Significance
Cryptic binding sites are absent or occluded in unbound proteins but present in ligand-bound structures. They can provide druggable pockets in cases where the main functional site of the protein cannot be targeted with sufficient potency or specificity. We show that unbound structures of proteins with cryptic sites have strong binding hot spots and above-average flexibility around the incipient pockets. Binding hot spots can be easily identified computationally, and protein flexibility can often be assessed by the analysis of X-ray structures of the unbound protein, including structures with low resolution. The approach presented here helps to assess the potential druggability of cryptic sites prior to running expensive screening experiments and reveals information about how cryptic sites form.
Keywords: protein–ligand interaction, drug discovery, druggability, binding hot spot, protein flexibility
Abstract
Molecular dynamics (MD) simulations of proteins reveal the existence of many transient surface pockets; however, the factors determining what small subset of these represent druggable or functionally relevant ligand binding sites, called “cryptic sites,” are not understood. Here, we examine multiple X-ray structures for a set of proteins with validated cryptic sites, using the computational hot spot identification tool FTMap. The results show that cryptic sites in ligand-free structures generally have a strong binding energy hot spot very close by. As expected, regions around cryptic sites exhibit above-average flexibility, and close to 50% of the proteins studied here have unbound structures that could accommodate the ligand without clashes. Nevertheless, the strong hot spot neighboring each cryptic site is almost always exploited by the bound ligand, suggesting that binding may frequently involve an induced fit component. We additionally evaluated the structural basis for cryptic site formation, by comparing unbound to bound structures. Cryptic sites are most frequently occluded in the unbound structure by intrusion of loops (22.5%), side chains (19.4%), or in some cases entire helices (5.4%), but motions that create sites that are too open can also eliminate pockets (19.4%). The flexibility of cryptic sites frequently leads to missing side chains or loops (12%) that are particularly evident in low resolution crystal structures. An interesting observation is that cryptic sites formed solely by the movement of side chains, or of backbone segments with fewer than five residues, result only in low affinity binding sites with limited use for drug discovery.
Most proteins binding known ligands have surface pockets that are already formed in their ligand-free state. For example, in single-chain enzymes, in 83% of cases, the substrate binds in the largest cleft present in the unbound structure (1). However, in the unbound structures of certain proteins, no binding site is easily detectable, and yet the proteins are capable of ligand binding by exposing clefts or pockets in the presence of a ligand. Thus, such binding sites, termed “cryptic,” require conformational changes to become apparent (2–6). Many biologically relevant drug targets lack appropriately sized pockets in their unbound structures to support the strong binding of drug-sized ligands (3, 7). Consequently, there is growing interest in identifying cryptic sites. It has been suggested that such sites can provide previously undescribed pockets on proteins that are already considered tractable targets, or can make it possible to target proteins that are currently considered undruggable (3). In particular, identification of ligand binding pockets is crucial for the inhibition of protein–protein interactions using small molecules (8). Discovering cryptic allosteric sites is also important if the main functional site of the protein cannot be targeted with sufficient specificity (9), or if targeting the distal site gives a different pharmacological profile (10). Experimental identification of cryptic sites generally involves screening libraries of small molecules or fragments (9, 11), site-directed tethering (9, 12), or the use of antibodies (13). Since these approaches require substantial effort and frequently have negative outcomes, computational tools have also been developed, in most cases based on long molecular dynamics (MD) simulations (3, 4), possibly paired with fragment docking (6). More recently, Cimermancic et al. (5) reported CryptoSite, a prediction tool based on machine learning, with features involving attributes of sequence, structure, and dynamics, which detected cryptic sites with remarkable accuracy.
Despite the current interest in cryptic site discovery, a number of fundamental questions remain unanswered. First, since existence of cryptic sites requires both the ability to bind ligands and substantial local flexibility, to what extent is this complex property predictable by analysis of available structures of an unbound protein? Second, what features distinguish those cryptic sites that are potentially useful, either as target sites for drugs or for the biological function of the protein? The opening of numerous transient pockets has been observed in long MD simulations (3, 4, 14, 15), and the ability to perform such calculations has generated substantial enthusiasm for studying cryptic sites (3, 4). However, which of these transient pockets will be druggable, and how many druggable cryptic sites do we expect to find on a protein? A third group of questions concerns the nature of cryptic sites. While some studies have reported the spontaneous opening of pockets in MD simulations (3, 4, 14, 16), others suggested that cryptic sites result from interactions with ligands (5, 6). This latter mechanism was recently demonstrated by a simulation study of three proteins with known cryptic pockets (6). Using a variety of energy functions and sampling methods, Oleinikovas et al. (6) observed that starting the simulations in the open (i.e., bound-like) conformation but without the ligands, the pockets promptly closed in all molecular dynamics simulation runs, irrespective of approach. However, adding fragment-sized molecules as probes occasionally resulted in the opening of cryptic sites. The need for ligands to promote the formation of pockets appears to contradict the results of earlier MD simulations (3, 4, 14). Thus, it is of interest to study whether cryptic sites are typically formed by conformational selection, induced-fit, or a “mixed” mechanism (6), and to what extent different sites use different mechanisms.
The goal of this paper is to address the above questions through analysis of a representative set of X-ray structures of proteins with validated cryptic binding sites. This set was originally selected for training and testing the CryptoSite cryptic site prediction protocol (5), and hence will be referred to as the CryptoSite set. Starting with 504,647 candidate pairs of ligand-bound structures with their unbound counterparts, Sali and coworkers (5) used pocket detection algorithms to retain only pairs with a small pocket score in the unbound form and a substantially larger score in the bound form. Manual inspection of the structures resulted in a dataset of 93 bound–unbound pairs in which each unbound structure had a site considered cryptic due to its low pocket score, and each bound structure had a biologically relevant ligand bound at the site (5). While the original CryptoSite set included only one unbound structure in each pair, to study the information provided by different unbound structures of a given protein, for each bound structure in the set, we added all unbound structures with at least 95% sequence identity that were available in the Protein Data Bank (PDB). Structures with any other ligand within 5 Å of the cryptic site were excluded. The number of such additional unbound structures varied from zero to 498 per protein (SI Appendix, Fig. S1A), resulting in an extended CryptoSite dataset that includes 4,950 structures rather than the original 186.
Our analysis focuses on two properties of the proteins: binding energy hot spots, and local protein flexibility. Hot spots are relatively small regions on the protein surface that can contribute a disproportionate amount to the ligand binding free energy (17–20). We have shown previously that no significant binding can occur without binding hot spots (20–22), and thus it is reasonable to look for hot spots near cryptic sites. The concept of hot spots is related to fragment binding; it has been well-established, both experimentally and computationally, that hot spots are characterized by their ability to bind a variety of fragment-sized ligands (23, 24). We also expect that protein structures have sufficient flexibility around cryptic sites to enable formation of the ligand binding pocket (5, 16). By exploring the bound complex structure and multiple unbound structures of each protein in the extended CryptoSite set, we can compare conformational variations near and far from the cryptic site. The results shed light on the types and magnitudes of conformational changes that occur spontaneously, and others that are most likely promoted by ligand binding. As will be shown, the high level of flexibility at these sites frequently leads to local disorder, including missing side chains or loops and possibly weak self-interactions between the protein surface and unstructured regions. Such disorder is particularly evident in low resolution X-ray structures, and we show that, combined with the analysis of hot spots, detecting disorder may provide information on potential cryptic sites. Therefore, even very low resolution X-ray data, which are frequently ignored and not even refined, can be useful for drug discovery.
In addition to reporting the above method for identifying cryptic binding sites from ligand-free protein crystal structures, we also study the possible reasons why a cryptic site is closed in the unbound structure, including side chains, loops, or unstructured segments protruding into the site, loops being too open and not forming a pocket, loops not visible in the X-ray structure, interdomain sites affected by hinge or other motion, and moving secondary structure elements, most frequently helices, being either too close or too far. For each of the 93 proteins, we discuss the conformational changes by accounting for both the hot spot structures and the original papers that describe the structures. Using this information, we attempt to place each system on the mechanistic continuum between conformational selection and induced fit, and, although this is not always possible, we believe that the analysis provides a number of interesting findings.
Results
Hot Spots near Cryptic Sites.
