Significance
We describe unexpected cooperation between two cytokines that are important in regulating the growth of cancers, namely, type I interferons (IFNs) and interleukin 6 (IL6). It is well established that IL6 is vital for the ability of many tumor types to prosper, and the work in the current paper reveals that the signaling pathway driven by IFN, which is also evident in many cancers, increases the expression of IL6 through a direct effect on the IL6 gene. The findings may help to identify new antitumor targets for therapy.
Keywords: STAT2, IRF9, IL6, ISGF3, p65
Abstract
In response to IFNβ, the IL6 gene is activated, modestly at early times by ISGF3 (IRF9 plus tyrosine-phosphorylated STATs 1 and 2), and strongly at late times by U-ISGF3 (IRF9 plus U-STATs 1 and 2, lacking tyrosine phosphorylation). A classical IFN-stimulated response element (ISRE) at −1,513 to −1,526 in the human IL6 promoter is required. Pretreating cells with IFNβ or increasing the expression of U-STAT2 and IRF9 exogenously greatly enhances IL6 expression in response to the classical NF-κB activators IL1, TNF, and LPS. U-STAT2 binds tightly to IRF9, the DNA binding subunit of ISGF3, and also to the p65 subunit of NF-κB. Therefore, as shown by ChIP analyses, U-STAT2 can bridge the ISRE and κB elements in the IL6 promoter. In some cancer cells, the protumorigenic activation of STAT3 will be enhanced by the increased synthesis of IL6 that is facilitated by high expression of U-STAT2 and IRF9.
Type I interferons (IFNα and IFNβ) mediate major innate immune responses to viruses and other infectious agents. Following the binding of type I IFNs to their dimeric receptor, IFNAR1/2, the JAK1 and TYK2 tyrosine kinases are activated, catalyzing phosphorylation of the receptors and receptor-bound STATs 1 and 2. Tyrosine-phosphorylated STAT1/2 heterodimers bind to IRF9, forming IFN-stimulated gene factor 3 (ISGF3), which then binds to IFN-stimulated response elements (ISREs) in the promoters of IFN-induced genes (ISGs) to initiate their transcription (1). STAT2 contributes its strong transactivation domain to ISGF3 (2), IRF9 contributes the principal DNA binding domain, and STAT1 stabilizes the complex and also provides additional DNA contacts (3). Notably, the STAT1, STAT2, and IRF9 genes are all ISGs that are strongly induced by type I IFNs.
Although the expression of most ISGs is mediated by this canonical signaling pathway, the components of the pathway have additional, noncanonical functions (4). U-STATs 1 and 2, lacking tyrosine phosphorylation, are still able to combine with IRF9 to form U-ISGF3, which sustains the transcription of about a quarter of the initially induced ISGs during the late phase of the response to type I IFNs (5). Even without STAT1, type I IFNs still trigger prolonged ISG expression in a U-STAT2–IRF9-dependent manner (6), and tyrosine phosphorylated STAT2 forms a homodimer that combines with IRF9 to form a complex that can still stimulate ISG expression (7). In addition to their functions within ISGF3, STAT2 and IRF9 interact with components of other signaling pathways to stimulate transcription. For example, IFNβ and TNFα synergistically induce DUOX2 expression, which helps to mediate the late phase of the antiviral response to Sendai virus in a STAT2- and IRF9-dependent, but STAT1-independent manner (8). STAT2 and IRF9 also help to drive the expression of Gene G in NB4 cells in response to retinoic acid plus IFNα, independently of STAT1 (9). Recently, STAT2 has also been found to negatively regulate IFNγ signaling, by forming an unproductive complex with STAT1 (10).
IL6 is a pathogenic cytokine whose up-regulation leads to chronic inflammatory diseases, for example, rheumatoid arthritis (11). Chronic IL6-dependent signaling is also associated with tumorigenesis in numerous mouse models as well as in human disease (12). Several studies show that IL6 is required for tumor initiation in response to activated oncoproteins such as K-RAS and EGFR (13–15). IL6 is a major inducer of STAT3 phosphorylation and activation, thus providing a dominant prosurvival benefit to cancer cells. Clinical studies reveal that elevated serum levels of IL6 are associated with advanced tumor stages in various cancers and poor survival (16). Therefore, the timing and location of IL6 production needs to be strictly regulated. Activated NF-κB family members have a major role in inducing the expression of IL6 (17). A recent study unexpectedly found that a deficiency of STAT2 inhibited colorectal carcinogenesis, due to a lower level of inflammatory cytokines (18), and mice lacking STAT2, but not lacking STAT1 or IFNARs, are hypersensitive to LPS (19). These findings indicate that STAT2 participates in inflammatory responses through mechanisms that have not yet been discovered.
We find that IFNβ stimulates robust IL6 secretion, which is sufficient to overcome the ability of IFNβ to inhibit cell growth. Priming cells with IFNβ synergistically enhances IL6 induction in response to treatments that activate NF-κB, in a process that depends upon the recruitment of STAT2, IRF9, and NF-κB to ISRE and κB elements in the IL6 promoter. Our new appreciation that STAT2 and IRF9 contribute to STAT3 activation in cancer cells, by helping to up-regulate IL6 expression, identifies potential targets to inhibit this process.
Results
IFNβ Induces IL6 Expression Without Activating NF-κB.
