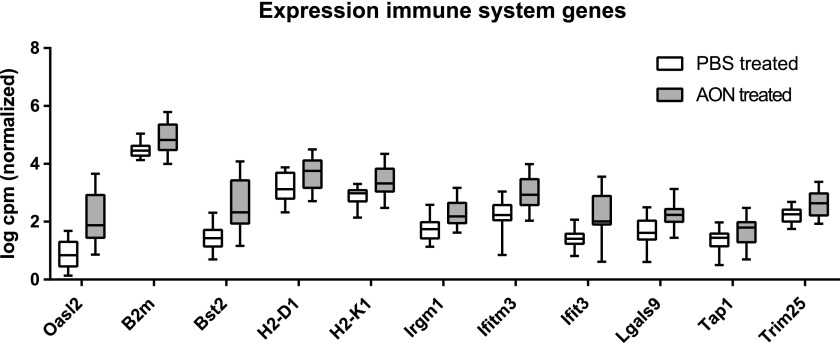
Examination of the biological processes with DAVID showed the most significant alteration in immune system associated genes. This process included 11 differentially expressed genes that were most strongly upregulated in striatum (Fig. 3). We validated five of these genes through ddPCR (Fig. 2B and Supplementary Table S2), which confirmed upregulation of the immune system associated genes Oasl2, Ifit3, Bst2, Trim25, and Lgals3bp in striatum of AON treated mice in both the FVB test and C57Bl/6 validation cohort. Together, these upregulated genes point to an innate immune response and complement activation, which constitutes the immediate defense in response to potential pathogens. In the context of the brain, this would suggest microglia involvement. For this reason, we next performed histological analysis of microglial activation.
FIG. 3.
Immune system associated genes are upregulated in brain after AON injection. Expression of 11 significantly altered genes in brain belonging to immune system process (GO:0002376). Based on RNA sequencing analysis of cortex, cerebellum, and striatum from nine PBS treated versus nine AON treated animals.
Microglia activation and increased astrocyte density
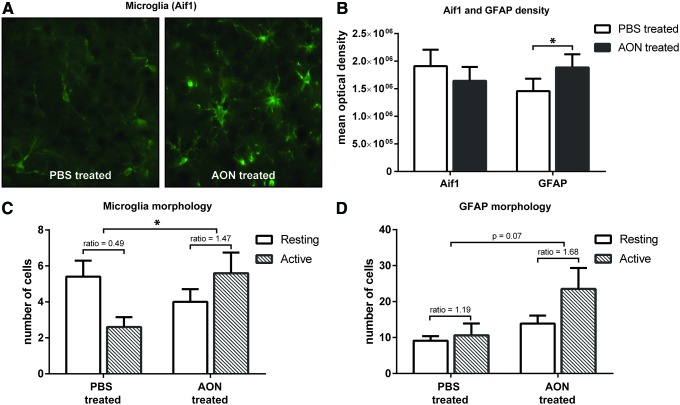
Based on the transcriptomic analysis, administration of the AON resulted in an immunologic response most strongly in the striatum of the mice. To further assess this innate immune response, immunohistological staining was performed on brain sections for the microglia marker allograft inflammatory factor 1 (Aif1) and for the astrocyte marker GFAP. Microglia are brain resident macrophages that are widely expressed in the brain and constitute a major component of the innate arm of the brain immune system [39,40]. In the RNA sequencing analysis, Aif1 expression was significantly increased in the AON treated mouse brain. Mean optical density measurements in striatum did not reveal a significant change in the Aif1 levels in AON treated mice (Fig. 4B), indicating no significant increase in the number of microglia cells. However, when more closely examining the number of activated and resting state cells, an increase in activated microglia was observed in striatum of AON treated mice (Fig. 4C), similar to previously reported for CpG-containing AONs [41].
FIG. 4.
