Significance
Widespread conversion of natural habitats to human land use creates evolutionarily novel environments and causes declines of native species. Stemming biodiversity loss requires an understanding of why some species persist while others decline in these novel habitats. We analyzed survey data of amphibian species from around the globe to determine whether closely related species respond similarly to habitat conversion. We find that species that persist in converted habitats tend to come from the same clades within the amphibian tree of life and that by favoring these widely distributed clades, habitat conversion leads to nonrandom extirpations and loss of evolutionary history. Our results show that the identity of winners and losers during the Anthropocene can be tightly linked to their evolutionary history.
Keywords: habitat loss, land use, Anthropocene biogeography, biodiversity, phylogenetic generalized linear mixed model
Abstract
Habitat conversion is driving biodiversity loss and restructuring species assemblages across the globe. Responses to habitat conversion vary widely, however, and little is known about the degree to which shared evolutionary history underlies changes in species richness and composition. We analyzed data from 48 studies, comprising 438 species on five continents, to understand how taxonomic and phylogenetic diversity of amphibian assemblages shifts in response to habitat conversion. We found that evolutionary history explains the majority of variation in species’ responses to habitat conversion, with specific clades scattered across the amphibian tree of life being favored by human land uses. Habitat conversion led to an average loss of 139 million years of amphibian evolutionary history within assemblages, high species and lineage turnover at landscape scales, and phylogenetic homogenization at the global scale (despite minimal taxonomic homogenization). Lineage turnover across habitats was greatest in lowland tropical regions where large species pools and stable climates have perhaps given rise to many microclimatically specialized species. Together, our results indicate that strong phylogenetic clustering of species’ responses to habitat conversion mediates nonrandom structuring of local assemblages and loss of global phylogenetic diversity. In an age of rapid global change, identifying clades that are most sensitive to habitat conversion will help prioritize use of limited conservation resources.
Human populations have established nonnative habitats across the Earth that represent novel environments for biodiversity (1–3). Conversion of natural habitats occurs on short time scales—typically over few generations—relative to the often-vast time scales of species’ niche evolution (4). As ecological communities are increasingly exposed to rapid habitat conversion, many species decline in altered habitats, whereas others persist or even thrive. This variation in response to habitat conversion is poorly understood and may result, in part, from both the environmental conditions experienced by assemblages and the shared evolutionary history of species. If closely related species exhibit similar responses to altered environments, then habitat conversion could precipitate losses of entire clades from local assemblages (5).
Habitat conversion favors some species that are fortuitously preadapted to human-altered habitats while threatening others (6). An explicitly phylogenetic perspective is crucial because extinction risk and sensitivity to habitat conversion are ultimately determined by species traits, which often reflect the shared evolutionary histories of vulnerable species (7–11). For example, frugivorous and insectivorous birds, cool-adapted and terrestrial-developing amphibians, and small-bodied bats often have reduced abundances in converted habitats (10, 12–17). However, the highly intercorrelated nature of many species traits means that clearly identifying specific traits that underlie tolerance is challenging, thereby limiting prediction. Phylogenetic relatedness acts as a useful proxy by integrating across all phylogenetically conserved traits, including those that may govern responses to habitat conversion.
The cumulative outcome of environment-by-trait sorting and species interactions in altered habitats generates changes in local diversity (18). Diversity is usually considered solely from the taxonomic (i.e., species identity) perspective, without considering the phylogenetic relationships among species. Patterns of taxonomic α-diversity are highly idiosyncratic, however, with different studies reporting increased, decreased, or unchanged species richness following habitat conversion (3, 19). Local responses can vary, in part, depending on the type of habitat alteration, with converted habitats that structurally contrast with natural habitats (e.g., intensive monocultures) often causing substantial decreases in local richness (3, 20). The magnitude of change in phylogenetic α-diversity (the total evolutionary history contained within a local community) can mirror or deviate from that of taxonomic α-diversity (21), depending on the shape of the community phylogeny and the relatedness of species in each habitat (22). We currently know little, however, about the phylogenetic relatedness of animal species that persist in converted habitats or how these species are distributed across the tree of life (but see recent studies of birds and bats at local and landscape scales; refs. 17, 21, and 23).
At landscape scales, negligible changes in species richness can obscure substantial shifts in species composition between natural and converted habitats (24), which can arise from the concurrent loss of species that depend on natural habitats and the gain of species that can exploit converted habitats (20). Taxonomic β-diversity measures the degree to which sites differ in species composition. If habitat conversion consistently benefits the same set of species across broad spatial scales, then loss of β-diversity through taxonomic homogenization can occur, as assemblages in distant areas become increasingly similar (sensu refs. 25 and 26). In turn, phylogenetic β-diversity measures the degree to which evolutionary history is shared across sites, such that phylogenetically unique species contribute more to community differentiation than species with many close relatives. Importantly, even if distinct areas of the globe share no species in common, phylogenetic homogenization can still occur if habitat conversion repeatedly benefits different species in the same clade. Measures of taxonomic and phylogenetic β-diversity can each be partitioned into turnover, which measures the loss and gain of unique species or lineages, and nestedness, which measures the differentiation among sites resulting from changes in α-diversity. By linking patterns of taxonomic diversity to evolutionary history, measures of phylogenetic β-diversity can provide new insights into how communities are being restructured at multiple spatial scales in the face of habitat conversion (21, 27–29).
In addition to the evolutionary history of species, regional environmental conditions can mediate community responses to habitat conversion. For example, negative effects of habitat loss and fragmentation on α-diversity appear to be most pronounced in warm, tropical climates (30), possibly owing to large increases in temperatures and thermal stress in fragments and the surrounding converted habitats (31–34). Similarly, we expect that taxonomic and phylogenetic β-diversity between natural and converted habitats vary with latitude and elevation because regional climate will affect the magnitude of microclimatic differences between natural and converted habitats. For example, differences in temperature between forest and pastures increase with decreasing elevation (and increasing regional temperatures) in Central America (15), thereby modifying steepness of habitat suitability gradients for ectotherms. High degrees of climatic niche specialization among tropical clades (e.g., narrow thermal tolerance breadths) could further intensify environmental filtering and lineage turnover in converted habitats in the tropics (35–37). Finally, the degree of landscape modification in a region may determine the consequences of local habitat conversion on both α- and β-diversity by enriching the regional pool for species that are tolerant of habitat conversion, while eliminating sensitive taxa or lineages.
Here, we quantified patterns of taxonomic and phylogenetic diversity of amphibians affected by habitat conversion across a range of converted-habitat types and biogeographic contexts. We compiled amphibian assemblage data from natural and converted habitats from published studies, and we mapped responses onto a large-scale phylogeny to address the following questions: (i) Do closely related species exhibit similar responses to habitat conversion, or are contrasting habitat affiliations interspersed randomly throughout the tree of life? (ii) Are patterns of phylogenetic α- and β-diversity concordant with those of taxonomic α- and β-diversity in converted habitats, and do these patterns depend upon regional environmental context? (iii) Are amphibian assemblages being homogenized, as evidenced by decreased β-diversity, in converted habitats along taxonomic and phylogenetic axes of biodiversity? By addressing these questions, we present a global analysis linking changes in amphibian abundance and diversity in converted habitats to the shared evolutionary history of species.
Results
Species Responses.