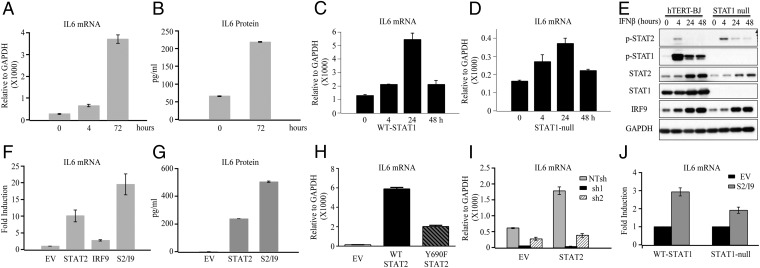
A previous study found that IFNα induced the autocrine secretion of IL6 in myeloma cells (20), and the plasma level of IL6 was found to be elevated rapidly in patients following injection of IFNα (21). We now show that IFNβ induces the expression of IL6 mRNA and protein in human mammary epithelial (HME) cells (Fig. 1 A and B). Of note, IL6 induction after 72 h was much higher than after 4 h. However, we observed that several NF-κB–dependent genes, including IL1 and TNFα, were not significantly induced by IFNβ in HME cells (SI Appendix, Fig. S1A). IFNβ stimulation did not change the phosphorylation of IκBα or the p65 subunit of NF-κB or the level of IκBα between 5 min and 4 h (SI Appendix, Fig. S1B), indicating that the expression of IL6 induced in response to IFNβ. We found a canonical ISRE element in the distal human IL6 promoter, between −1,513 and −1,526. When the human and mouse IL6 promoters were compared, except for the region surrounding the κB element, the most conserved region is between −1,513 and −1,565 (SI Appendix, Fig. S1C), implying that this ISRE might be important for transcriptional regulation by IFNβ. Previously, we found that phosphorylated ISGF3 and U-ISGF3 are responsible for ISG induction during the early and late phases of the response to type I IFNs, respectively (5). The amount of IL6 induced in response to IFNβ increased gradually from 4 to 72 h (SI Appendix, Fig. S1A), along with decreases in phosphorylated STAT1 and STAT2 and increases in U-STAT1, U-STAT2, and IRF9 (SI Appendix, Fig. S1D). In contrast to the induction of canonical ISGs, whose expression is driven by phosphorylated ISGF3 only (for example, CXCL10) or by both phosphorylated ISGF3 and U-ISGF3 (for example, DDX58), the IFNβ-dependent induction of IL6 was much stronger in response to U-ISGF3 than to phosphorylated ISGF3 (SI Appendix, Fig. S1E).
Fig. 1.
IFNβ induces IL6 expression without activating NF-κB. (A and B) HME cells were treated or not with IFNβ (200 units/mL) for 4 or 72 h. IL6 expression was analyzed by q-PCR and ELISA. (C and D) hTERT–BJ and hTERT–STAT1-null fibroblasts were treated with IFNβ (200 units/mL) for 4, 24, or 48 h or were untreated. q-PCR analysis of IL6 expression and (E) Western analyses of STAT2, STAT1, and IRF9 are shown. (F and G) The amount of IL6 induced by U-STAT2 alone, IRF9 alone, or U-STAT2 and IRF9 together were analyzed by q-PCR and ELISA. EV, control HME cells made with empty vector. (H) The amounts of IL6 mRNA induced by IRF9 alone (EV), WT STAT2 and IRF9 together, and Y690F–STAT2 and IRF9 together were analyzed by q-PCR. (I) HME control and HME cells with increased expression of U-STAT2 were infected with lentiviruses containing an shRNA targeting IRF9 or a nontargeted shRNA control (NT shRNA). IL6 mRNA was analyzed by q-PCR. (J) The levels of expression of U-STAT2 and IRF9 were increased in BJ human foreskin fibroblasts and STAT1-null fibroblasts, respectively, followed by q-PCR analysis of IL6 mRNA expression.
High Expression of STAT2 and IRF9 Are Sufficient for IL6 Induction.
STAT1 is an important component of ISGF3, but in some cases, type I IFNs stimulate the expression of some ISGs even in the absence of STAT1. However, STAT2 and IRF9 are still required (6, 22, 23). To clarify this point, we examined whether STAT1 is required for IFNβ-induced IL6 expression. When wild-type or STAT1-null fibroblasts were stimulated with IFNβ, the expression of IL6 was increased by twofold in the STAT1-null fibroblasts and by about fourfold in the wild-type cells after 24 h (Fig. 1 C and D), indicating that STAT1 has a supporting role but is not required for IFNβ-induced IL6 expression. Consistently, the induction of canonical ISGs, such as STAT2 and DDX58, was less in IFNβ-treated STAT1-null cells than in wild-type cells (Fig. 1E and SI Appendix, Fig. S1F). We also noted different IL6 induction kinetics in HME and BJ cells. The induction of DDX58, a canonical U-ISGF3 target gene, was also decreased after 48 h compared with 24 h of IFNβ treatment in BJ cells (SI Appendix, Fig. S1F), but the induction of STAT1, IRF9, and STAT2 were not decreased at 48 h (Fig. 1E), similarly to the induction of IL6. We appreciate that the ISG induction pattern is different in different cell lines, perhaps because the function and formation of U-ISGF3 is not only dependent on the level of expression, but also on other factors, including posttranslational modifications.
To exclude the impact of unknown factors that might be activated by IFNβ stimulation, we expressed U-STAT2 and IRF9 exogenously in HME cells. Increased expression of U-STAT2 alone strongly increased IL6 mRNA levels, by about 10-fold, while increased expression of IRF9 alone increased IL6 mRNA levels by about 2-fold, consistent with the lack of intrinsic transcriptional function for IRF9 and its ability to help U-STAT2 to enter the nucleus (24, 25). The combination of U-STAT2 and IRF9 increased IL6 mRNA by about 19-fold (Fig. 1F). ELISA results are consistent with the changes in IL6 mRNA levels (Fig. 1G). To further test whether tyrosine phosphorylation of STAT2 is required for the induction of IL6, the STAT2 phosphorylation-deficient mutant Y690F was coexpressed with IRF9 in HME cells. IL6 expression was enhanced, compared with the effect of increased expression of IRF9 alone (Fig. 1H). However, the level of induction was not as high as with wild-type STAT2. Since the C terminus of STAT2 participates in stabilizing the STAT2–IRF9 interaction (3), the Y690F mutation might impact the stability of the STAT2–IRF9 dimer. The expression of additional NF-κB–dependent genes induced by U-STAT2 was examined by using an Illumina Gene Expression Array (SI Appendix, Fig. S2). To study the role of IRF9 further, we reduced its expression in HME cells expressing a high level of exogenous U-STAT2. Knockdown of IRF9 blocked IL6 induction by U-STAT2, showing that IRF9 is also required for IL6 induction in response to IFNβ (Fig. 1I and SI Appendix, Fig. S3A). Increased expression of U-STAT2 plus IRF9 increased IL6 expression by about 2-fold in STAT1-null fibroblasts and by about 3-fold in wild-type fibroblasts (Fig. 1J), similarly to the effects on DDX58 expression (SI Appendix, Fig. S3B). Collectively, these data indicate that levels of expression of exogenous U-STAT2 and IRF9 that are comparable to the levels induced at late times in response to IFNβ are sufficient to induce IL6 expression, and that STAT1 is not essential for this process.