Assessment of astrocyte and microglia markers in striatum. Histological analysis of the microglia marker Aif1 and astrocyte marker GFAP was performed on three coronal striatum sections per mouse. (A) Representative example of microglia staining with Aif1 of a PBS and AON treated mouse at 40 × magnification. (B) Quantification of the optical density revealed no observable change in Aif1 levels, but did show a significantly increased level of GFAP. (C) Despite no increased number of Aif1 positive cells, the ratio of active to resting state microglia was significantly increased in striatum of AON treated mice. (D) Scoring of GFAP morphology showed a general increase in astrocytes in AON treated mice and a trend toward increased ratio of active over resting state cells. Based on three sections per mouse from five versus five mice. *P < 0.05 with Student's t-test. GFAP, glial fibrillary acidic protein. Color images available online at www.liebertpub.com/nat
RNA sequencing results did not show significant alteration of GFAP transcript levels in AON treated mice. However, staining of GFAP in AON treated animals did reveal a small significant increase in mean optical GFAP density in striatum (Fig. 4B). GFAP is a marker for reactive astrocytes, which is a common cell type of the CNS with a wide range of functions. Astrocytes express TLRs and are capable of producing several cytokines and chemokines [42]. Reactive astrogliosis is often a response to cytokines such as TGF-α or through indirect activation by neurons, microglia, or endothelial cells [43]. When examining the activity state of astrocytes in the striatum, an increase in the number of active glia cells was observed in AON treated mice (Fig. 4D). To determine whether the immune stimulatory effect was not restricted to a single AON sequence, additional immunohistological stainings of brain tissue from a previous mouse study (described in [24]) were used. This AON was also a 2′-O-methyl PS-AON and was complementary to both the murine and human huntingtin mRNA, but showed no effect on the murine huntingtin transcript [24]. Similar to the AON used in the present study, this second AON resulted in increased astrocyte and microglia activation in the striatum of AON treated mice (Supplementary Fig. S3), suggesting that the immune stimulation reported in this study is not restricted to a single AON sequence.
Discussion
AONs are increasingly being used for treatment of CNS disorders and have recently received FDA approval for treatment of SMA [44]. In this study we set out to determine the side effects of a uniformly modified 2′-O-methyl PS-AON in the mouse brain. This type of AON is typically implemented in steric blocking applications, such as exon inclusion or exon skipping [3].
The AON did not contain CpG motifs, which are known to be potent immunostimulatory agents [45]. A bolus ICV injection led to widespread distribution and cellular uptake throughout the mouse brain, in line with previous reports using similar AON chemistries [4,6,26]. Despite using the same AON, we observed different effects on body weight in our FVB test and C57Bl/6 validation cohort of mice. Mice in the validation cohort showed a reduction in body weight already after 100 μg ICV administration of AON, while our test cohort showed no discernible difference in body weight at a dose of 615 μg AON. This disparity may be the result of differences between the FVB and C57Bl/6 mouse background or the differences in dosing schedule between the two cohorts.
Through RNA sequencing analysis, we found that the AON led to upregulation of several transcripts associated with the immune system. The largest upregulation was observed for Oasl2, Bst2, Ifit3, and H2-Q4. These genes were upregulated in all brain regions tested (striatum, cortex, and cerebellum) and generally showed a slightly larger fold change in the striatum compared to cortex and cerebellum. Given comparable AON uptake in striatum and cortex, this suggests that the striatum is more sensitive to the immunostimulatory effects of the AON. The top differentially expressed genes were also found upregulated in the C57Bl/6 validation cohort, indicating that the immune system activation is not dependent on the genetic background of the mice.
It has been found that particular AON sequences, even without CpG motifs, can result in an atypical inflammatory response [46]. It is therefore important to consider that the immune stimulatory effects we describe in this study can potentially be confounded by the specific sequence of the AON used. However, we have confirmed the microglia and astrocyte activation for a different AON sequence of identical chemistry (Supplementary Fig. S3), indicating that the results described in this study are not unique to one sequence.