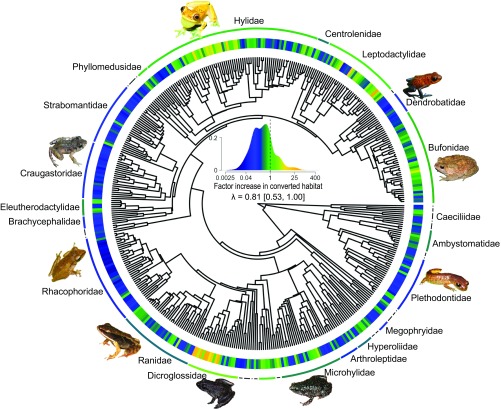
Species varied widely in their responses to habitat conversion. Based on a Bayesian implementation of a phylogenetic generalized linear mixed model (GLMM), 354 of the 438 species included in the analysis decreased in abundance (posterior mean response to habitat conversion <0), whereas 84 species increased in abundance when natural habitats were converted to pastures, plantations, agriculture, or clearcuts (posterior mean response >0). These responses to habitat conversion exhibited strong phylogenetic signal (Pagel’s λ = 0.81) [95% highest posterior density (HPD) interval: 0.53–1.00; Fig. 1]. Among the clades most affiliated with converted habitats were the Asian grass frogs of the genus Fejervarya (Dicroglossidae), which are common in rice agriculture (mean expected abundance increase of 13.2× in converted habitats), and neotropical treefrogs, such as species of Dendropsophus (Hylidae; mean expected increase of 2.4×), that often breed in still waters in cattle pastures. Clades strongly affiliated with natural habitats included genera with numerous species that forego an aquatic larval stage and often breed on vegetation or in leaf litter, such as salamanders in the genus Bolitoglossa (Plethodontidae; mean expected decrease of 4.3× in converted habitats), paleotropical frogs in Pseudophilautus (Rhacophoridae; expected decrease of 27.7×), as well as neotropical frogs in Craugastor (Craugastoridae; expected decrease of 9.3×) and Pristimantis (Strabomantidae; expected decrease of 6.2×). Furthermore, some large clades exhibited mixed responses or contained many generalist species with no strong response (Fig. 1), such as New World frogs of the family Leptodactylidae (mean expected abundance change of 1.0×, ranging from a decrease of 22.8× to an increase of 36.9×), the widely distributed true frogs of the family Ranidae (mean decrease of 2.7×, ranging from a decrease of 13.8× to an increase of 3.9×), and the true toads of Bufonidae (mean decrease of 1.7×, ranging from a decrease of 8.9× to an increase of 6.1×).
Fig. 1.
Species’ responses to habitat conversion are conserved in the amphibian phylogeny. Phylogenetic signal (λ and 95% HPD) was estimated by using a Bayesian implementation of a phylogenetic GLMM. The color gradient indicates effect sizes from the model; blue shades indicate species that were most abundant in natural habitats, green indicates species that were similarly abundant in natural and converted habitats, and yellow shades indicate species that were most abundant in converted habitats. All families represented by five or more species are labeled, and the color of arcs represent mean family responses. Among the clades most affiliated with converted habitats were neotropical hylids, such as species from the genus Dendropsophus, as well as species of Fejevarya (Dicroglossidae) from southeastern Asia. Among the clades that were most sensitive to habitat conversion were species from the neotropical families Craugastoridae and Dendrobatidae, as well as species of Rhacophoridae from southern Asia.
Impacts of Habitat Conversion on Taxonomic and Phylogenetic α-Diversity.
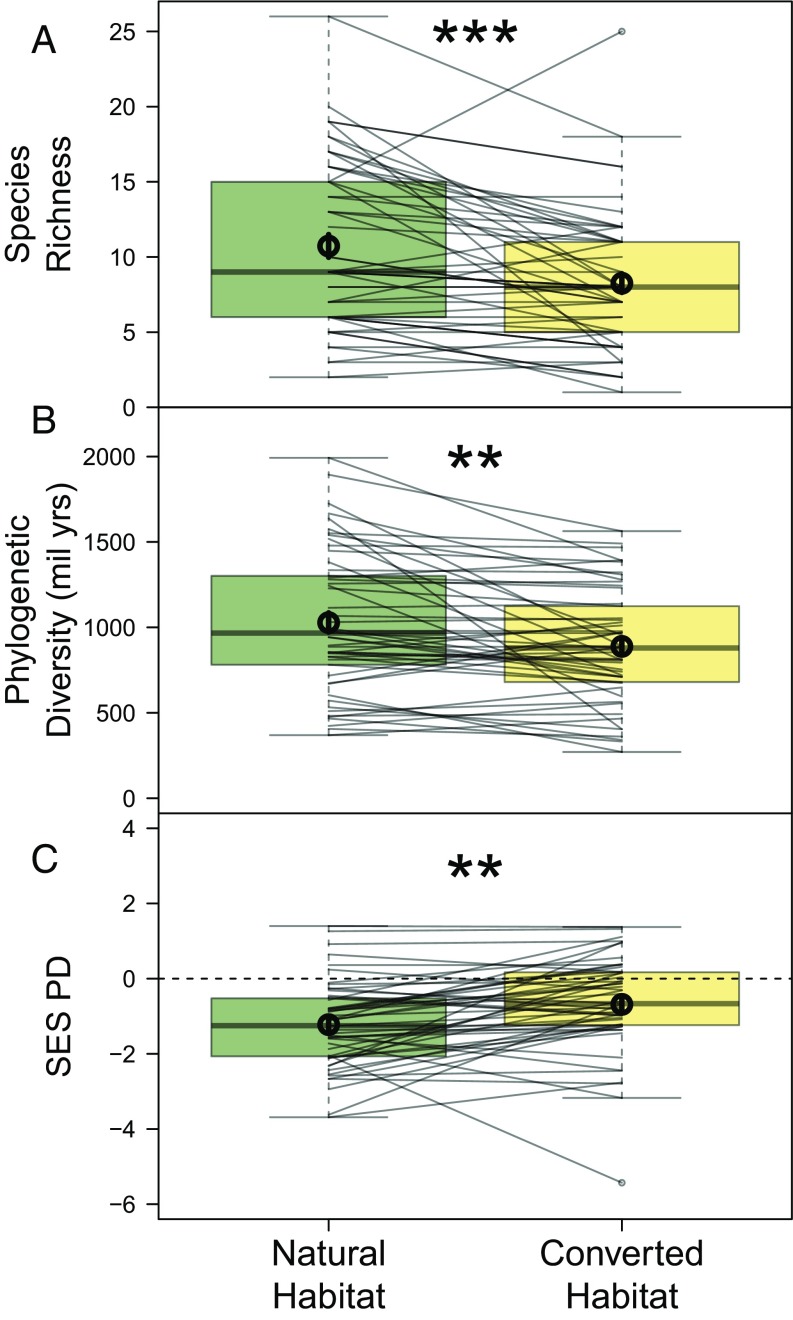
Within studies, habitat conversion on average resulted in a 22% decrease in species richness, constituting a loss of 2.4 species (95% CI = [1.2, 3.6], df = 1, χ2 = 17.1, P < 0.001; Fig. 2A) from a mean of 10.7 species within natural habitat assemblages. Despite this average trend, responses from individual studies varied considerably, with 17% exhibiting no change and 17% having greater species richness in converted habitats. Phylogenetic diversity (PD) loss followed similar patterns. On average, habitat conversion resulted in the local loss of 139 My of amphibian evolutionary history (95% CI = [63, 215], df = 1, χ2 = 10.9, P = 0.001, proportion significant (sig.) from sample of posterior trees = 1; Fig. 2B), such that PD decreased by 13.5% from a mean of 1.03 billion years of evolutionary history within natural-habitat assemblages (study-level assemblages, including species in both natural and converted-habitats, contained 1.21 billion years of evolutionary history on average). Once again, however, individual studies varied widely, with 4% of study comparisons displaying no change and 30% showing increased PD in converted habitats.
Fig. 2.
Mean α-diversity declines in converted habitats. Both taxonomic (A) and phylogenetic (B) α-diversity decline with habitat conversion, yet PD declines less than expected based on species richness loss (C; SES PD is PD standardized by species richness), indicating that species affiliated with converted habitats come from across the phylogenetic tree. Box plots depict medians, interquartile ranges, and full ranges and are overlaid with a circle representing the mean (error bars representing 95% CIs are included, but are too small to see). Thin lines connect values within the same study. Significant differences are shown by asterisks. **P < 0.01; ***P < 0.001.
After controlling for differences in species richness, both natural and converted habitats had less PD than expected if communities were assembled randomly with respect to phylogeny. Given the observed species richness, the average natural-habitat site had 211 million fewer years of evolutionary history than expected at random (P < 0.001, proportion sig. from sample of posterior trees = 1), whereas the average converted-habitat site had 120 million fewer years (P < 0.001, proportion sig. from sample of posterior trees = 1). Interestingly, after controlling for species richness, converted habitats had relatively greater PD than natural habitats. This result suggests that natural habitats, such as forest, support a larger number of closely related species, whereas converted habitats more thoroughly sample the total phylogenetic space, even though they often have fewer species to do so (df = 1, χ2 = 9.2, P = 0.002, proportion sig. trees = 1; Fig. 2C).