IFNβ Synergistically Enhances IL6 Induction in Response to Inflammatory Factors in an ISRE-Dependent Manner.
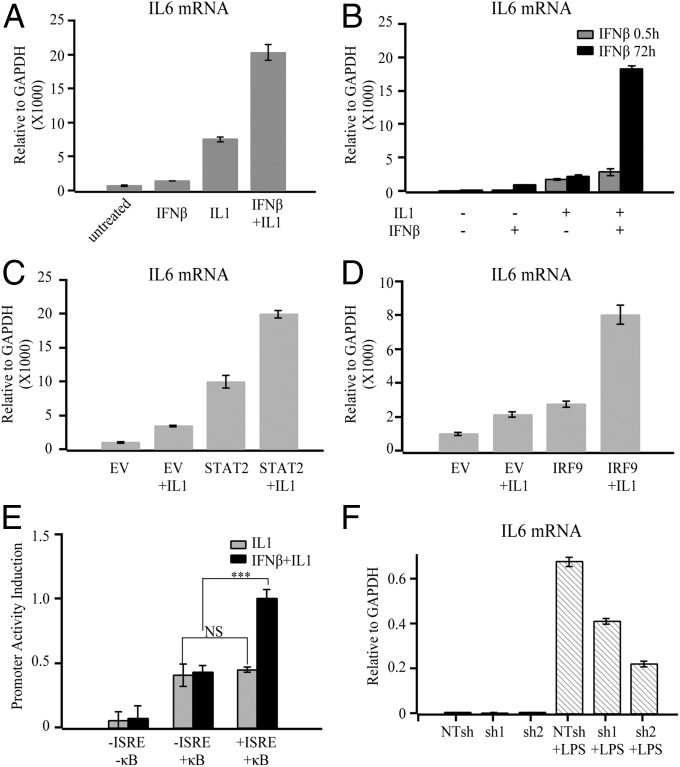
To understand the broader impact of IFNβ-mediated IL6 expression, we examined the effect of IRF9 and U-STAT2 on the expression induced by agents that activate NF-κB. HME cells primed with IFNβ for 4 h expressed increased amounts of IL6 mRNA following treatment with proinflammatory factors, including LPS, IL1, and TNFα (Fig. 2A and SI Appendix, Fig. S4A). To evaluate the relative effects of phosphorylated ISGF3 (P-ISGF3) and U-ISGF3, HME cells were primed with IFNβ for 30 min or 72 h. At 30 min, STAT1 and STAT2 tyrosine phosphorylation was maximum (P-ISGF3) and it started to decline between 1 and 2 h (SI Appendix, Fig. S1B); at 4 h, the treated cells contained both P-ISGF3 and U-ISGF3 (SI Appendix, Fig. S1B); at 72 h, U-ISGF3 was the dominant form, rather than P-ISGF3. As expected, priming with U-ISGF3 was much more synergistic with IL1 in inducing IL6 expression than priming with phosphorylated ISGF3 (Fig. 2B). In addition, exogenously expressed U-STAT2 or IRF9 strongly increased the expression of IL6 induced by IL1 (Fig. 2 C and D). To determine whether the ISRE in the IL6 promoter is responsible for the synergistic induction of IL6, we tested luciferase reporters with different IL6 promoter fragments (SI Appendix, Fig. S4B). The increases in IL6 promoter activity in response to IL1 were comparable between the promoters with and without the ISRE (Fig. 2E). Notably, IFNβ priming increased the activity of the IL6 promoter with the ISRE, but not the one without it, indicating that this element is responsible for the IFNβ-mediated responses. Similar results were obtained with TNFα-stimulated cells (SI Appendix, Fig. S4C). Collectively, these results indicate that the IL6 ISRE is required for IFNβ priming to enhance the induction of IL6 in response to proinflammatory factors that activate NF-κB. Also, knockdown of IRF9 impaired IL6 expression induced by LPS in normal human fibroblasts (Fig. 2F and SI Appendix, Fig. S4D), but the induction of other NF-κB–dependent genes (IL1 and IL8) was not inhibited (SI Appendix, Fig. S4 E and F).
Fig. 2.
IFNβ synergistically enhances IL6 expression in response to induction by inflammatory factors in an IL6–ISRE-dependent manner. (A) HME cells primed or not with IFNβ (200 units/mL) for 4 h were treated with IL1 (20 ng/mL) for 3 h. IL6 mRNA expression levels were assayed by q-PCR. (B) HME cells were treated or not with IFNβ for 0.5 or 72 h, then stimulated or not with IL1 for 3 h. IL6 mRNA was assayed by q-PCR. (C and D) HME cells expressing control vector, STAT2, or IRF9 were treated with IL1 for 3 h, and q-PCR analysis of IL6 expression was performed. EV, control HME cells made with empty vector. (E) Stable pools of HME cells with different IL6 reporter constructs were primed or not with IFNβ (500 units/mL) for 4 h and then stimulated with IL1 for 24 h. Promoter activity was analyzed by luciferase assay. (F) BJ cells were infected with lentiviruses containing IRF9 shRNA or scrambled shRNA. The cells were then treated with LPS for 3 h, and q-PCR analysis of IL6 mRNA expression was performed.
U-STAT2 and IRF9 Increase p65 Occupancy on the IL6 Promoter.