The most strongly upregulated gene in response to the AON, Oasl2, belongs to the 2′,5′-oligoadenylate synthetase gene family, of which a total of 11 have been identified in mouse, compared to 4 in human [47]. In the mouse brain, Oasl2 is the most highly expressed of the enzyme family [47] and is an antiviral enzyme which through oligoadenylate synthetase activity activates ribonucleases, resulting in degradation of viral RNA. Oasl is one of the proteins upregulated in response to type I and II IFN cytokines and confers resistance to viral infection [48]. In response to virus infection, IFN induced upregulation of Oas activates the ribonuclease L, which leads to RNA degradation [49].
The second most upregulated gene observed in AON treated mice, the transmembrane protein Bst2, is also an IFN induced protein. Similar to Oasl2, Bst2 is a component of the innate defense response to viruses [50]. Bst2 is also known as tetherin, because it was identified as a protein that caused retention of virions on infected cell surfaces [51]. Bst2 is not only able to tether these virions based on recognition of pathogen-associated molecular patterns (PAMPs) but also is able to act as a pattern-recognition receptor capable of signaling to the immune system through NFκB-dependent pro-inflammatory gene expression [52]. Indeed, double-stranded RNA is a known PAMP recognized by TLR3, which upon activation results in NF-κB and IFN-beta production [53]. Surprisingly, however, Bst2 showed negligible ability to induce NF-κB response [52], suggesting that this downstream signaling pathway may not have been activated in the mouse brain in this study. In line with this, we did not observe significant alteration in expression levels of NFKB1 between PBS and AON treated mice. The increase in Bst2 expression in this study (Log2FC = 2) is not substantially different from what was previously observed in transcriptional analysis of a mouse centrally injected with lipopolysaccharide (LPS), where Bst2 showed a Log2FC of 2.82 [54]. This observation indicates that the immune activation in response to the AON can be quite substantial at the dosages tested in this study, although expression changes of other immune response genes are more mildly affected in our study compared to LPS injection [54].
Another strongly upregulated gene in brain of the AON treated mice was Ifit3. Similar to Oasl2 and Bst2, Ifit3 is an antiviral protein that is upregulated in response to IFN signaling [55]. Ifit3 restricts DNA and RNA virus replication, and evidence suggests that the Ifit family of proteins plays an important role in destruction of invasive RNA [56]. Ifit proteins are able to bind viral RNA and sequester viral proteins in the cytoplasm [57]. The observation that several IFN induced genes are upregulated in response to the AON suggests that the innate immune system is activated in response to the AON, resulting in IFN signaling. In line with this, we observed microglia activation based on significant Aif1 transcript upregulation in RNA sequencing analysis, as well as increased active state microglia cells in histological examination. These observations are in concordance with previous reports, where doses higher than 100 μg of 2′-O-methyl PS-AON led to significant Aif1 transcript upregulation [6]. Similarly, the observed increase in astrocyte activation in this study is indicative of a neuroprotective response in response to the AON [58]. Although the role of reactive astrocytes is still not well understood, it was recently found that A1 reactive astrocytes are induced by cytokines originating from activated neuroinflammatory microglia [59]. The observed astrocyte activation in striatum of AON treated mice may thus be a downstream result of microglial activation.
The mechanisms underlying the immune activation by RNA and AONs have been previously investigated, and a large portion of the immune activation originates from TLR stimulation (reviewed in [60]). This activation can be antagonized upon 2′-O-methylation of RNA [61,62], which strongly reduces the immunostimulatory effects of the AON. Nonetheless, even in absence of CpG motifs and having 2′-methoxyethyl modifications, AONs are still capable of inducing an immune response [63]. In particular TLR3, 7, and 8 were shown to recognize single-stranded RNA [64–66]. Although the majority of TLRs sense stimulatory components on the cell surface, TLR3, 7, and 8 sense nucleic acids in endosomal compartments of the cell [65]. These three TLRs are expressed in microglia of mice [67], while astrocytes in contrast only express low levels of TLR2, 4, 5, and 9 in resting state [68]. This argues for an initiatory role of the microglia, which are able to sense the AONs through their TLRs, resulting in cytokine production and downstream activation of the astrocytes. However, it is also known that human neurons express TLR3 [69] and TLR8 [70] and are able to initiate an antiviral response characterized by cytokines (TNF-α, IL-6), chemokines (CCL5, CXCL-10), and antiviral molecules 2′5′OAS and IFN-beta [69].