The hot spots of unbound structures were determined by using both the FTMap and FTFlex programs (22, 25, 26). FTMap distributes small organic probe molecules of different size, shape, and polarity on the surface of the protein to be studied, finds the most favorable positions for each probe type, clusters the probes, and ranks the clusters on the basis of their average energy. Regions that bind several different probe clusters are called consensus sites (CSs) and are the predicted binding hot spots. While FTMap considers the protein as rigid, the related algorithm FTFlex allows side chain conformers of residues around selected hot spots to vary, facilitating pocket opening (26). The results of FTMap have been extensively validated (20, 22, 25, 27–34) and shown to provide reliable information on the location of binding sites (35) and on the druggability of a site: i.e., its ability to bind drug-like small molecules with sufficient affinity for pharmacological activity (21).
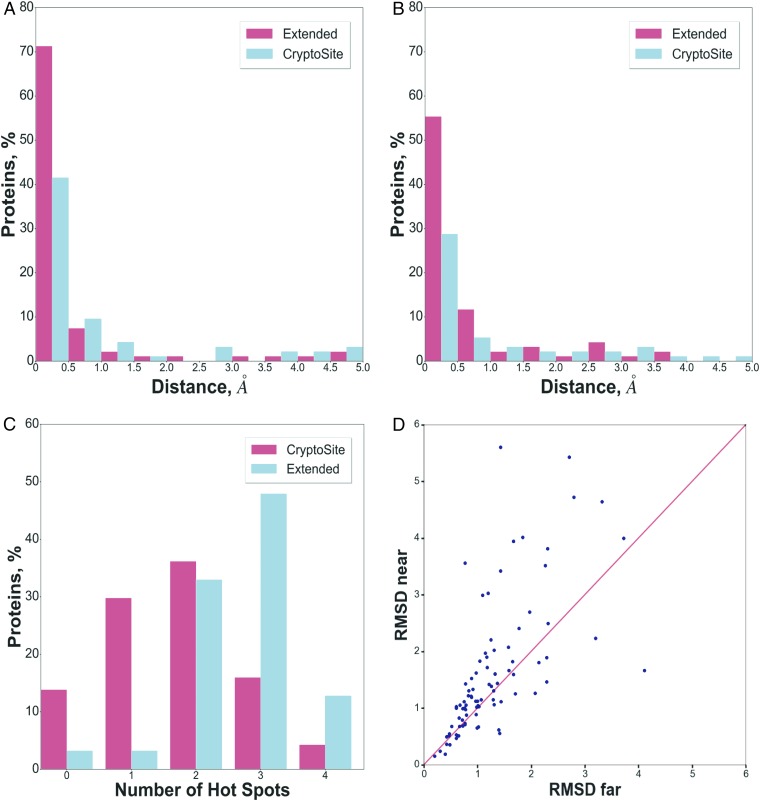
FTMap and FTFlex were applied to all 4,857 unbound and 93 bound structures in the extended CryptoSite set. All ligands and water molecules were removed before mapping as FTMap includes a continuum water approximation. For each of the 93 proteins represented in this set, we show mapping results for the original unbound and bound structures in the CryptoSite set, as well as for a number of unbound structures in the extended set that have strong hot spots close to the location of the cryptic site (SI Appendix, Table S1). As described previously (21), a necessary condition for a site to bind a small molecule with at least micromolar affinity is to have a hot spot with 13 or more probe clusters. As shown in Fig. 1A, 67% of the unbound structures in the original CryptoSite dataset have a hot spot that satisfies this condition and is located within 5 Å of the cryptic site. If the additional unbound structures in the extended set are also considered, the percentage of proteins that have a hot spot with ≥13 probe clusters within 5 Å of the cryptic site increases to 88%. While the 5-Å distance may appear to be large, we note that, when constructing the CryptoSite set, Cimermancic et al. (5) defined cryptic binding sites by selecting residues with at least one atom <5 Å away from any ligand atom, and we adopt this definition. Moreover, 79% of unbound structures in the extended set actually have a hot spot with ≥13 probe clusters within 1 Å of the cryptic site (Fig. 1A). We have shown that a binding site is potentially druggable (although not necessarily using a drug-sized molecule) if it binds at least 16 probe clusters (21). According to Fig. 1B, 50% of the unbound structures in the original CryptoSite set have such hot spots within 5 Å of the cryptic site. This value increases to 81% of the 93 proteins if all unbound structures in the extended set are considered. As will be shown, the hot spots do not necessarily overlap with the cryptic sites, but they are exploited by the ligands in the bound structures and thus contribute to binding.
Fig. 1.
Distribution of strong hot spots and rmsd values near the location of the cryptic sites for the unbound structures in the extended and original CryptoSite sets. (A) Hot spots with 13 or more probe clusters. (B) Hot spots with 16 or more probe clusters, strong enough to potentially support druggability. (C) Distribution of the number of strong hot spots with 16 or more probe clusters in the unbound structures of the CryptoSite set, and distribution of the maximum number of such hot spots in the unbound structures of the extended CryptoSite set. (D) Average pairwise rmsd values between the hot spot regions of unbound structures near the cryptic site, versus average pairwise rmsd values between the hot spot regions of the unbound structures far from the cryptic site. For each protein, the rmsd values were calculated between all pairs of unbound structures in the extended CryptoSite set.
Since strong proximal hot spots are necessary for ligand binding, the number of cryptic sites that are potentially druggable on a protein is limited by the number of such hot spots. This observation is important because long MD simulations have been reported to result in numerous pockets that were open sufficiently long to be detectable (3, 14). For example, for IL-2 and β-lactamase, Bowman and Geissler (3) reported over 50 pockets that were open and most likely accessible more than 10% of the time. Such sites were distributed across the surface of the proteins and were proposed to provide viable drug target sites. However, as shown in Fig. 1C, the number of strong hot spots with more than 16 probe clusters on a given protein never exceeds four, even considering all of the different unbound structures in the extended CryptoSite set. Furthermore, in addition to the cryptic site, most of the proteins also have known (i.e., noncryptic) binding sites with ligands at one, two, or sometimes three of these strong hot spots. Thus, based on the analysis of protein structures with validated cryptic sites, we conclude that, among the pockets opened by conformational changes or MD simulations, only one or two may qualify as genuine cryptic sites, capable of binding a ligand with substantial affinity. As will be further discussed, the druggability of a cryptic site also depends on its type: i.e., the mechanism of its opening.
We note that Sali and coworkers (5) compared the accuracy of their CryptoSite cryptic site prediction method with that of our FTFlex algorithm with respect to 14 proteins and found the two methods to be comparable overall based on area under the curve (AUC) values (0.77 for FTFlex versus 0.83 for CryptoSite). The agreement was very good (10 out of 14 cases) for pockets that, even in the unbound state, present a small surface concavity that can fit the small-molecule fragments used as probes by FTMap and FTFlex. CryptoSite was found more accurate than FTFlex when a cryptic site was fully buried or when it resided in a large protein. In the majority of cases involving large proteins for which disagreement between the methods was found, the cryptic site was located between two domains. For FTFlex and FTMap, we generally used a domain-separating algorithm (36) and mapped the domains separately (22). In many proteins, domain separation substantially improves the detection of hot spots at cryptic sites (SI Appendix, Table S1). However, Cimermancic et al. (5) did not separate the domains before testing FTFlex and thus most likely underestimated the accuracy of the method. Overall, based on the similar AUC values and considering the improved results due to domain separation, the potential difference in the accuracy of FTFlex and CryptoSite is very small. In fact, our focus here is not on the performance of different methods for identifying such sites, but rather to gain insight into the structural features and changes that result in cryptic sites. We note, however, that, although both FTMap and FTFlex fail to find relevant hot spots in any unbound structure for 12 of the 93 proteins in the extended CryptoSite set, in most cases, it is easy to understand why this happens. Some cryptic sites bind only tiny ligands and are too small, even in the bound structure, to fit our probes; others are inherently located between two chains and therefore are not found by mapping of only one chain, or have nearby sites that bind ligands with much higher affinity than the cryptic site and hence attract most of the probes used for the mapping.
Protein Flexibility Around Cryptic Sites.