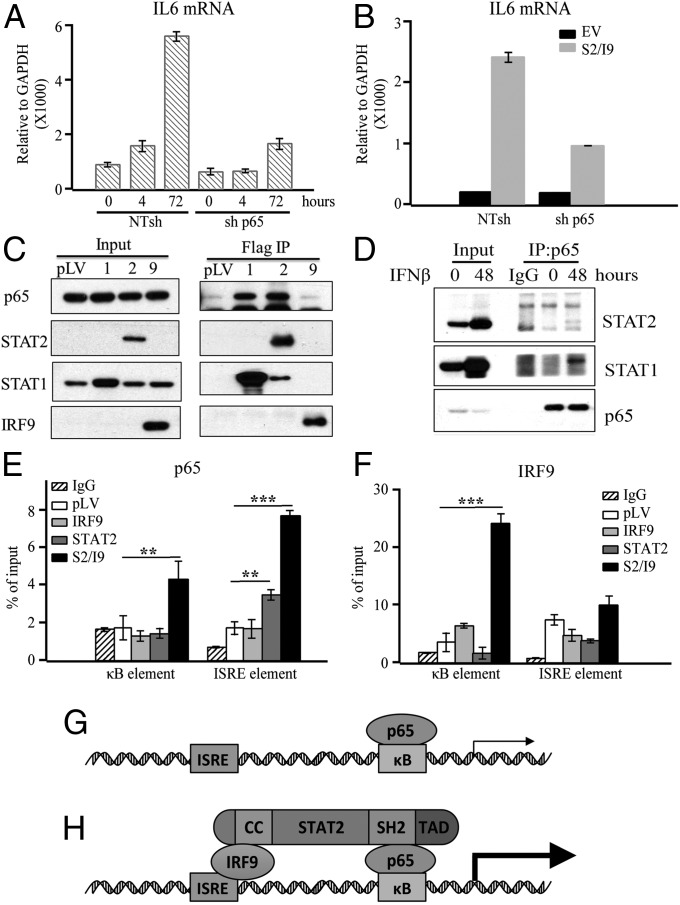
The induced expression of IL6 in response to U-STAT2 and IRF9 is similar to the induction of a subset of IFN-induced genes in response to U-ISGF3, but quite different from the induction of classical ISGs in response to phosphorylated ISGF3 (SI Appendix, Fig. S1E); therefore, the IL6 gene is not a classical ISG. Transcription of the IL6 gene is well known to be driven by NF-κB, but NF-κB is not activated by IFNβ in HME cells (SI Appendix, Fig. S1B). Exogenous expression of U-STAT2 and IRF9, either individually or together, did not lead to the translocation of p65 from the cytosol to the nucleus (SI Appendix, Fig. S5 A and B). We conclude that the activation of IL6 expression in response to IFNβ or to increased expression of U-STAT2 plus IRF9 is not due to its ability to activate NF-κB. Since EGF, upon binding to EGFR, activates NF-κB through a mechanism that depends on TLR4 phosphorylation (26), HME cells will have basal activation of NF-κB because EGF is present in the medium used for these cells. To study the role of NF-κB further, we knocked the expression of its p65 subunit down, finding that the ability of IFNβ, or increased expression of U-STAT2 and IRF9 to enhance IL6 expression, was severely inhibited (Fig. 3 A and B).
Fig. 3.
U-STAT2 plus IRF9 increased p65 occupancy at the IL6 promoter. (A) HME cells were infected with lentiviruses containing p65 shRNA or NTshRNA control and the cells were treated with IFNβ for the indicated times. q-PCR analysis of IL6 mRNA levels are shown. (B) Control HME cells with empty vector (EV) or with increased expression of U-STAT2 and IRF9 were infected with lentiviruses containing p65 shRNA or NT shRNA control. q-PCR analysis of IL6 expression was performed. (C) Whole-cell lysates from HME cells expressing STAT1, STAT2, and IRF9 were used for immunoprecipitation of Flag-tagged proteins. (D) Whole-cell lysates from HME cells treated or not with IFNβ (50 units/mL) for 48 h were analyzed by immunoprecipitation, using anti-p65. (E) The occupancy of p65 at the IL6 promoter in HME cells expressing a high level of IRF9 alone, U-STAT2 alone, or U-STAT2 and IRF9 together were assayed by ChIP. (F) ChIP analysis of the occupancy of IRF9 at the IL6 promoter in the above cell lines. (G) p65 drives IL6 transcription when it occupies the κB element. (H) STAT2 bridges and stabilizes a complex with IRF9 and p65, which increases IL6 transcription. The CC domain of STAT2 binds to IRF9, the SH2 domain probably binds to p65, and the TAD domain enhances the transcriptional response. **P ≤ 0.01, ***P ≤ 0.001.
We tested the ability of p65 to bind to U-STAT1 or U-STAT2. Immunoprecipitation from HME cells expressing exogenous U-STAT1, U-STAT2, or IRF9 showed that p65 binds strongly to U-STAT1 and U-STAT2, but not to IRF9 (Fig. 3C). This result is consistent with the finding that exogenous expression of IRF9 alone did not induce IL6 expression strongly (Fig. 1F). Furthermore, we found an interaction between U-STAT1, U-STAT2, and p65 in HME cells following stimulation with IFNβ (Fig. 3D). However, consistent with results above showing that U-STAT2 or IRF9 did not cause nuclear translocation of p65 in HME cells, exogenous expression of each protein also did not change the interaction between p65 and IκBα (SI Appendix, Fig. S5C). Taken together, these results strongly support a model in which the interaction of p65 and the U-STAT2–IRF9 complex is required for enhanced induction of IL6 expression.
To examine how a U-STAT2–IRF9–p65 complex might regulate IL6 transcription, we performed ChIP experiments, using HME cells expressing exogenous U-STAT2 alone, IRF9 alone, or U-STAT2 plus IRF9. Exogenous expression of U-STAT2 alone increased the occupancy of p65 on the ISRE element of the IL6 gene by about twofold, but there was little change at the κB element (Fig. 3E), indicating that a U-STAT2–IRF9–p65 complex can be recruited to the IL6 promoter by binding to the ISRE. STAT2 has a constitutively active nuclear export signal and is localized mainly in the cytosol in the absence of IFN. In quiescent cells, the translocation of U-STAT2 to the nucleus depends on IRF9, which has a strong nuclear localization sequence (24). As a consequence, cells coexpressing U-STAT2 and IRF9 led to high levels of these two proteins in the nucleus, and a dramatically increased occupancy of p65 at both the ISRE and κB elements (Fig. 3E). IRF9 recruitment to the IL6 promoter was not changed by increasing the expression of U-STAT2 (Fig. 3F). Surprisingly, the occupancy of IRF9 at the κB site was strongly enhanced by coexpression of U-STAT2 and IRF9 but not by the expression of IRF9 alone, indicating that the U-STAT2–IRF9–p65 complex also binds to the κB element (Fig. 3F). Furthermore, consistent with the results of q-PCR analyses (Fig. 1F), the recruitment of p65 to the IL6 promoter was not increased significantly in HME cells expressing exogenous IRF9 (Fig. 3E). To further test our hypothesis, we knocked the expression of p65 down and examined the occupancy of IRF9 on the IL6-κB and IL6-ISRE elements, finding a decrease in the recruitment of IRF9 to the IL6-κB element but not the IL6-ISRE element (SI Appendix, Fig. S6A). Knockdown of p65 did not affect the levels of IRF9 (SI Appendix, Fig. S6B). Collectively, these results suggest that U-STAT2 serves as a bridge within the IL6 promoter, connecting IRF9 bound to the ISRE with p65 bound to the κB element.