Of note, the TLRs and general immune response are known to differ between mouse and human [71,72]. For instance, CpG stimulatory sequences do not elicit TLR9 dependent TNF-α dependent toxicity in humans after pulmonary administration, while they do in mice [73]. This difference was hypothesized to be due to differential TLR9 expression patterns between rodents and primates, as monocyte/macrophage cells do not express TLR9 in primates [73]. Furthermore, small interfering RNA (siRNA) was shown to be a ligand for the murine TLR3, whereas the human TLR3 was unresponsive to the siRNA [74]. It may therefore be difficult to extrapolate the murine immune response for AONs to the human situation.
However, also the use of monkeys as a model for AON tolerability has presented difficulties, as monkeys have a greater sensitivity to complement activation in response to second generation AONs compared to humans [75,76]. As such, mouse models may overestimate the contribution of AON sequence specific effects, while experiments in monkeys appear to overestimate the AON induced complement activation aspect.
In conclusion, we show in this study that a 2′-O-methyl PS-AON of 19 nucleotides in length results in mild activation of the innate immune response when administered to the mouse brain. The immune activation is characterized by increased activation of microglia and astrocytes, likely resulting in IFN signaling. The resulting transcriptional changes in the brain are most strongly identified by upregulation of Oasl2 and Bst2.
Supplementary Material
Acknowledgments
The authors thank Bharath Sampadi for performing Ingenuity analysis, Melvin Evers for advice on study design, Frank Rigo for providing antibody reagents, Peter-Bram ‘t Hoen and Szymon Kielbasa for advice on RNA sequencing analysis, and Ólafur Magnússon (Decode Genetics) for performing the RNA sequencing.
Author Disclosure Statement
The authors declare no competing financial interests.
References
- 1.Lundin KE, Gissberg O. and Smith CI. (2015). Oligonucleotide therapies: the past and the present. Human Gene Ther 26:475–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khvorova A. and Watts JK. (2017). The chemical evolution of oligonucleotide therapies of clinical utility. Nat Biotechnol 35:238–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evers MM, Toonen LJ. and van Roon-Mom WM. (2015). Antisense oligonucleotides in therapy for neurodegenerative disorders. Adv Drug Deliv Rev 87:90–103 [DOI] [PubMed] [Google Scholar]
- 4.Casaca-Carreira J, Temel Y, Larrakoetxea I. and Jahanshahi A. (2017). Distribution and penetration of intracerebroventricularly administered 2′OMePS oligonucleotide in the mouse brain. Nucleic Acid Ther 27:4–10 [DOI] [PubMed] [Google Scholar]
- 5.Juliano RL. (2016). The delivery of therapeutic oligonucleotides. Nucleic Acids Res 44:6518–6548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rigo F, Chun SJ, Norris DA, Hung G, Lee S, Matson J, Fey RA, Gaus H, Hua Y, et al. (2014). Pharmacology of a central nervous system delivered 2′-O-methoxyethyl-modified survival of motor neuron splicing oligonucleotide in mice and nonhuman primates. J Pharmacol Exp Ther 350:46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiriboga CA. (2017). Nusinersen for the treatment of spinal muscular atrophy. Expert Rev Neurother 17:955–962 [DOI] [PubMed] [Google Scholar]
