Abstract
Huntington's disease (HD) is an inherited brain disorder characterized by progressive motor, cognitive, and behavioral dysfunctions. It is caused by abnormally large trinucleotide cytosine–adenine–guanine (CAG) repeat expansions on exon 1 of the Huntingtin gene. CAG repeat length (CAG-RL) inversely correlates with an earlier age of onset. Region-based studies have shown that HD gene mutation carrier (HDgmc) individuals (CAG-RL ≥36) present functional connectivity alterations in subcortical (SC) and default mode networks. In this analysis, we expand on previous HD studies by investigating associations between CAG-RL and connectivity in the whole brain, as well as between CAG-dependent connectivity and motor and cognitive performances. We used group-independent component analysis on resting-state functional magnetic resonance imaging scans of 261 individuals (183 HDgmc and 78 healthy controls) from the PREDICT-HD study, to obtain whole-brain resting state networks (RSNs). Regression analysis was applied within and between RSNs connectivity (functional network connectivity [FNC]) to identify CAG-RL associations. Connectivity within the putamen RSN is negatively correlated with CAG-RL. The FNC between putamen and insula decreases with increasing CAG-RL, and also shows significant associations with motor and cognitive measures. The FNC between calcarine and middle frontal gyri increased with CAG-RL. In contrast, FNC in other visual (VIS) networks declined with increasing CAG-RL. In addition to observed effects in SC areas known to be related to HD, our study identifies a strong presence of alterations in VIS regions less commonly observed in previous reports and provides a step forward in understanding FNC dysfunction in HDgmc.
Key words: : CAG repeat length, group-independent component analysis, prodromal Huntington's disease, resting-state fMRI, within- and between-networks functional connectivity
Introduction
Huntington's disease (HD) is a rare, neurodegenerative, heritable disorder caused by a trinucleotide cytosine–adenine–guanine (CAG) repeat expansion in exon 1 of the Huntington gene (HTT) (Paulsen et al., 2014), leading to a mutant form of the Huntington protein (i.e., Huntingtin). This devastating disorder is characterized by abnormal movements, cognitive impairments, and psychiatric abnormalities (Paulsen et al., 2008, 2014). HD affects people from different backgrounds and ethnicities worldwide with prevalence estimates from 0.4 per 100,000 habitants to 5.7 per 100,000 habitants (Baig et al., 2016; Pringsheim et al., 2012). A greater number of CAG repeats is associated with earlier HD onset (Langbehn et al., 2010; Lee et al., 2012; Torres Ramírez et al., 2006) and algorithms using CAG repeat length (CAG-RL) and age have been widely used to stage the severity of prodromal HD (An et al., 2012; Ha and Fung, 2012; Killoran et al., 2013; Langbehn et al., 2010; Paulsen et al., 2008; Reilmann et al., 2014; Zhang et al., 2010).
HD diagnosis is based on the presence of significant motor abnormalities (primarily chorea). In general, HD gene mutation carriers are undiagnosed and diagnosed individuals with CAG repeats ≥36. Premanifest HD states are stage as at risk individuals with CAG mutation without symptoms or signs. Prodromal HD states are individuals with CAG mutation presenting symptoms or signs, but insufficient to reaching clinical diagnosable levels (Misiura et al., 2017; Reilmann et al., 2014). Manifest HD states are individuals with CAG mutation showing significant involuntary movements to reach clinical diagnoses. Healthy controls (HCs) are individuals with Huntingtin gene without CAG mutation.
It has been shown that the premanifest and prodromal HD states correlate with cognitive decline, psychiatric symptoms, and brain abnormalities that can be detected up to 15 years before diagnosis (Ha and Fung, 2012; Paulsen et al., 2008; Ross et al., 2014).
Pathological studies have demonstrated that after diagnosis, HD progression is characterized by neuronal loss in the caudate, putamen, and layers IV and V of the cortex (Halliday et al., 1998; Vonsattel et al., 1985); brain imaging studies both pre- and postdiagnosis HD show volumetric reductions in putamen, external globus pallidus, caudate nucleus, and ventral midbrain in the region of the substantia nigra (Aylward et al., 2004; Buckner et al., 2008; Kipps et al., 2005; van den Bogaard et al., 2011), as well as in the cerebral cortex (Ciarochi et al., 2016; Rosas et al., 2008). Task performances related to functional magnetic resonance imaging (fMRI) suggest that activation abnormalities can be found before manifest signs or symptoms appear (Kloppel et al., 2015; Paulsen et al., 2004; Reading et al., 2004; Wolf et al., 2011).
Most recently, resting-state fMRI (rs-fMRI) measures have gained interest to explore whole-brain functional connectivity (FC). These measures of intrinsic FC may be sensitive to progression of prodromal HD (Dogan et al., 2013; Dumas et al., 2013; Harrington et al., 2015; Koenig et al., 2014; Poudel et al., 2014; Werner et al., 2014). Specific findings, however, vary across studies, partly due to the varying participant samples studied and methods used. The finding of reduced connectivity is common, particularly in subcortical (SC) regions and motor areas (Aylward et al., 2004; van den Bogaard et al., 2011; Wolf and Kloppel, 2013); however, some findings indicating hyperconnectivity in HD or prodromal HD have been found, suggesting compensatory relationships (Kloppel et al., 2015; Wolf et al., 2012). Although many studies utilized a seed-based approach, findings are somewhat difficult to directly compare since connectivity findings likely vary by seed location. The work by Quarantelli and associates (2013) and Wolf and colleagues (2011) showed dysfunctions in the default mode network (DMN) as well as reduced connectivity consistent with known disease atrophy in the basal ganglia and its connections (Unschuld et al., 2012). A study by Harrington and colleagues (2015) used prodromal staging estimates to determine whether network abnormalities were associated with the disease burden of HD in prodromal stages. Their results confirmed a largely disease burden-dependent functional reorganization of whole-brain networks in prodromal HD. These findings showed a robust association, emphasizing the value of ongoing research in this area. Consistent among all studies is the potential sensitivity of the imaging methods to elucidate unknown underlying pathways and to detect and track HD trajectories in all stages.
To date, there is still need to identify and understand HD structural and functional brain changes before phenotype manifestation and after clinical diagnosis. By analyzing rs-fMRI data, we can examine brain regions' coactivations without the bias of a task performance. In this study, our aim was to expand on previous HD literature by unrestrictedly examining connectivity in the whole brain and its associations with CAG-RLs. Our motivation for this study is to identify all brain networks with brain alterations related to CAG-RLs. Whole-brain connectivity was explored using a large rs-fMRI sample from the PREDIT-HD study composed of HD gene mutation carriers (HDgmc) and HC participants, and the data-driven approach group-independent component analysis (GICA) (Calhoun and Adali, 2012; Calhoun et al., 2001). Whole-brain parcellation was obtained by GICA, which provides spatially independent brain regions or resting-state networks (RSNs) and their spatial maps and timecourses. We first explore within network (voxel-wise) connectivity in a given RSN. Next, we explore between-network connectivity among all RSNs. Functional network connectivity (FNC) measures the level of coactivation of RSN timecourses (Jafri et al., 2008) and is estimated as the pairwise correlations between two RSN timecourses. We examined CAG repeat effects on the connectivity within a given RSN and CAG repeat effects on the FNC among all the RSNs. Finally, we investigate any relationships between all significant FNC alterations affected by CAG repeat and seven clinical HD measures that evaluate motor and cognitive control (CC) performances. A subset of the resting-state data has been previously analyzed (48 prHD and 16 HCs) using graph-theory approach, identifying the gradual loss of hub status in prHD (Harrington et al., 2015).
Methods
Participants
In this study, we use a large rs-fMRI sample from the PREDIT-HD study composed of HD gene mutation carriers (HDgmc) and HCs. The PREDICT-HD study (Paulsen, 2010; Paulsen et al., 2008) is an international multicenter longitudinal observational study focused on identifying HD biological and refined clinical markers. Since 2001, PREDICT-HD study has collected HDgmc participants who had previously been tested for the presence of the CAG expanded gene and were known to be at certain genetic risk for HD (Paulsen et al., 2008), and matched HC participants. PREDICT-HD participants data included repeated neuropsychological testing, clinical measures, motor function, structural, and fMRI scanning. The PREDICT-HD study was approved by all participating sites' institutional review boards, and all individuals provided written informed consent.