In addition to typically requiring a strong hot spot close to the location of a cryptic site, the opening of pockets also requires sufficient structural plasticity, as has been demonstrated by molecular dynamics (MD) simulations of multiple proteins (3, 4, 14, 15). Here, we explore whether the availability of several X-ray structures of a protein in the unbound state can be used to reveal conformational changes indicating above-average flexibility around cryptic sites. This is not a simple task since proteins may have very flexible or even unstructured regions unrelated to ligand binding, such as large variations in terminal fragments or loops. To restrict the analysis to potential ligand binding sites, we considered only conformational differences of residues in the vicinity of hot spots. Residues within a 9-Å neighborhood were selected around the center of each hot spot, and the average pairwise rmsd for the atoms of these nearby residues between all unbound structures of the same protein in the extended data set was calculated. This analysis was applied to the 88 proteins from the original CryptoSite set that have additional structures in the extended set. To assess the degree of flexibility in regions close to versus far from cryptic sites, we separated the calculated average rmsd values into those surrounding hot spots that were close to the cryptic site (i.e., where any probe atom was closer than 5 Å from any atom of the ligand superimposed from the bound structure), versus hot spots not meeting this criterion that were classified as far from the cryptic site. For each protein, Fig. 1D shows the mean pairwise all-atom rmsd values for the hot spots close to the cryptic site versus the mean pairwise rmsd values averaged over all other hot spots of the same protein far from the cryptic site. According to these calculations, for 69% of the 88 proteins with multiple unbound structures, the rmsd values near the cryptic site were larger than the ones at the far sites, indicating increased structural variability closer to the cryptic site. The overall mean rmsd values, averaged over the 88 proteins, were 1.00 ± 1.04 Å and 0.69 ± 0.56 Å, respectively, for the near and far rmsd values. While the overall variances were large when all proteins were considered, we emphasize that we compared near and far rmsd values for each protein separately, and hence we used the Wilcoxon signed-ranks test, a nonparametric analog of the pairwise t test. The null hypothesis was that the rmsd values near the cryptic site were the same as the ones at the far sites, and the hypothesis was rejected at P < 0.01, indicating that increase in flexibility, while modest, is significant.
The Structural Origins of Cryptic Sites.
The results described so far suggest that formation of a druggable cryptic site requires both the existence of a nearby hot spot and some level of local flexibility. We used the extended CryptoSite set and the mapping results to study two further questions. First, what are the conformational differences between bound and unbound structures: i.e., what causes the site to be cryptic? Second, are these changes induced by ligand binding, or are they spontaneous and thus also present in some unbound structures, with the open pocket stabilized by subsequent ligand binding through conformational selection?
To measure the conformational differences between bound and different unbound structures, we selected the residues within 9 Å around the location of the cryptic site in each bound structure and, for each protein, calculated the local backbone, side chain, and all atom rmsd for the selected residues between the bound structure and the unbound structure in the CryptoSite set (SI Appendix, Table S2). Relatively small backbone rmsd values can be found if either a number of residues around the site are missing in the unbound, bound, or both structures, or if the backbone conformational changes are really small. As will be further discussed, after the removal of cases with missing residues, we identified 18 proteins in which the cryptic sites are created by the movement of side chains, without substantial change in the backbone conformation (SI Appendix, Tables S1 and S2).
We also calculated a number of measures that show if any of the unbound structures would or would not clash with the ligand, the latter to identify cryptic sites that could be the results of conformational selection. The first of these measures is the minimum distance between any atom of in the ligand-bound protein in the CryptoSite set and any atom of its bound ligand, which shows the baseline interatomic distance we need for the specific protein to avoid any receptor–ligand clashes (column D in SI Appendix, Table S2). The values are between 2.5 and 3 Å as expected for interatomic distances. The next measure is the minimal distance between any (nonhydrogen) atom of the unbound protein in the CryptoSite set and any atom of the ligand superimposed from the bound structure (column E in SI Appendix, Table S2). According to this column, the minimal interatomic distance in the unbound structure is generally smaller than the value in the bound structure, revealing that the proteins in the selected unbound conformation would clash with the superimposed ligand. However, the unbound structures in the extended set show substantial variation, and, for many proteins, there exist unbound structures substantially closer to the bound form than the one in the CryptoSite set (SI Appendix, Fig. S1B). The third measure is the same minimum distance but maximized over all unbound structures in the extended CryptoSite set (column F in SI Appendix, Table S2). This measure shows if there exists an unbound structure that would not clash with the superimposed ligand. Finally, to show the diversity of unbound structures in the extended CryptoSite set, we present the smallest value of the same distance over all unbound structures (column G in SI Appendix, Table S2). According to these results, in 45 cases in at least one of the unbound structures in the extended set, the minimal value of the receptor–ligand interatomic distance would be larger than 90% of the interatomic distance seen in the real receptor–ligand pair (column F in SI Appendix, Table S2). Thus, despite the limited set of structures in the PDB, it appears that, for 48.3% of the 93 proteins, there exists at least one unbound structure that could accommodate the ligand without any conformational change. For example, in the catalytic subunit of PKA (item 3 in SI Appendix, Table S1), the smallest interatomic distance between the ligand and one of the unbound structures is 2.73 Å, slightly higher than the shortest distance in the actual receptor–ligand complex (row 3 of SI Appendix, Table S2), implying that binding could occur without any ligand-induced conformational change. In contrast, for the Niemann–Pick C2 protein, the minimal distance between any atom of the protein and any atom of the superimposed ligand does not exceed 0.59 Å in any of the unbound structures; thus, the ligand could not bind without substantial conformational changes (item 5 in SI Appendix, Tables S1 and S2).
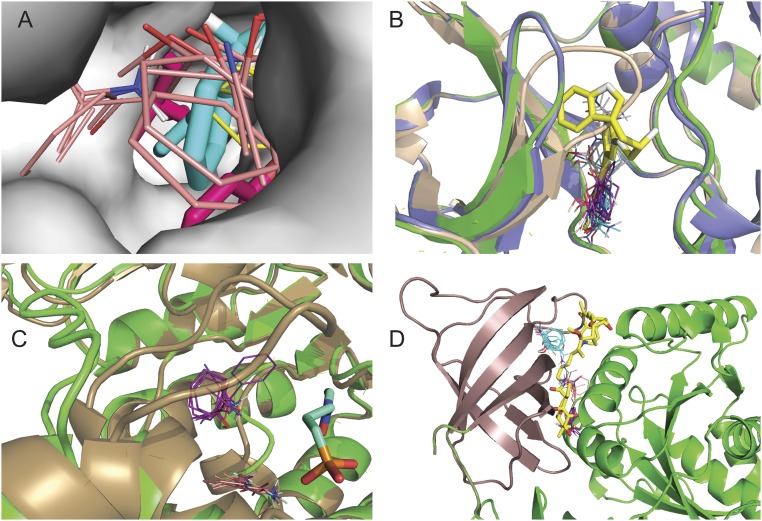
We also studied the types of conformational changes that occur between unbound and bound structures (Table 1). Among the 93 structures of the CryptoSite set, in 18 protein pairs, the backbone rmsd is less than 1.0 Å, and there are no missing residues (SI Appendix, Tables S1 and S2). Thus, in these 18 cases, there are no significant main-chain motions in the vicinity of the binding site, and the cause of the pocket being cryptic is that one or more side chains protrude into the site in the unbound structure. This type of conformational change is designated by an “S” (side chain). An example is provided by myosin II heavy chain (item 2 in SI Appendix, Table S1). In the unbound structure (2AKA, chain A), the side chains of Leu262 and Tyr634 protrude into the very narrow binding site (Fig. 2A). Mapping yields hot spots at the entrance of the pocket, 2.11 Å from the ligand superimposed from the bound structure 1YV3 (chain A). Any of the currently known unbound structure of myosin II would clash with the ligand (column E of SI Appendix, Table S2), and hence it is likely that the site is formed by induced fit. However, in many other cases, we found that the site-occluding side chains can move out of the cryptic site spontaneously. Examples include the chitinase B1 enzyme (item 1 in SI Appendix, Tables S1 and S2). In this case, the minimum receptor–ligand distance is essentially the same (2.60 Å) in some of the unbound structures as the minimum receptor–ligand distance in the bound structure (2.64 Å), indicating that unbound forms exist that can accommodate the ligand without conformational change. Another example is ribonuclease A (item 40 in SI Appendix, Table S1), in which residue His110 has two side chain conformers, but only one of them protrudes into the pocket. Accordingly, in one of the unbound structures, the minimum receptor–ligand distance is 4.08 Å, much larger than the minimum receptor–ligand distance, 2.65 Å, in the bound structure (SI Appendix, Table S2).