- 8.Hua Y, Sahashi K, Hung G, Rigo F, Passini MA, Bennett CF. and Krainer AR. (2010). Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev 24:1634–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kordasiewicz HB, Stanek LM, Wancewicz EV, Mazur C, McAlonis MM, Pytel KA, Artates JW, Weiss A, Cheng SH, et al. (2012). Sustained therapeutic reversal of Huntington's disease by transient repression of huntingtin synthesis. Neuron 74:1031–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith RA, Miller TM, Yamanaka K, Monia BP, Condon TP, Hung G, Lobsiger CS, Ward CM, McAlonis-Downes M, et al. (2006). Antisense oligonucleotide therapy for neurodegenerative disease. J Clin Invest 116:2290–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller TM, Pestronk A, David W, Rothstein J, Simpson E, Appel SH, Andres PL, Mahoney K, Allred P, et al. (2013). An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurol 12:435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma VK, Sharma RK. and Singh SK. (2014). Antisense oligonucleotides: modifications and clinical trials. MedChemComm 5:1454–1471 [Google Scholar]
- 13.Levin AA. (1999). A review of the issues in the pharmacokinetics and toxicology of phosphorothioate antisense oligonucleotides. Biochim Biophys Acta 1489:69–84 [DOI] [PubMed] [Google Scholar]
- 14.Brown DA, Kang SH, Gryaznov SM, DeDionisio L, Heidenreich O, Sullivan S, Xu X. and Nerenberg MI. (1994). Effect of phosphorothioate modification of oligodeoxynucleotides on specific protein binding. J Biol Chem 269:26801–26805 [PubMed] [Google Scholar]
- 15.Guvakova MA, Yakubov LA, Vlodavsky I, Tonkinson JL. and Stein CA. (1995). Phosphorothioate oligodeoxynucleotides bind to basic fibroblast growth factor, inhibit its binding to cell surface receptors, and remove it from low affinity binding sites on extracellular matrix. J Biol Chem 270:2620–2627 [DOI] [PubMed] [Google Scholar]
- 16.Galbraith WM, Hobson WC, Giclas PC, Schechter PJ. and Agrawal S. (1994). Complement activation and hemodynamic changes following intravenous administration of phosphorothioate oligonucleotides in the monkey. Antisense Res Dev 4:201–206 [DOI] [PubMed] [Google Scholar]
- 17.Bjersing JL, Eriksson K, Tarkowski A, and Collins LV. (2004). The arthritogenic and immunostimulatory properties of phosphorothioate oligodeoxynucleotides rely on synergy between the activities of the nuclease-resistant backbone and CpG motifs. Inflammation 28:39–51 [DOI] [PubMed] [Google Scholar]
- 18.Elepfandt P, Rupprecht S, Schoning-Burkhardt B, Volk HD. and Woiciechowsky C. (2002). Oligodeoxynucleotides induce brain inflammation in rats when infused intracerebroventricularly. Neurosci Lett 322:107–110 [DOI] [PubMed] [Google Scholar]
- 19.Meng W, Yamazaki T, Nishida Y. and Hanagata. N. (2011). Nuclease-resistant immunostimulatory phosphodiester CpG oligodeoxynucleotides as human Toll-like receptor 9 agonists. BMC Biotechnol 11:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henry S, Stecker K, Brooks D, Monteith D, Conklin B. and Bennett CF. (2000). Chemically modified oligonucleotides exhibit decreased immune stimulation in mice. J Pharmacol Exp Therapeut 292:468–479 [PubMed] [Google Scholar]
- 21.Monteith DK, Henry SP, Howard RB, Flournoy S, Levin AA, Bennett CF. and Crooke ST. (1997). Immune stimulation—a class effect of phosphorothioate oligodeoxynucleotides in rodents. Anticancer Drug Des 12:421–432 [PubMed] [Google Scholar]
- 22.Zhao Q, Temsamani J, Iadarola PL, Jiang Z. and Agrawal S. (1996). Effect of different chemically modified oligodeoxynucleotides on immune stimulation. Biochem Pharmacol 51:173–182 [DOI] [PubMed] [Google Scholar]
- 23.Peng Ho S, Livanov V, Zhang W, Li J. and Lesher T. (1998). Modification of phosphorothioate oligonucleotides yields potent analogs with minimal toxicity for antisense experiments in the CNS. Brain Res Mol Brain Res 62:1–11 [DOI] [PubMed] [Google Scholar]
- 24.Casaca-Carreira J, Toonen LJ, Evers MM, Jahanshahi A, van-Roon-Mom WM. and Temel Y. (2016). In vivo proof-of-concept of removal of the huntingtin caspase cleavage motif-encoding exon 12 approach in the YAC128 mouse model of Huntington's disease. Biomed Pharmacother 84:93–96 [DOI] [PubMed] [Google Scholar]