While assemblages in both natural and converted habitats showed patterns of overall phylogenetic clustering (Fig. S1), mean phylogenetic distance (MPD) was not different between habitats (df = 1, χ2 = 2.4, P = 0.12, proportion sig. trees = 0). Mean nearest taxon distance (MNTD), however, was diminished in natural-habitat assemblages (df = 1, χ2 = 13.8, P < 0.001, proportion sig. trees = 1), indicating that the greater species richness in natural habitats results, in part, from the presence of a greater number of recently diverged species than in converted habitats. In all cases, the levels of decline in taxonomic and phylogenetic α-diversity between natural and converted habitats were statistically indistinguishable among the different types of habitat conversion (i.e., agriculture vs. pasture, etc.; for all tests: df = 3, all χ2 < 3.7, all P > 0.29; Fig. S2). All biogeographic variables (e.g., regional temperature, precipitation, etc.) were nonsignificant (P > 0.05) in analyses of interhabitat differences in α-diversity across studies. There was no evidence of spatial nonindependence of model residuals (Fig. S3) or directional biases associated with sampling effort (Fig. S4).
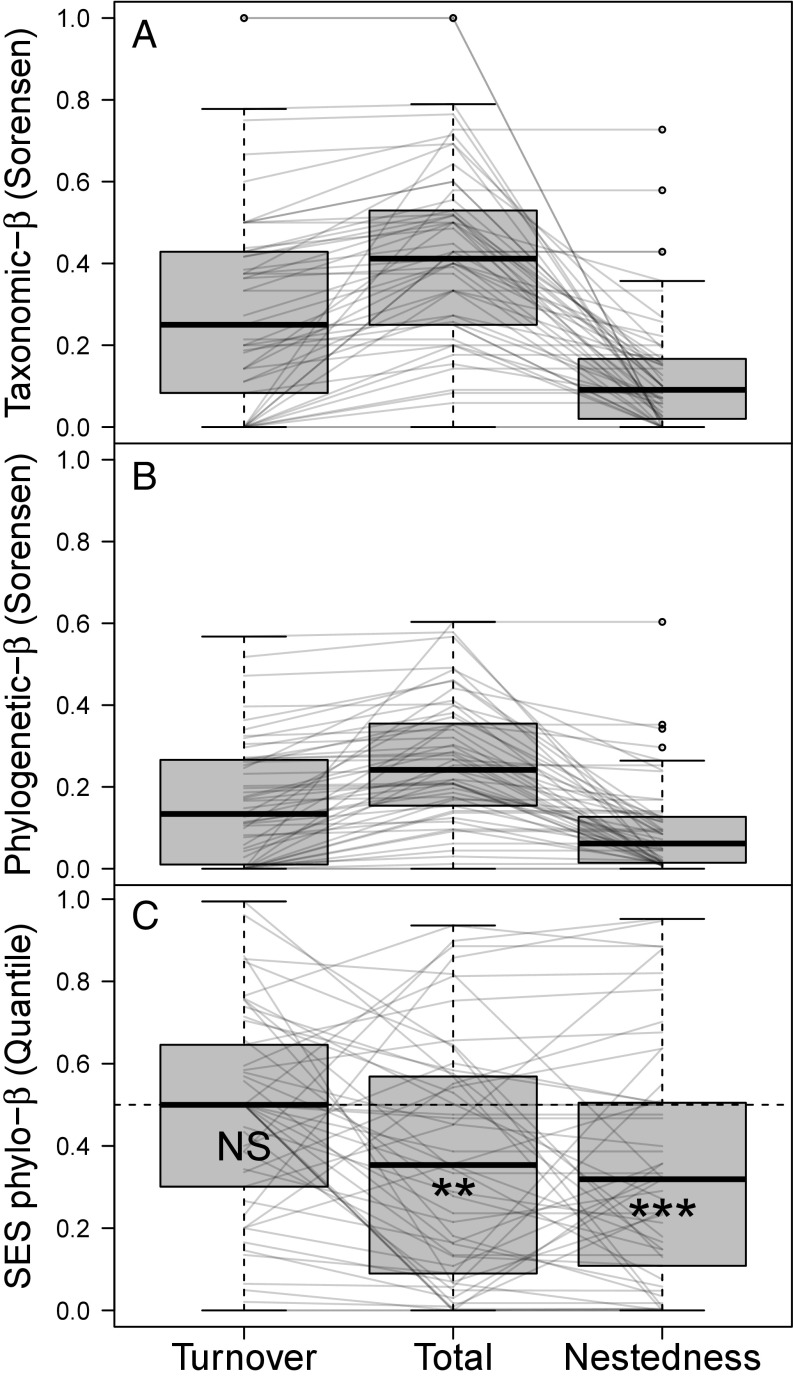
Impacts of Habitat Conversion on Taxonomic and Phylogenetic β-Diversity Within Study Regions.
Overall taxonomic and phylogenetic β-diversity between natural and converted habitats varied widely throughout the dataset, in some cases representing entirely nonoverlapping species assemblages, and in other cases, assemblages were identical. However, despite this variation in total taxonomic β-diversity, turnover—the loss and gain of unique species—tended to play a larger role than nestedness—the differentiation attributable to changes in species richness (turnover comprised 61% of total β-diversity, 95% CI: [51%, 71%]; Fig. 3A). In contrast, total phylogenetic β-diversity was, on average, composed of roughly equal portions of turnover and nestedness (turnover comprised 56%, 95% CI: [45%, 66%]; Fig. 3B). As a result, study sites were on average less phylogenetically differentiated between natural and converted habitats than expected based on their taxonomic differentiation, if species composition were random with respect to phylogeny (Fig. 3C). This effect did not vary strongly by converted-habitat type (Fig. S5) and was largely attributable to the nestedness component of phylogenetic β-diversity. Lower-than-expected nestedness indicated that, while phylogenetic α-diversity differed between habitats within a study, this difference was less than expected given changes in taxonomic diversity, signaling that species additions in natural habitats come predominantly from shallow positions in the tree.
Fig. 3.
Average β-diversity between natural and converted habitats (Sorensen dissimilarity index). (A and B) Total β-diversity is partitioned into turnover and nestedness components for both taxonomic (A) and phylogenetic (B) β-diversity. (C) However, comparing observed values to a null model distribution shows that there is less total phylogenetic differentiation between natural and converted habitats than expected based on changes in taxonomic species composition. This is primarily attributed to especially low levels of phylogenetic nestedness between habitats, indicating that within regions, relatively closely related species are found in both natural and converted habitats. Thin lines connect values within the same study. In C, significant deviations from neutral expectation are shown by asterisks. **P < 0.01; ***P < 0.001. NS, not significant.
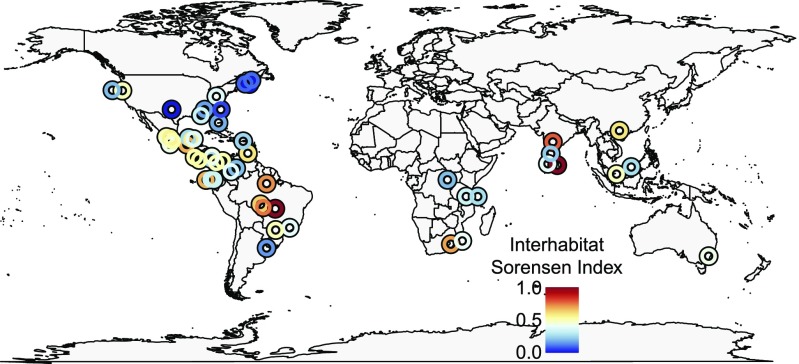
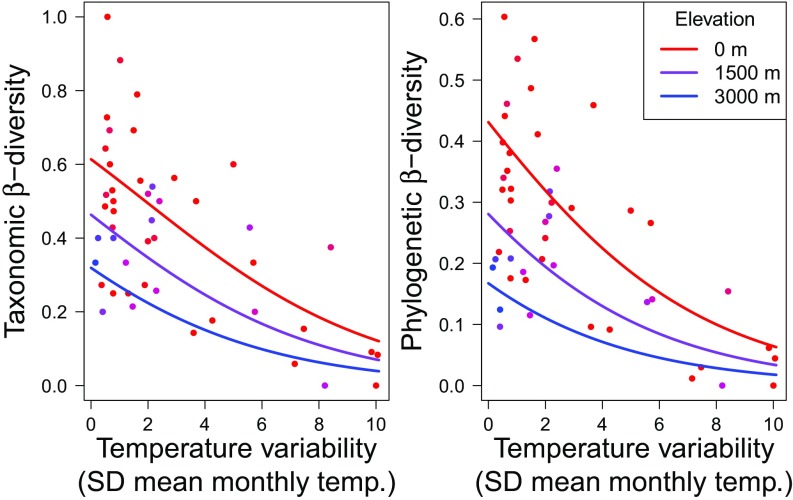
The greatest between-habitat community dissimilarity typically occurred in tropical regions, whereas communities in natural and converted habitats tended to be quite similar in temperate zones (Fig. 4). When considering environmental variables, the best-fit model indicated that differentiation between natural and converted habitats increased with decreasing temperature variability and elevation (Fig. 5 and Tables S1 and S2). For example, in the most temperate regions at high elevations, >95% of species were shared between habitats, whereas in the most equatorial climate zones at sea level, only 40% of species were shared. This effect remained significant even when species richness was included as a covariate, suggesting that observed patterns cannot be attributed solely to the effect of latitudinal species richness gradients.