U-STAT2 and IRF9 Promote the Survival of Cancer Cells by Enhancing IL6 Expression and STAT3 Activation.
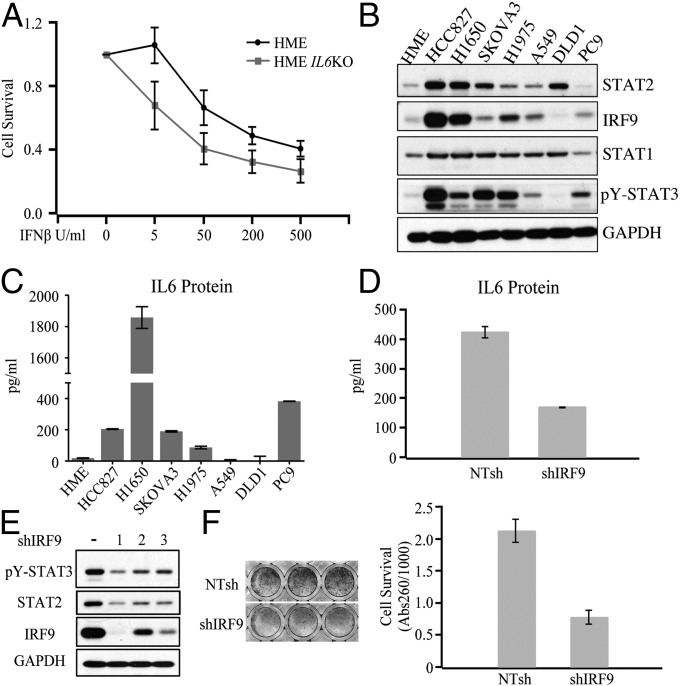
IL6 is famous for its ability to inhibit apoptosis and activate proliferation (27, 28). To study the role of the IL6 that is induced in response to IFNβ directly, we knocked the IL6 gene out in HME cells. The IL6-deficient cells were more sensitive to growth inhibition induced by IFNβ, at both low and high doses (Fig. 4A), suggesting that the IL6 induced by IFNβ helps to protect the cells from the growth inhibitory effects of IFNβ. Paracrine or autocrine IL6, derived from stromal or cancer cells, accounts for substantial STAT3 activation in cancers of the lung, liver, breast, head and neck, and other organs (29, 30). NF-κB, constitutively activated in virtually all oncogenic abnormalities, is required for the autocrine secretion of IL6 in some cancers (31). To analyze correlations with autocrine IL6 expression, we examined the levels of STAT2, IRF9, and tyrosine-phosphorylated STAT3 in cancer cells (Fig. 4B). Cells with relatively high levels of IRF9 or U-STAT2 (HCC827, H1975, H1650, and SKOVA3) also have more STAT3 activation. In contrast, DLD1 and A549 cells, which have relatively low expression of IRF9 and U-STAT2, also have less STAT3 activation. Accordingly, HCC827, H1975, H1650, and SKOVA3 cells have higher levels of autocrine IL6 expression than do DLD1 and A549 cells (Fig. 4C). Of note, the level of U-STAT1 is not related to IL6 secretion or to the level of tyrosine- phosphorylated STAT3 in these cells, confirming that STAT2 and IRF9 but not STAT1 are principally required for the production of autocrine IL6. To confirm that the U-STAT2–IRF9 complex is responsible for autocrine IL6 secretion and STAT3 activation in cancer cells, we knocked the expression of IRF9 down in HCC827 cells, finding that IL6 secretion was reduced by 60% (Fig. 4D). Reduced expression of IRF9 dramatically inhibited both STAT3 tyrosine phosphorylation and STAT2 expression (Fig. 4E). Consistently, knockdown of IRF9 strongly inhibited the proliferation of lung cancer cells (Fig. 4F) but not normal cells (SI Appendix, Fig. S7).
Fig. 4.
U-STAT2 and IRF9 promote cancer cell survival by enhancing IL6 expression and STAT3 phosphorylation. (A) HME and HME IL6 KO cells were treated with IFNβ (0, 5, 50, 200, or 500 units/mL) for 10 d. Cell survival was tested by staining with crystal violet. (B) Analysis of IRF9, STAT2, STAT1, and p-STAT3 levels by the Western method. (C) Levels of secreted IL6 were analyzed by ELISA. (D) ELISA analysis of IL6 levels in HCC827 cells with knockdown of IRF9 or control cells. (E) HCC827 cells were infected with lentiviruses containing an shRNA targeting IRF9 or shRNA control, and p-STAT3 and STAT2 levels were assayed by the Western method. (F) The same cells used in E were seeded into 48-well plates and cell survival was analyzed by crystal violet staining after 6 d.
Encouraged by these in vitro data, we investigated whether the ability of U-STAT2 and IRF9 to facilitate IL6 expression is associated with survival in cancer patients. High levels of STAT2, IRF9, and IL6 correlated with poor overall survival in lung adenocarcinoma patients, and STAT1 and IFNB expression did not (SI Appendix, Fig. S8). In contrast, a high level of IFNAR1 correlated with good survival, and high levels of IFNA1, IFNA2, and IFNA10 with poor survival. We also examined the data for gastric, breast, and ovarian cancers, finding that IL6 expression did not correlate significantly with the overall survival of these patients, and neither did STAT2 or IRF9. IL6 has been shown to facilitate resistance to erlotinib, afatinib, and cisplatin in nonsmall cell lung cancer (32–34), providing a rationale for the lung cancer-specific high levels of STAT2 and IRF9. Together, these results provide evidence that U-STAT2 and IRF9 cooperate with p65 to facilitate lung cancer cell growth or survival by facilitating the expression of IL6 and the activation of STAT3.
Discussion
Mechanistic Aspects.