- 25.Evers MM, Tran HD, Zalachoras I, Meijer OC, den Dunnen JT, van Ommen GJ, Aartsma-Rus A. and van Roon-Mom WM. (2014). Preventing formation of toxic N-terminal huntingtin fragments through antisense oligonucleotide-mediated protein modification. Nucleic Acid Ther 24:4–12 [DOI] [PubMed] [Google Scholar]
- 26.Toonen LJA, Rigo F, van Attikum and H.van Roon-Mom WMC. (2017). Antisense oligonucleotide-mediated removal of the polyglutamine repeat in spinocerebellar ataxia type 3 mice. Mol Ther Nucleic acids 8:232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shitaka Y, Tran HT, Bennett RE, Sanchez L, Levy MA, Dikranian K. and Brody DL. (2011). Repetitive closed-skull traumatic brain injury in mice causes persistent multifocal axonal injury and microglial reactivity. J Neuropathol Exp Neurol 70:551–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown GC. and Neher JJ. (2010). Inflammatory neurodegeneration and mechanisms of microglial killing of neurons. Mol Neurobiol 41:242–247 [DOI] [PubMed] [Google Scholar]
- 29.VanGuilder HD, Bixler GV, Brucklacher RM, Farley JA, Yan H, Warrington JP, Sonntag WE. and Freeman. WM. (2011). Concurrent hippocampal induction of MHC II pathway components and glial activation with advanced aging is not correlated with cognitive impairment. J Neuroinflamm 8:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson MD. and Oshlack A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson MD, McCarthy DJ. and Smyth GK. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toonen LJ, Schmidt I, Luijsterburg MS, van Attikum H. and van Roon-Mom WM. (2016). Antisense oligonucleotide-mediated exon skipping as a strategy to reduce proteolytic cleavage of ataxin-3. Sci Rep 6:35200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goelz MF, Mahler J, Harry J, Myers P, Clark J, Thigpen JE. and Forsythe DB. (1998). Neuropathologic findings associated with seizures in FVB mice. Lab Anim Sci 48:34–37 [PubMed] [Google Scholar]
- 34.Altschul SF, Gish W, Miller W, Myers EW. and Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215:403–410 [DOI] [PubMed] [Google Scholar]
- 35.Eskildsen S, Justesen J, Schierup MH. and Hartmann R. (2003). Characterization of the 2′-5′-oligoadenylate synthetase ubiquitin-like family. Nucleic Acids Res 31:3166–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Justesen J, Hartmann R. and Kjeldgaard NO. (2000). Gene structure and function of the 2′-5′-oligoadenylate synthetase family. Cell Mol Life Sci 57:1593–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kramer A, Green J, Pollard J, Jr., and Tugendreich S. (2014). Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30:523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang da W, Sherman BT. and Lempicki RA. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57 [DOI] [PubMed] [Google Scholar]
- 39.Prinz M. and Priller J. (2014). Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat Rev Neurosci 15:300–312 [DOI] [PubMed] [Google Scholar]
- 40.Filiano AJ, Gadani SP, and Kipnis J. (2015). Interactions of innate and adaptive immunity in brain development and function. Brain Res 1617:18–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dalpke AH, Schafer MK, Frey M, Zimmermann S, Tebbe J, Weihe E. and Heeg K. (2002). Immunostimulatory CpG-DNA activates murine microglia. J Immunol 168:4854–4863 [DOI] [PubMed] [Google Scholar]
- 42.Carpentier PA, Begolka WS, Olson JK, Elhofy A, Karpus WJ. and Miller SD. (2005). Differential activation of astrocytes by innate and adaptive immune stimuli. Glia 49:360–374 [DOI] [PubMed] [Google Scholar]
- 43.Hol EM. and Pekny M. (2015). Glial fibrillary acidic protein (GFAP) and the astrocyte intermediate filament system in diseases of the central nervous system. Curr Opin Cell Biol 32:121–130 [DOI] [PubMed] [Google Scholar]