Table 1.
Types of cryptic sites in the CryptoSite set of 93 proteins
| Type* | Origin of the site being cryptic | No.† |
| LC | Loops protruding into the site, making it closed to ligand binding | 21 |
| LO | Loops too open in the unbound structure, making the pocket not well-formed | 18 |
| S | Side chains protruding into the site in the unbound structure | 18 |
| LM | Loops missing in the unbound structure, leading to loss of the pocket | 11 |
| I | Interdomain cryptic site, affected by hinge or other motion of the two domains | 11 |
| U | Unstructured regions in the unbound structure, in most cases N or C termini | 3 |
| SC | Secondary structure elements too close, closing on the pocket | 5 |
| SO | Secondary structure elements too far, making the pocket too open | 2 |
| SM | Secondary structure elements missing in the unbound or bound structure | 2 |
| CT | Very large conformational transition (calmodulin) | 1 |
| F | FTMap fails: Pocket is too weak to bind probes | 1 |
Notation used in SI Appendix, Table S1.
Number of cryptic sites of the particular type among the 93 proteins.
Fig. 2.
Mapping results for various types of cryptic sites. (A) Mapping of the myosin II heavy chain. The bound structure (1YV3, chain A, shown as gray surface) has a very narrow pocket that binds a blebbistatin molecule (cyan sticks). The unbound structure (2AKA, chain A) is superimposed as magenta sticks to show that the side chains of L262 and Y634 protrude into the pocket. Mapping of this structure using FTFlex placed the hot spots CS0 (beige, 17 probe clusters) and CS4 (yellow, eight probe clusters) at the entrance of the pocket. (B) Mapping of the unbound structure of the catalytic subunit of the cAMP-dependent protein kinase PKA (PDB ID code 2GFC, chain A) shown as tan schematic. The hot spots, obtained after domain splitting, are CS0 (cyan, 18 probe clusters), CS1 (magenta, 16 probe clusters), and CS4 (gray, 13 probe clusters). An inhibitor (yellow) is superimposed for reference. We also superimpose the bound structure (PDB ID code 2JDS, chain A, green) and an alternative unbound structure (PDB ID code 4DFZ, chain E, blue) to show that the loop 51 to 56 moves out of the pocket, freeing up the site for the ligand. (C) Mapping of the unbound structure of 1-deoxy-d-xylulose-5-phosphate reductoisomerase (PDB ID code 1K5H, chain C, show in green), with the loop 208 to 215 far from the ligand (cyan), superimposed from the bound structure (PDB ID code 2EGH, shown in tan color). Since the site is too open, FTMap finds only the hot spots CS0 (magenta, 14 probe clusters) and CS3 (orange, 11 probe clusters). The loop in 2EGH moves toward the ligand and forms a lid on the binding pocket. (D) Mapping of the unbound structure of the elongation factor TU (1EXM, chain A) after splitting the protein into two domains (shown as brown and green). FTMap found the hot spots CS0 (cyan, 18 probe clusters), CS1 (magenta, 17 probe clusters), and CS3 (tan, 12 probe clusters) on the domain shown in brown. The ligand, superimposed from the bound structure (1HA3 chain B), is shown as yellow sticks.
An interesting aspect of cryptic sites caused by side chain motion is that the binding affinity of such sites is usually low, in the micromolar range. For 10 of these 18 proteins with small backbone conformational change, literature values for the binding affinities of the ligands that occupy the cryptic site show that ligand binding is very weak (SI Appendix, Table S1). Although we could not find data for the remaining eight proteins, in no case is there evidence of strong binding (Kd < 300 nM) by any ligand. This observation suggests that cryptic sites in regions of the protein that have a relatively rigid backbone, such that pocket opening and closing involves only side-chain motions, do not tend to bind ligands with high affinity.
The most frequent origin of cryptic sites is actually not side chain movement but loops protruding into the pocket. The CryptoSite set includes 21 such cases, denoted as “LC” (loops closed). For example, in the unbound structure of the catalytic subunit of PKA (2GFC, chain A) mentioned above, the loop comprising residues 51 to 56 restricts the site, which, in this closed conformation, has no hot spots (Fig. 2B). In the bound structure (2JDS, chain A), and also in many unbound structures such as chain E of 4DFZ, the loop is farther from the site, leaving it fully open. Thus, the protein can assume the conformation required for ligand binding without the presence of any ligand. Another example is the already discussed Niemann–Pick C2 protein (item 5 in SI Appendix, Table S1). In the unbound structure 1NEP, the loop comprising residues 96 to 103 is closer to the ligand binding site than in the bound structure 2HKA, chain C, and the side chains of Phe66 and Tyr100 would clash with the ligand. We note that 2HKA has three protein molecules in the unit cell. Chain A of the same structure has no bound ligand, but the 96 to 103 loop is substantially further from the site, which shows local flexibility. However, F66 still protrudes into the site and would clash with the ligand. Thus, there is no proof that the site can become fully open without ligand binding.
Loop movement can have an opposite effect: in 18 of the 93 unbound structures in the CryptoSite set (16%), the cause of a diminished site is that one or more loops are too open [denoted as “LO” (loops open)], and the pocket is not well formed. One example is 1-deoxy-d-xylulose-5-phosphate reductoisomerase (item 35 in SI Appendix, Table S1). The loop of residues 208 to 215 is extremely flexible, and, in the unbound structure 1K5H, chain C, it moves outward, whereas, in the bound structure 2EGH, chain B, it closes down on the small ligand 2-phosphoglycolic acid (Fig. 2C). Since, in chain C of 1K5H, the pocket is too open, FTMap finds only two relatively weak hot spots, CS0 (14) and CS3 (11), near the cryptic site. (FTMap numbers the strongest consensus site CS0, the next strongest CS1, etc. The number in parentheses is the number of probe clusters the consensus site contains, which provides a measure of the strength of the hot spot.) In contrast, in chain B of the same unbound protein, part of the loop is unstructured, and residues 212 through 215 are not visible in the X-ray structure. Nevertheless, the remaining part of the loop is sufficient to close on the site, resulting in two strong hot spots: CS0 (19) and CS1 (18) (Fig. 2C). We note that, despite the loop being too open in the unbound structure 1K5H, simply superimposing the ligand still leads to clashes with other nearby residues (SI Appendix, Table S2); thus, ligand binding involves both the closing of the loop and moving some side-chain atoms out of the site.
In 11 proteins (indicated by “I”), the sites are cryptic because they are located at the interface between two domains and are affected by hinge-type or other interdomain motions. After separating the domains and mapping them individually, FTMap found strong hot spots on one or both proteins as the interdomain surfaces became accessible to the FTMap probes, but these hot spots could not always be detected without the domain separation. One example is elongation factor TU (item 25 in SI Appendix, Table S1). In the bound structure (1HA3, chain B), the large ligand binds between two domains. In the unbound structure (1EXM, chain A), the two domains move closer together, and the pocket becomes too narrow for ligands or FTMap probes to enter. However, after separating the protein into its two domains (shown as brown and green in Fig. 2D), FTMap found three hot spots on the C-terminal domain shown in brown. There is evidence that binding of the antibiotic aurodox at this cryptic site occurs by induced fit (SI Appendix, Table S1). We note that domain separation was also found to be useful in mapping sites located between domains where the main cause of the site’s absence in the unbound structure was either a side chain or a loop protruding into the pocket. In such cases, we used the S and LC classification, respectively, rather than I, as the interdomain location of the site is not the main reason it is cryptic.