Our working model for how U-STAT2, IRF9, and p65 collaborate on the IL6 promoter to enhance expression is shown in Fig. 3 G and H. We propose that STAT2 bridges the ISRE and κB elements by binding simultaneously to IRF9 and p65, bringing the potent transactivation domain of STAT2 into play to help activate the transcriptional machinery. As a component of U-ISGF3, STAT1 can participate in ISRE-dependent activation of IL6 expression, but it is not required since the U-STAT2–IRF9 complex functions well even without STAT1. However, STAT1 may assist by inducing the expression of U-STAT2 and IRF9 as target genes of phosphorylated ISGF3. Since the ISRE and κB elements are not close to one another in the linear DNA sequence of the promoter, it is likely that a loop is formed to facilitate their interaction. The high concentrations of U-STAT2 and IRF9 that help to drive IL6 expression are achieved late in response to type I IFNs, since the STAT1, STAT2, and IRF9 genes are all ISGs that are activated strongly by tyrosine-phosphorylated ISGF3 during the initial response to these IFNs. In many cancers, these three proteins, which comprise U-ISGF3, are expressed at a high level constitutively, as a result of exposure of the tumors to type I or III IFNs, or as a consequence of other inducing mechanisms, such as cell crowding (35). Although U-STAT2 plays a predominant role in driving IL6 expression, it remains possible that tyrosine-phosphorylated STAT2 also contributes. As shown in Fig. 1A, IL6 was induced 4 h after treatment with IFNβ, when tyrosine phosphorylation of STAT2 was apparent, but increased STAT2 expression was not.
The expression of IL6 in response to type I IFN requires that free NF-κB p65 is already present, but IFNβ does not liberate NF-κB from IκB in the cells we have studied. IL6 is not the only gene that is regulated by type I IFNs in an NF-κB–dependent manner, as both ISGF3 and NF-κB are required to induce expression of the βR1 gene in response to IFNβ (36). Signals generated in response to type I IFN interact with NF-κB–dependent signals in bone marrow-derived cells. In lymphoblasts, IFNα-induced activation of NF-κB is due to IκBα degradation, catalyzed by the IFN-dependent activation of PI3K and AKT (37). We show that IFNβ induces the expression of a small subset of NF-κB–dependent genes, including IL6, without activating the release of NF-κB from IκB or increasing the phosphorylation of p65. However, p65 is required for U-STAT2 plus IRF9 or IFNβ to induce IL6 expression.
We show that enhanced induction of IL6 by U-ISGF3 in combination with NF-κB stimulators depends on the presence of an ISRE in the IL6 promoter. In another study, Listeria monocytogenes and IFNβ were shown to induce the expression of the NOS2 and IL6 genes synergistically (38). Tyrosine-phosphorylated ISGF3 and NF-κB cooperatively regulate NOS2 transcription by recruiting STAT1 and p65 to a distant enhancer that is ∼30 kb upstream of the transcription start site (TSS), but not to putative ISREs located at −940 to −952 and −911 to −924. The cooperation of phosphorylated ISGF3 and NF-κB was considered to occur early in the response to type I IFNs, in agreement with our finding that priming by phosphorylated ISGF3 enhances IL6 induction in response to IL1 (Fig. 2B). In addition, synergistic induction of the IL6 gene in response to IFNγ and TLR-dependent signaling is associated with STAT1 occupancy at an enhancer that is ∼25 kb upstream of the TSS, followed by the priming of histone acetylation (39). Both studies show that STAT1 is recruited to the IL6 distal enhancer, thus increasing the formation of initiation complexes that include RNA polymerase II at the TSS. However, in our study, the enhanced activation of IL6 expression is much stronger in cells with high levels of U-ISGF3 than at early times in cells with phosphorylated ISGF3 (Fig. 2B). An ISRE at −1,513 to −1,526 in the IL6 promoter, rather than a much more distal enhancer, is responsible for the enhanced induction of IL6 expression in response to the U-ISGF3 that is formed late in the response to IFNβ, in combination with activators of NF-κB.
As an important coordinator of immune responses, NF-κB interacts with many other transcription factors. STAT3 and STAT1 interact with p65 physically. For example, STAT1α binds to p65, decreasing NF-κB nuclear localization and inhibiting target gene activation (40). We confirmed the interaction between U-STAT1 and p65 (Fig. 3 C and D) but it is not clear whether this interaction is direct. However, we did not detect the binding of p65 to IRF9, even though IRF9 is required for STAT2-induced NF-κB–dependent gene expression (Fig. 1I). U-STAT2 binds to IRF9 through its coil–coil domain (41), but we do not yet know which domain of U-STAT2 is responsible for its interaction with p65. However, we do know that the SH2 domain of STAT3 is essential for its interaction with NF-κB (38), suggesting that this domain of STAT2 might bind to p65 similarly.
The STAT1–NF-κB and STAT3–NF-κB complexes are likely to form in the cytosol, and we know that U-STAT2 is required for the translocation of p65 into the nucleus in bone marrow-derived macrophages (19). The EGF that is present in the culture medium for HME cells activates NF-κB (42, 43). However, we did not find a significant difference in the amount of IκBα bound to p65 when we compared HME cells with and without the expression of exogenous U-STAT2 (SI Appendix, Fig. S5C). We conclude that U-STAT2 and IRF9 do not affect the translocation of p65 into the nucleus in HME cells, in contrast to the situation in macrophages. IRF9 and STAT2 are required for enhanced IL6 expression but do not augment the expression of IL1 or IL8 in response to LPS (SI Appendix, Fig. S4 E and F), further supporting our conclusion that the ability of U-STAT2 and IRF9 to enhance NF-κB–dependent gene expression is promoter specific and probably dependent upon the presence of an ISRE element in the promoters of genes that show this cooperation.
We now find that IRF9 occupies a κB element that is very close to the transcription start site of the IL6 gene, and that p65 occupies a putative ISRE at the promoter in cells expressing high levels of both U-STAT2 and IRF9, but not in cells expressing a high level of IRF9 only. Combined with our finding that p65 interacts with U-STAT2 but not IRF9, the data strongly support our hypothesis that STAT2 functions as a bridge connecting p65 and IRF9 on the IL6 promoter. On the other hand, a recent study found that TNF induced the binding of NF-κB to an ISRE, driving the expression of some ISGs in hepatocytes (44). These findings suggest that the U-STAT2–IRF9–p65 complex might be assembled independently of κB elements, and that activation of NF-κB–dependent signaling might increase the formation of this complex, which might occupy ISREs at ISG promoters to drive a modest level of transcription. In summary, the U-STAT2–IRF9–p65 complex drives the transcription of a subset of NF-κB–dependent genes in response to type I and type III IFNs and a subset of ISGs in response to activators of NF-κB.