For the analysis, we selected each participant's first available rs-fMRI scan. The data consisted of a total of 261 participants (183 HDgmc [53 M/130 F] and 78 HCs [25 M/53 F]). HDgmc participants were between the ages of 19 and 79 (mean age = 42.48 years and standard deviation = 12.82 years) and HC individuals were between the ages of 25 and 69 (mean age = 48.59 years and standard deviation = 11.35 years). Based on their CAG-RL, the participants can be divided into two groups, the HDgmc group with CAG-RL ranging from 37 to 58 and the HC group with CAG-RL ranging from 15 to 35. Twenty-three participants from the HDgmc group were diagnosed before or during the scan's visit. Table 1 displays a breakdown of the demographic and genetic information of the sample showing significant group differences.
Table 1.
Demographic, Genetic, and Clinical Information for Huntington Disease Gene Mutation Carriers and Healthy Control Participants
| N | HDgmc | HCs | p Value | |
|---|---|---|---|---|
| Gender (F/M) | 261 | 130/53 | 53/25 | |
| Age (years) | 261 | 42.48 ± 12.82, (19:79) | 48.59 ± 11.35, (25:62) | 0.0003* |
| CAG-repeats | 261 | 42.33 ± 3.09, (37:58) | 21.04 ± 4.43, (15:35) | 2.8495e-123* |
| Education | 256 | 15.20 ± 2.39, (10–20) | 15.72 ± 2.08, (12–20) | 0.1036 |
| Motor total | 254 | 7.53 ± 9.65, (0–54) | 3.676 ± 4.80, (0–30) | 0.0012* |
| SDMT raw score | 251 | 51.74 ± 10.82, (20–84) | 56.58 ± 9.25, (21–78) | 0.0008* |
| Stroop color | 250 | 77.15 ± 14.34, (31–112) | 83.79 ± 14.38, (36–137) | 0.0009 * |
| Stroop interference | 252 | 45.77 ± 11.77, (16–76) | 48.58 ± 9.69, (20–70) | 0.0695 |
| Stroop word | 252 | 97.19 ± 19.49, (41–141) | 104.08 ± 16.77, (37–143) | 0.0080* |
| TMTA | 251 | 24.80 ± 9.74, (11–57) | 21.46 ± 7.82, (10–69) | 0.0095* |
| TMTB | 252 | 58.09 ± 27.68, (20–222) | 52.74 ± 24.52, (12–20) | 0.1478 |
Significant group differences, p < 0.05.
N, total number of participants; HDgmc, Huntington disease gene mutation carriers; HC, healthy control, mean ± SD; SD, standard deviation, range values (min:max); CAG, cytosine–adenine–guanine; TMTA, trail making test A; TMTB, trail making test part B; SDMT, symbol digit modalities test.
Clinical variables
Seven clinical variables assessing motor and cognitive abilities (Huntington-Study-Group, 1996) were selected from the PREDICT-HD data set (Misiura et al., 2017; Paulsen et al., 2014). The motor measure was total motor score from the Unified Huntington's Disease Rating Scale defined as the sum of all individual motor abnormality ratings (oculomotor, bradykinesia, rigidity, dystonia, and chorea) with higher scores indicating more severe motor impairments. The cognitive measures were the symbol digit modalities test raw score, the total raw scores from the color naming task, word reading and interference conditions of the Stroop color and word test, trail making test part A seconds to completion, and trail making test part B seconds to completion (Table 1).
Data acquisition and imaging parameters
The PREDICT-HD study acquired rs-fMRI scans in seven centers. All sites used identical Siemens Magnetom TrioTim scanners running on Syngo MR B17 software. The acquisition parameters were tuned by a magnetic resonance physicist at The University of Iowa, and the protocol was distributed to partner sites to ensure consistent acquisition across sites and scanners. Before rs-fMRI data collection started, individuals were asked to lie still, keep their eyes open, and not to fall asleep. T2*-weighted functional gradient-echo echo-planar images were acquired with the following parameters: voxel size = 2.0 × 2.0 × 4.0 mm3; repetition time = 2800 msec; echo time = 29 msec; flip angle (FA) = 80°; field of view = 256 × 256mm2; matrix = 128 × 128, slice thickness = 4 mm, gap = 0 or 0.5 mm, and number of slices = 31 interleaved axial oblique. Resting-state scans lasted 6 min, 15 sec (132 volumes).
Data preprocessing
The rs-fMRI data were preprocessed using SPM5 (www.fil.ion.ucl.ac.uk/spm/). Volume alignment was performed using INRIalign (http://sop.inria.fr/epidaure/Collaborations/IRMf/INRIAlign.html), and slice-timing correction was set to 31 slices using slice 15 as a reference frame. Next, the data were spatially normalized to the Montreal Neurological Institute (Friston, 1994) space, resliced to 3 mm × 3 mm × 3 mm voxels, and smoothed using a Gaussian kernel with a full-width at half maximum of 6 mm. A group mask was computed as the intersection of all individuals' first time point retaining voxels greater than the mean of the distribution −0.6 times the standard deviation. We verified that no participant exhibited severe motion on any of the six motion parameters; most participants have maximum translation values around 2 mm, and only two participants have their maximum translation values between 2.5 and 4.5 mm. The maximum rotation value was 0.02°. No participants were removed due to severe movements.
GICA and postprocessing
We used group-level spatial ICA (GICA) to obtain whole-brain parcellation into functional networks without the need to select seed regions. Spatial ICA applied to rs-fMRI data identifies RSNs; the spatial sources are spatial maps that are maximally independent in their linearly mixed fMRI signals or timecourses. After removing the first four time points from each participant's scan to mitigate T1 relaxation effects, GICA was applied to the aligned, smoothed, and normalized data from all subjects as a group, using the GIFT toolbox [http://mialab.mrn.org/software/gift (Calhoun et al., 2001)]. The rs-fMRI data were compressed using two stages of principal component analysis (PCA) (Rachakonda et al., 2016). For the first data reduction, we retained 128 principal components. For the second stage, to capture more anatomical and functional brain regions (Allen et al., 2011; Erhardt et al., 2011), 100 independent components (ICs) were estimated using the expectation maximization algorithm to avoid otherwise prohibitive memory requirements (Roweis, 1998). The Infomax ICA algorithm (Bell and Sejnowski, 1995) was repeated 10 times in ICASSO (www.cis.hut.fi/projects/ica/icasso) and aggregate spatial maps were estimated as the modes of the component clusters. Participants-specific spatial maps and timecourses were estimated using the GICA1 back reconstruction method based on PCA compression and projection (Calhoun et al., 2001; Erhardt et al., 2011). Out of the 100 ICs (C = 100) that were estimated, 46 ICs were identified as meaningful RSNs by evaluating the high to low frequency power ratio in the spectra of components in the RSN timecourse, and the location of maxima in the spatial map (Allen et al., 2011; Meda et al., 2008; Robinson et al., 2009). The other 54 ICs were discarded from further analysis because they were considered to be related to artifacts, white matter, ventricular, or cerebrospinal fluid spatial maps, or had irregular timecourse spectra power.
Before computing whole-brain FC, the additional postprocessing of RSN timecourses included linear, quadratic, and cubic detrending, despiking using 3DDespike (Cox, 1996), and filtering using a fifth-order Butterworth low-pass filter with a high frequency cutoff of 0.15 Hz (Allen et al., 2011; Cordes et al., 2001). For each participant, the FNC values for each pair of the 46 RSNs were calculated as the Pearson's pairwise correlation between timecourses for that pair. A total of 1035 [= 46 RSN × (46 RSNs − 1)/2] FNC pairs were obtained.