Fig. 4.
Map of taxonomic β-diversity (Sorensen dissimilarity index) between natural and converted habitats. Points are jittered to show overlapping locations.
Fig. 5.
Total taxonomic and phylogenetic β-diversity between natural and converted habitats within each study decreases with increasing annual temperature variation and at higher elevations. Lines represent best-fit expectations at different elevations above sea level.
Global β-Diversity in Natural and Converted Habitats.
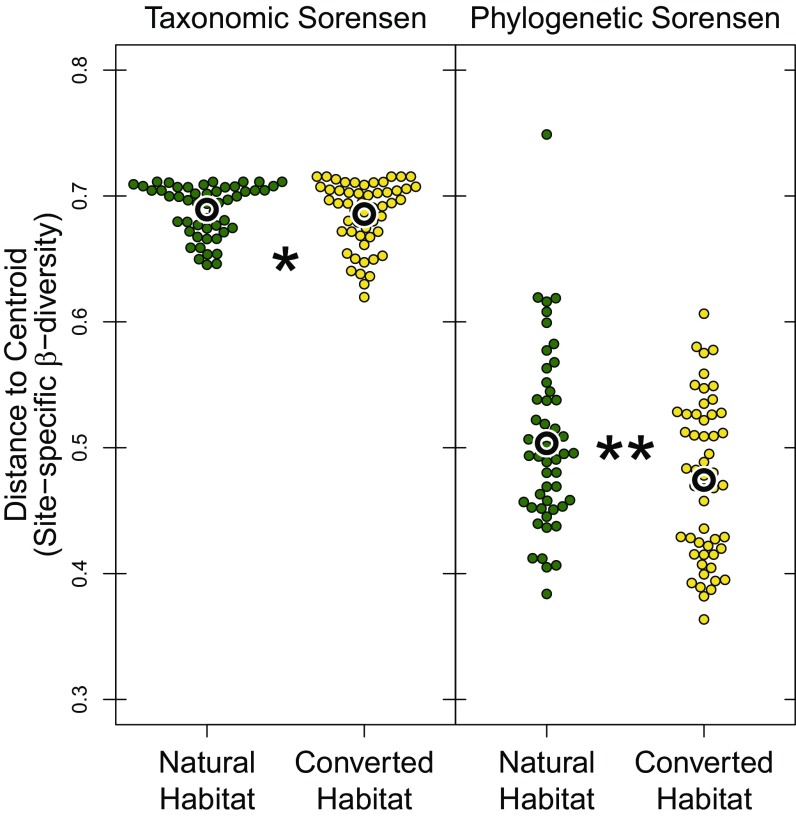
Biotic homogenization at the global scale was apparent in terms of taxonomic diversity and PD, although the effect size, especially for taxonomic homogenization, was small (taxonomic: df = 1, χ2 = 4.2, P = 0.04; phylogenetic: df = 1, χ2 = 10.0, P = 0.002; proportion sig. trees = 1; Fig. 6). Assemblages in converted habitats were 0.6% more taxonomically similar to themselves than were assemblages in natural habitats (based on mean pairwise Sorensen dissimilarity index), but shared 4% more of their phylogenetic history than did assemblages in natural habitats. The greater loss of phylogenetic β-diversity in converted habitats persisted when directly controlling for taxonomic differences (SES phylo β, χ2 = 35.6, P < 0.001, proportion sig. trees = 1), or when considering only the true turnover component of taxonomic (df = 1, χ2 = 5.1, P = 0.024) and phylogenetic (df = 1, χ2 = 8.0, P = 0.005, proportion sig. trees = 1) β-diversity. Total γ-diversity declined as a result of concurrent decreases in phylogenetic α- and β-diversity. Our global sample of 438 species comprised a total of 18.3 billion years of unique evolutionary history, of which 16.3 billion years (89% of total) occurred in natural habitats and only 12.7 billion years (69%) occurred in converted habitats. In our sample, habitat conversion resulted in a decline of 3.6 billion years, diminishing PD by 22%.
Fig. 6.
Converted habitats across the globe have less β-diversity than natural habitats. Taxonomic β-diversity is slightly lower in converted habitats, although habitat identity explains very little of the total variation (marginal R2 = 0.007). Phylogenetic β-diversity differences between habitats are greater (marginal R2 = 0.05), suggesting that, across the globe, some clades that are able to use converted habitats are widely distributed, even if the constituent species are not. Significant differences are shown by asterisks. *P < 0.05; **P < 0.01.
Discussion
As global environmental change reshapes and diminishes biodiversity, it is imperative to predict not only the magnitude of species loss but also which groups will decline and which will persist (38, 39). We found that phylogenetic relatedness explained most of the variation in amphibian species responses to anthropogenic habitat conversion, one of the primary threats to amphibian biodiversity (40). As a result, habitat conversion is selecting for new species pools within each region of the globe, which often have reduced taxonomic diversity and PD. Even though phylogenetic α-diversity did not decline as much as expected given decreases in species richness, species replacement across sites was dominated by a reoccurring subset of phylogenetically clustered taxa that are robust to habitat conversion. Ultimately, the shared evolutionary history of species contributes to the nonrandom structuring of local assemblages following habitat conversion and to the homogenization of amphibian biotas globally.
The strong phylogenetic signal in sensitivity to habitat conversion presumably stems from phylogenetically conserved traits (10, 41). Notably, species-rich clades of frogs with terrestrial development tended to be especially sensitive to habitat conversion. For example, clades that were strongly dependent upon natural habitats included direct developers (species that forego a free-living larval stage) such as lungless salamanders of the family Plethodontidae; the neotropical frog genus Pristimantis (Strabomantidae), which includes >500 described species; and the frog genus Pseudophilautus (Rhacophoridae), which is endemic to the Western Ghats–Sri Lanka biodiversity hotspot and is experiencing high rates of extinction (42, 43). In contrast, clades characterized by aquatic larval development, especially those breeding in still waters, were often more tolerant of habitat conversion, such as the neotropical hylids and members of the families Leptodactylidae, Bufonidae, and Dicroglossidae. These species likely benefit from the creation of novel stillwater bodies in human-modified habitats (e.g., cattle ponds, irrigation ponds, or rice agriculture). Aquatic breeding is the ancestral form of reproduction in class Amphibia, and terrestrial development has evolved multiple times across the amphibian phylogeny (44).
Terrestrial development may often restrict local distributions to humid forests, as the global distributions of these taxa are often constrained to wet tropical regions (44). Terrestrial-breeding species typically have reduced clutch sizes, which likely evolved as a tradeoff to the production of large eggs that are less prone to desiccation on land. However, terrestrial eggs still require humid conditions for development, such as moist leaf litter found in forests (45). Because these strongly forest-affiliated taxa tend to have small clutch sizes, they may be especially vulnerable to landscape-level habitat conversion and fragmentation effects (46). In addition, terrestrial-developing species frequently have small bodies with high surface-to-volume ratios and low heat tolerances (14, 47, 48), increasing the risk of desiccation and thermal stress in warm, open-canopy habitats (32, 49, 50). These or other phylogenetically conserved traits likely contribute to the observed changes to amphibian assemblages following habitat conversion (10, 41).
Habitat conversion erodes multiple dimensions of amphibian biodiversity. At the local scale, mean taxonomic and phylogenetic α-diversity decreased in converted habitats by 22 and 13.5%, respectively. Furthermore, the amount of PD in both habitat types was less than expected based on species richness. This pattern, in concert with the phylogenetic signal in habitat affiliation across the global phylogeny (Fig. 1), suggests that species that co-occur in each habitat are more related than expected by chance, likely resulting from environmental filtering via conserved niches (ref. 4; but see ref. 51). In contrast, bird assemblages were phylogenetically overdispersed in forest habitats in Costa Rica, whereas agriculture-affiliated species were more phylogenetically clustered (21), patterns that perhaps reflect different roles of density-dependent interactions in structuring bird and amphibian assemblages in forest. The magnitude of decrease was similar for both taxonomic and phylogenetic α-diversity of amphibians, indicating that while there are typically fewer species in converted habitats, these species represent clades scattered throughout the phylogeny. The additional species found in natural habitats tended to be more closely related to one another at the tips (lower MNTD), whereas relatedness in each habitat was roughly equivalent when considering deeper phylogenetic time scales (similar MPD). These patterns of community tree shape may be attributable, in part, to the ancestral state of aquatic breeding strategies, which tend to be relatively successful in converted habitats, vs. the more recent emergence of terrestrial development, which appears less successful in converted habitats (10, 44).
