Abstract
Second-generation (Gen 2) Antisense oligonucleotides (ASOs) show increased nuclease stability and affinity for their RNA targets, which has translated to improved potency and therapeutic index in the clinic. Gen 2 ASOs are typically modified using the phosphorothioate (PS) backbone modification, which enhances ASO interactions with plasma, cell surface, and intracellular proteins. This facilitates ASO distribution to peripheral tissues and also promotes cellular uptake after injection into animals. Previous work identified that Stabilin receptors specifically internalize PS-ASOs in the sinusoidal endothelial cells of the liver and the spleen. By modulating expression of specific proteins involved in the trafficking and maturation of the endolysosomal compartments, we show that Rab5C and EEA1 in the early endosomal pathway, and Rab7A and lysobisphosphatidic acid in the late endosomal pathway, are important for trafficking of PS-ASOs and facilitate their escape from endolysosomal compartments after Stabilin-mediated internalization. In conclusion, this work identifies key rate-limiting proteins in the pathway for PS-ASO translocation and escape from the endosome.
Keywords: : endosome maturation, endosome escape, antisense oligonucleotide, Stabilin, ASO
Introduction
Oligonucleotide therapeutics have made rapid progress in clinical settings, [1] but the precise pathways by which these agents enter cells and escape from endolysosomal compartments to exert antisense effects are just starting to be understood [1,2]. Antisense oligonucleotides (ASOs) are short (∼16–20 bp), chemically modified nucleic acids, which utilize Watson–Crick base pairing for hybridizing to their cognate RNA in cells. Second-generation ASOs are typically modified using the phosphorothioate (PS) backbone and 2′ modifications of the ribose sugar, which enhance ASO stability in biological fluids [3]. PS-ASOs can exert their biological effects through a variety of antisense mechanisms based on the chemical composition of the oligonucleotide. For example, DNA gapmer PS-ASOs promote degradation of their target RNA by RNase H-mediated hydrolysis while uniformly modified MOE PS-ASOs can modulate RNA splicing [4]. Understand the biological mechanisms responsible for binding, internalizing, and trafficking PS-ASOs are currently areas of active investigation. Recent work showed that Annexin A2 facilitates transport of PS-ASOs from early endosomes to late endosomes (LE) and is involved in the release of ASOs from late endosomal compartments in several common cell lines, such as A431, HEK293, and HepG2 [5].
PS-ASOs were recently shown to be rapidly internalized by clathrin-mediated endocytosis by a class of membrane-bound scavenging receptor proteins known as the Stabilin receptors in the liver sinusoidal endothelial cells, a site for the clearance of many blood-borne matrix and synthetic molecules [6]. Stabilin receptors (Stabilin-1 and Stabilin-2) bind to and internalize a host of negatively charged polymers, ranging from smaller, ∼20 nucleotide PS-ASOs [6] (both Stabilin receptors), to large glycosaminoglycans, such as ∼30 kDa unfractionated heparin and 3–400 kDa hyaluronan (Stabilin-2 only) [7,8]. Stabilin-2, also known as the Hyaluronic-Acid Receptor for Endocytosis (HARE), has two distinctly expressed isoforms in native tissue,190-kDa and 315-kDa [9]. The 190-kDa isoform is a truncated version of the full 315-kDa isoform, but retains full functionality for binding and endocytosis [10]. Both isoforms have been shown to be able to internalize PS-ASOs equally; however, for this work, we decided to utilize recombinant cells overexpressing the 190-kDa isoform. The use of this cell line is relevant based on the 1:1 expression ratio of both Stabilin-2 isoforms in native tissues [11].
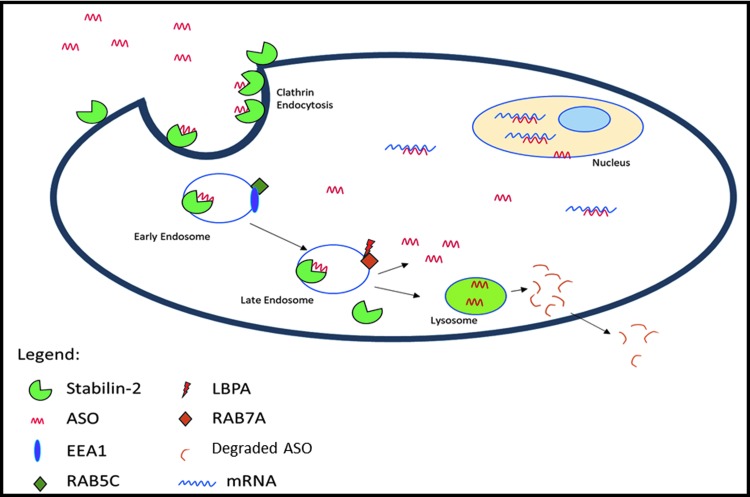
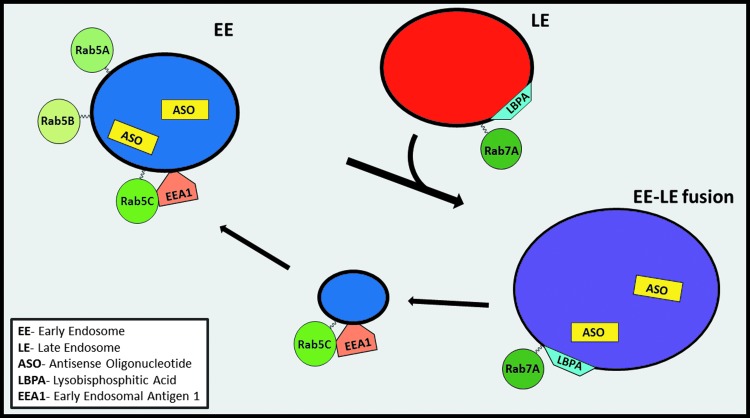
Our initial efforts of this work focused on early endosomal antigen 1 (EEA1), which is a peripheral membrane associated protein found in the early endosome, containing zinc-dependent binding motifs [12]. This protein was selected based on its role in early endosomal transport by localizing to the early endosome with Rab5 and phosphatidylinositol (3)P-kinase [13]. Rab5 has three distinct isoforms, Rab5A, B, and C, which all bind to EEA1, however, Rab5C has distinct functional differences compared to Rab5A and Rab5B. Rab5 isoforms have shown preferential differences to specific protein targets, along with identification of differing roles in functionality such as EGFR degradation [14]. Rab5C binding to EEA1 confers directionality of the endosomes from early to late stages, by serving as a regulatory element for SNARE protein family members in EE fusion and maturation to LEs [13,15]. The role of Rab7A in altering PS-ASO activity was also evaluated, as this protein is an activator in endosomal maturation, lysosome biogenesis, fusion, and morphological changes of LEs, as well as a variety of interacting partners [16–18]. One LE-specific resident of particular interest is lysobisphosphatidic acid (LBPA) (Fig. 1). LBPA is an uncommon lipid that is enriched in the LE [19], and has shown to be involved in controlling cholesterol levels in LEs [20]. Recent work has suggested that LPBA is an essential component for PS-ASO escape in the LE [21]. Taken together, manipulation of these endosomal molecules may increase the understanding of PS-ASO activity after internalization by Stabilin-1 and -2.
FIG. 1.
Proposed mechanism of late endosomal PS-ASO escape by internalization through Stabilin receptors. Key interactor molecules highlighted for escape in both early and late endosomes. PS-ASO, phosphorothioate–antisense oligonucleotide.