RSN categorization
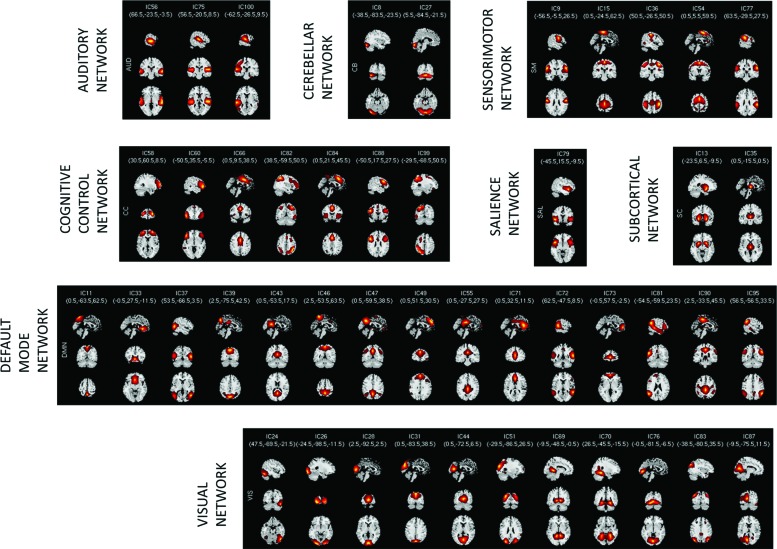
Based on their anatomical, functional properties, neurosynth labeling (http://neurosynth.org/), and similarities to other RSNs found on previous rs-fMRI studies (Allen et al., 2011, 2014; Smith et al., 2009), the 46 RSNs were categorized into 8 functional domains: auditory (AUD), cerebellar (CB), CC, default mode network (DMN), salience (SAL), SC, sensorimotor (SM), and visual (VIS). Each of the 46 RSNs was described based on where the strongest signal occurred.
The AUD networks are represented by three components in the middle temporal gyrus (IC 56) and superior temporal gyrus (ICs 75 and 100). The CB networks are represented by two components in the cerebellum (ICs 8 and 27). The CC networks are represented by seven components: middle frontal gyrus (ICs 58 and 88), inferior frontal gyrus (60), presupplementary motor area (ICs 66 and 84), and posterior parietal cortex (ICs 82 and 99). The DMN is captured by 15 components: precuneus medial (ICs 11 and 39), medial prefrontal cortex (ICs 33, 71, and 73), temporal parietal junction (ICs 37, 72, and 81), posterior cingulate cortex (ICs 43, 55, and 90), precuneus (ICs 46 and 47), superior frontal gyrus (IC 49), and angular gyrus (95). The SAL network is represented by a single component insular cortex (IC 79). The SC networks are represented by two important components in HD studies: putamen (IC 13) and thalamus (IC 35). The SM networks are described by five components postcentral gyrus (ICs 9, 15, and 36), supplementary motor area (IC54), and supramarginal gyrus (IC 77). And, the VIS networks were represented by 11 components: inferior occipital gyrus (ICs 24, 26, and 76), calcarine lobule (ICs 28 and 87), cuneus (IC 31), lingual gyrus (ICs 44 and 69), middle occipital gyrus (ICs 51 and 83), and fusiform gyrus (IC 70).
Statistical analyses
We used the multivariate analysis of covariance (MANCOVAN) toolbox within GIFT [http://mialab.mrn.org/software/gift (Allen et al., 2011)] to find significant associations between the participant's connectivity within and between RSN and the covariates of interest: age, gender, CAG-RL, age × CAG-RL, education, head translation, head rotation, and sites. For each individual, the motion parameters head translation and rotation were computed as the mean of the sum of the absolute difference between time frames. Age, CAG-RL, education, head translation, and head rotation were treated as continuous variables, whereas gender and the seven sites were treated as discrete variables. The gender variable was coded as 1 for females and 0 for males. Regression models considering all the covariates of interest are referred to as the full models.
Within-network connectivity
Full regression models were built for each of the 46 spatial maps with the covariates of interest: age, gender, CAG repeat, age × CAG-RL, education, head translation, head rotation, and sites as independent variables and spatial maps voxel's intensities as dependent variables. Next, MANCOVAN provided a reduced model obtained by backward elimination removing the insignificant variables from the full model (Allen et al., 2011). The age × CAG-RL interaction and education variables were dropped from the 46 full models. We report on those that included the CAG-RL variable in their reduced models.
Between network connectivity
A full regression model was built for the FNC values with the following covariates of interest: age, gender, CAG-RL, age × CAG-RL, education, head translation, head rotation, and sites as independent variables and the 1035 FNCs pairs as the dependent variable. Next, MANCOVAN provided a reduced model obtained by backward elimination. The nonsignificant variables age × CAG-RL interaction and education variables were dropped from the full model. The FNC reduced model retained the covariates age, gender, CAG-RL, head translation, head rotation, and sites.
Multivariate analysis allows us to identify all important covariates associated with the spatial maps and FNC values. However, regression models with several covariates become difficult to interpret. Therefore, we further explored within and between RSNs connectivity performing univariate testing, selecting one covariate of interest at a time and fixing the other variables. Since we are interested in identifying CAG repeat effects on whole-brain FC, linear relationships between CAG repeats and the 46 spatial maps and 1035 FNC pairs values were tested. A correction for multiple comparisons was incorporated using the Matlab function mafdr based on the Storey's false discovery rate algorithm (Storey, 2002), using a significant adjusted alpha level of 0.05 to identify all the adjusted p values that survived false discovery rate correction.
Finally, we investigated any associations between the FNCs showing significant CAG effects and seven clinical measures. Regression analysis was performed using the cognitive and motor clinical measures (already listed). The FNC pairs showing significant CAG effects were the main covariates of interest, and age, gender, motion parameters, and sites were also included as covariates in the seven regression models.
Results
Figure 1 shows the most representative voxels of the 46 spatial maps overlaid on a standard template, and plotted as t-statistics thresholded at t > mean + 4 × standard deviation.
FIG. 1.
Spatial maps of the 46 ICs identified as RSNs, grouped into 8 domains based on their anatomical and functional properties: AUD, CB, CC, DMN, SAL, SC, SM, and VIS. All spatial maps are overlaid on a standard template and plotted as t-statistics (t) thresholded at t > mean + 4 × standard deviation. RSN, resting state network; IC, independent component; AUD, auditory; CB, cerebellar; CC, cognitive control; DMN, default mode network; SAL, salience; SC, subcortical; SM, sensorimotor; VIS, visual. Color images available online at www.liebertpub.com/brain
Within FNC results
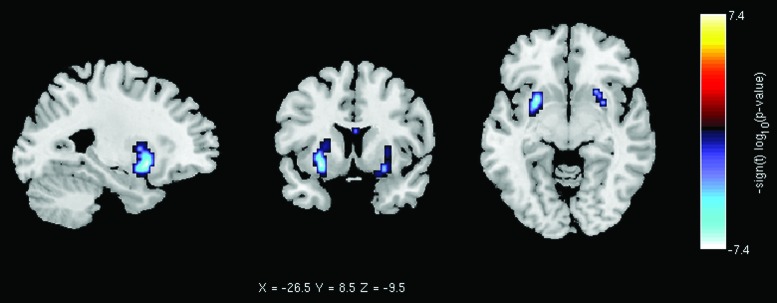
At the voxel level, the univariate CAG repeat test results (Fig. 2) show that a cluster of 85 voxels in the putamen spatial map (IC13, SC network, out of a total of 59,642 voxels per spatial map) survived false discovery rate correction. A significant anticorrelation between the CAG-RL and the putamen connectivity was obtained. All significant t and p values are displayed in Figure 2. This significant result shows a within network decrease in connectivity in the putamen as the number of CAG repeats increases. The covariates CAG, age, gender, rotation, and sites were retained in the reduced regression model for the putamen spatial map.
FIG. 2.