Taxonomic and phylogenetic β-diversity provided complementary perspectives on the effects of habitat conversion at regional and global scales. Within study regions, species turnover drove changes in taxonomic composition across habitats to a greater extent than nestedness, suggesting that species in natural habitats were often replaced by disturbance-tolerant or agriculture-affiliated species (20, 52). Changes in phylogenetic composition were driven similarly by turnover and nestedness (Fig. 3B), reflecting shallow PD gradients as well as the gain and loss of unique lineages across habitats (Fig. 3C). However, because habitat affiliations were clustered at the tips but broadly distributed across deeper branches (Fig. 1), species stemming from evolutionarily divergent clades often co-occurred in both habitats. These changes in α-diversity and regional composition culminated in phylogenetic homogenization at the global scale. Globally, habitat conversion led to a sevenfold greater decrease in phylogenetic β-diversity than taxonomic β-diversity (Fig. 6). The greater magnitude of decline in phylogenetic β-diversity indicates that, even though different regions infrequently share species, the same widely distributed clades tend time and again to benefit from the conversion of natural habitats. The observed losses of phylogenetic α- and β-diversity were compounded at the global scale, resulting in a 22% decrease in total γ-diversity in converted habitats across our sample.
The geographic context in which habitat conversion occurs likely interacts with the evolutionary origins of the species themselves to influence winners and losers in local assemblages (30, 53). Such a dynamic could mean that habitat conversion has strikingly different results across biogeographic regions. Indeed, among amphibian assemblages, both taxonomic and phylogenetic β-diversity between natural and converted habitats increased with decreasing temperature variation toward the tropics and with decreasing elevation (Fig. 5). These results extend the hypothesis that specialization among tropical clades gives rise to greater β-diversity in the tropics (35, 54, 55). Just as “mountain passes are higher in the tropics,” boundaries between natural and converted habitats also likely exhibit greater contrast in the tropics. At high latitudes, differences in temperature between natural (typically forest) and converted habitats are relatively small compared with the thermal extremes experienced over the course of a year. This should lead to less thermal habitat partitioning in open- vs. closed-canopy habitats among temperate-zone amphibians and, therefore, smaller community shifts when habitats are disturbed. Furthermore, that increasing elevation moderates differentiation between natural- and converted-habitat assemblages suggests that converted habitats represent an especially-selective filter in the lowland tropics. Thermal niche tracking, for example, can lead tropical ectotherms to shift habitat affiliation from cool forests in the lowlands to warm, converted habitats at higher elevations (15, 56).
The observed changes in multiple dimensions of amphibian diversity were robust to phylogenetic uncertainty and reflect a broad geographic set of community samples. These results, nevertheless, should be viewed in light of several assumptions and potential sources of variation. First, geographic sampling biases in the literature resulted in the underrepresentation of studies from regions such as northern Africa and Asia. Despite these geographic gaps, the results here include a large sample of species that represent amphibian trait variation across the globe (Fig. S6). Second, by analyzing observational studies that substitute space for time, we assume that patterns of β-diversity between natural and converted habitats reflect a signature of habitat conversion rather than solely preexisting factors, such as differences in topography among sites; for example, flat lands are often preferred for agriculture (57). However, our analysis of β-diversity across many studies, along with spatial replication and interspersion of natural and converted sites within studies, should mitigate uncertainty when identifying habitat conversion as a factor influencing β-diversity (Fig. S7). Finally, community tree topology and associated variation in diversification rates among clades can affect the magnitude of PD loss resulting from nonrandom local extinctions (58). Different community tree topologies, therefore, may have contributed to the substantial variation in PD loss that we observed across studies (Fig. 2). Accounting for community tree shape, especially when there is strong phylogenetic signal in extinction risk, will be necessary when developing strategies to reduce loss of evolutionary history from assemblages.
Identifying patterns of PD in converted habitats can have core ramifications for conservation. For instance, the distribution of habitat associations in the phylogeny determines our ability to maintain assemblages that are phylogenetically representative of native, natural-habitat species pools. For amphibians, a large proportion of species and lineages are present in human-altered landscapes, occurring in forest remnants and areas of agriculture, which presents important opportunities for maintaining multiple dimensions of amphibian diversity outside of protected areas. At the same time, many species are highly dependent on natural habitats and often have restricted geographic ranges (10) that do not overlap with current protected areas, requiring targeted habitat protection to close a substantial conservation gap for these sensitive clades (59, 60).
An approach that attempts to balance both increased habitat protection (e.g., through community-managed forest reserves; ref. 61) and diversified agricultural systems (e.g., through payments for environmental services; ref. 62) would likely be necessary to maintain diversity across the amphibian tree of life. Maintenance of remnant natural vegetation, such as small groves of trees, in human-modified landscapes can maximize species richness by allowing some forest-dependent species to persist and can increase overall community similarity to protected reserves (63). For example, the forest-affiliated strawberry poison frog (Oophaga pumilio) is able to persist in pastures, but it does so almost exclusively by occupying the buttresses of scattered, single trees rather than occurring in open-pasture habitat (20, 64). There are necessary tradeoffs, however, in allocating resources toward wildlife-friendly agriculture or reserve protection that have consequences for PD (23). Conservation planning should, therefore, explicitly account for these tradeoffs and integrate strategies when possible (e.g., through optimizing land sharing and sparing strategies; refs. 65 and 66) to maximize taxonomic and PD of focal groups.
In an era of rapid global change, we are challenged with stemming the loss of biodiversity, which requires an understanding of why some species persist while others decline. Bringing a phylogenetic approach to bear on community responses to habitat conversion will help identify lineages that are sensitive or robust to this pervasive threat. Human activities that convert natural habitats into systems for producing food and fiber have inadvertently pruned the amphibian tree of life into a smaller, less diverse topiary by favoring a relatively few tolerant clades. By identifying which clades are most sensitive to habitat conversion, we can better guide prioritization of conservation assessments and fill conservation gaps through targeted reserve acquisition and diversified agricultural practices, particularly as many new species are described each year and are data-deficient (60, 67). Together, our results show that strong phylogenetic dependence of species’ responses to habitat conversion helps explain nonrandom structuring of local assemblages and global patterns of biotic homogenization. The identity of winners and losers during the Anthropocene are, therefore, inextricably linked to their evolutionary history.
Materials and Methods
Data Collection.
We searched the literature using Institute for Scientific Information Web of Science and Google Scholar to compile amphibian species counts in natural habitats and in nearby converted habitats, as reported from published field surveys (detailed search methods are provided in SI Materials and Methods and ref. 10). We extracted observed species abundances from data tables and figures for all studies that sampled amphibian assemblages in natural and converted habitats in the same landscape, that used standardized sampling methods, and that reported sampling effort and replication. For most studies in the dataset (45 studies; 94%), “natural habitats” refers to forest; however, we included three studies for which the natural habitat type is undisturbed wetland or grassland. Removing these nonforest studies from the dataset did not affect the results. To examine potential differences among converted-habitat types, we categorized converted habitats as agriculture, pastures, tree plantations, or clear cuts. The dataset included 52,887 individual observations of 438 species reported from 48 studies on 5 continents (available in Datasets S1 and S2).
Phylogenetic Tree.
We used a time-calibrated phylogenetic tree generated by Pyron (68). This tree contains 3,309 taxa, representing ∼42% all known amphibian diversity. Of the 438 taxa in the community dataset, 369 were present in the phylogeny. To analyze the full community dataset, we used the PASTIS species imputation method to generate a posterior distribution of trees while constraining placement of the remaining 69 species within the relevant genus of each species (ref. 69; additional details are provided in SI Materials and Methods). We sampled 100 of these trees to account for species placement and phylogenetic uncertainty in downstream analyses. To examine the influence of choice of imputation method, we alternatively placed the missing species into the full Pyron (68) tree by randomly inserting branches along the subtree representing the genus of each species. The probability of insertion at any particular branch was proportional to the length of that branch, and the tip addition was equally probable along any point of the focal branch. We repeated this procedure 100 times to generate a series of pseudoposterior trees for analysis. Random placement of species into phylogenies can lead to downward-biased estimates of phylogenetic signal (70); so, we note that estimates presented here represent lower bounds of true phylogenetic signal. Because taxa were constrained by genus, the most variable portion of the phylogeny existed only in extremely shallow evolutionary time. As a result, the average correlation coefficient between cophenetic distances across all pairwise comparisons of posterior and pseudoposterior trees was extremely high (mean ρ = 0.9953, range 0.9842–0.9993).