Materials and Methods
Cell lines
Stabilin-2 (190-HARE) and empty vector cell lines were generated as previously described [9,10]. Briefly, the cDNA of interest was cloned into pcDNA5/FRT/V5–6xHIS-TOPO (Life Technologies) and stably transfected in HEK293 Flp-In cells within the FRT site to eliminate off-target effects of gene disruption (Life Technologies). Cell lines were grown and maintained in DMEM containing 8% FBS supplemented with 50 μg/mL Hygromycin B.
Oligonucleotide synthesis and delivery
Phosphorothioate PS-ASOs were synthesized using a standard phosphoramidite chemistry by using a DNA synthesizer at Integrated DNA Technologies (Iowa City, IA) or at Ionis Pharmaceuticals (Carlsbad, CA) against Malat-1. PS-ASOs were delivered to cultured cells utilizing standardized gymnotic conditions. PS-ASOs were added to standard Dulbecco's modified Eagle's medium (DMEM) without serum, and diluted to 1 μM. One hundred microliters of medium was added to 900 μL of DMEM with 8% FBS to reach a final concentration of 0.1 μM of PS-ASO. The PS-ASO used for this study was GCTTCAGTCATGACTTCCTT, which targets the long noncoding RNA Malat-1 and previously verified [6]. The PS-ASO was fully modified using the PS backbone and the bold letters indicate 2′-Methoxyethyl (MOE) RNA nucleotides. The biotinylated ASO (bASO) used for this study was GCTTCAGTCATGACTTCCTT (Ionis AA7141) with a modified 5′ terminus containing a piperidine linker to accept biotin-NHS. Details of the synthesis and purification procedures are in supplemental materials (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/nat).
siRNA delivery
Stabilin-2 190-kDa cells were plated in 24-well dishes and grown to 50% confluency followed by transfection of 5 pmol siRNAs in duplicate wells, giving a final concentration of 2.5 pmol siRNA per well. siRNAs were used against specific protein targets and transfection was facilitated by RNAiMAX Lipofectamine (Life Technologies). siRNAs were ordered from Silencer Select (Ambion) for each protein target in the pathway. The sense sequences are: EEA1–5′-GCUAAGUUGCAUUCCGAAtt-3′, Rab7A- 5′-GAGCUGACUUUCUGACCAAtt-3′, Rab5A- 5′-GCAAGCAAGUCCUAACAUUtt-3′, Rab5B- 5′-GGAGCGAUAUCACAGCUUAtt-3′, and Rab5C- 5′-GGACAGGAGCGGUAUCACA-3′. Negative/scrambled control was prestocked with the catalog number 4390843.
Purification of RNA and qPCR analysis
Cells were plated in 24-well dishes and treated with siRNAs above when cells were at 50% confluency. After 24 h of incubation with siRNAs, cells were treated with 0.1 μM PS-ASO against Malat-1, and incubated for 24 h. After incubation of PS-ASOs, cells were lysed using a lysis buffer consisting of PBS +0.5% NP40 + protease inhibitors (aprotin-100 μM, leupeptin-20 μM, pepstatin-20 μM, PMSF-400 μM). RNA was isolated from cell lysates using TRIzol RNA isolation protocol (Ambion) following the manufacturer's direction. RNA concentration was measured using a NanoDrop 2000 spectrophotometer. cDNA reverse transcription was performed using Improm-II RTase reagents (Promega). qPCR was run using ∼50 ng cDNA per reaction with 2 × SYBR qPCR master mix, and primers for Malat-1 (Forward–5′-AAAGCAAGGTCTCCCCACAAG-3′, Reverse- 5′-TGAAGGGTCTGTGCTAGATCAAAA-3′) (Eurofins), and beta-actin control (Forward- 5′-AAGTCAGTGTACAGGTAAGCC-3′, Reverse- 5′-GTCCCCCAACTTGAGATGTATG-3′) (IDT). Samples were run in at least triplicate for statistical analysis on a Bio-Rad CFX Connect® qPCR machine, using the following cycling protocols: 10 min at 50°C, 5 min at 95°C, and 40 cycles of 15 s at 95°C and 1 min 15 s at 60°C. Primer specificity was verified by using a melt curve analysis after PCR analysis was performed. Cq values were determined using Bio-Rad the CFX manager software. RNA levels of the controls were compared with that of Malat-1 (PS-ASO target) by determination of the relative quantity from the beta-actin control; afterward the relative quantities were normalized to non-PS-ASO-treated controls for each sample to generate reported gene expression levels for each RNA target. Statistical analysis was done by using a Student's t-test for pairwise comparison using Microsoft Excel.
Overexpression of LBPA by DHA induction
Lysobisphosphatidic Acid (LBPA) cannot be directly induced into mammalian cells; therefore, the lipid precursor docosahexaenoic acid (DHA) was used. Working stock solutions of DHA (50.0 mM in ethanol) and lipid-free bovine serum albumin (BSA; 5 mM in water) were mixed at a 4:1 ratio, respectively. The solution was diluted to a working solution of 10.0 μM DHA that has been reported to a working concentration for overexpression of DHA (and by conversion of LBPA) without leading to cytotoxic effects on the cells. Cells were first treated with siRNAs for RAB7A, EEA1, or scrambled siRNA for 24 h to provide proper target knockdown. The cells were then incubated in 10.0 μM DHA-BSA DMEM. Given the cells inability to survive long term in BSA media, a 6-h incubation for DHA media was done with or without PS-ASO contained in the media. This should not have any impact on PS-ASO activity as these molecules have shown transport to LE/lysosome at 3–6 h [6]. After 6 h of DHA-BSA ± PS-ASO media, cells were harvested and qPCR analysis was done for Malat-1 expression (see “Purification of RNA and qPCR analysis section”).
Rab5C overexpression and Western blot analysis
The Rab5 plasmid (OriGene NM_201434) was transiently transfected into 190-kDa HARE-overexpressing cells using Lipofectamine 2000 under predetermined optimal conditions. Cells were treated for 24, 48, and 72 h to determine if transfection was possible for qPCR analysis with siRNA and PS-ASO treatment to follow. Cell lysates were separated by 10% SDS-PAGE using 30 μg of protein, blotted, and probed with a mouse monoclonal anti-DDK primary antibody (OriGene), followed by an anti-mouse fluorescence secondary antibody (LiCor). After overexpression levels were verified by western blot, a transient transfection of Rab5C using Lipofectamine 2000 was performed and incubated for 24 h before siRNAs were added, followed by PS-ASO incubation. A qPCR analysis was performed targeting Malat-1.
Rab5C (S35 N) point mutant
The Rab5C plasmid from OriGene was subject to site-directed mutagenesis (serine to asparagine substitution) using Q5 (New England Biolabs) reagents. Primers used were: Forward–5′-GTAGGCAAAAACAGCCTCGTCCTCCGCTTTG-3′, Reverse–5′-CGAGGCTGTTTTTGCCTACCGCAGACTCC-3′ (Eurofins). Rab5C plasmid was PCR amplified using Q5-HiFi Polymerase (NEB) for 18 cycles. Samples were Dpn1 digested for 1 h to remove methylated template DNA, and heat shock transformed into competent Escherichia coli cells. Cells were plated, grown, and screened for correct sequence. Rab5CS35 N plasmids were transfected into 190-HARE cells and incubated for 24 h, followed by siRNA treatment. After 24 h of siRNA treatment, PS-ASOs (0.1 μM) were added and incubated for 24 h, followed by qPCR targeting Malat-1 as described above.