Putamen RSN spatial map (IC 13), within-network functional connectivity, CAG-RL univariate test results. Voxels in blue are those whose average intensity significantly decays as the number of CAG-RLs increases. Results are displayed as −sign(t) × log10(p value), and only presenting the p values that survived FDR correction. CAG-RL, cytosine–adenine–guanine repeat length; FDR, false discovery rate. Color images available online at www.liebertpub.com/brain
Between FNC results
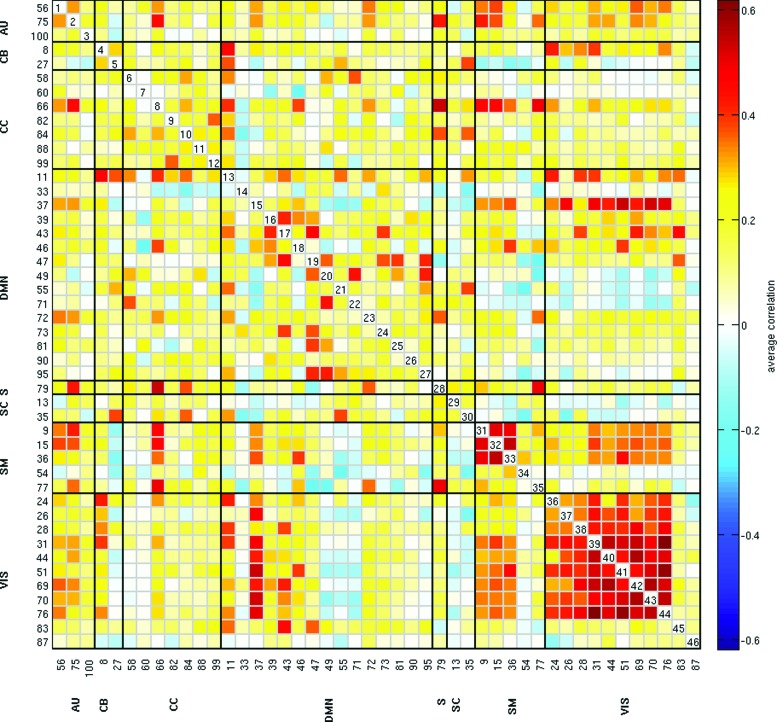
Figure 3 shows the FNC matrix computed as the Pearson's pairwise correlation between the timecourses of the 46 RSNs averaged across individuals. From this figure we can observe that the VIS domain is the only domain showing highly positive correlations between its RSNs. We also see evidence of anticorrelation among some RSNs from the DMN and CC, SM and VIS domains.
FIG. 3.
Unthresholded FNC matrix, showing all pairwise correlations between RSNs timecourses averaged across subjects. Positive correlations are in the yellow to red range, whereas negative correlations are light to dark blue. FNC, functional network connectivity. Color images available online at www.liebertpub.com/brain
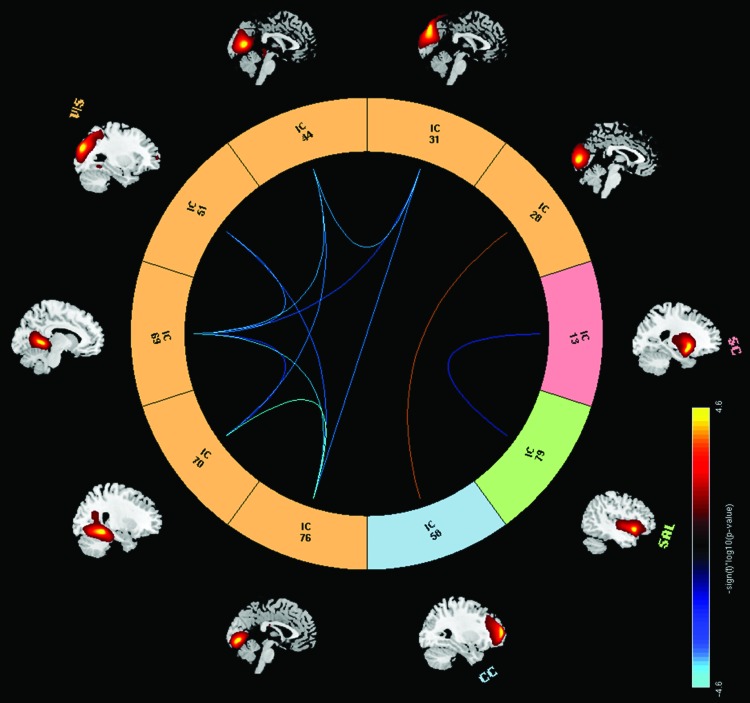
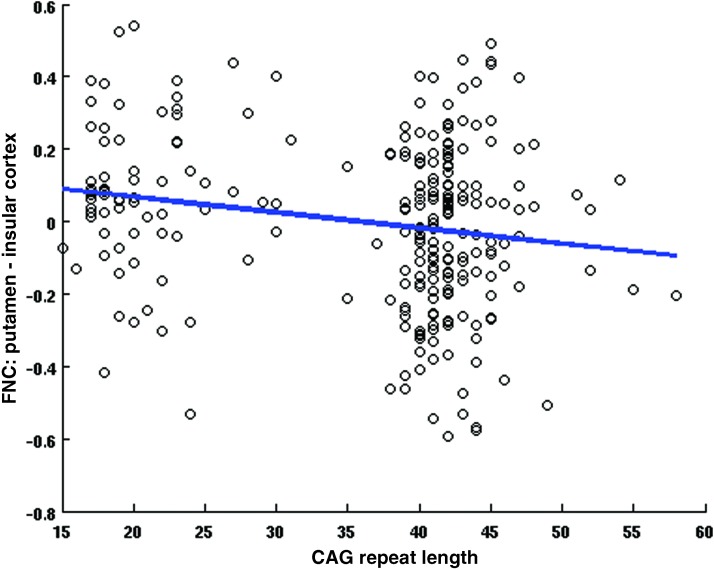
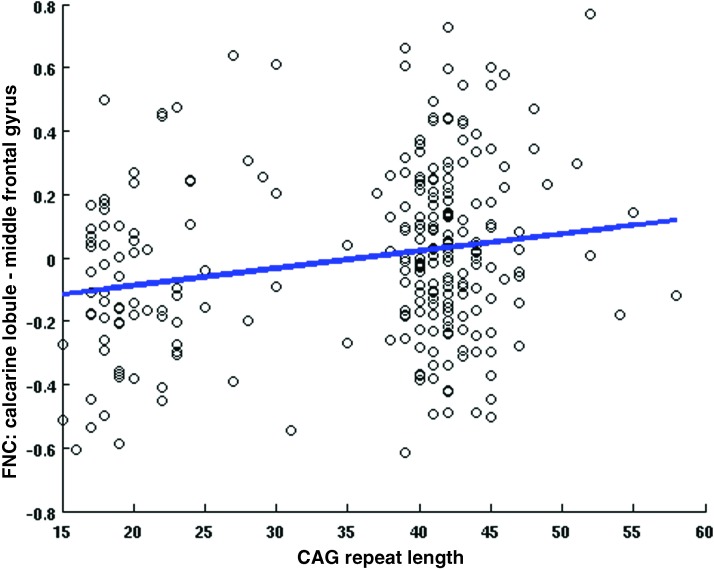
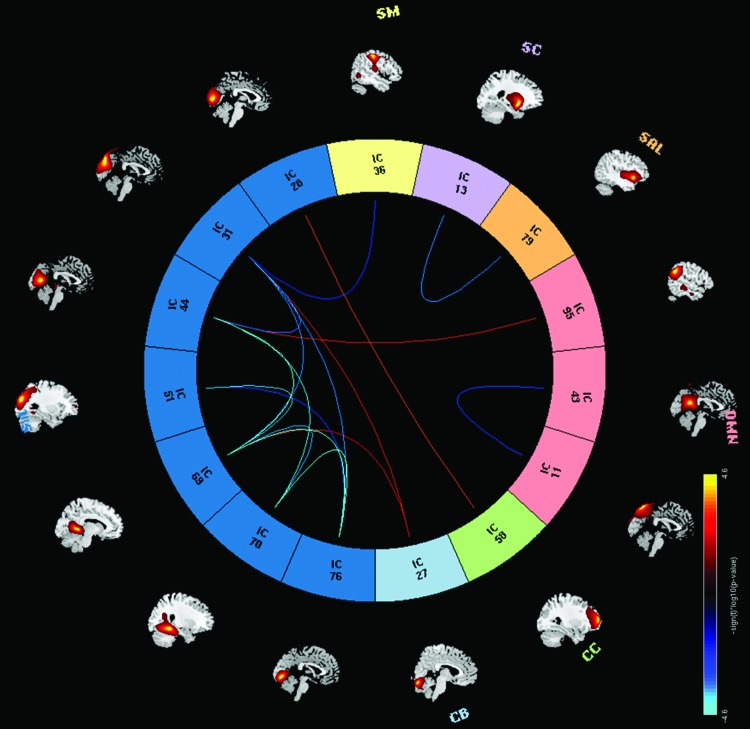
At the network level, we applied multivariate analysis to the Fisher transformed FNC correlation values and the selected covariates of interest. From the univariate CAG test results shown in Figure 4, we observe mostly negative (11) pairs compared with 1 positive pair of linear relationships between CAG-RL and FNCs whose p values survived false discovery rate correction thresholded at a significance level of 0.05. These results imply that as the number of CAG-RLs increases, the FNC between 11 RSNs pairs from the SC, SAL, and VIS domains decreases. As the number of CAG-RLs increases, the FNC between two RSNs from the VIS and CC increases. The β and p values for the 12 pairs of RSNs are listed in Table 2. Figures 5 and 6 show the scatter plots of two pairs of RSNs (1) putamen and insular cortex and (2) calcarine lobule and middle frontal gyrus showing negative, and positive relationships with the number of CAG-repeats.