Phylogenetic Signal in Response to Habitat Conversion.
To estimate the degree to which response to habitat conversion was conserved in the amphibian phylogeny, we used a Bayesian implementation of a phylogenetic GLMM (71, 72) that integrated over uncertainty in tree topology (73). For all amphibian species observed at a site, the model estimated their abundance with Poisson error using a log link. Fixed effects included an intercept and a response to land use (coded as natural vs. converted habitats). We fit the model with random intercept terms for species and two random slopes in response to habitat conversion. The first random slope was drawn from a normal distribution with a variance term estimated from the data (quantifying the degree to which responses to the environment are independent of phylogeny). The second was drawn from a multivariate normal distribution characterized by a variance scalar (estimated from the data) and the expected correlation matrix under Brownian motion derived from the phylogeny. The relative magnitude of these two variance terms were used to estimate the degree of phylogenetic signal in responses to habitat conversion, expressed as Pagel’s λ (also termed phylogenetic heritability, H2; ref. 74). Finally, the model additionally included a random error term to account for variation not conserved within species or between habitats.
Quantifying Effects of Habitat Conversion on Taxonomic and Phylogenetic α-Diversity.
We quantified taxonomic α-diversity as observed species richness based on species occurrences in natural and converted habitats reported in each study, and we quantified phylogenetic α-diversity as Faith’s PD (75)—the sum of total branch lengths of assemblages within each habitat. To examine sensitivity of results to different measures of α-diversity, we also derived estimates of taxonomic and phylogenetic α-diversity using extrapolation of accumulation curves (76). Because results using extrapolation and observed α-diversity were qualitatively identical (Fig. S8), we reported results obtained from observed species occurrences. To examine the change in PD, while controlling for changes in taxonomic diversity, we first extracted the local phylogenetic tree for all species (in both natural and converted habitats) from a given study. We then shuffled the tip labels 1,000 times, each time drawing the observed number of species in each habitat, to generate null distributions of PDs. We standardized the observed values of PD by subtracting the mean and dividing by the SD of the null distribution, generating a standardized effect size Z-score (SES PD). For both observed PD and the SES PD, we calculated values for each of the 100 posterior trees and took the mean to use as a response variable in downstream analyses. To examine phylogenetic divergence among species within each habitat, we calculated MPD, which measures divergence across the community tree and is mathematically equivalent to phylogenetic species variability (77), and MNTD, which measures divergence at shallow portions of the tree.
To examine changes in α-diversity with habitat conversion, we subtracted α-diversity values (species richness, PD, SES PD, MPD, and MNTD) in each study’s converted habitat(s) from that study’s value in natural habitat. We compared alternative nested linear mixed-effect models (LMMs) to determine which best described the data. The first model included an independent effect of each of the four converted habitat types, the second had only an intercept (estimating the overall effect of converted habitats, regardless of type), and a null model included an intercept term set to 0 (postulating no difference in α-diversity between natural and converted habitats). All models included the study as a random intercept because some studies contained comparisons with multiple converted habitats. We used likelihood ratio tests evaluated against a χ2 distribution to calculate significance levels for each term.
To better understand the relationship between taxonomic and phylogenetic α-diversity and environmental drivers, we regressed α-diversity against mean annual temperature, the SD of monthly temperatures, mean annual precipitation, elevation, and mean human footprint for each study location. Temperature and precipitation measures were extracted from the WorldClim dataset (78) within 5-km buffers centered on each study area, representing local climates. The human footprint index—a composite of human population density, land use, infrastructure, and accessibility—was extracted within 25-km buffers centered on each study area and represents the overall influence of human activities within the study region (79). We examined all single-variable models with an error structure as above and used likelihood ratio tests to determine variable significance.
Quantifying Effects of Habitat Conversion on Taxonomic and Phylogenetic β-Diversity Within Study Regions.
Based on species occurrence in natural and converted habitats, we calculated pairwise taxonomic and phylogenetic β-diversity metrics within each study region. We partitioned total dissimilarity (Sorensen index) between habitats into turnover—the gain and loss of unique species (or branches for phylogenetic β-diversity)—and nestedness—the contribution of richness (or PD) gradients to total dissimilarity—(80, 81) using functions within the betapart package in R. For phylogenetic β-diversity standardized by taxonomic β-diversity, we shuffled tip labels 1,000 times on the study-specific phylogeny, while holding constant the species composition in each habitat type. We then calculated the standardized effect size (SES) as both a Z score and as a rank of the observed value in the null distribution of phylogenetic β-diversities. We repeated the calculations on each of our 100 posterior trees and then took the mean value for downstream analysis.
We assessed whether interhabitat β-diversity was primarily made up of turnover or nestedness by analyzing the proportion of total β-diversity that was made up of turnover, using a GLM with binomial error and a nonfixed dispersion parameter to facilitate testing proportional responses. We tested against the null hypothesis that the proportion of turnover was 0.5 (i.e., neither turnover nor nestedness predominated as a driver of β-diversity) using likelihood ratio tests. Similarly, we evaluated whether phylogenetic β-diversity was different from a neutral expectation based on taxonomic β-diversity. We used the same GLM procedure described above but evaluated the rank at which the observed β-diversity value fell in the null distribution. A value of 0.5 indicates that PD perfectly matched the expectation, whereas values close to zero indicate that there was much less phylogenetic turnover between habitats than expected (i.e., species that differed between habitats were more closely related than expected). Because GLMMs implemented within R currently do not permit the evaluation of over/under dispersion by using a “quasibinomial” distribution, we fit GLMs with individual studies as replicates, taking the mean value of within-study β-diversity between natural and converted habitats when multiple converted-habitat types were evaluated. We also regressed β-diversity against the environmental variables described above to test the hypothesis that community dissimilarity between natural and converted habitats is affected by regional environmental context.
Global β-Diversity Within Natural and Human Modified Habitats.
To understand whether habitat conversion leads to taxonomic or phylogenetic homogenization across the globe, we quantified β-diversity among all study assemblages, separately for natural and converted habitats. We first calculated pairwise β-diversity across all sites, using taxonomic, phylogenetic, and SES phylogenetic β-diversity metrics. To quantify taxonomic and phylogenetic β-diversity within each habitat, we then calculated each site’s distance from its habitat’s multivariate median (82) using the betadisper function in package vegan. We compared these β-diversity values between habitats using a LMM with a random intercept of study (values were sufficiently far from 0 and 1 to not violate the Gaussian error assumption). Because pairwise SES phylogenetic β-diversities do not represent dissimilarities, we instead converted pairwise dissimilarities to a single measure by taking each site’s average dissimilarity to every other site within its habitat class and then proceeded to analyze these values with a LMM as above.
Supplementary Material
Acknowledgments
We thank M. Veiman and M. Donnelly for logistical support. This work was supported by a University of Toronto Ecology and Evolutionary Biology Postdoctoral Fellowship (to L.O.F.). A.J.N. and M.E.T. were supported by Florida International University (FIU) Evidence Acquisition Fellowships; M.E.T. was supported a Fulbright Student Research Award; and A.J.N. was supported by an FIU Dissertation Year Fellowship during data collection.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 3739.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714891115/-/DCSupplemental.