Labeling and endocytosis of hyaluronan and ASOs
Approximately 100 μg of hyaluronan (HA) with an average size of 130 kDa was labeled with a Bolton-hunter adduct on the reducing end of the polymer using the method of Raja et al. [22]. The Bolton-hunter adduct is iodinated with 0.3 mCi carrier-free 125I-Na (MPBio) and the HA is quantified using the carbazole assay [23]. For the HA endocytosis assays, cells were plated at least 2 days before the assay and allowed to grow up to about 80% confluence in 24-well polystyrene tissue culture plates. Endocytosis medium was prepared with DMEM supplemented with 0.05% BSA and 1 μg/mL 125I-HA alone or with 100 μg/mL unlabeled HA to serve as a nonspecific uptake control. Cells were incubated with the HA ligands for 2 h, washed three times with ice-cold Hank's Balance Salt Solution (HBSS) and cell lysates were prepared with 0.3 mL 0.3 M NaOH. Counts per minute (CPM) of each lysate was measured with a Wallac 1470 Wizard Gamma Counter and total protein was measured by the Bradford assay.
Specific endocytosis of HA was calculated by subtracting the 125I-HA + HA samples from the 125I-HA samples. For the ASO endocytosis assays, 125I-streptavidin (SA) was prepared by the method of Harris et al. [7] and mixed in a 1:4 ratio with bASO. An equivalent amount of 100 nM 125I-SA-bASO or 125I-SA alone was incubated with prepared cells for 6 h, washed, and prepared in the same manner as the HA uptake described above. Specific endocytosis of ASO was measured by subtracting the 125I-SA alone values from the 125I-SA-bASO values. Nonspecific binding of 125I-HA + HA was less than 3% 125I-HA and 125I-SA was less than 1% of the 125I-SA-bASO. Each experiment was performed at least twice with at least three replicates per sample.
Statistics
All experiments were performed at least three times with three replicates per experiment. The mean of each experiment was calculated with standard deviation. Groups of experimental data were compared using the Student's t test with a p-value equal to or less than 0.05 considered significantly different.
Results
ASO chemistry and design
A second-generation PS-ASO targeting the long noncoding RNA Malat-1 was used for our experiments. Gen 2 PS-ASOs have a central DNA region that is flanked on both ends with a 2′-O-methoxyethyl RNA (MOE) modification. The DNA gap promotes degradation of the targeted RNA by RNaseH1-mediated hydrolysis. RNaseH1 is a ubiquitously expressed endonuclease, which selectively degrades the RNA strand of a DNA–RNA heteroduplex. The 2′ MOE modifications work to increase metabolic stability of these PS-ASOs, while also increasing the affinity of target binding. The knockdown of Malat-1, as measured by quantitative RT-PCR (qRT-PCR), was used as a surrogate for PS-ASO escape from the endosomal compartments as RNA knockdown can only occur when PS-ASO is released from these compartments [24,25].
Early endosome versus LE targets
Once internalized by the Stabilin receptors into clathrin-coated vesicles, PS-ASOs are trafficked to the lysosome for degradation [6]. However, at some point in the endosomal trafficking pathway, a fraction of the internalized PS-ASO has to escape these vesicles and enter the cytoplasm and/or the nucleus to induce RNase H-mediated degradation of Malat-1. Early endosome antigen 1 (EEA1), a peripheral hydrophilic membrane protein found in early endosomal membrane fractions as well as in the cytosol, was selected as a possible candidate for screening [12]. EEA1 is involved in early endosomal vesicle transport for fusion with LEs through SNARE proteins, as well as early endosome sorting [26,27]. With regard to LEs, the small GTPase Rab7A, which plays an important role in a variety of LE functions, including motility, biogenesis, cargo transport, among many others, was chosen as a potential candidate affecting PS-ASO release [17,19,28].
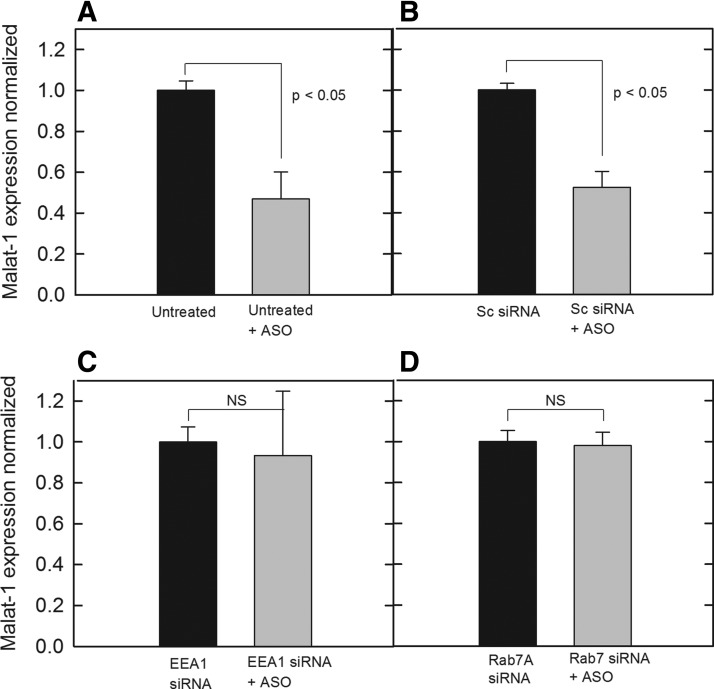
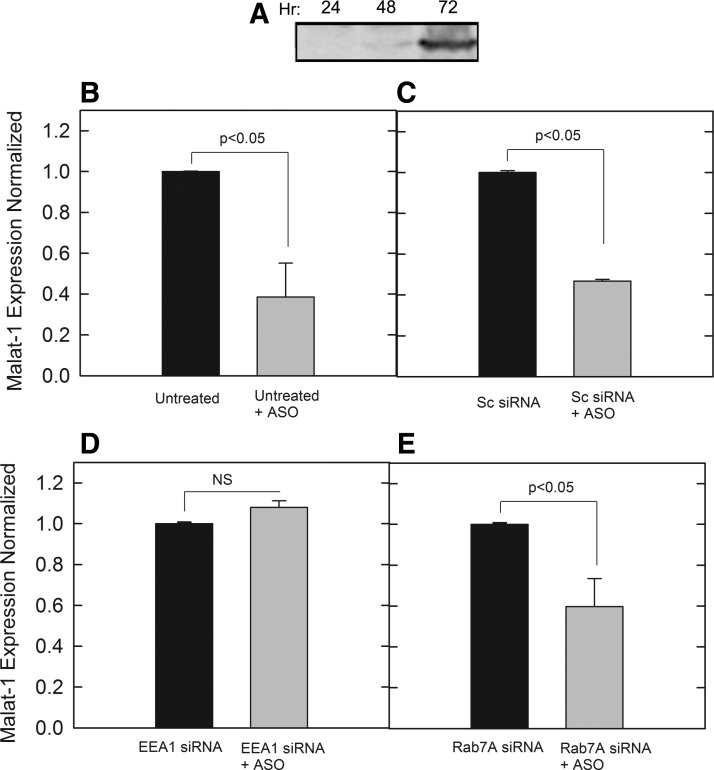
After verifying siRNA functionality for target RNA knockdown (Supplementary Fig. S2), cells were incubated in 5.0 pmol siRNA for EEA1 or Rab7A, followed by PS-ASO incubation for 24 h. This was done to determine if knockdown of specific proteins in the early or LE affected Malat-1 expression as a measure of PS-ASO release. From our results, cells left untreated (Fig. 2A) or treated with a scrambled negative control siRNA (Fig. 2B) showed significant PS-ASO activity for reducing Malat-1 RNA expression. However, when EEA1 siRNA (Fig. 2C) or Rab7A siRNA (Fig. 2D) was added to cells, expression levels of Malat-1 were not affected and no different than paired samples that were not treated with PS-ASO. Given that EEA1 and Rab7A are in two separate maturation stages of the endosomal pathway, this suggested that PS-ASO activity is a result of endosomal escape before transport to the lysosome. The results also provided evidence that these proteins could be working in conjunction through the same mechanism to influence PS-ASO release.