The Role of Synergistic Activation of IL6 Expression in Cancer.
Considering the important role of IL6 in the tumor microenvironment, we investigated the ability of U-STAT2 and IRF9 to regulate IL6 secretion in cancer cells, finding that decreasing the expression of IRF9 and U-STAT2 inhibited IL6 production and STAT3 activation in lung cancer HCC827 cells (Fig. 4 D and E), where STAT3 activation is dependent on autocrine IL6 (45). As a consequence, reducing the level of IRF9 repressed the growth of these cells (Fig. 4F), which contrasts with the function of ISGF3 as an activator of antiproliferative gene expression. Of note, decreasing IRF9 expression did not inhibit STAT3 activation and cell growth in normal fibroblasts (SI Appendix, Fig. S7). This is not the first time that ISGF3 has been found to promote cancer survival and metastasis. Our previous work showed that U-ISGF3 regulates about a quarter of IFNβ-induced ISGs (5), and that this subset is virtually identical to the set of IFN-induced genes in the interferon-related DNA damage resistance signature (IRDS) (46). In triple negative breast cancer cells, but not in ER-positive cells, the IRDS subset of ISGs is induced by a RIG-1 (DDX58)-dependent antiviral pathway when the cancer cells contact stromal fibroblasts (47). Additional functions of ISGF3 in cancer cell survival are not yet clear. Considering the instability of the cancer genome, ISGF3 might also play an important role in overcoming cell death induced by genomic instability in the process of tumorigenesis. We did not observe basal STAT2 tyrosine phosphorylation in the cancer cells that we studied. Therefore, U-STAT2–IRF9 and U-ISGF3 might be the principal mediators of collaboration with activators of NF-κB in cancer cells, and we did observe that high levels of U-STAT2 and IRF9 correlate with poor prognoses in lung adenocarcinoma. The U-STAT2–IRF9 complex, U-ISGF3, or both, may also interact with transcription factors other than NF-κB to induce the expression of genes that facilitate the growth of cancer cells.
Materials and Methods
Detailed information on cell culture, reagents, constructs in lentiviral vectors, constructs of the IL6 promoter, luciferase assay, real-time PCR, immunoprecipitations, ChIP assays, transfections and luciferase assays, ELISAs, cell survival assay, Kaplan–Meier analysis, and statistical analysis is available in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was funded by National Institutes of Health Grant P01 CA062220 (to G.R.S.), National Natural Science Foundation of China Grant 31571439, and Ocean University of China “Zhufeng Talented Professionals” start-up fund (to J.Y.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714102115/-/DCSupplemental.
References
- 1.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharya S, et al. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 3.Martinez-Moczygemba M, Gutch MJ, French DL, Reich NC. Distinct STAT structure promotes interaction of STAT2 with the p48 subunit of the interferon-alpha-stimulated transcription factor ISGF3. J Biol Chem. 1997;272:20070–20076. doi: 10.1074/jbc.272.32.20070. [DOI] [PubMed] [Google Scholar]
- 4.Majoros A, et al. Canonical and non-canonical aspects of JAK-STAT signaling: Lessons from interferons for cytokine responses. Front Immunol. 2017;8:29. doi: 10.3389/fimmu.2017.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheon H, et al. IFNβ-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J. 2013;32:2751–2763. doi: 10.1038/emboj.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaszczyk K, et al. STAT2/IRF9 directs a prolonged ISGF3-like transcriptional response and antiviral activity in the absence of STAT1. Biochem J. 2015;466:511–524. doi: 10.1042/BJ20140644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bluyssen HA, Levy DE. Stat2 is a transcriptional activator that requires sequence-specific contacts provided by stat1 and p48 for stable interaction with DNA. J Biol Chem. 1997;272:4600–4605. doi: 10.1074/jbc.272.7.4600. [DOI] [PubMed] [Google Scholar]
- 8.Fink K, et al. IFNβ/TNFα synergism induces a non-canonical STAT2/IRF9-dependent pathway triggering a novel DUOX2 NADPH oxidase-mediated airway antiviral response. Cell Res. 2013;23:673–690. doi: 10.1038/cr.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lou YJ, et al. IRF-9/STAT2 [corrected] functional interaction drives retinoic acid-induced gene G expression independently of STAT1. Cancer Res. 2009;69:3673–3680, and erratum (2009) 69:4553. doi: 10.1158/0008-5472.CAN-08-4922. [DOI] [PubMed] [Google Scholar]
- 10.Ho J, et al. STAT2 is a pervasive cytokine regulator due to its inhibition of STAT1 in multiple signaling pathways. PLoS Biol. 2016;14:e2000117. doi: 10.1371/journal.pbio.2000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka T, Narazaki M, Kishimoto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb Perspect Biol. 2014;6:a016295. doi: 10.1101/cshperspect.a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neurath MF, Finotto S. IL-6 signaling in autoimmunity, chronic inflammation and inflammation-associated cancer. Cytokine Growth Factor Rev. 2011;22:83–89. doi: 10.1016/j.cytogfr.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Brooks GD, et al. IL6 trans-signaling promotes KRAS-driven lung carcinogenesis. Cancer Res. 2016;76:866–876. doi: 10.1158/0008-5472.CAN-15-2388. [DOI] [PubMed] [Google Scholar]
- 14.Ancrile B, Lim KH, Counter CM. Oncogenic Ras-induced secretion of IL6 is required for tumorigenesis. Genes Dev. 2007;21:1714–1719. doi: 10.1101/gad.1549407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao SP, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salgado R, et al. Circulating interleukin-6 predicts survival in patients with metastatic breast cancer. Int J Cancer. 2003;103:642–646. doi: 10.1002/ijc.10833. [DOI] [PubMed] [Google Scholar]