- 44.Aartsma-Rus A. (2017). FDA approval of nusinersen for spinal muscular atrophy makes 2016 the year of splice modulating oligonucleotides. Nucleic Acid Ther 27:67–69 [DOI] [PubMed] [Google Scholar]
- 45.Dalpke AH, Zimmermann S, Albrecht I. and Heeg K. (2002). Phosphodiester CpG oligonucleotides as adjuvants: polyguanosine runs enhance cellular uptake and improve immunostimulative activity of phosphodiester CpG oligonucleotides in vitro and in vivo. Immunology 106:102–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burel SA, Machemer T, Ragone FL, Kato H, Cauntay P, Greenlee S, Salim A, Gaarde WA, Hung G, et al. (2012). Unique O-methoxyethyl ribose-DNA chimeric oligonucleotide induces an atypical melanoma differentiation-associated gene 5-dependent induction of type I interferon response. J Pharmacol Exp Ther 342:150–162 [DOI] [PubMed] [Google Scholar]
- 47.Kakuta S, Shibata S. and Iwakura Y. (2002). Genomic structure of the mouse 2′,5′-oligoadenylate synthetase gene family. J Interferon Cytokine Res 22:981–993 [DOI] [PubMed] [Google Scholar]
- 48.Chebath J, Benech P, Revel M. and Vigneron M. (1987). Constitutive expression of (2′–5′) oligo A synthetase confers resistance to picornavirus infection. Nature 330:587–588 [DOI] [PubMed] [Google Scholar]
- 49.Silverman RH. (2007). Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol 81:12720–12729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tokarev A, Skasko M, Fitzpatrick K. and Guatelli J. (2009). Antiviral activity of the interferon-induced cellular protein BST-2/tetherin. AIDS Res Hum Retroviruses 25:1197–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neil SJ, Zang T. and Bieniasz PD. (2008). Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430 [DOI] [PubMed] [Google Scholar]
- 52.Galão RP, Le Tortorec A, Pickering S, Kueck T. and Neil SJ. (2012). Innate sensing of HIV-1 assembly by tetherin induces NFκB-dependent proinflammatory responses. Cell Host Microbe 12:633–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsumoto M, Kikkawa S, Kohase M, Miyake K. and Seya T. (2002). Establishment of a monoclonal antibody against human Toll-like receptor 3 that blocks double-stranded RNA-mediated signaling. Biochem Biophys Res Commun 293:1364–1369 [DOI] [PubMed] [Google Scholar]
- 54.Bonow RH, Aid S, Zhang Y, Becker KG. and Bosetti F. (2009). The brain expression of genes involved in inflammatory response, the ribosome, and learning and memory is altered by centrally injected lipopolysaccharide in mice. Pharmacogenomics J 9:116–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmeisser H, Mejido J, Balinsky CA, Morrow AN, Clark CR, Zhao T, and Zoon KC. (2010). Identification of alpha interferon-induced genes associated with antiviral activity in Daudi cells and characterization of IFIT3 as a novel antiviral gene. J Virol 84:10671–10680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou X, Michal JJ, Zhang L, Ding B, Lunney JK, Liu B. and Jiang Z. (2013). Interferon induced IFIT family genes in host antiviral defense. Int J Biol Sci 9:200–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diamond MS. and Farzan M. (2013). The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol 13:46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pekny M, Wilhelmsson U. and Pekna M. (2014). The dual role of astrocyte activation and reactive gliosis. Neurosci Lett 565:30–38 [DOI] [PubMed] [Google Scholar]
- 59.Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, Bennett ML, Münch AE, Chung W-S, et al. (2017). Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541:481–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dalpke A. and Helm M. (2012). RNA mediated Toll-like receptor stimulation in health and disease. RNA Biol 9:828–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robbins M, Judge A, Liang L, McClintock K, Yaworski E. and MacLachlan I. (2007). 2′-O-methyl-modified RNAs act as TLR7 antagonists. Mol Ther 15:1663–1669 [DOI] [PubMed] [Google Scholar]