FIG. 4.
FNC pairs for the CAG-RL univariate test results, blue curves show significant negative correlations between pairs of RSNs from SC–SAL and VIS–VIS domains and CAG-RL. Red curve shows significant positive correlation between a pair of RSNs from CC–VIS domains. Results are displayed as −sign(t) × log10(p value), and only presenting the p values that survived FDR correction. Color images available online at www.liebertpub.com/brain
Table 2.
List of Resting State Network Pairs Showing Cytosine–Adenine–Guanine Effects
| RSN pairs | ICs | Domains | β | p Values | β | p Values |
|---|---|---|---|---|---|---|
| Posterior cingulate cortex and precuneus med | 43-11 | DMN–DMN | −0.0068 | 0.5e-03 | ||
| Putamen and insular cortex | 13-79 | SC–SAL | −0.0052 | 0.7e-03 | −0.0058 | 0.2e-03 |
| Cuneus and postcentral gyrus | 31-36 | VIS–SM | −0.0070 | 0.7e-03 | ||
| Lingual gyrus and cuneus | 44-31 | VIS–VIS | −0.0085 | 0.2e-03 | −0.0082 | 0.3e-03 |
| Lingual gyrus and cuneus | 69-31 | VIS–VIS | −0.0088 | 0.4e-03 | −0.0093 | 0.2e-03 |
| Lingual gyrus and lingual gyrus | 69-44 | VIS–VIS | −0.0096 | 0.1e-03 | −0.0104 | 0.1e-03 |
| Lingual gyrus and middle occipital gyrus | 69-51 | VIS–VIS | −0.0071 | 0.1e-03 | −0.0077 | 0.1e-03 |
| Fusiform gyrus and lingual gyrus | 70-44 | VIS–VIS | −0.0074 | 0.2e-03 | −0.0079 | 0.1e-03 |
| Fusiform gyrus and lingual gyrus | 70-69 | VIS–VIS | −0.0073 | 0.6e-03 | −0.0081 | 0.2e-03 |
| Inferior occipital gyrus and cuneus | 76-31 | VIS–VIS | −0.0080 | 0.3e-03 | −0.0083 | 0.2e-03 |
| Inferior occipital gyrus and middle occipital gyrus | 76-51 | VIS–VIS | −0.0069 | 0.4e-03 | −0.0068 | 0.6e-03 |
| Inferior occipital gyrus and lingual gyrus | 76-69 | VIS–VIS | −0.0083 | 0.1e-03 | −0.0091 | 0.3e-04 |
| Inferior occipital gyrus and fusiform gyrus | 76-70 | VIS–VIS | −0.0084 | 0.1e-03 | −0.0090 | 0.2e-04 |
| Calcarine lobule and middle frontal gyrus | 28-58 | VIS–CC | 0.0066 | 0.3e-03 | 0.0066 | 0.4e-03 |
| Cuneus and cerebellum | 31-27 | VIS–CB | 0.0083 | 0.7e-03 | ||
| Lingual gyrus and angular gyrus | 44-95 | VIS–DMN | 0.0046 | 0.9e-03 | ||
| Lingual gyrus and cerebellum | 69-75 | VIS–CB | 0.0065 | 0.1e-02 |
Columns 4 and 5 display β and FDR-corrected p values when considering all HC and HDgmc individuals with CAG, columns 6 and 7 display β and FDR-corrected p values when only considering all HC and HDgmc individuals with CAG repeat length ≥40.
RSN, resting state network; IC, independent component; FDR, false discovery rate; VIS, visual; DMN, default mode network; SAL, salience; SC, subcortical; CC, cognitive control; CB, cerebellar.
FIG. 5.
CAG effects, FNC between the putamen (IC13) and insular cortex (IC79) versus CAG-RL. The data points are in black circles and best regression fit is indicated by the blue line. Color images available online at www.liebertpub.com/brain
FIG. 6.
CAG effects, FNC between calcarine lobule (IC28) middle frontal gyrus (IC58) versus CAG-RL. The data points are in black circles and best regression fit is indicated by the blue line. Color images available online at www.liebertpub.com/brain
Since our sample includes HDgmc individuals in the reduced penetrance range [i.e., CAG-RL = 36–39 (McNeil et al., 1997; Quarrell et al., 2007)], we also analyzed the FNC correlation values by removing the HDgmc individuals with <40 CAG-RLs. A total of 21 HDgmc (35 < CAG-repeat <40) individuals were removed, leaving 162 HDgmc (40 ≤ CAG-repeat ≤58) and 78 HC individuals. By removing HDgmc individuals in the reduced penetrance range, the remaining sample data are divided into two well-separated groups, HDgmc with full HD penetrance and HC individuals.
Application of the multivariate regression and planned follow-up univariate tests showed that findings were strengthened with removal of the participants with incomplete penetrance. Results replicated previously identified negative correlations between RSNs from SC, SAL, CC, and VIS domains, and showed additional negative associations between RSNs from CB, VIS, and SM domains and within RSNs from the DMN and VIS domains. Figure 7 shows negative and positive CAG effects on 17 pairs of RSNs; correlation values from the DMN, SC, SAL, CB, SM, and VIS domains survived FDR correction. These results imply that as CAG-RL increases, FNC decreases between 13 pairs of RSNs. Conversely, as CAG-RL increases, the FNC between four pairs of RSNs increases (Table 2). Findings successfully detected FNC differences between HC and HDgmc individuals with 40 or more CAG-repeats. Results may suggest that weakened connectivity is more pronounced in individuals in the full penetrance range (McNeil et al., 1997; Quarrell et al., 2007).
FIG. 7.
FNC pairs for the CAG-RL univariate test results [subsample, HC (15 ≤ CAG-RL ≤ 35) and HDgmc (40 ≤ CAG-repeat ≤ 58)], blue curves show significant negative correlations between pairs of RSNs from SC–SAL and VIS–VIS domains and number of CAG repeats. Red curve shows significant positive correlation between a pair of RSN from CC–VIS domains. Results are displayed as −sign(t) × log10(p value), and only presenting the p values that survived FDR correction. CAG-RL, CAG repeat length; HCs, healthy controls; HDgmc, Huntington disease gene mutation carriers. Color images available online at www.liebertpub.com/brain
Relationship with neuropsychological measures
Tables 3 and 4 display a summary of the 12 FNC pairs β and p values from the linear regression. The signs of the β values indicate the direction of the linear relationships between the 12 FNC pairs and the clinical variables. From these results we can observe significant positive and negative associations between 8 of the 12 FNC pairs and the 7 clinical measures. FNC pairs showing negative associations with the motor and cognitive scores indicate that as FNC decreases, clinical correlates worsen, as higher scores on those tests indicate greater impairment. Similarly, FNC pairs showing positive associations with the clinical measures indicate that as FNC increases, cognitive performances also improve. The FNC pair putamen–insular cortex was the only pair showing associations with all of the motor and cognitive measures.
Table 3.