- 17.Hoesel B, Schmid JA. The complexity of NF-κB signaling in inflammation and cancer. Mol Cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamero AM, et al. STAT2 contributes to promotion of colorectal and skin carcinogenesis. Cancer Prev Res (Phila) 2010;3:495–504. doi: 10.1158/1940-6207.CAPR-09-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alazawi W, et al. Stat2 loss leads to cytokine-independent, cell-mediated lethality in LPS-induced sepsis. Proc Natl Acad Sci USA. 2013;110:8656–8661. doi: 10.1073/pnas.1221652110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jourdan M, et al. IFN-alpha induces autocrine production of IL-6 in myeloma cell lines. J Immunol. 1991;147:4402–4407. [PubMed] [Google Scholar]
- 21.Ito N, et al. Induction of interleukin-6 by interferon alfa and its abrogation by a serine protease inhibitor in patients with chronic hepatitis C. Hepatology. 1996;23:669–675. doi: 10.1053/jhep.1996.v23.pm0008666316. [DOI] [PubMed] [Google Scholar]
- 22.Hahm B, Trifilo MJ, Zuniga EI, Oldstone MB. Viruses evade the immune system through type I interferon-mediated STAT2-dependent, but STAT1-independent, signaling. Immunity. 2005;22:247–257. doi: 10.1016/j.immuni.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 23.Sarkis PT, Ying S, Xu R, Yu XF. STAT1-independent cell type-specific regulation of antiviral APOBEC3G by IFN-alpha. J Immunol. 2006;177:4530–4540. doi: 10.4049/jimmunol.177.7.4530. [DOI] [PubMed] [Google Scholar]
- 24.Banninger G, Reich NC. STAT2 nuclear trafficking. J Biol Chem. 2004;279:39199–39206. doi: 10.1074/jbc.M400815200. [DOI] [PubMed] [Google Scholar]
- 25.Reich NC. STATs get their move on. JAKSTAT. 2013;2:e27080. doi: 10.4161/jkst.27080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De S, et al. Erlotinib protects against LPS-induced endotoxicity because TLR4 needs EGFR to signal. Proc Natl Acad Sci USA. 2015;112:9680–9685. doi: 10.1073/pnas.1511794112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirano T, Ishihara K, Hibi M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene. 2000;19:2548–2556. doi: 10.1038/sj.onc.1203551. [DOI] [PubMed] [Google Scholar]
- 28.Ogata A, et al. IL-6 triggers cell growth via the Ras-dependent mitogen-activated protein kinase cascade. J Immunol. 1997;159:2212–2221. [PubMed] [Google Scholar]
- 29.Schafer ZT, Brugge JS. IL-6 involvement in epithelial cancers. J Clin Invest. 2007;117:3660–3663. doi: 10.1172/JCI34237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Y, Xu F, Lu T, Duan Z, Zhang Z. Interleukin-6 signaling pathway in targeted therapy for cancer. Cancer Treat Rev. 2012;38:904–910. doi: 10.1016/j.ctrv.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 31.Staudt LM. Oncogenic activation of NF-kappaB. Cold Spring Harb Perspect Biol. 2010;2:a000109. doi: 10.1101/cshperspect.a000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee HJ, et al. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell. 2014;26:207–221. doi: 10.1016/j.ccr.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 33.Kim SM, et al. Activation of IL-6R/JAK1/STAT3 signaling induces de novo resistance to irreversible EGFR inhibitors in non-small cell lung cancer with T790M resistance mutation. Mol Cancer Ther. 2012;11:2254–2264. doi: 10.1158/1535-7163.MCT-12-0311. [DOI] [PubMed] [Google Scholar]
- 34.Duan S, et al. IL-6 signaling contributes to cisplatin resistance in non-small cell lung cancer via the up-regulation of anti-apoptotic and DNA repair associated molecules. Oncotarget. 2015;6:27651–27660. doi: 10.18632/oncotarget.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolosenko I, et al. Cell crowding induces interferon regulatory factor 9, which confers resistance to chemotherapeutic drugs. Int J Cancer. 2015;136:E51–E61. doi: 10.1002/ijc.29161. [DOI] [PubMed] [Google Scholar]
- 36.Rani MR, et al. A role for NF-kappa B in the induction of beta-R1 by interferon-beta. J Biol Chem. 2001;276:44365–44368. doi: 10.1074/jbc.C100417200. [DOI] [PubMed] [Google Scholar]
- 37.Rani MR, Hibbert L, Sizemore N, Stark GR, Ransohoff RM. Requirement of phosphoinositide 3-kinase and Akt for interferon-beta-mediated induction of the beta-R1 (SCYB11) gene. J Biol Chem. 2002;277:38456–38461. doi: 10.1074/jbc.M203204200. [DOI] [PubMed] [Google Scholar]
- 38.Yang J, et al. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev. 2007;21:1396–1408. doi: 10.1101/gad.1553707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiao Y, et al. Synergistic activation of inflammatory cytokine genes by interferon-γ-induced chromatin remodeling and toll-like receptor signaling. Immunity. 2013;39:454–469. doi: 10.1016/j.immuni.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krämer OH, et al. Acetylation of Stat1 modulates NF-kappaB activity. Genes Dev. 2006;20:473–485. doi: 10.1101/gad.364306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rengachari S, et al. 2017. Structural basis of STAT2 recognition by IRF9 reveals molecular insights into ISGF3 function. bioRxiv:10.1101/131714.
- 42.Habib AA, et al. The epidermal growth factor receptor engages receptor interacting protein and nuclear factor-kappa B (NF-kappa B)-inducing kinase to activate NF-kappa B. Identification of a novel receptor-tyrosine kinase signalosome. J Biol Chem. 2001;276:8865–8874. doi: 10.1074/jbc.M008458200. [DOI] [PubMed] [Google Scholar]
- 43.Sun L, Carpenter G. Epidermal growth factor activation of NF-kappaB is mediated through IkappaBalpha degradation and intracellular free calcium. Oncogene. 1998;16:2095–2102. doi: 10.1038/sj.onc.1201731. [DOI] [PubMed] [Google Scholar]
- 44.Wang W, et al. Convergent transcription of interferon-stimulated genes by TNF-α and IFN-α augments antiviral activity against HCV and HEV. Sci Rep. 2016;6:25482. doi: 10.1038/srep25482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song L, Rawal B, Nemeth JA, Haura EB. JAK1 activates STAT3 activity in non-small-cell lung cancer cells and IL-6 neutralizing antibodies can suppress JAK1-STAT3 signaling. Mol Cancer Ther. 2011;10:481–494. doi: 10.1158/1535-7163.MCT-10-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weichselbaum RR, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci USA. 2008;105:18490–18495. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boelens MC, et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell. 2014;159:499–513. doi: 10.1016/j.cell.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.