There are several less frequently occurring causes of sites being not fully formed in the unbound structure. As will be further discussed, pockets may become undetectable due to very flexible loops that are missing in the X-ray structure [11 cases, denoted as “LM” (loops missing)]. Entire secondary structure elements, usually smaller helices, may also be missing in the unbound or bound structure [two cases, denoted as “SM” (secondary structure element missing)]. The site may also disappear if the unbound protein has unstructured regions, most frequently near the N or C terminus [three cases, indicated by “U” (unstructured)]. Another source of cryptic sites is that secondary structure elements, primarily helices, are too close to each other in the unbound protein, closing the pocket [five cases, indicated as “SC” (secondary structures closed)]. A well-known example of this type is TEM β-lactamase (item 92 in SI Appendix, Table S1). Two proteins have the opposite problem: i.e., they have secondary structure elements that are too far from each other in the unbound structure, resulting in a flat surface, but come closer and create a pocket in the bound protein [indicated by “SO” (secondary structure open)]. Finally, there are two proteins in the CryptoSite set that have cryptic sites for other reasons. Calmodulin (item 28 in SI Appendix, Table S1) is subject to major backbone conformational change upon binding the drug trifluoperazine (local rmsd is 9.1 Å), but, interestingly, the strong hot spot at the cryptic site is retained in the unbound structure. Finally, FTMap failed to find any hot spots even in the bound structure of human liver glycogen phosphorylase (item 57 in SI Appendix, Table S1). However, this site binds a small ligand (uric acid) with very low affinity (Kd = 550,000 nM) and does not attract the FTMap probes despite being open even in the unbound structure SI Appendix, Table S2).
As discussed above, cryptic sites caused exclusively by side chain motion are unlikely to bind ligands with high affinity. This observation is important because many of the sites generated by molecular dynamics are in this category. For the proteins in the CryptoSite set, we found evidence of high affinity binding (Kd < 300 nM) only when the conformational change involved a backbone segment with at least five residues. Thus, it seems that localized movement of very small segments cannot create large enough pockets for ligand binding. Loop motion or missing or unstructured loops are seen in 53 of the 93 proteins in the CryptoSite set. Loop reorganization was found spontaneous in 23 (43%) of these proteins, and, in 13 (57%) of these cases, compounds with nanomolar binding affinity can be found in the literature (SI Appendix, Table S1). Two proteins with well-known cryptic sites, interleukin-2 (IL-2) (37, 38) and TEM β-lactamase (39), are discussed in more detail, and we also present a prospective application of the knowledge gained in this work to identify a cryptic site on the cyclin-dependent kinase 2 (CDK2) that was subsequently validated experimentally (SI Appendix).
Regions of Weak Electron Density in X-Ray Structures as Predictors of Cryptic Sites.
Hardy and Wells (9) have demonstrated that, in X-ray structures, it is not uncommon to see adventitious binding of small compounds to protein cavities, and, while these observations are typically ignored, such “crystallization artifacts” recorded in the Protein Data Bank (40) may provide a large repository of information on serendipitous allosteric sites (9). Here, we focus on a different aspect of X-ray structures, frequently indicating cryptic binding sites: namely, that some protein atoms, either just a side chain or several residues of a loop, may be missing due to the high level of local flexibility. The CryptoSite set includes 11 unbound structures with missing loops that also have a nearby hot spot, predicting a cryptic site. The following examples further demonstrate that such regions of weak electron density can provide useful information as to the existence of cryptic sites on important drug target proteins and thereby increase the value of low resolution X-ray data that are frequently not even considered worth refining.
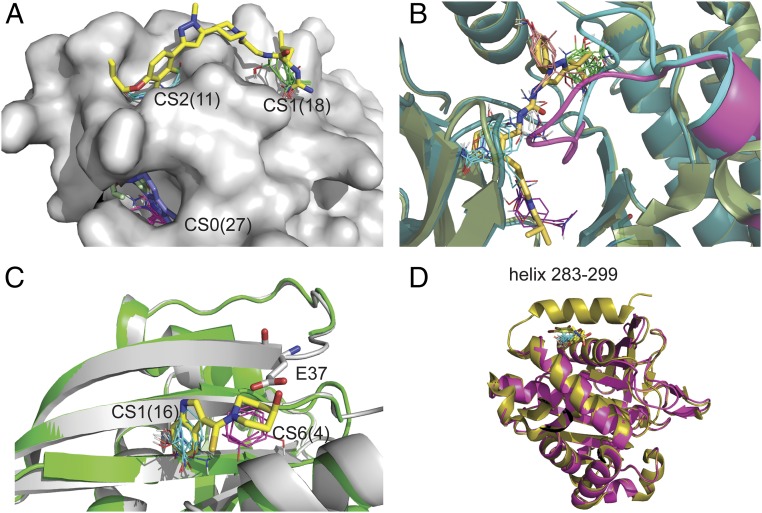
Our first example is interleukin-2 (IL-2). The protein has a known cryptic site with an inhibitor bound to two strong hot spots in the IL-2/IL-2Rα interface (6) (SI Appendix). It was noted by Hyde and coworkers (37, 38) that several X-ray structures of IL-2 miss some residues in the loop 75 to 82 on a different side of the protein. For example, residues 75 to 80 are not visible in the unbound structure 1PY2 shown in Fig. 3A, and this gap region has two hot spots, one of them very strong: CS0 (27) and CS3 (10). It has been shown that this site binds a ligand (blue sticks in Fig. 3A) that allosterically regulates the binding of the inhibitor (shown as yellow sticks) at the main site. Thus, the region identified by the missing loop adjacent to a strong hot spot proved to be a cryptic site that allosterically affected ligand binding at the IL-2/IL-2Rα interface site.
Fig. 3.
Missing regions in X-ray structures as predictors of cryptic sites. (A) Mapping of the bound IL-2 structure 1PY2 with inhibitor (yellow) shown for reference. The loop of residues 75 to 80 is not visible in the X-ray structure, and the strongest hot spot, CS0 (magenta, 27 probe clusters), is at the gap created by the missing loop, which indicates a cryptic site. The site binds an allosteric ligand (blue) in the structure 1NBP. Other structures also show that the region at this cryptic site is very flexible. For example, in the inhibitor-bound structure 1M48, residues 79 to 82 are missing, but F78 (colored light green) protrudes into the pocket. (B) Substrate binding region of P38 protein kinase. Shown are the unbound structures 3D83 (light green), with the DFG loop shown in magenta, and 2ZB0 (dark blue), with the DFG loop shown in cyan. Both loops are in DFG-in conformation and would clash with the ligand (shown as yellow sticks), superimposed from the bound structure 2YIW, which is in DFG-out conformation. Mapping the ligand-bound DFG-out structures 1KV1, in which residues 171 to183 are missing, yields the hot spots CS0 (green, 23 probe clusters), CS1 (magenta, 12 probe clusters), CS3 (orange, nine probe clusters), and CS4 (white, eight probe clusters). In contrast, mapping the DFG-in structure 2ZB0 yields only the hot spot CS0 (cyan, 16 probe clusters). As shown, mapping both DFG-in and DFG-out conformations, the hot spots map out the entire ligand binding site. (C) Inhibitors of the KRAS/SOS interaction. Shown are chain A (gray) and chain C (green) of the unbound structure 3GFT. Residues 36 and 37 are not visible in chain C. Mapping of chain A identifies the hot spot CS1(cyan, 16 probe clusters) in a hydrophobic pocket that binds the indole group of an inhibitor. The mapping of chain C finds the additional hot spot CS6 (magenta, 4) in a secondary site that enables the binding of the slightly higher affinity inhibitor (yellow sticks) in the structure 4EPW. However, in chain A, the side chain of Glu37 (shown as white sticks) protrudes into this secondary site and would clash with the inhibitor. (D) Protein tyrosine phosphatase 1B (PTP1B). Shown are the unbound structures 1SUG (gold) and 2HNP (magenta), the latter missing the C-terminal helix. 1SUG was mapped after removal of residues 283 to 299, resulting in the hot spot CS1 (cyan, 17 probe clusters), overlapping with the allosteric inhibitor superimposed from the structure 1T48 (yellow sticks).
The second example is the P38 MAP kinase. As with most kinases, P38 MAP kinase has several potential allosteric sites, including a cryptic lipid binding site in the C-terminal domain (item 64 in SI Appendix, Table S1). Here, we focus on the flexible “DFG” activation loop in the N-terminal domain, comprising residues 170 to 185, part of which is missing in several X-ray structures (41). Fig. 3B shows that mapping one such bound structure, 1KV1, in which residues 171 to 183 are missing, yields several hot spots that trace out the shapes of the bound ligands whereas mapping unbound structures in the DFG-in loop conformation (e.g., 3D83 or 2ZB0) finds only the hot spot at the ATP binding site. Thus, in this example, mapping conformations with missing loops provided information on a functionally relevant cryptic binding site.