FIG. 2.
Knockdown of specific targets by siRNA treatment followed by treatment of 0.1 μM PS-ASO against Malat-1 with relative quantities was normalized. Following RNA extraction, cDNA synthesis was performed and normalized to ∼50 ng/reaction. Expression of Malat-1was assessed by SYBR green qPCR amplification with beta-actin controls in cells with (A) no siRNA treatment, (B) a negative control scrambled siRNA, (C) siRNA for EEA1, (D) siRNA for RAB7A. Bars represent normalized Malat-1 expression as mean ± standard deviation, n = 3. Statistical analysis was performed using two-tailed Student's t-test P < 0.05. EEA1, early endosomal antigen 1. NS, not statistically significant.
Rab5 isoforms alter PS-ASO activity
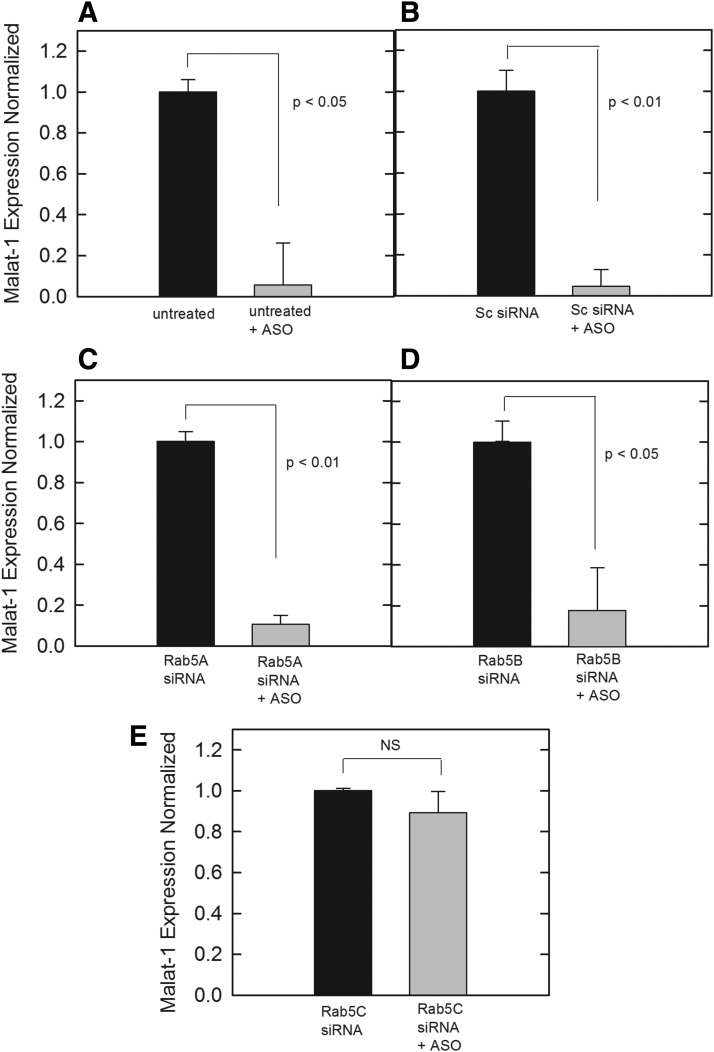
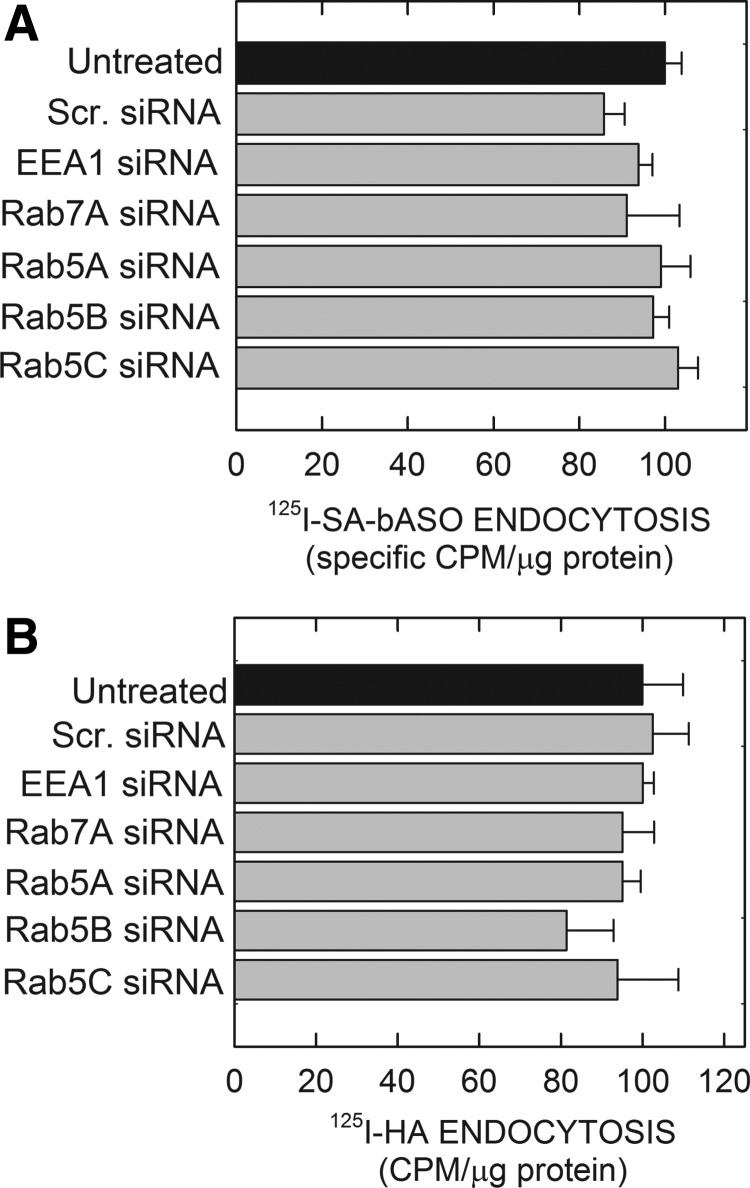
A potential molecular pathway that regulates early endosomal trafficking and is known to interact with EEA1 is Rab5 [27], which is expressed in cells in three distinct isoforms: Rab5A, Rab5B, and Rab5C. These isoforms share a sequence homology of roughly 90%; however, Rab5A and 5B are functionally different from Rab5C due to each isoform's respective ability to interact with different effector proteins, such as Rab5C, being the only isoform shown to interact with Rac1 [14,29]. Each isoform of Rab5 was disrupted in our 190-HARE-overexpressing cells by siRNA knockdown, which was verified by qPCR analysis (Supplementary Fig. S3). After siRNA induction, cells were treated with 0.1 μM of PS-ASO against Malat-1 to determine if any knockdown of a specific isoform influenced PS-ASO release. Untreated siRNA controls (Fig. 3A), as well as scrambled siRNA (negative) controls (Fig. 3B) were carried out to demonstrate that the PS-ASO was functional. Rab5A and Rab5B followed a similar pattern to the controls, indicating no functional role in altering PS-ASO activity (Fig. 3C, D). However, when Rab5C expression was inhibited by siRNA, no knockdown of Malat-1 was observed, indicating that Rab5C is working in conjunction with EEA1 to facilitate PS-ASO escape from endosomes and/or PS-ASO trafficking from early to LEs (Fig. 3E). To ensure that our methods were not perturbing Stabilin-2-mediated endocytosis or other mechanisms of ASO internalization, we treated 190-HARE cells with all of the siRNAs individually followed by the addition of radiolabeled PS-ASO. The effect of siRNA treatment with these cells did not affect PS-ASO uptake (Fig. 4A) nor did it affect other ligands for Stabilin-2 such as hyaluronan (Fig. 4B).