- 62.Sioud M, Furset G. and Cekaite L. (2007). Suppression of immunostimulatory siRNA-driven innate immune activation by 2′-modified RNAs. Biochem Biophys Res Commun 361:122–126 [DOI] [PubMed] [Google Scholar]
- 63.Senn JJ, Burel S. and Henry SP. (2005). Non-CpG-containing antisense 2′-methoxyethyl oligonucleotides activate a proinflammatory response independent of Toll-like receptor 9 or myeloid differentiation factor 88. J Pharmacol Exp Ther 314:972–979 [DOI] [PubMed] [Google Scholar]
- 64.Diebold SS, Kaisho T, Hemmi H, Akira S. and Reis e Sousa C. (2004). Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303:1529–1531 [DOI] [PubMed] [Google Scholar]
- 65.Xagorari A. and Chlichlia K. (2008). Toll-like receptors and viruses: induction of innate antiviral immune responses. Open Microbiol J 2:49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Butchi NB, Pourciau S, Du M, Morgan TW. and Peterson KE. (2008). Analysis of the neuroinflammatory response to TLR7 stimulation in the brain: comparison of multiple TLR7 and/or TLR8 agonists. J Immunol 180:7604–7612 [DOI] [PubMed] [Google Scholar]
- 67.Olson JK. and Miller SD. (2004). Microglia initiate central nervous system innate and adaptive immune responses through multiple TLRs. J Immunol 173:3916–3924 [DOI] [PubMed] [Google Scholar]
- 68.Bowman CC, Rasley A, Tranguch SL. and Marriott I. (2003). Cultured astrocytes express toll-like receptors for bacterial products. Glia 43:281–291 [DOI] [PubMed] [Google Scholar]
- 69.Lafon M, Megret F, Lafage M. and Prehaud C. (2006). The innate immune facet of brain: human neurons express TLR-3 and sense viral dsRNA. J Mol Neurosci 29:185–194 [DOI] [PubMed] [Google Scholar]
- 70.Ma Y, Haynes RL, Sidman RL. and Vartanian T. (2007). TLR8: an innate immune receptor in brain, neurons and axons. Cell Cycle 6:2859–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bryant CE. and Monie TP. (2012). Mice, men and the relatives: cross-species studies underpin innate immunity. Open Biol 2:120015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ketloy C, Engering A, Srichairatanakul U, Limsalakpetch A, Yongvanitchit K, Pichyangkul S. and Ruxrungtham K. (2008). Expression and function of Toll-like receptors on dendritic cells and other antigen presenting cells from non-human primates. Vet Immunol Immunopathol 125:18–30 [DOI] [PubMed] [Google Scholar]
- 73.Campbell JD, Cho Y, Foster ML, Kanzler H, Kachura MA, Lum JA, Ratcliffe MJ, Sathe A, Leishman AJ, et al. (2009). CpG-containing immunostimulatory DNA sequences elicit TNF-alpha-dependent toxicity in rodents but not in humans. J Clin Invest 119:2564–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weber C, Muller C, Podszuweit A, Montino C, Vollmer J. and Forsbach A. (2012). Toll-like receptor (TLR) 3 immune modulation by unformulated small interfering RNA or DNA and the role of CD14 (in TLR-mediated effects). Immunology 136:64–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shen L, Frazer-Abel A, Reynolds PR, Giclas PC, Chappell A, Pangburn MK, Younis H. and Henry SP. (2014). Mechanistic understanding for the greater sensitivity of monkeys to antisense oligonucleotide-mediated complement activation compared with humans. J Pharmacol Exp Ther 351:709–717 [DOI] [PubMed] [Google Scholar]
- 76.Henry SP, Jagels MA, Hugli TE, Manalili S, Geary RS, Giclas PC. and Levin AA. (2014). Mechanism of alternative complement pathway dysregulation by a phosphorothioate oligonucleotide in monkey and human serum. Nucleic Acid Ther 24:326–335 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.