Our third example is KRAS, an extremely important cancer drug target (42, 43). A structure of unbound human KRAS, 3GFT, has seven chains in the asymmetric unit. Mapping chain A, the one with the fewest missing residues and most likely with the least disorder, yielded only one hot spot with 13 probe clusters in an isolated pocket in the KRAS–SOS interface. The pocket can accommodate only very small ligands with low binding affinity (44). However, chain C of the same structure is less ordered and is missing residues 36 and 37. Mapping this chain placed a stronger hot spot at the same location and also found a second hot spot nearby (Fig. 3C). The movement of residues 36 and 37 to reveal the stronger hot spot ensemble contributes to the binding of larger and slightly higher affinity inhibitors (43). However, since the site is created by moving only two side chains rather than a longer loop, based on the analysis presented in the previous section, we can predict that it will have only moderate affinity, and this is confirmed by the difficulty of developing drug candidates that bind at this location (43, 44).
A fourth example demonstrating the potential use of the information provided by weak electron density focuses on protein tyrosine phosphatase 1B (PTP1B), another well-validated drug target. Although a number of high affinity inhibitors binding at the active site of PTP1B have been identified, they are charged, are very large, and have limited selectivity (45). Therefore, a cryptic allosteric site found close to the C terminus has potential significance (46, 47). The unbound structures 2F6V and 1SUG (with resolutions of 1.7 Å and 1.95 Å, respectively) have a well-resolved C-terminal helix, 285 to 299, which covers this allosteric site. However, the helix is completely missing in other unbound structures, such as 2HNP, with a resolution of 2.85 Å, and 1T49, demonstrating a high level of flexibility at the C terminus. After removing residues 283 to 299 from 1SUG, mapping the truncated protein yielded a strong hot spot, CS1 (17), which overlaps with an inhibitor that binds in this cryptic site, superimposed from the structure 1T48 (46) (Fig. 3D). In contrast, mapping high resolution unbound structures without removing the helix yielded only CS9 (3) (item 76 in SI Appendix, Table S1). Thus, lower resolution structures can provide clues on regions that are very flexible, but are “frozen” in some particular state that covers the cryptic site, and hence are not detected in higher resolution X-ray structures.
Discussion
We have explored the structural origin of cryptic sites in a representative set of 93 proteins in which the structure of the sites in unbound and ligand-bound structures differs substantially. The set was expanded by adding, for each protein, all suitable unbound structures with at least 95% sequence identity that were available in the Protein Data Bank. Mapping of all unbound structures revealed that 88% of the proteins in the set had a strong hot spot within 5 Å of the ligand superimposed from a bound structure. As described earlier, such binding hot spots can contribute a disproportionally large amount to the binding free energy of any ligand. In many cases, the ligand exploits this adjacent hot spot when bound at the cryptic site, suggesting the possibility that formation of the cryptic site may involve an induced fit mechanism in which the ligand first binds at the nearby hot spot and then induces a conformational change in the protein to form the final complex. The conformational adaptation is facilitated by the fact that regions around cryptic sites have moderately but significantly higher flexibility than around other hot spots. However, we have also shown that, for almost 50% of the proteins in the CryptoSite set, there exists an unbound structure in the PDB that would not clash with the ligand even without any further conformational change, thus potentially enabling a conformational selection mechanism. Nevertheless, the binding to nearby hot spots is still likely to be important for stabilizing the bound state.
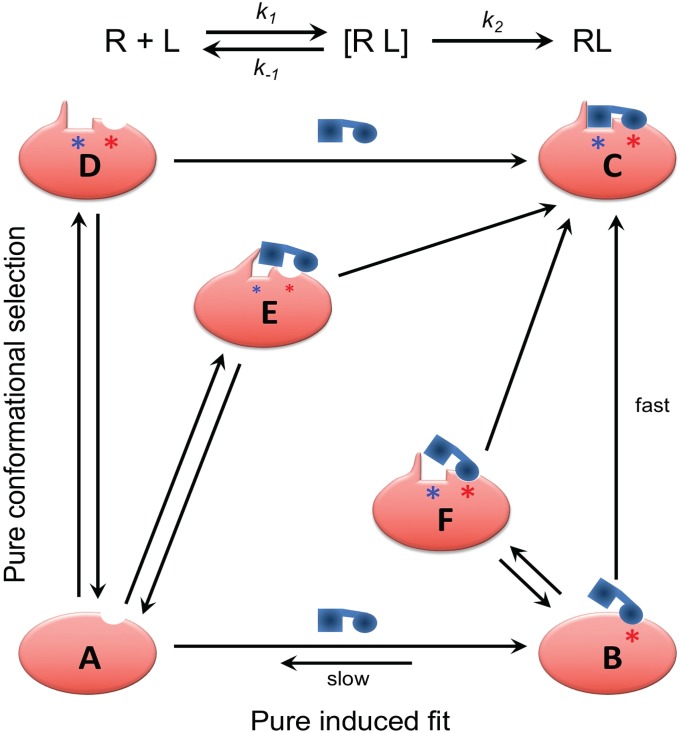
Kinetically, the distinction between an induced fit binding mechanism and binding by conformational selection is that, in the former, the initial encounter of the ligand with the unbound protein is “sticky.” That is, even when the initial encounter is with a conformation of the protein in which the cryptic ligand binding site is occluded, dissociation of the encounter complex is slow compared with conformational conversion to the final, stable complex. The relatively long lifetime of the initial encounter complex gives time for the ligand-induced conformational change in the protein to take place within the lifetime of this complex, resulting in induced fit binding (Fig. 4). For the alternative, conformational selection mechanism, collisional encounters between the ligand and forms of the protein in which the cryptic site is occluded will be nonproductive, with the ligand rapidly dissociating. Only when the initial encounter occurs with one of the small fraction of protein molecules in which the cryptic site is fully formed will reaction proceed to give a stable complex (Fig. 4). Induced fit and conformational selection must properly be considered as extremes in a continuum of binding mechanisms, however (48–50). Even in cases in which initial binding is to the cryptic conformation of the protein, exploiting a site-adjacent hot spot, there may well be kinetic competition between dissociation of this initial encounter complex and conversion to the final complex, depending on the rate of the conformational changes that form the adjacent site. Conversely, even when some spontaneous conformation enables the initial binding of the ligand to occur, it is likely that, in many cases, further conformation adjustments between protein and ligand are involved in formation of the final complex (48–50) (Fig. 4). Our finding that almost half of the proteins in the CryptoSite set have a reported unbound structure in which the cryptic site is sufficiently open to accommodate the ligand without significant steric clashes indicates that, in this subset of cases, an open site can form in the absence of ligand. This result suggests that these proteins are candidates to bind ligand by a conformational selection mechanism. However, whether conformational selection represents the main pathway for ligand binding in these cases depends in part on what fraction of the protein contains an open site at equilibrium, as well as the kinetics of the protein’s conformational change relative to the dissociation rate of the encounter complex formed with other conformational states of the protein in which the cryptic site is not open. Therefore, observation of X-ray structures of the unbound protein that contain an open cryptic site do not, by themselves, prove that binding is by conformational selection. The presence of a strong hot spot close to a cryptic site suggests a likelihood that the initial binding of ligand to this hot spot in the cryptic conformation of the protein can be strong enough that the initial encounter complex will be relatively stable, so that reaction continues on to form the final stable complex in an induced fit binding mechanism. Our observation that a majority of cryptic sites in the current study have a strong hot spot adjacent to the binding site suggests that an induced fit mechanism is likely in many of these cases. Most clear cut are those 31 cases in which a strong hot spot exists, and there is no reported structure of the unbound protein that contains an open cryptic site. In these cases, which encompass a substantial fraction of the CryptoSite set, an induced fit mechanism seems highly likely. Conversely, there are three members in which there is no strong hot spot adjacent to the cryptic site and many examples of unbound structures with an open site, suggesting that binding occurs by conformational selection.
Fig. 4.