Regression Model of Functional Network Connectivity Measures Against Clinical Variables
| β Values | Motor total | SDMT raw score | Stroop color | Stroop interference | Stroop word | TMTA | TMTB |
|---|---|---|---|---|---|---|---|
| PUT-SAL (IC13-IC79) | −8.5551* | 12.7881* | 14.2768* | 7.9351* | 18.0495* | −8.6295* | −22.1525* |
| CL-MFG (IC28-IC58) | −1.6353 | −1.9635 | 4.0175 | 2.6179 | 2.3895 | 2.1512 | −0.9772 |
| LG-CUN (IC44-IC31) | −1.6258 | 7.1899* | 7.5365* | 5.2879* | 9.4050* | −5.9208* | −15.9820* |
| LG-CUN (IC69-IC31) | −1.0621 | 3.0239 | 3.0295 | 2.3755 | 3.4514 | −3.1940 | −3.8722 |
| LG-LG (IC69-IC44) | −0.8166 | 2.2792 | 2.4269 | 1.0300 | 4.6044 | −3.4466* | −5.9391 |
| LG-MOG (IC69-IC51) | −3.2192 | 4.9289 | 6.2551 | 5.3194 | 6.7466 | −3.9303 | −8.6091 |
| FFG-LG (IC70-IC44) | −4.5853* | 8.2284* | 7.2251* | 4.8544 | 11.4753* | −5.5388* | −14.3846* |
| FFG-LG (IC70-IC69) | −4.9955* | 7.0897* | 6.5760 | 5.3009* | 8.3072 | −6.1042* | −16.0205* |
| IOG-CUN (IC76-IC31) | −2.3820 | 5.9267* | 6.0445 | 5.7123* | 2.8964 | −3.9717* | −10.1489 |
| IOG-MOG (IC76-IC51) | −4.1871* | 7.1561 | 8.8066 | 8.0540 | 11.2357 | −7.0222* | −19.6900* |
| IOG-LG (IC76-IC69) | −2.1169 | 4.3739 | 5.0721 | 3.7829 | 4.0027 | −4.5592* | −8.8428 |
| IOG-FFG (IC76-IC70) | −2.5963 | 5.6224* | 5.0340 | 3.5055 | 4.4640 | −2.1396 | −3.7288 |
p Values that passed a significance level of 0.05 using FDR correction. Putamen (PUT, IC13), salience (SAL, IC79), calcarine lobule (CL, IC28), middle frontal gyrus (MFG, IC58), lingual gyrus (LG, ICs 44, 69), cuneus (CUN, IC31), middle occipital gyrus (MOG, IC51), fusiform gyrus (FFG, IC70), and inferior occipital gyrus (IOG, IC76).
Table 4.
Beta p Values from Table 3
| β p Values | |||||||
|---|---|---|---|---|---|---|---|
| Motor total | SDMT raw score | Stroop color | Stroop interference | Stroop word | TMTA | TMTB | |
| PUT-SAL (IC13-IC79) | 0.0002* | 3.27E-06* | 0.0003* | 0.0082* | 0.0003* | 0.0005* | 0.0019* |
| CL-MFG (IC28-IC58) | 0.3945 | 0.4116 | 0.2409 | 0.3107 | 0.5834 | 0.3117 | 0.8735 |
| LG-CUN (IC44-IC31) | 0.2981 | 0.0001* | 0.0055* | 0.0098* | 0.0064* | 0.0004* | 0.0011* |
| LG-CUN (IC69-IC31) | 0.4555 | 0.0772 | 0.2197 | 0.2001 | 0.2697 | 0.0355 | 0.3797 |
| LG-LG (IC69-IC44) | 0.5520 | 0.1717 | 0.3121 | 0.5670 | 0.1282 | 0.0196* | 0.1663 |
| LG-MOG (IC69-IC51) | 0.0775 | 0.0247 | 0.0462 | 0.0239 | 0.0899 | 0.0443 | 0.1273 |
| FFG-LG (IC70-IC44) | 0.0082* | 9.39E-05* | 0.0174* | 0.0337 | 0.0028* | 0.0033* | 0.0082* |
| FFG-LG (IC70-IC69) | 0.0025* | 0.0004* | 0.0236 | 0.0154* | 0.0244 | 0.0007* | 0.0022* |
| IOG-CUN (IC76-IC31) | 0.1279 | 0.0018* | 0.0272 | 0.0054* | 0.4062 | 0.0190* | 0.0384 |
| IOG-MOG (IC76-IC51) | 0.0206* | 0.0012 | 0.0060 | 0.0008 | 0.0056 | 0.0003* | 0.0005* |
| IOG-LG (IC76-IC69) | 0.1996 | 0.0294 | 0.0782 | 0.0805 | 0.2736 | 0.0106* | 0.0868 |
| IOG-FFG (IC76-IC70) | 0.1223 | 0.0057* | 0.0840 | 0.1094 | 0.2271 | 0.2394 | 0.4787 |
All the p values that passed FDR correction using a 0.05 significant level are marked with an asterisk. Putamen (PUT, IC13), salience (SAL, IC79), carcarine lobule (CL, IC28), middle frontal gyrus (MFG, IC58), lingual gyrus (LG, ICs 44, 69), cuneus (CUN, IC31), middle occipital gyrus (MOG, IC51), fusiform gyrus (FFG, IC70), and inferior occipital gyrus (IOG, IC76).
Discussion
Findings from this whole-brain analysis, using a data-driven approach and a large rs-fMRI data set including HD gene mutation carriers and HC individuals, showed multiple robust RSN abnormalities. Consistent with known volumetric and pathological research, the within network connectivity in the putamen was largely reduced in HDgmc and the functional connectivities between the putamen and other networks were similarly aberrant. The identified reduced connectivity between the putamen and the anterior insular cortex RSNs from the cortical and SAL domains is consistent with previous publications (Harrington et al., 2015; McColgan et al., 2015). One of the most frequently cited psychiatric changes in HDgmc is apathy or lack of motivation to act (Krishnamoorthy and Craufurd, 2011; Rosenblatt and Leroi, 2000). The SAL network (Seeley et al., 2009), with its rich connections between the anterior insular and limbic system, may be reflective of this observation. Such network abnormalities in prodromal HD may also be associated with deficits in proprioception or awareness of bodily perceptions as well as the inability of persons with prodromal HD to select the most relevant or important aspect of work or social demands (Brossman et al., 2012). Additional studies can determine whether network abnormalities of the putamen and insula may be relevant for those prodromal individuals with apathy that impacts quality of life (Uddin, 2015). The decline in FNC between the occipital gyrus and the fusiform gyrus finding is also consistent with previous results (Dogan et al., 2013; McColgan et al., 2015). This pair of RSNs has been implicated in the differentiation of familiar faces from new faces (Rossion et al., 2003). One of the tasks most sensitive to deficiencies across both the PREDICT-HD and TRACK-HD studies is emotion recognition, which requires the detailed examination of facial expression denoting feelings in others. Despite excellent research designed to enhance understanding of emotion recognition abnormalities in prodromal HD (Henley et al., 2012; Novak et al., 2012; van Asselen et al., 2012), further study of network circuitry and its dependence on the complexities of emotional intelligence is critical for many neurological and psychiatric diseases and disorders.
When removing HDgmc participants with CAG-RL in the intermediate penetrance range (dividing the sample data into two well-separated HDgmc and HC groups), negative and positive correlations with CAG-RL were identified in a pair of RSNs from the DMN and a pair of RSNs from the DMN and VIS networks. Findings regarding the DMN in prodromal HD studies have been mixed (Dumas et al., 2013; Koenig et al., 2014; Quarantelli et al., 2013; Werner et al., 2014). Our findings suggest that brain abnormalities in participants with reduced penetrance CAG alleles may alter research outcomes; this may help understand previous mixed results. Alternatively, Harrington and associates (2015) suggest that network topology disturbances appear to be related to network disorganization rather than reduced connectivity. This hypothesis remains to be tested. Most notably, both publications of connectivity in prodromal HD using rich club and network analyses show findings addressing both global efficiency and network-based statistics, demonstrating that an integration of approaches may be preferable to fully understand the abnormality of connectivity before HD manifestation.