FIG. 3.
Malat-1 expression was analyzed through qPCR based on RAB5 knockdowns by siRNA, followed by treatment with 0.1 μM PS-ASO against Malat-1. cDNA was normalized to ∼50 ng/reaction. Expression of Malat-1assessed by SYBR green qPCR amplification with beta-actin controls in cells (A) with no siRNA treatment, (B) treated with a negative control scrambled siRNA, (C) treated with siRNA for RAB5A, (D) treated with siRNA for RAB5B, (E) treated with siRNA for RAB5C. Bars represent normalized Malat-1 expression as mean ± standard deviation, n = 3. Statistical analysis was performed using two-tailed Student's t-test P < 0.05.
FIG. 4.
siRNA targeted knockdown of individual proteins had no effect on endocytosis of I125 radiolabeled ligands. Cells were grown to 50% confluency and then treated with siRNAs as indicated. Twenty-four hours posttreatment, cells were washed and treated with radiolabeled (A) PS-ASO or (B) HA for 6 h and 2 h, respectively. The background control for PS-ASO uptake was free 125I-SA, which was not conjugated to the PS-ASO and the control for the HA uptake were cells incubated with 100-fold excess unlabeled HA. Bars represent CPM normalized to protein concentration with mean ± standard deviation, n = 3. Scr. siRNA is a negative control for siRNA induction. Statistics used two-tailed Student's t-test, P < 0.05.
DHA-supplemented medium alters PS-ASO activity
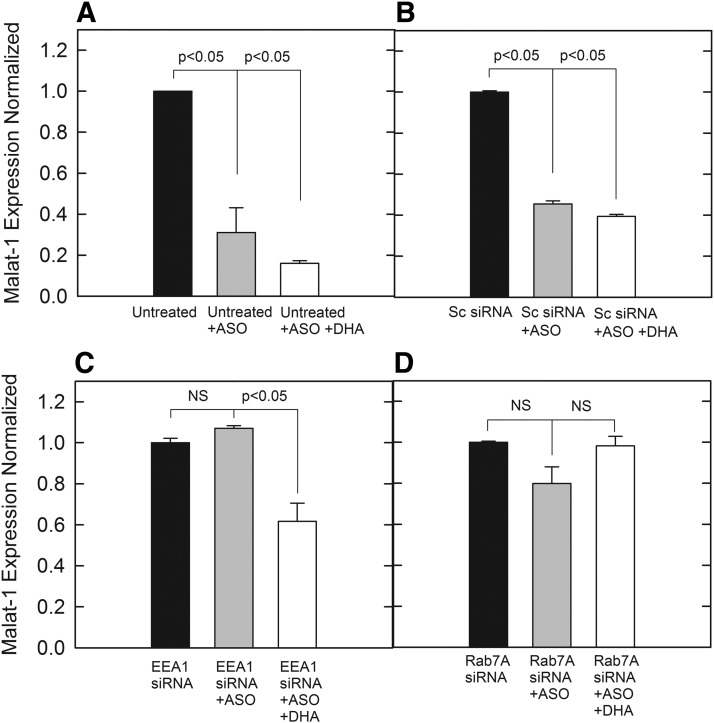
A recent report indicated that LPBA is a molecular component of endosomal escape in cell lines that are amenable for PS-ASO activity through free uptake [21]. LBPA has previously been reported to be enriched in late endosomal vesicles, whereas also being reported to be required to assist is endosomal escape for PS-ASOs within the LE [21,30]. LPBA cannot be simply incorporated into cells in its active form. A method for LPBA overexpression in mammalian cells is to incorporate 10.0 μM docosahexaenoic acid (DHA) as a starting substrate that has been shown to be converted to LBPA [31]. Replicating this method in our cell lines proved to be successful and an increase in LBPA was detected by an anti-LBPA antibody as previously reported [21] (Supplementary Fig. S4A, B). The 190-HARE-overexpressing cells were supplemented with 10.0 μM DHA conjugated with BSA in the medium, followed by treatment with siRNA for endosomal protein knockdown, followed by PS-ASO treatment. In our non-siRNA untreated controls, as well as scrambled siRNA (negative) controls, a similar trend of PS-ASOs facilitating Malat-1 knockdown was noticed, which was on par, or further knocked down by DHA medium supplementation (Fig. 5A, B). However, for EEA1 knockdown, the incorporation of DHA was able to rescue PS-ASO activity, shown by Malat-1 knockdown (Fig. 5C). Interestingly, when Rab7A was knocked down, the supplementation of LBPA was unable to rescue the effects of PS-ASO activity, pointing to Rab7A as the limiting factor in late endosomal PS-ASO transport (Fig. 5D). The knockdown of Rab7A in addition to the isoforms of Rab5 and EEA1 did not affect degradation of Stabilin-2 ligands. Since we could not measure degradation of the PS-ASO molecule efficiently, we monitored 125I-hyaluronan degradation under each siRNA treatment and found that degradation for all conditions was not affected (Supplementary Fig. S5). Therefore, the integrity of the early and late endosomal pathway was intact despite manipulations in EEA1, Rab7A, and LBPA levels.
FIG. 5.
Malat-1 expression analyzed through qPCR based on PS-ASO against Malat-1 in conjunction with siRNA treatment and DHA media supplementation for LBPA overexpression analysis. cDNA was normalized to ∼50 ng/reaction. Expression of Malat-1assessed by SYBR green qPCR amplification with beta-actin controls. (A) Expression of cells with no siRNA treatment, (B) Expression of cells treated with a negative control scrambled siRNA, (C) Expression of cells treated with siRNA for EEA1, (D) Expression of cells treated with siRNA for RAB7A. Bars represent normalized Malat-1 expression with mean ± standard deviation, n = 3. Statistical analysis was performed using two-tailed Student's t-test P < 0.05.