Continuum of mechanisms for binding to cryptic sites. In an induced fit binding mechanism, the initial encounter of the ligand with the unbound protein, A, is “sticky”: that is, dissociation of the initial encounter complex B, with rate k-1, is slow compared with conversion to the final, stable complex C, with rate constant k2, allowing time for the ligand-induced conformational change in the protein to take place within the lifetime of the encounter complex. For the alternative, conformational selection mechanism, most encounters between the ligand and protein are nonproductive, with the ligand rapidly dissociating. Only when the initial encounter occurs with one of the small fraction of protein molecules in which the cryptic site (shown by a blue asterisk) is fully formed (D) will reaction proceed to give a stable complex. Intermediate situations can be conceived where dissociation of the initial encounter complex occurs at a rate comparable with that for partitioning forward to form a stable complex. One hybrid mechanism involves an initial encounter with a form of the protein, E, in which the cryptic site is only partially formed through spontaneous conformational fluctuations, requiring the presence of the ligand to promote conversion to the final complex. Another is when ligand binds to a noncompetent conformation of the protein in a relatively sticky manner but must then wait for one or more adjacent subpockets to open through stochastic conformational fluctuations (F) before forming the final complex. We propose that mechanisms toward the induced fit end of this mechanistic spectrum (Bottom Right) are more likely when there is a strong hot spot (shown by a red asterisk) adjacent to the cryptic site even in protein conformers in which the cryptic site itself is occluded in all known unbound structures, a situation that describes over 50% of the examples present in the CryptoSite protein set.
We additionally observed that ligand binding is generally weak in surface pockets that are exclusively created by side chain motion. While there are different ways to interpret this finding, we speculate that perhaps it is because only relatively small binding cavities can be rendered cryptic solely by occluding them with side-chain atoms whereas to close a large cleft or cavity of the kind that binds ligands strongly typically requires some main-chain motions, too. This observation is potentially important because many of the transitional pockets that open in MD simulations involve only side chains (3, 14). Our analysis suggests that such pockets generally have limited ligand binding potential and, hence, are unlikely to be relevant for drug discovery.
We also studied the mechanisms of forming the pocket at the various cryptic sites. It was shown that, in addition to the cases involving only side chain motion, these include moving flexible loops and, in a few cases, entire secondary structural elements out of the pocket to form the ligand binding site. These changes frequently occur at domain–domain interfaces and may be emphasized by slight motions of the two domains relative to each other. Since mapping employs fragment-sized small molecules as probes, these are generally able to bind in the pocket despite its reduced size, particularly if the domains are separated and individually mapped. In a number of proteins, the cryptic site is the result of the opposite mechanism: i.e., a pocket is too open in the unbound structure, but side chains, loops, or entire secondary structure elements move closer to each other to form a better defined pocket. Since the success of mapping depends more on highly local surface properties than on the shape of the overall pocket, it generally can find hot spots at such sites.
In some cases, the high level of local flexibility leads to side chains or entire residues missing in the X-ray structure. Most frequently, weak electron density occurs in loops that become disordered, or in unstructured terminal regions. Although other X-ray structures of the same protein (e.g., determined in a different crystal form or at higher resolution) may be more complete, the missing fragments are predictive of structural uncertainty. This observation emphasizes that these structures can be very informative for drug discovery, even when more complete structures are also available. We have also shown that, in such cases, it may be useful to map the higher resolution structures, but after removing the uncertain regions, to identify cryptic sites.
Methods
Constructing the Extended CryptoSite Set.
For each protein in the CryptoSite set (5), we extracted from the Protein Data Bank all structures within 95% of sequence identity, defined as the percentage of identical residues based on sequence alignment by BLAST. The blastcut feature of PDB (www.rcsb.org/pdb/statistics/clusterStatistics.do) was used to extract homologous sequences. Five proteins did not have any other structure with 95% sequence identity in the PDB. Structural variations were analyzed in the ensembles of unbound structures for the remaining 88 proteins. This was done by local alignment using the steps as follows. (i) For each protein, the sequences of all selected structures were aligned using the CLUSTALW multiple alignment algorithm (51). (ii) Residues of the bound structure with any atom closer than 9 Å from the geometric center of the ligand were selected to define the extended cryptic site. (iii) The α-carbon atoms of the residues of this extended cryptic site in the bound structure and the α-carbon atoms of the same residues in the other structures were used for local structural alignment. Alignment was based on the least square algorithm using quaternions. (iv) We tested the aligned structures for the presence of any ligand atom (excluding water and metal ions) within 5 Å from any atom of the ligand in the bound structure. Any assumed unbound structure with such a ligand was removed, and the remaining structures formed the ensemble of unbound structures in the extended CryptoSite set for each bound structure.
Analysis of Hot Spots and Flexibility.
Before mapping, proteins were split into domains using the Protein Domain Parser, a structure-based method that relies on the number of contacts between regions of the protein to separate domains (36). Each unbound structure in the CryptoSite set was mapped using the FTMap program (22), both as the structure of the entire protein and as a set of separate domains. The hot spots were ranked based on the number of probe clusters, starting with consensus site 0 (CS0) with the largest number of probe clusters. Hot spots that had any probe atoms closer than 5 Å from any atom of the ligand superimposed from the bound structure were classified as being near the cryptic site and the others as far from the cryptic site. The rank of hot spots near the consensus sites is listed in SI Appendix, Table S1 as line 1 for each protein, with the number in parentheses indicating the number of probe clusters in the consensus site. The hot spots of the unbound structures in the extended set were locally aligned to the hot spots of the original unbound structure in the CryptoSite set as described in the previous section. Local all-atom rmsd values, calculated for each pair of aligned hot spots, were used to characterize local flexibility near and far from the cryptic site. For each protein, the additional unbound structures in the extended set were mapped using the FTMap program, again both with and without domain split. The resulting hot spots were classified as near or far from the cryptic site. Results for some unbound structures with strong hot spots near the cryptic site are also reported (SI Appendix, Table S1).
Supplementary Material
Acknowledgments
This investigation was supported by Grant R35-GM118078 from the National Institute of General Medical Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.K.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711490115/-/DCSupplemental.