The limitations of this study are scans were acquired with slightly anisotropic resolution (2 mm × 2 mm × 4 mm) to follow existent protocols and to make them compatible with older scans. The scans were also preprocessed using an older SPM version, SPM5. We have verified that the processing steps used in this analysis are robust and quite similar to the output from the current SPM version, SPM12. Another limitation is the assumption that FC among networks is consistent throughout the whole scan period. In future work, we plan to look at time-varying or dynamic connectivity, which may provide additional FC information (Calhoun et al., 2014). In particular, we hypothesize that time-dependent interactions among networks may have been buried and consequently not captured by our static correlation analysis.
Conclusions
This whole-brain connectivity analysis of rs-fMRI data captured CAG-RL associations with intra- and internetwork connectivity among Huntington disease gene mutation carrier (HDgmc) and HC individuals. Significant correlations between CAG-RL and FC from SC, SAL, CC, and VIS domains were identified. In particular, our results complement previously identified putamen degradation. An important contribution, the identified CAG-RL-dependent reduced connectivity between putamen and insular cortex, and its association with motor and cognitive performance could reflect an HD FNC abnormality measure.
In summary, the results from the characterization of FNC changes in individuals with CAG mutation suggest that much can be learned from ongoing study in this area. Application of these methods will be essential to improving anatomical and functional correlates of the earliest HD manifestations. The RSNs identified by GICA as well as those captured in seed-based studies might be used to develop a composite of RSNs that can facilitate comparisons across HD studies. Consistency across studies is critical for ensuring that stages of HD are considered in every research study to provide characterization of the earliest changes associated with HD. Our future work will focus on exploring differences between time-varying FC among HDgmc and HC individuals.
Acknowledgments
This work was supported by grants from the National Institutes of Health (5R01NS040068, 1U01NS082083, 5R01NS054893, P20GM103472, and R01REB020407) and the CHDI Foundation (A-5008). We thank the Predict-HD Investigators and coordinators of the Huntington Study Group; PREDICT-HD sites: University of Iowa, Cleveland Clinic, University College London, Indiana University, Washington University in St. Louis, University California San Francisco; the study participants; the National Research Roster for Huntington Disease Patients and Families; the Huntington's Disease Society of America. We acknowledge the assistance of Hans J. Johnson and Mark Lowe in imaging acquisition.
Author Disclosure Statement
No competing financial interest exists.
References
- Allen EA, Damaraju E, Plis SM, Erhardt EB, Eichele T, Calhoun VD. 2014. Tracking whole-brain connectivity dynamics in the resting state. Cereb Cortex 24:663–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EA, Erhardt EB, Damaraju E, Gruner W, Segall JM, Silva RF, et al. 2011. A baseline for the multivariate comparison of resting-state networks. Front Syst Neurosci 5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An MC, Zhang N, Scott G, Montoro D, Wittkop T, Mooney S, et al. 2012. Genetic correction of Huntington's disease phenotypes in induced pluripotent stem cells. Cell Stem Cell 11:253–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylward EH, Sparks BF, Field KM, Yallapragada V, Shpritz BD, Rosenblatt A, et al. 2004. Onset and rate of striatal atrophy in preclinical Huntington disease. Neurology 63:66–72 [DOI] [PubMed] [Google Scholar]
- Baig SS, Strong M, Quarrell OWJ. 2016. The global prevalence of Huntington's disease: a systematic review and discussion. Neurodegener Dis Manag 6:331–343 [DOI] [PubMed] [Google Scholar]
- Bell AJ, Sejnowski TJ. 1995. An information-maximization approach to blind separation and blind deconvolution. Neural Comput 7:1129–1159 [DOI] [PubMed] [Google Scholar]
- Brossman B, Williams JK, Downing N, Mills JA, Paulsen JS. 2012. Development of the Huntington disease work function scale. J Occup Environ Med 54:1300–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Andrews-Hanna JR, Schacter DL. 2008. The brain's default network: anatomy, function, and relevance to disease. Ann N Y Acad Sci 1124:1–38 [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Adali T. 2012. Multisubject independent component analysis of fMRI: a decade of intrinsic networks, default mode, and neurodiagnostic discovery. IEEE Rev Biomed Eng 5:60–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ. 2001. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp 14:140–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Miller R, Pearlson G, Adalı T. 2014. The chronnectome: time-varying connectivity networks as the next frontier in fMRI data discovery. Neuron 84:262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarochi JA, Calhoun VD, Lourens S, Long JD, Johnson HJ, Bockholt HJ, et al. 2016. Patterns of co-occurring gray matter concentration loss across the Huntington disease prodrome. Front Neurol 7:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes D, Haughton VM, Arfanakis K, Carew JD, Turski PA, Moritz CH, et al. 2001. Frequencies contributing to functional connectivity in the cerebral cortex in “resting-state” data. Am J Neuroradiol 22:1326–1333 [PMC free article] [PubMed] [Google Scholar]
- Cox RW. 1996. AFNI: software for analysis and visualization of functional magnetic resonance neuroima. Comput Biomed Res 29:162–173 [DOI] [PubMed] [Google Scholar]
- Dogan I, Eickhoff SB, Schulz JB, Shah NJ, Laird AR, Fox PT, et al. 2013. Consistent neurodegeneration and its association with clinical progression in Huntington's disease: a coordinate-based meta-analysis. Neurodegener Dis 12:23–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas EM, van den Bogaard SJ, Hart EP, Soeter RP, van Buchem MA, van der Grond J, et al. 2013. Reduced functional brain connectivity prior to and after disease onset in Huntington's disease. NeuroImage Clin 2:377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt EB, Rachakonda S, Bedrick EJ, Allen EA, Adali T, Calhoun VD. 2011. Comparison of multi-subject ICA methods for analysis of fMRI data. Hum Brain Mapp 32:2075–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston K. 1994. Functional and effective connectivity in neuroimaging: a synthesis. Hum Brain Mapp 2:56–78 [Google Scholar]
- Ha AD, Fung VS. 2012. Huntington's disease. Curr Opin Neurol 25:491–498 [DOI] [PubMed] [Google Scholar]
- Halliday GM, McRitchie DA, Macdonald V, Double KL, Trent RJ, McCusker E. 1998. Regional specificity of brain atrophy in Huntington's disease. Exp Neurol 154:663–672 [DOI] [PubMed] [Google Scholar]
- Harrington DL, Rubinov M, Durgerian S, Mourany L, Reece C, Koenig K, et al. 2015. Network topology and functional connectivity disturbances precede the onset of Huntington's disease. Brain 138:2332–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henley SM, Novak MJ, Frost C, King J, Tabrizi SJ, Warren JD. 2012. Emotion recognition in Huntington's disease: a systematic review. Neurosci Biobehav Rev 36:237–253 [DOI] [PubMed] [Google Scholar]
- Huntington-Study-Group. 1996. Unified Huntington's disease rating scale: reliability and consistency. Mov Disord 11:136–142 [DOI] [PubMed] [Google Scholar]
- Jafri MJ, Pearlson GD, Stevens M, Calhoun VD. 2008. A method for functional network connectivity among spatially independent resting-state components in schizophrenia. NeuroImage 39:1666–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killoran A, Biglan KM, Jankovic J, Eberly S, Kayson E, Oakes D, Young AB, Shoulson I. 2013. Characterization of the Huntington intermediate CAG repeat expansion phenotype in PHAROS. Neurology 80:2022–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipps CM, Duggins AJ, Mahant N, Gomes L, Ashburner J, McCusker EA. 2005. Progression of structural neuropathology in preclinical Huntington's disease: a tensor based morphometry study. J Neurol Neurosurg Psychiatry 76:650–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloppel S, Gregory S, Scheller E, Minkova L, Razi A, Durr A, et al. 2015. Compensation in preclinical Huntington's disease: evidence from the track-on HD study. EBioMedicine 2:1420–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig KA, Lowe MJ, Harrington DL, Lin J, Durgerian S, Mourany L, et al. 2014. Functional connectivity of primary motor cortex is dependent on genetic burden in prodromal Huntington disease. Brain Connect 4:535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy A, Craufurd D. 2011. Treatment of apathy in Huntington's disease and other movement disorders. Curr Treat Options Neurol 13:508–519 [DOI] [PubMed] [Google Scholar]