References
- 1.Foley JA, et al. Global consequences of land use. Science. 2005;309:570–574. doi: 10.1126/science.1111772. [DOI] [PubMed] [Google Scholar]
- 2.Haddad NM, et al. Habitat fragmentation and its lasting impact on earth’s ecosystems. Sci Adv. 2015;1:e1500052. doi: 10.1126/sciadv.1500052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newbold T, et al. Global effects of land use on local terrestrial biodiversity. Nature. 2015;520:45–50. doi: 10.1038/nature14324. [DOI] [PubMed] [Google Scholar]
- 4.Wiens JJ, et al. Niche conservatism as an emerging principle in ecology and conservation biology. Ecol Lett. 2010;13:1310–1324. doi: 10.1111/j.1461-0248.2010.01515.x. [DOI] [PubMed] [Google Scholar]
- 5.Purvis A, Agapow P-M, Gittleman JL, Mace GM. Nonrandom extinction and the loss of evolutionary history. Science. 2000;288:328–330. doi: 10.1126/science.288.5464.328. [DOI] [PubMed] [Google Scholar]
- 6.McKinney ML, Lockwood JL. Biotic homogenization: A few winners replacing many losers in the next mass extinction. Trends Ecol Evol. 1999;14:450–453. doi: 10.1016/s0169-5347(99)01679-1. [DOI] [PubMed] [Google Scholar]
- 7.Lee TM, Jetz W. Unravelling the structure of species extinction risk for predictive conservation science. Proc Biol Sci. 2011;278:1329–1338. doi: 10.1098/rspb.2010.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson AD, et al. Drivers and hotspots of extinction risk in marine mammals. Proc Natl Acad Sci USA. 2012;109:3395–3400. doi: 10.1073/pnas.1121469109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keinath DA, et al. A global analysis of traits predicting species sensitivity to habitat fragmentation. Glob Ecol Biogeogr. 2017;26:115–127. [Google Scholar]
- 10.Nowakowski AJ, Thompson ME, Donnelly MA, Todd BD. Amphibian sensitivity to habitat modification is associated with population trends and species traits. Glob Ecol Biogeogr. 2017;26:700–712. [Google Scholar]
- 11.Todd BD, Nowakowski AJ, Rose JP, Price SJ. Species traits explaining sensitivity of snakes to human land use estimated from citizen science data. Biol Conserv. 2017;206:31–36. [Google Scholar]
- 12.Karp DS, Ziv G, Zook J, Ehrlich PR, Daily GC. Resilience and stability in bird guilds across tropical countryside. Proc Natl Acad Sci USA. 2011;108:21134–21139. doi: 10.1073/pnas.1118276108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newbold T, et al. Ecological traits affect the response of tropical forest bird species to land-use intensity. Proc Biol Sci. 2013;280:20122131. doi: 10.1098/rspb.2012.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nowakowski AJ, et al. Tropical amphibians in shifting thermal landscapes under land-use and climate change. Conserv Biol. 2017;31:96–105. doi: 10.1111/cobi.12769. [DOI] [PubMed] [Google Scholar]
- 15.Frishkoff LO, Hadly EA, Daily GC. Thermal niche predicts tolerance to habitat conversion in tropical amphibians and reptiles. Glob Change Biol. 2015;21:3901–3916. doi: 10.1111/gcb.13016. [DOI] [PubMed] [Google Scholar]
- 16.Trimble MJ, van Aarde RJ. Amphibian and reptile communities and functional groups over a land-use gradient in a coastal tropical forest landscape of high richness and endemicity. Anim Conserv. 2014;17:441–453. [Google Scholar]
- 17.Frank HK, Frishkoff LO, Mendenhall CD, Daily GC, Hadly EA. Phylogeny, traits, and biodiversity of a neotropical bat assemblage: Close relatives show similar responses to local deforestation. Am Nat. 2017;190:200–212. doi: 10.1086/692534. [DOI] [PubMed] [Google Scholar]
- 18.Mayfield MM, et al. What does species richness tell us about functional trait diversity? Predictions and evidence for responses of species and functional trait diversity to land‐use change. Glob Ecol Biogeogr. 2010;19:423–431. [Google Scholar]
- 19.Thompson ME, Nowakowski AJ, Donnelly MA. The importance of defining focal assemblages when evaluating amphibian and reptile responses to land use. Conserv Biol. 2016;30:249–258. doi: 10.1111/cobi.12637. [DOI] [PubMed] [Google Scholar]
- 20.Kurz DJ, Nowakowski AJ, Tingley MW, Donnelly MA, Wilcove DS. Forest-land use complementarity modifies community structure of a tropical herpetofauna. Biol Conserv. 2014;170:246–255. [Google Scholar]
- 21.Frishkoff LO, et al. Loss of avian phylogenetic diversity in neotropical agricultural systems. Science. 2014;345:1343–1346. doi: 10.1126/science.1254610. [DOI] [PubMed] [Google Scholar]
- 22.Vellend M, Cornwell WK, Magnuson-Ford K, Mooers AØ. Biological Diversity: Frontiers in Measurement and Assessment. Oxford Univ Press; Oxford: 2011. Measuring phylogenetic biodiversity; pp. 194–207. [Google Scholar]
- 23.Edwards DP, Gilroy JJ, Thomas GH, Uribe CAM, Haugaasen T. Land-sparing agriculture best protects avian phylogenetic diversity. Curr Biol. 2015;25:2384–2391. doi: 10.1016/j.cub.2015.07.063. [DOI] [PubMed] [Google Scholar]
- 24.Supp SR, Ernest SK. Species-level and community-level responses to disturbance: A cross-community analysis. Ecology. 2014;95:1717–1723. doi: 10.1890/13-2250.1. [DOI] [PubMed] [Google Scholar]
- 25.Clavel J, Julliard R, Devictor V. Worldwide decline of specialist species: Toward a global functional homogenization? Front Ecol Environ. 2011;9:222–228. [Google Scholar]
- 26.Godet L, Gaüzere P, Jiguet F, Devictor V. Dissociating several forms of commonness in birds sheds new light on biotic homogenization. Glob Ecol Biogeogr. 2015;24:416–426. [Google Scholar]
- 27.Graham CH, Fine PV. Phylogenetic beta diversity: Linking ecological and evolutionary processes across space in time. Ecol Lett. 2008;11:1265–1277. doi: 10.1111/j.1461-0248.2008.01256.x. [DOI] [PubMed] [Google Scholar]
- 28.Helmus MR, Savage K, Diebel MW, Maxted JT, Ives AR. Separating the determinants of phylogenetic community structure. Ecol Lett. 2007;10:917–925. doi: 10.1111/j.1461-0248.2007.01083.x. [DOI] [PubMed] [Google Scholar]
- 29.Sol D, Bartomeus I, González-Lagos C, Pavoine S. Urbanisation and the loss of phylogenetic diversity in birds. Ecol Lett. 2017;20:721–729. doi: 10.1111/ele.12769. [DOI] [PubMed] [Google Scholar]
- 30.Mantyka-Pringle CS, Martin TG, Rhodes JR. Interactions between climate and habitat loss effects on biodiversity: A systematic review and meta-analysis. Glob Change Biol. 2012;18:1239–1252. [Google Scholar]
- 31.Nowakowski AJ, Jimenez BO, Allen M, Diaz-Escobar M, Donnelly MA. Landscape resistance to movement of the poison frog, Oophaga pumilio, in the lowlands of northeastern Costa Rica. Anim Conserv. 2013;16:188–197. [Google Scholar]
- 32.Nowakowski AJ, Veiman-Echeverria M, Kurz DJ, Donnelly MA. Evaluating connectivity for tropical amphibians using empirically derived resistance surfaces. Ecol Appl. 2015;25:928–942. doi: 10.1890/14-0833.1. [DOI] [PubMed] [Google Scholar]
- 33.Arroyo-Rodríguez V, Saldaña-Vázquez RA, Fahrig L, Santos BA. Does forest fragmentation cause an increase in forest temperature? Ecol Res. 2016;32:81–88. [Google Scholar]
- 34.Huey RB, et al. Why tropical forest lizards are vulnerable to climate warming. Proc Biol Sci. 2009;276:1939–1948. doi: 10.1098/rspb.2008.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janzen DH. Why mountain passes are higher in the tropics. Am Nat. 1967;101:233–249. [Google Scholar]
- 36.Pyron RA, Wiens JJ. Large-scale phylogenetic analyses reveal the causes of high tropical amphibian diversity. Proc Biol Sci. 2013;280:20131622. doi: 10.1098/rspb.2013.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonetti MF, Wiens JJ. Evolution of climatic niche specialization: A phylogenetic analysis in amphibians. Proc Biol Sci. 2014;281:20133229. doi: 10.1098/rspb.2013.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dirzo R, et al. Defaunation in the Anthropocene. Science. 2014;345:401–406. doi: 10.1126/science.1251817. [DOI] [PubMed] [Google Scholar]
- 39.Murray KA, Verde Arregoitia LD, Davidson A, Di Marco M, Di Fonzo MM. Threat to the point: Improving the value of comparative extinction risk analysis for conservation action. Glob Change Biol. 2014;20:483–494. doi: 10.1111/gcb.12366. [DOI] [PubMed] [Google Scholar]
- 40.Stuart SN, et al. Threatened Amphibians of the World. Lynx Edicions; Barcelona: 2008. [Google Scholar]
- 41.Almeida-Gomes M, Rocha CFD. Habitat loss reduces the diversity of frog reproductive modes in an Atlantic forest fragmented landscape. Biotropica. 2015;47:113–118. [Google Scholar]
- 42.Frost DR. Amphibian Species of the World: An Online Reference. American Museum of Natural History; New York: 2017. [Google Scholar]
- 43.IUCN 2016. The IUCN Red List of Threatened Species (International Union for Conservation of Nature and Natural Resources, Cambridge, UK), Version 2016-3.