Rab5C overexpression leads to increased PS-ASO activity
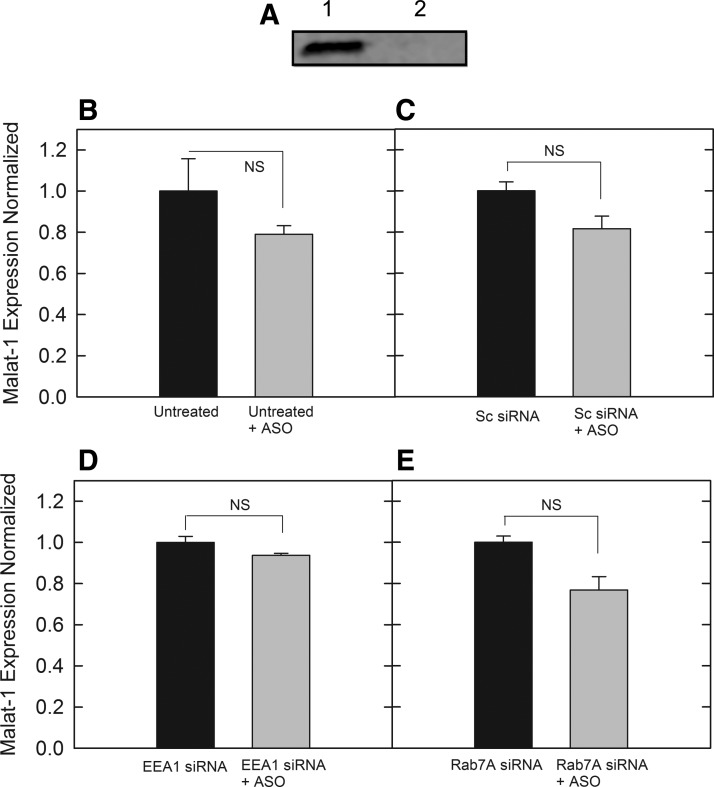
Given the importance of Rab5C to modulate PS-ASO activity by activating EEA1 for endosomal fusion, we examined the effect of overexpressing Rab5C. A Rab5C plasmid was transiently transfected into cells for 24, 48, and 72 h to determine if production would be substantial in our 190-HARE cells over 72 h. As shown by western blot analysis, overexpression of Rab5C peaked at 72 h posttransfection (Fig. 6A). It was verified that 72 h was sufficient for Rab5C expression in cells. The 190-HARE cells were transfected with Rab5C, followed by an incubation with siRNAs for EEA1 and Rab7A at 24 h postinduction. This was followed by PS-ASO treatment 24 h post siRNA induction, and was incubated for a final 24 h followed by cell lysis. When Malat-1 levels were analyzed in our control cells, PS-ASO activity was unchanged and similar to previous controls (Fig. 6B, C). When EEA1 was knocked down, overexpressing Rab5C was not able to overcome the absence of its interacting partner, as Malat-1 levels were that of non-PS-ASO-treated control cells (Fig. 6D). Interestingly, in the Rab7A knockdown cells, PS-ASO activity was restored (Fig. 6E), as higher levels of Rab5C are able to increase ability of Rab5-Rab7 exchange and Rab5 becomes displaced from the endosome concomitantly by the addition of Rab7A [32].
FIG. 6.
Malat-1 expression based on PS-ASO against Malat-1 in conjunction with RAB5C overexpression and siRNA treatment. qPCR analysis performed by SYBR green qPCR amplification with beta-actin controls with cDNA normalized to ∼50 ng/reaction. (A) Western blot of transient transfection using varied Lipofectamine concentrations over multiple time points, (B) Expression of cells with no siRNA treatment, (C) Expression of cells treated with a negative control scrambled siRNA, (D) Expression of cells treated with siRNA for EEA1, (E) Expression of cells treated with siRNA for RAB7A. Bars represent normalized Malat-1 expression with mean ± standard deviation, n = 3. Statistical analysis was performed using two-tailed Student's t-test P < 0.05.
Dominant negative Rab5C expression alters PS-ASO activity
To determine if Rab5C activity was a crucial driving force for PS-ASO activity, a S35 N substitution was created in the GTP-binding site leading to a dominant negative protein. The dominant negative Rab5C was then transiently overexpressed in 190-HARE cells and verified by western blot analysis (Fig. 7A). This confirmed that the expression of the inactive mutant is sufficient over the time course of treatment similar to the Rab5C overexpression experiment. The cells were then treated in a similar fashion to our previous Rab5C-overexpressing cells with siRNA knockdowns for EEA1 and Rab7A, followed by incubation of PS-ASOs against Malat-1. The results from the untreated and negative control (scrambled) siRNA samples indicated that the dominant negative Rab5C ablated PS-ASO activity as Malat-1 expression was not inhibited (Fig. 7B, C). Similarly, when siRNA treatments were induced for EEA1 and Rab7A, followed by PS-ASO induction, there was no significant change in Malat-1 expression (Fig. 7D, E). These results provide evidence that Rab5C activity is important for PS-ASO escape from endosomal compartments.
FIG. 7.
Dominant negative RAB5C transfection of 190-HARE-overexpressing cells. qPCR analysis performed by SYBR green qPCR amplification with beta-actin controls with cDNA normalized to ∼50 ng/reaction. (A) Western blot showing inactive mutant expression 72 h posttransfection (lane 1) and untransfected cells (lane 2), (B) Expression of cells with no siRNA treatment, (C) Expression of cells treated with a negative control scrambled siRNA, (D) Expression of cells treated with siRNA for EEA1, (E) Expression of cells treated with siRNA for RAB7A. Bars represent normalized Malat-1 expression with mean ± standard deviation, n = 3. Statistical analysis was performed using two-tailed Student's t-test P < 0.05.
Discussion
In this study, we report that specific trafficking molecules in the early and late endosomal pathway affect PS-ASO activity starting with initial uptake by the Stabilin-2 (190-HARE) receptor (Fig. 8). We discovered that knockdown of EEA1 and Rab7A reduces PS-ASO efficacy without affecting bulk ASO internalization in Stabilin-2-overexpressing HEK cells. Somewhat surprisingly, the knockdown of Rab5C or overexpressing the dominant negative Rab5C also reduced the ASO activity.
FIG. 8.
Schematic representation of interacting partners from early endosome–late endosomal fusion. Rab5C interacts with EEA1 to allow for endosomal maturation and fusion with a late endosome. The late endosome facilitates Rab7A to interact with LBPA, which is involved in PS-ASO escape. Rab5C and EEA1 are returned back to the early endosome by a vesicle, as they remain membrane tethered. LBPA, lysobisphosphatidic acid.
Nearly two decades ago, it was discovered that early endosomes are quite heterogeneous depending on receptors, cargo, and intracellular components. A single early endosomal vesicle may be a mosaic of different biochemical domains that split off and mature as single entities [33]. It is possible that RabC may cluster to form an enriched domain to sequester Stabilin-2 and its cargo for trafficking to the LE. Rab GTPases undergo specific spatial organization and distribution, allowing for vesicle motility, formation, and fusion [34]. Specific compartmentalization of enriched membrane domains induces cooperativity, facilitating spatial segregation for different cargos. The identification of Rab5 as an activator of EEA1 through a zinc finger binding domain has been well established [35]; however, the identification of the specificity for the Rab5C isoform with regard to regulation of PS-ASO transport helps provide insight into the mechanism behind PS-ASO escape and activity. Although previous works have identified possible mechanisms for endosomal escape of PS-ASOs, this article provides specific molecular manipulations and interactions for the mechanism underlying the proposed escape pathway [2,36,37].
As PS-ASOs are trafficked through the endosomal pathway after receptor mediated endocytosis, they are subsequently transported through the LE/multivesicular body to the lysosome for degradation. During transport to the LE, Rab7A interacts with the phospholipid LBPA, which is a component in other late endosomal processes such as protein trafficking and cholesterol transport [38,39]. LBPA has been previously shown to have a role in the escape of PS-ASOs [21] and other cargos such as vesicular stomatitis virus nucleocapsids and anthrax toxin from late endosomal compartments [40]. LBPA-containing MVB/LEs represent a subpopulation containing ubiquitinated membrane proteins [41] suggesting that a higher proportion of PS ASOs are transported to LBPA-containing vesicles after Stabilin-mediated internalization. In addition, recent work has identified that the cytosolic autophagy protein, Alix, alters levels of LBPA [5,21]. Alix has also been reported to be involved in the regulation of the endolysosomal system [42]. These results demonstrate that modulating LBPA and Rab7A affects activity of PS-ASOs and suggests that escape of PS-ASOs occurs as a late endosomal event before trafficking to the lysosome.