- Langbehn DR, Hayden MR, Paulsen JS. 2010. CAG-repeat length and the age of onset in Huntington disease (HD): a review and validation study of statistical approaches. Am J Med Genet 153B:397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Ramos EM, Lee JH, Gillis T, Mysore JS, Hayden MR, et al. 2012. CAG repeat expansion in Huntington disease determines age at onset in a fully dominant fashion. Neurology 78:690–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColgan P, Seunarine KK, Razi A, Cole JH, Gregory S, Durr A, et al. 2015. Selective vulnerability of Rich Club brain regions is an organizational principle of structural connectivity loss in Huntington's disease. Brain 138:3327–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil SM, Novelletto A, Srinidhi J, Barnes G, Kornbluth I, Altherr MR, et al. 1997. Reduced penetrance of the Huntington's disease mutation. Hum Mol Genet 6:775–779 [DOI] [PubMed] [Google Scholar]
- Meda SA, Giuliani NR, Calhoun VD, Jagannathan K, Schretlen DJ, Pulver A, et al. 2008. A large scale (N = 400) investigation of gray matter differences in schizophrenia using optimized voxel-based morphometry. Schizophr Res 101:95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misiura MB, Lourens S, Calhoun VD, Long J, Bockholt J, Johnson H, et al. 2017. Cognitive control, learning, and clinical motor ratings are most highly associated with basal ganglia brain volumes in the premanifest Huntington's disease phenotype. J Int Neuropsychol Soc 23:159–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak MJ, Warren JD, Henley SM, Draganski B, Frackowiak RS, Tabrizi SJ. 2012. Altered brain mechanisms of emotion processing in pre-manifest Huntington's disease. Brain 135:1165–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen JS. 2010. Early detection of Huntington disease. Future Neurol 5 [Epub ahead of print]; DOI: 10.2217/fnl.09.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen JS, Langbehn DR, Stout JC, Aylward E, Ross CA, Nance M, et al. 2008. Detection of Huntington's disease decades before diagnosis: the Predict-HD study. J Neurol Neurosurg Psychiatry 79:874–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen JS, Long JD, Ross CA, Harrington DL, Erwin CJ, Williams JK, et al. 2014. Prediction of manifest Huntington's disease with clinical and imaging measures: a prospective observational study. Lancet Neurol 13:1193–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen JS, Zimbelman JL, Hinton SC, Langbehn DR, Leveroni CL, Benjamin ML, et al. 2004. fMRI biomarker of early neuronal dysfunction in presymptomatic Huntington's Disease. Am J Neuroradiol 25:1715–1721 [PMC free article] [PubMed] [Google Scholar]
- Poudel GR, Egan GF, Churchyard A, Chua P, Stout JC, Georgiou-Karistianis N. 2014. Abnormal synchrony of resting state networks in premanifest and symptomatic Huntington disease: the IMAGE-HD study. J Psychiatry Neurosci 39:87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringsheim T, Wiltshire K, Day L, Dykeman J, Steeves T, Jette N. 2012. The incidence and prevalence of Huntington's disease: a systematic review and meta-analysis. Mov Disord 27:1083–1091 [DOI] [PubMed] [Google Scholar]
- Quarantelli M, Salvatore E, Giorgio SM, Filla A, Cervo A, Russo CV, et al. 2013. Default-mode network changes in Huntington's disease: an integrated MRI study of functional connectivity and morphometry. PLoS One 8:e72159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarrell OWJ, Rigby AS, Barron L, Crow Y, Dalton A, Dennis N, et al. 2007. Reduced penetrance alleles for Huntington's disease: a multi‐centre direct observational study. J Med Genet 44:e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachakonda S, Silva RF, Liu J, Calhoun VD. 2016. Memory efficient PCA methods for large group ICA. Front Neurosci 10:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading SA, Dziorny AC, Peroutka LA, Schreiber M, Gourley LM, Yallapragada V, et al. 2004. Functional brain changes in presymptomatic Huntington's disease. Ann Neurol 55:879–883 [DOI] [PubMed] [Google Scholar]
- Reilmann R, Leavitt BR, Ross CA. 2014. Diagnostic criteria for Huntington's disease based on natural history. Mov Disord 29:1335–1341 [DOI] [PubMed] [Google Scholar]
- Robinson S, Basso G, Soldati N, Sailer U, Jovicich J, Bruzzone L, et al. 2009. A resting state network in the motor control circuit of the basal ganglia. BMC Neurosci 10:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas HD, Salat DH, Lee SY, Zaleta AK, Pappu V, Fischl B, et al. 2008. Cerebral cortex and the clinical expression of Huntington's disease: complexity and heterogeneity. J Neurol 131:1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt A, Leroi I. 2000. Neuropsychiatry of Huntington's disease and other basal ganglia disorders. Psychosomatics 41:24–30 [DOI] [PubMed] [Google Scholar]
- Ross CA, Aylward EH, Wild EJ, Langbehn DR, Long JD, Warner JH, et al. 2014. Huntington disease: natural history, biomarkers and prospects for therapeutics. Nat Rev Neurol 10:204–216 [DOI] [PubMed] [Google Scholar]
- Rossion B, Schiltz C, Crommelinck M. 2003. The functionally defined right occipital and fusiform “face areas” discriminate novel from visually familiar faces. NeuroImage 19:877–883 [DOI] [PubMed] [Google Scholar]
- Roweis S. 1998. EM algorithms for PCA and SPCA. Neural Inform Process Syst 10:616–626 [Google Scholar]
- Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. 2009. Neurodegenerative diseases target large-scale human brain networks. Neuron 62:42–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, et al. 2009. Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci U S A 106:13040–13045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD. 2002. A direct approach to false discovery rates. J R Stat Soc 64:479–498 [Google Scholar]
- Torres Ramírez L, Mori Quispe N, Mendoza Cabanillas M, Montoya Henriquez J, Cuentas Jara M, Domínguez Calderón J, et al. 2006. Clinical molecular study of Huntington disease in patients from Valle de Cañete-Peru. [In Spanish]. Diagnóstico (Perú) 45:102–108 [Google Scholar]
- Uddin LQ. 2015. Salience processing and insular cortical function and dysfunction. Nat Rev Neurosci 16:55–61 [DOI] [PubMed] [Google Scholar]
- Unschuld PG, Joel SE, Liu X, Shanahan M, Margolis RL, Biglan KM, et al. 2012. Impaired cortico-striatal functional connectivity in prodromal Huntington's Disease. Neurosci Lett 514:204–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Asselen M, Julio F, Januario C, Campos EB, Almeida I, Cavaco S, et al. 2012. Scanning patterns of faces do not explain impaired emotion recognition in Huntington disease: evidence for a high level mechanism. Front Psychol 3:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bogaard SJA, Dumas EM, Acharya TP, Johnson H, Langbehn DR, Scahill RI, et al. 2011. Early atrophy of pallidum and accumbens nucleus in Huntington's disease. J Neurol 258:412–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP. 1985. Neuropathological classification of Huntington's disease. J Neuropathol Exp Neurol 44:559–577 [DOI] [PubMed] [Google Scholar]
- Werner CJ, Dogan I, Sass C, Mirzazade S, Schiefer J, Shah NJ, et al. 2014. Altered resting-state connectivity in Huntington's disease. Hum Brain Mapp 35:2582–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf RC, Kloppel S. 2013. Clinical significance of frontal cortex abnormalities in Huntington's disease. Exp Neurol 247:39–44 [DOI] [PubMed] [Google Scholar]
- Wolf RC, Sambataro F, Vasic N, Wolf ND, Thomann PA, Landwehrmeyer GB, et al. 2011. Longitudinal functional magnetic resonance imaging of cognition in preclinical Huntington's disease. Exp Neurol 231:214–222 [DOI] [PubMed] [Google Scholar]
- Wolf RC, Sambataro F, Vasic N, Wolf ND, Thomann PA, Saft C, et al. 2012. Default-mode network changes in preclinical Huntington's disease. Exp Neurol 237:191–198 [DOI] [PubMed] [Google Scholar]
- Zhang N, An MC, Montoro D, Ellerby LM. 2010. Characterization of human Huntington's disease cell model from induced pluripotent stem cells. PLoS Curr 2:RRN1193. [DOI] [PMC free article] [PubMed] [Google Scholar]