- 44.Gomez-Mestre I, Pyron RA, Wiens JJ. Phylogenetic analyses reveal unexpected patterns in the evolution of reproductive modes in frogs. Evolution. 2012;66:3687–3700. doi: 10.1111/j.1558-5646.2012.01715.x. [DOI] [PubMed] [Google Scholar]
- 45.Crump ML. Anuran reproductive modes: Evolving perspectives. J Herpetol. 2015;49:1–16. [Google Scholar]
- 46.Quesnelle PE, Lindsay KE, Fahrig L. Low reproductive rate predicts species sensitivity to habitat loss: A meta-analysis of wetland vertebrates. PLoS One. 2014;9:e90926. doi: 10.1371/journal.pone.0090926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheffers BR, et al. Thermal buffering of microhabitats is a critical factor mediating warming vulnerability of frogs in the Philippine biodiversity hotspot. Biotropica. 2013;45:628–635. [Google Scholar]
- 48.Catenazzi A, Lehr E, Vredenburg VT. Thermal physiology, disease, and amphibian declines on the eastern slopes of the Andes. Conserv Biol. 2014;28:509–517. doi: 10.1111/cobi.12194. [DOI] [PubMed] [Google Scholar]
- 49.Tracy CR, Christian KA, Tracy CR. Not just small, wet, and cold: Effects of body size and skin resistance on thermoregulation and arboreality of frogs. Ecology. 2010;91:1477–1484. doi: 10.1890/09-0839.1. [DOI] [PubMed] [Google Scholar]
- 50.Nowakowski AJ, et al. Thermal biology mediates responses of amphibians and reptiles to habitat modification. Ecol Lett. 2018;21:345–355. doi: 10.1111/ele.12901. [DOI] [PubMed] [Google Scholar]
- 51.Mayfield MM, Levine JM. Opposing effects of competitive exclusion on the phylogenetic structure of communities. Ecol Lett. 2010;13:1085–1093. doi: 10.1111/j.1461-0248.2010.01509.x. [DOI] [PubMed] [Google Scholar]
- 52.Schneider-Maunoury L, et al. Abundance signals of amphibians and reptiles indicate strong edge effects in neotropical fragmented forest landscapes. Biol Conserv. 2016;200:207–215. [Google Scholar]
- 53.Frishkoff LO, et al. Climate change and habitat conversion favour the same species. Ecol Lett. 2016;19:1081–1090. doi: 10.1111/ele.12645. [DOI] [PubMed] [Google Scholar]
- 54.Buckley LB, Jetz W. Linking global turnover of species and environments. Proc Natl Acad Sci USA. 2008;105:17836–17841. doi: 10.1073/pnas.0803524105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zuloaga J, Kerr JT. Over the top: Do thermal barriers along elevation gradients limit biotic similarity? Ecography. 2017;40:478–486. [Google Scholar]
- 56.Larsen TH. Upslope range shifts of Andean dung beetles in response to deforestation: Compounding and confounding effects of microclimatic change. Biotropica. 2012;44:82–89. [Google Scholar]
- 57.Grau HR, Aide M. Globalization and land-use transitions in Latin America. Ecol Soc. 2008;13:16. [Google Scholar]
- 58.Heard SB, Mooers AØ. Phylogenetically patterned speciation rates and extinction risks change the loss of evolutionary history during extinctions. Proc Biol Sci. 2000;267:613–620. doi: 10.1098/rspb.2000.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nori J, et al. Amphibian conservation, land-use changes and protected areas: A global overview. Biol Conserv. 2015;191:367–374. [Google Scholar]
- 60.Nowakowski AJ, Angulo A. Targeted habitat protection and its effects on the extinction risk of threatened amphibians. FrogLog. 2015;116:8–10. [Google Scholar]
- 61.Porter-Bolland L, et al. Community managed forests and forest protected areas: An assessment of their conservation effectiveness across the tropics. For Ecol Manage. 2012;268:6–17. [Google Scholar]
- 62.Morse W, et al. Consequences of environmental service payments for forest retention and recruitment in a Costa Rican biological corridor. Ecol Soc. 2009;14:23. [Google Scholar]
- 63.Mendenhall CD, et al. Countryside biogeography of neotropical reptiles and amphibians. Ecology. 2014;95:856–870. doi: 10.1890/12-2017.1. [DOI] [PubMed] [Google Scholar]
- 64.Robinson D, Warmsley A, Nowakowski AJ, Reider KE, Donnelly MA. The value of remnant trees in pastures for a neotropical poison frog. J Trop Ecol. 2013;29:345–352. [Google Scholar]
- 65.Mendenhall CD, Shields-Estrada A, Krishnaswami AJ, Daily GC. Quantifying and sustaining biodiversity in tropical agricultural landscapes. Proc Natl Acad Sci USA. 2016;113:14544–14551. doi: 10.1073/pnas.1604981113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fischer J, et al. Land sparing versus land sharing: Moving forward. Conserv Lett. 2014;7:149–157. [Google Scholar]
- 67.Catenazzi A. State of the world’s amphibians. Annu Rev Environ Resour. 2015;40:91–119. [Google Scholar]
- 68.Pyron RA. Biogeographic analysis reveals ancient continental vicariance and recent oceanic dispersal in amphibians. Syst Biol. 2014;63:779–797. doi: 10.1093/sysbio/syu042. [DOI] [PubMed] [Google Scholar]
- 69.Thomas GH, et al. PASTIS: An R package to facilitate phylogenetic assembly with soft taxonomic inferences. Methods Ecol Evol. 2013;4:1011–1017. [Google Scholar]
- 70.Rabosky DL. No substitute for real data: A cautionary note on the use of phylogenies from birth-death polytomy resolvers for downstream comparative analyses. Evolution. 2015;69:3207–3216. doi: 10.1111/evo.12817. [DOI] [PubMed] [Google Scholar]
- 71.Frishkoff LO, de Valpine P, M’Gonigle LK. Phylogenetic occupancy models integrate imperfect detection and phylogenetic signal to analyze community structure. Ecology. 2017;98:198–210. doi: 10.1002/ecy.1631. [DOI] [PubMed] [Google Scholar]
- 72.Ives AR, Helmus MR. Generalized linear mixed models for phylogenetic analyses of community structure. Ecol Monogr. 2011;81:511–525. [Google Scholar]
- 73.de Villemereuil P, Wells JA, Edwards RD, Blomberg SP. Bayesian models for comparative analysis integrating phylogenetic uncertainty. BMC Evol Biol. 2012;12:102. doi: 10.1186/1471-2148-12-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hadfield JD, Nakagawa S. General quantitative genetic methods for comparative biology: Phylogenies, taxonomies and multi-trait models for continuous and categorical characters. J Evol Biol. 2010;23:494–508. doi: 10.1111/j.1420-9101.2009.01915.x. [DOI] [PubMed] [Google Scholar]
- 75.Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. [Google Scholar]
- 76.Chao A, et al. Rarefaction and extrapolation of phylogenetic diversity. Methods Ecol Evol. 2015;6:380–388. [Google Scholar]
- 77.Tucker CM, et al. A guide to phylogenetic metrics for conservation, community ecology and macroecology. Biol Rev Camb Philos Soc. 2017;92:698–715. doi: 10.1111/brv.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25:1965–1978. [Google Scholar]
- 79.Sanderson EW, et al. The human footprint and the last of the wild: The human footprint is a global map of human influence on the land surface, which suggests that human beings are stewards of nature, whether we like it or not. Bioscience. 2002;52:891–904. [Google Scholar]
- 80.Baselga A. Partitioning the turnover and nestedness components of beta diversity. Glob Ecol Biogeogr. 2010;19:134–143. [Google Scholar]
- 81.Leprieur F, et al. Quantifying phylogenetic beta diversity: Distinguishing between ‘true’ turnover of lineages and phylogenetic diversity gradients. PLoS One. 2012;7:e42760. doi: 10.1371/journal.pone.0042760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anderson MJ, Ellingsen KE, McArdle BH. Multivariate dispersion as a measure of beta diversity. Ecol Lett. 2006;9:683–693. doi: 10.1111/j.1461-0248.2006.00926.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.