References
- 1.Laskowski RA, Luscombe NM, Swindells MB, Thornton JM. Protein clefts in molecular recognition and function. Protein Sci. 1996;5:2438–2452. doi: 10.1002/pro.5560051206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Durrant JD, McCammon JA. Molecular dynamics simulations and drug discovery. BMC Biol. 2011;9:71. doi: 10.1186/1741-7007-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowman GR, Geissler PL. Equilibrium fluctuations of a single folded protein reveal a multitude of potential cryptic allosteric sites. Proc Natl Acad Sci USA. 2012;109:11681–11686. doi: 10.1073/pnas.1209309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman GR, Bolin ER, Hart KM, Maguire BC, Marqusee S. Discovery of multiple hidden allosteric sites by combining Markov state models and experiments. Proc Natl Acad Sci USA. 2015;112:2734–2739. doi: 10.1073/pnas.1417811112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cimermancic P, et al. CryptoSite: Expanding the druggable proteome by characterization and prediction of cryptic binding sites. J Mol Biol. 2016;428:709–719. doi: 10.1016/j.jmb.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oleinikovas V, Saladino G, Cossins BP, Gervasio FL. Understanding cryptic pocket formation in protein targets by enhanced sampling simulations. J Am Chem Soc. 2016;138:14257–14263. doi: 10.1021/jacs.6b05425. [DOI] [PubMed] [Google Scholar]
- 7.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 8.Whitty A, Kumaravel G. Between a rock and a hard place? Nat Chem Biol. 2006;2:112–118. doi: 10.1038/nchembio0306-112. [DOI] [PubMed] [Google Scholar]
- 9.Hardy JA, Wells JA. Searching for new allosteric sites in enzymes. Curr Opin Struct Biol. 2004;14:706–715. doi: 10.1016/j.sbi.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Arkin MR, Whitty A. The road less traveled: Modulating signal transduction enzymes by inhibiting their protein-protein interactions. Curr Opin Chem Biol. 2009;13:284–290. doi: 10.1016/j.cbpa.2009.05.125. [DOI] [PubMed] [Google Scholar]
- 11.Ludlow RF, Verdonk ML, Saini HK, Tickle IJ, Jhoti H. Detection of secondary binding sites in proteins using fragment screening. Proc Natl Acad Sci USA. 2015;112:15910–15915. doi: 10.1073/pnas.1518946112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erlanson DA, Wells JA, Braisted AC. Tethering: Fragment-based drug discovery. Annu Rev Biophys Biomol Struct. 2004;33:199–223. doi: 10.1146/annurev.biophys.33.110502.140409. [DOI] [PubMed] [Google Scholar]
- 13.Lawson AD. Antibody-enabled small-molecule drug discovery. Nat Rev Drug Discov. 2012;11:519–525. doi: 10.1038/nrd3756. [DOI] [PubMed] [Google Scholar]
- 14.Eyrisch S, Helms V. Transient pockets on protein surfaces involved in protein-protein interaction. J Med Chem. 2007;50:3457–3464. doi: 10.1021/jm070095g. [DOI] [PubMed] [Google Scholar]
- 15.Ulucan O, Eyrisch S, Helms V. Druggability of dynamic protein-protein interfaces. Curr Pharm Des. 2012;18:4599–4606. doi: 10.2174/138161212802651652. [DOI] [PubMed] [Google Scholar]
- 16.Johnson DK, Karanicolas J. Druggable protein interaction sites are more predisposed to surface pocket formation than the rest of the protein surface. PLoS Comput Biol. 2013;9:e1002951. doi: 10.1371/journal.pcbi.1002951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeLano WL. Unraveling hot spots in binding interfaces: Progress and challenges. Curr Opin Struct Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- 18.Ciulli A, Williams G, Smith AG, Blundell TL, Abell C. Probing hot spots at protein-ligand binding sites: A fragment-based approach using biophysical methods. J Med Chem. 2006;49:4992–5000. doi: 10.1021/jm060490r. [DOI] [PubMed] [Google Scholar]
- 19.Metz A, et al. Hot spots and transient pockets: Predicting the determinants of small-molecule binding to a protein-protein interface. J Chem Inf Model. 2012;52:120–133. doi: 10.1021/ci200322s. [DOI] [PubMed] [Google Scholar]
- 20.Hall DR, Kozakov D, Whitty A, Vajda S. Lessons from hot spot analysis for fragment-based drug discovery. Trends Pharmacol Sci. 2015;36:724–736. doi: 10.1016/j.tips.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozakov D, et al. New frontiers in druggability. J Med Chem. 2015;58:9063–9088. doi: 10.1021/acs.jmedchem.5b00586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozakov D, et al. The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat Protoc. 2015;10:733–755. doi: 10.1038/nprot.2015.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mattos C, Ringe D. Locating and characterizing binding sites on proteins. Nat Biotechnol. 1996;14:595–599. doi: 10.1038/nbt0596-595. [DOI] [PubMed] [Google Scholar]
- 24.Hajduk PJ, Huth JR, Fesik SW. Druggability indices for protein targets derived from NMR-based screening data. J Med Chem. 2005;48:2518–2525. doi: 10.1021/jm049131r. [DOI] [PubMed] [Google Scholar]
- 25.Brenke R, et al. Fragment-based identification of druggable ‘hot spots’ of proteins using Fourier domain correlation techniques. Bioinformatics. 2009;25:621–627. doi: 10.1093/bioinformatics/btp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grove LE, Hall DR, Beglov D, Vajda S, Kozakov D. FTFlex: Accounting for binding site flexibility to improve fragment-based identification of druggable hot spots. Bioinformatics. 2013;29:1218–1219. doi: 10.1093/bioinformatics/btt102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landon MR, et al. Detection of ligand binding hot spots on protein surfaces via fragment-based methods: Application to DJ-1 and glucocerebrosidase. J Comput Aided Mol Des. 2009;23:491–500. doi: 10.1007/s10822-009-9283-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozakov D, Chuang GY, Beglov D, Vajda S. Where does amantadine bind to the influenza virus M2 proton channel? Trends Biochem Sci. 2010;35:471–475. doi: 10.1016/j.tibs.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozakov D, et al. Structural conservation of druggable hot spots in protein-protein interfaces. Proc Natl Acad Sci USA. 2011;108:13528–13533. doi: 10.1073/pnas.1101835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall DH, et al. Robust identification of binding hot spots using continuum electrostatics: Application to hen egg-white lysozyme. J Am Chem Soc. 2011;133:20668–20671. doi: 10.1021/ja207914y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buhrman G, et al. Analysis of binding site hot spots on the surface of Ras GTPase. J Mol Biol. 2011;413:773–789. doi: 10.1016/j.jmb.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Golden MS, et al. Comprehensive experimental and computational analysis of binding energy hot spots at the NF-κB essential modulator/IKKβ protein-protein interface. J Am Chem Soc. 2013;135:6242–6256. doi: 10.1021/ja400914z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landon MR, et al. Novel druggable hot spots in avian influenza neuraminidase H5N1 revealed by computational solvent mapping of a reduced and representative receptor ensemble. Chem Biol Drug Des. 2008;71:106–116. doi: 10.1111/j.1747-0285.2007.00614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wassman CD, et al. Computational identification of a transiently open L1/S3 pocket for reactivation of mutant p53. Nat Commun. 2013;4:1407. doi: 10.1038/ncomms2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ngan CH, et al. FTSite: High accuracy detection of ligand binding sites on unbound protein structures. Bioinformatics. 2012;28:286–287. doi: 10.1093/bioinformatics/btr651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexandrov N, Shindyalov I. PDP: Protein domain parser. Bioinformatics. 2003;19:429–430. doi: 10.1093/bioinformatics/btg006. [DOI] [PubMed] [Google Scholar]
- 37.Arkin MR, et al. Binding of small molecules to an adaptive protein-protein interface. Proc Natl Acad Sci USA. 2003;100:1603–1608. doi: 10.1073/pnas.252756299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hyde J, Braisted AC, Randal M, Arkin MR. Discovery and characterization of cooperative ligand binding in the adaptive region of interleukin-2. Biochemistry. 2003;42:6475–6483. doi: 10.1021/bi034138g. [DOI] [PubMed] [Google Scholar]
- 39.Horn JR, Shoichet BK. Allosteric inhibition through core disruption. J Mol Biol. 2004;336:1283–1291. doi: 10.1016/j.jmb.2003.12.068. [DOI] [PubMed] [Google Scholar]
- 40.Berman HM, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson KP, et al. Crystal structure of p38 mitogen-activated protein kinase. J Biol Chem. 1996;271:27696–27700. doi: 10.1074/jbc.271.44.27696. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Kaiser CE, Frett B, Li HY. Targeting mutant KRAS for anticancer therapeutics: A review of novel small molecule modulators. J Med Chem. 2013;56:5219–5230. doi: 10.1021/jm3017706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun Q, et al. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew Chem Int Ed Engl. 2012;51:6140–6143. doi: 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maurer T, et al. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc Natl Acad Sci USA. 2012;109:5299–5304. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barr AJ. Protein tyrosine phosphatases as drug targets: Strategies and challenges of inhibitor development. Future Med Chem. 2010;2:1563–1576. doi: 10.4155/fmc.10.241. [DOI] [PubMed] [Google Scholar]
- 46.Wiesmann C, et al. Allosteric inhibition of protein tyrosine phosphatase 1B. Nat Struct Mol Biol. 2004;11:730–737. doi: 10.1038/nsmb803. [DOI] [PubMed] [Google Scholar]
- 47.Li S, et al. The mechanism of allosteric inhibition of protein tyrosine phosphatase 1B. PLoS One. 2014;9:e97668. doi: 10.1371/journal.pone.0097668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daniels KG, Suo Y, Oas TG. Conformational kinetics reveals affinities of protein conformational states. Proc Natl Acad Sci USA. 2015;112:9352–9357. doi: 10.1073/pnas.1502084112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greives N, Zhou HX. Both protein dynamics and ligand concentration can shift the binding mechanism between conformational selection and induced fit. Proc Natl Acad Sci USA. 2014;111:10197–10202. doi: 10.1073/pnas.1407545111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou HX. From induced fit to conformational selection: A continuum of binding mechanism controlled by the timescale of conformational transitions. Biophys J. 2010;98:L15–L17. doi: 10.1016/j.bpj.2009.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002;Chap 2:Unit 2.3. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.