Stabilin-2 (190-HARE) internalizes a wide variety of negatively charged molecules ranging from sizes of polymers of several hundred kDa (Hyaluronan) down to oligos of a 20+ nucleotides (PS-ASOs) [6,43]. Although not much is known about Rab5C, it has previously been identified to operate independently of Rabs 5A and 5B in the trafficking and signaling of the EGF receptor [14,29]. EGF receptors are dependent on Rab5A for internalization and degradation, whereas, we did not see the same effect on the internalization for Stabilin-2. The fact that knocking down Rab5C ablated the function of the PS-ASO cargo suggests that Rab5C, and not Rabs 5A or 5B, is essential for downstream trafficking of this cargo. It remains unknown if Rab5C is only involved in trafficking of PS-ASO cargo or if other Stabilin-2 cargos use the same trafficking mechanism. We previously reported that Stabilin-2 is a signaling receptor [44] and that downstream signaling is cargo dependent [8], which may have further implications into how the receptor is trafficked depending on the bound cargo.
Efficacy of a therapeutic ASO targeted against a specific RNA for either systemic or tissue-specific knockdown of gene expression is dependent on several factors, including initial dose, site of injection (subcutaneous, intravascular, intrathecal, etc), clearance rates by liver and kidney, internalization by the target tissue, and escape from the endosomal pathway [45]. The physicochemical principles for designing an ASO to remain intact in biological fluids and for enhancing binding to its target RNA are relatively well understood. The difficulty lies in the delivery of the ASO to target tissues/cells and allowing an effectual number of the ASOs to pass biological barriers in the form of membranes and lysosomes.
We previously reported that both Stabilin-1 and Stabilin-2 receptors are specific endocytic receptors for nontargeted PS-ASOs with high affinity and endocytic activity. Although Stabilin receptors are highly expressed in specific tissues such as in the sinusoids of liver, lymph node, spleen, and bone marrow [11,46], it is becoming evident that their expression is more widespread than previously thought by more recent reports [47,48]. Both receptors are internalized by classic clathrin-mediated mechanisms [49] that result in the majority of ligand to be degraded in lysosomes for anabolic cellular functions. Any additional knowledge about how ASO-binding receptors are trafficked through the cells of targeted and nontargeted tissues is of therapeutic value as their mechanisms may be exploited to increase delivery efficiency.
Conclusion
After internalization by Stabilin receptors, PS-ASOs are transported in the classical Rab5-mediated endosomal route. As the transition from early to LEs occurs, EEA1 is activated by Rab5C to facilitate vesicle fusion. The majority of internalized PS-ASO is trafficked to the lysosome for degradation, with a small fraction escaping from the LE. Escape is facilitated by Rab7 and LBPA in the LE, allowing for the increased release of PS-ASO molecules into the cytosol and nucleus for exerting antisense effects.
Supplementary Material
Acknowledgments
The authors acknowledge the generosity of Ionis Pharmaceuticals for providing high-quality antisense oligonucleotides for this study and NIH for funding under grant R01HL130864 to E.N.H.
Author Disclosure Statement
P.P.S and W.B.W. are employees of Ionis Pharmaceuticals. The other authors report no competing financial interests.
References
- 1.Miller CM. and Harris EN. (2016). Antisense oligonucleotides: treatment strategies and cellular internalization. RNA Dis 3:1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crooke ST, Wang S, Vickers TA, Shen W. and Liang XH. (2017). Cellular uptake and trafficking of antisense oligonucleotides. Nat Biotechnol 35:230–237 [DOI] [PubMed] [Google Scholar]
- 3.Altmann K-H, Dean NM, Fabbro D, Freier SM, Geiger T, Hanera R, Hiisken DA, Martina P, Monia BPB, Miiller M, et al. (1996). Second generation of antisense oligonucleotides: from nuclease resistance to biological efficacy in animals. CHIMIA Int J Chem 50:168–176 [Google Scholar]
- 4.Kole R, Krainer AR. and Altman S. (2012). RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov 11:125–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang S, Sun H, Tanowitz M, Liang XH. and Crooke ST. (2016). Annexin A2 facilitates endocytic trafficking of antisense oligonucleotides. Nucleic Acids Res 44:7314–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller CM, Donner AJ, Blank EE, Egger AW, Kellar BM, Ostergaard ME, Seth PP. and Harris EN. (2016). Stabilin-1 and Stabilin-2 are specific receptors for the cellular internalization of phosphorothioate-modified antisense oligonucleotides (ASOs) in the liver. Nucleic Acids Res 44:2782–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris EN, Weigel JA. and Weigel PH. (2008). The human hyaluronan receptor for endocytosis (HARE/Stabilin-2) is a systemic clearance receptor for heparin. J Biol Chem 283:17341–17350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pandey MS, Baggenstoss BA, Washburn J, Harris EN. and Weigel PH. (2013). The hyaluronan receptor for endocytosis (HARE) activates NF-kappaB-mediated gene expression in response to 40-400-kDa, but not smaller or larger, hyaluronans. J Biol Chem 288:14068–14079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris EN, Kyosseva SV, Weigel JA. and Weigel PH. (2007). Expression, processing, and glycosaminoglycan binding activity of the recombinant human 315-kDa hyaluronic acid receptor for endocytosis (HARE). J Biol Chem 282:2785–2797 [DOI] [PubMed] [Google Scholar]
- 10.Harris EN, Weigel JA. and Weigel PH. (2004). Endocytic function, glycosaminoglycan specificity, and antibody sensitivity of the recombinant human 190-kDa hyaluronan receptor for endocytosis (HARE). J Biol Chem 279:36201–36209 [DOI] [PubMed] [Google Scholar]
- 11.Zhou B, Weigel JA, Fauss L. and Weigel PH. (2000). Identification of the hyaluronan receptor for endocytosis (HARE). J Biol Chem 275:37733–37741 [DOI] [PubMed] [Google Scholar]
- 12.Mu FT, Callaghan JM, Steele-Mortimer O, Stenmark H, Parton RG, Campbell PL, McCluskey J, Yeo JP, Tock EP. and Toh BH. (1995). EEA1, an early endosome-associated protein. EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine “fingers” and contains a calmodulin-binding IQ motif. J Biol Chem 270:13503–13511 [DOI] [PubMed] [Google Scholar]
- 13.Simonsen A, Lippe R, Christoforidis S, Gaullier JM, Brech A, Callaghan J, Toh BH, Murphy C, Zerial M. and Stenmark H. (1998). EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature 394:494–498 [DOI] [PubMed] [Google Scholar]
- 14.Chen PI, Kong C, Su X, and Stahl PD. (2009). Rab5 isoforms differentially regulate the trafficking and degradation of epidermal growth factor receptors. J Biol Chem 284:30328–30338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jovic M, Sharma M, Rahajeng J. and Caplan S. (2010). The early endosome: a busy sorting station for proteins at the crossroads. Histol Histopathol 25:99–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vonderheit A. and Helenius. A. (2005). Rab7 associates with early endosomes to mediate sorting and transport of Semliki forest virus to late endosomes. PLoS Biol 3:e233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bucci C, Thomsen P, Nicoziani P, McCarthy J, and van Deurs B. (2000). Rab7: a key to lysosome biogenesis. Mol Biol Cell 11:467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vitelli R, Santillo M, Lattero D, Chiariello M, Bifulco M, Bruni CB. and Bucci C. (1997). Role of the small GTPase Rab7 in the late endocytic pathway. J Biol Chem 272:4391–4397 [DOI] [PubMed] [Google Scholar]
- 19.Lebrand C, Corti M, Goodson H, Cosson P, Cavalli V, Mayran N, Faure J. and Gruenberg J. (2002). Late endosome motility depends on lipids via the small GTPase Rab7. EMBO J 21:1289–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chevallier J, Chamoun Z, Jiang G, Prestwich G, Sakai N, Matile S, Parton RG. and Gruenberg J. (2008). Lysobisphosphatidic acid controls endosomal cholesterol levels. J Biol Chem 283:27871–27880 [DOI] [PubMed] [Google Scholar]
- 21.Wang S, Sun H, Tanowitz M, Liang XH. and Crooke ST. (2017). Intra-endosomal trafficking mediated by lysobisphosphatidic acid contributes to intracellular release of phosphorothioate-modified antisense oligonucleotides. Nucleic Acids Res 45:5309–5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raja RH, LeBoeuf RD, Stone GW. and Weigel PH. (1984). Preparation of alkylamine and 125I-radiolabeled derivatives of hyaluronic acid uniquely modified at the reducing end. Anal Biochem 139:168–177 [DOI] [PubMed] [Google Scholar]
- 23.Bitter T. and Muir HM. (1962). A modified uronic acid carbazole reaction. Anal Biochem 4:330–334 [DOI] [PubMed] [Google Scholar]
- 24.Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, Wentworth BM, Bennett CF. and Thornton CA. (2012). Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature 488:111–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, et al. (2013). The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 73:1180–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawe DC, Chawla A, Merithew E, Dumas J, Carrington W, Fogarty K, Lifshitz L, Tuft R, Lambright D. and Corvera S. (2002). Sequential roles for phosphatidylinositol 3-phosphate and Rab5 in tethering and fusion of early endosomes via their interaction with EEA1. J Biol Chem 277:8611–8617 [DOI] [PubMed] [Google Scholar]
- 27.Wilson JM, de Hoop M, Zorzi N, Toh BH, Dotti CG. and Parton RG. (2000). EEA1, a tethering protein of the early sorting endosome, shows a polarized distribution in hippocampal neurons, epithelial cells, and fibroblasts. Mol Biol Cell 11:2657–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansson M, Lehto M, Tanhuanpaa K, Cover TL. and Olkkonen VM. (2005). The oxysterol-binding protein homologue ORP1 L interacts with Rab7 and alters functional properties of late endocytic compartments. Mol Biol Cell 16:5480–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen PI, Schauer K, Kong C, Harding AR, Goud B. and Stahl PD. (2014). Rab5 isoforms orchestrate a “division of labor” in the endocytic network; Rab5C modulates Rac-mediated cell motility. PLoS One 9:e90384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guerra F. and Bucci C. (2016). Multiple Roles of the Small GTPase Rab7. Cells 5:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Besson N, Hullin-Matsuda F, Makino A, Murate M, Lagarde M, Pageaux JF, Kobayashi T. and Delton-Vandenbroucke I. (2006). Selective incorporation of docosahexaenoic acid into lysobisphosphatidic acid in cultured THP-1 macrophages. Lipids 41:189–196 [DOI] [PubMed] [Google Scholar]
- 32.Poteryaev D, Datta S, Ackema K, Zerial M. and Spang A. (2010). Identification of the switch in early-to-late endosome transition. Cell 141:497–508 [DOI] [PubMed] [Google Scholar]
- 33.Sonnichsen B, De Renzis S, Nielsen E, Rietdorf J. and Zerial M. (2000). Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol 149:901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zerial M. and McBride H. (2001). Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2:107–117 [DOI] [PubMed] [Google Scholar]
- 35.Mishra A, Eathiraj S, Corvera S. and Lambright DG. (2010). Structural basis for Rab GTPase recognition and endosome tethering by the C2H2 zinc finger of Early Endosomal Autoantigen 1 (EEA1). Proc Natl Acad Sci U S A 107:10866–10871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth CM. (2005). Molecular and cellular barriers limiting the effectiveness of antisense oligonucleotides. Biophys J 89:2286–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen KD, Kopeckova P. and Kopecek J. (2002). Antisense oligonucleotides delivered to the lysosome escape and actively inhibit the hepatitis B virus. Bioconjug Chem 13:975–984 [DOI] [PubMed] [Google Scholar]
- 38.Kobayashi T, Stang E, Fang KS, de Moerloose P, Parton RG. and Gruenberg J. (1998). A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature 392:193–197 [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi T, Beuchat MH, Lindsay M, Frias S, Palmiter RD, Sakuraba H, Parton RG. and Gruenberg J. (1999). Late endosomal membranes rich in lysobisphosphatidic acid regulate cholesterol transport. Nat Cell Biol 1:113–118 [DOI] [PubMed] [Google Scholar]
- 40.van der Goot FG. and Gruenberg J. (2006). Intra-endosomal membrane traffic. Trends Cell Biol 16:514–521 [DOI] [PubMed] [Google Scholar]
- 41.Russell MR, Nickerson DP. and Odorizzi G. (2006). Molecular mechanisms of late endosome morphology, identity and sorting. Curr Opin Cell Biol 18:422–428 [DOI] [PubMed] [Google Scholar]
- 42.Trioulier Y, Torch S, Blot B, Cristina N, Chatellard-Causse C, Verna JM. and Sadoul R. (2004). Alix, a protein regulating endosomal trafficking, is involved in neuronal death. J Biol Chem 279:2046–2052 [DOI] [PubMed] [Google Scholar]
- 43.Pempe EH, Xu Y, Gopalakrishnan S, Liu J. and Harris EN. (2012). Probing structural selectivity of synthetic heparin binding to stabilin protein receptors. J Biol Chem 287:20774–20783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kyosseva SV, Harris EN. and Weigel PH. (2008). The hyaluronan receptor for endocytosis mediates hyaluronan-dependent signal transduction via extracellular signal-regulated kinases. J Biol Chem 283:15047–15055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geary RS, Norris D, Yu R, and Bennett CF. (2015). Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv Drug Deliv Rev 87:46–51 [DOI] [PubMed] [Google Scholar]
- 46.Hare AK. and Harris EN. (2015). Tissue-specific splice variants of HARE/Stabilin-2 are expressed in bone marrow, lymph node, and spleen. Biochem Biophys Res Commun 456:257–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park SY, Yun Y, Lim JS, Kim MJ, Kim SY, Kim JE. and Kim IS. (2016). Stabilin-2 modulates the efficiency of myoblast fusion during myogenic differentiation and muscle regeneration. Nat Commun 7:10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim GW, Park SY. and Kim IS. (2016). Novel function of stabilin-2 in myoblast fusion: the recognition of extracellular phosphatidylserine as a “fuse-me” signal. BMB Rep 49:303–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen B, Longati P, Elvevold K, Nedredal GI, Schledzewski K, Olsen R, Falkowski M, Kzhyshkowska J, Carlsson F, et al. (2005). Stabilin-1 and stabilin-2 are both directed into the early endocytic pathway in hepatic sinusoidal endothelium via interactions with clathrin/AP-2, independent of ligand binding. Exp Cell Res 303:160–173 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.