Significance
Heart disease is associated with the development of fibrosis, a type of scarring that impedes cardiac function. The primary cellular source of cardiac fibrosis is the resident cardiac fibroblast. We found that cardiac fibroblasts from human heart failure patients or a mouse model of heart disease express excessive amounts of the SPRR2B protein. We provide evidence that SPRR2B is a signal-responsive regulatory subunit of the p53 ubiquitination complex that stimulates the destruction of p53 and the accumulation of pathological fibroblasts. This study defines a unique mechanism of cell cycle control that is dysfunctional in heart disease and may drive the development pathological fibrosis.
Keywords: fibroblast, heart, p53, proliferation, SPRR2B
Abstract
Heart disease is associated with the accumulation of resident cardiac fibroblasts (CFs) that secrete extracellular matrix (ECM), leading to the development of pathological fibrosis and heart failure. However, the mechanisms underlying resident CF proliferation remain poorly defined. Here, we report that small proline-rich protein 2b (Sprr2b) is among the most up-regulated genes in CFs during heart disease. We demonstrate that SPRR2B is a regulatory subunit of the USP7/MDM2-containing ubiquitination complex. SPRR2B stimulates the accumulation of MDM2 and the degradation of p53, thus facilitating the proliferation of pathological CFs. Furthermore, SPRR2B phosphorylation by nonreceptor tyrosine kinases in response to TGF-β1 signaling and free-radical production potentiates SPRR2B activity and cell cycle progression. Knockdown of the Sprr2b gene or inhibition of SPRR2B phosphorylation attenuates USP7/MDM2 binding and p53 degradation, leading to CF cell cycle arrest. Importantly, SPRR2B expression is elevated in cardiac tissue from human heart failure patients and correlates with the proliferative state of patient-derived CFs in a process that is reversed by insulin growth factor-1 signaling. These data establish SPRR2B as a unique component of the USP7/MDM2 ubiquitination complex that drives p53 degradation, CF accumulation, and the development of pathological cardiac fibrosis.
Heart disease is accompanied by hypertrophic growth and pathological remodeling of the myocardium and is the leading cause of death in the United States (1, 2). Activated cardiac fibroblasts (CFs), often called myofibroblasts, accumulate in heart disease and secrete extracellular matrix (ECM), driving the development of cardiac fibrosis (3, 4). Cardiac fibrosis is a form of scarring that replaces necrotic tissue and provides structural support to the expanding myocardium, but does so at the expense of cardiac function and ultimately leads to heart failure (HF) (1). In contrast, physiological hypertrophic growth in response to exercise improves cardiac function and attenuates the development of fibrosis. The mechanisms that control fibroblast accumulation and activation specifically in pathological remodeling are poorly defined.
Similarities between pathological fibroblast activation and unrestrained cell proliferation in cancerous tumors are evident (5); the p53 tumor suppressor has recently been linked to lung and cardiac fibrosis, and is also thought to impact CF plasticity during pressure overload and ischemic injury (6, 7). p53 activates cell cycle exit genes such as cyclin-dependent kinase inhibitor 1a (Cdkn1a) as well as proapoptotic genes such as BAX (8). p53 is a substrate for the E3 ubiquitin ligase MDM2, which targets p53 for proteasome-mediated degradation, relieving constraints on cell cycle. USP7 is an MDM2 deubiquitinase that stabilizes MDM2, whereas MDM2 might otherwise ubiquitinate itself, leading to self-destruction. The mechanisms controlling the balance between MDM2-autoubiquitination and MDM2-dependent p53 degradation are complex, context dependent, and incompletely understood (9).
We conducted an RNA sequencing (RNA-seq) screen to identify genes that are differentially expressed in mouse CFs during physiological (swim training) or pathological [transverse aortic constriction (TAC)] remodeling. Small proline-rich protein 2b (Sprr2b) was among the genes that were most profoundly up-regulated in pathological remodeling and lost in physiological remodeling. Here, we describe an interaction between Sprr2b and a nuclear complex of USP7, MDM2, and p53. Sprr2b promotes MDM2 accumulation and p53 degradation, and accelerates cell cycle progression specifically in CFs. We demonstrate that TGF-β1 and reactive oxygen species (ROS) signaling potentiates both Sprr2b gene expression and nonreceptor tyrosine kinase (NRTK)-dependent phosphorylation of the Sprr2b protein, facilitating USP7/MDM2 interaction and the degradation of p53. Importantly, we show that SPRR2B is significantly enriched within the cardiac interstitium in human HF. Indeed, SPRR2B expression positively correlates with proliferation of CFs isolated from human HF patients. Taken together, our study describes a unique stress-dependent mechanism regulating MDM2-dependent p53 degradation that controls pathological CF accumulation.
Results
Sprr2b Expression Is Induced in CFs During Pathologic Remodeling.
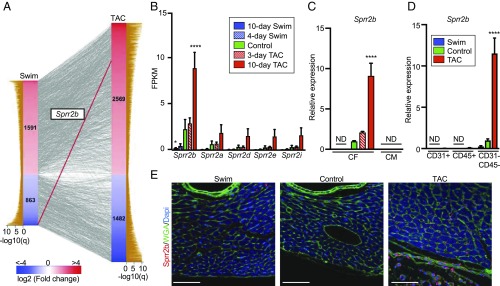
To identify genes that might underlie the divergent fibrotic response of the heart to pathological and physiological hypertrophic growth, we performed RNA-seq on CFs isolated from mice subjected to 10 days of left ventricle pressure overload or swim training (Fig. S1A and B) (10–14). This study identified 2,454 genes that changed in endurance exercise and 4,051 genes that changed in response to TAC (Fig. 1A). One of the most affected genes was Sprr2b (Fig. 1 A and B), which was 4.3-fold up-regulated in TAC relative to controls [fragments per kilobase per million reads (FPKM) 8.099 versus 1.890] and 9.1-fold down-regulated in swim relative to controls (FPKM 0.207 versus 1.890). We also identified five others members of the Sprr2 family (Sprr2a, -d, -e, -h, -i) that were only moderately up-regulated in TAC compared with swim (Fig. 1B and Fig. S1C). qRT-PCR confirmed the changes in Sprr2b expression during physiological and pathological remodeling and revealed that Sprr2b is specifically enriched in CFs and not detected in cardiomyocytes (CMs) (Fig. 1C). Separation of cardiac interstitial cells by FACS followed by qRT-PCR confirmed that Sprr2b is enriched following TAC in CD31−/CD45− mesenchymal cells/fibroblasts (Fig. 1D). Western blot confirmed that SPRR2B protein is also enriched in the CD31−/CD45− population (Fig. S1D). Immunostaining further demonstrated robust expression of SPRR2B following TAC, localizing to both the cytoplasm and nucleus of cardiac interstitial cells, but not CMs (Fig. 1E). In contrast, SPRR2B-expressing cells were rarely found in the heart of control animals and were undetectable in swim-trained animals.
Fig. 1.
RNA-seq reveals the Sprr2b gene is differentially regulated by exercise and disease specifically in CFs. (A) Comparison of genes expressed in CFs with fragments per kilobase per million reads (FPKM) > 1 for any condition that are significantly altered by exercise (swim, Left column) or pressure overload (TAC, Right column) compared with controls (q < 0.05). Genes that are altered in both treatment groups (n = 2,075) are indicated by connecting lines with Sprr2b highlighted in red showing inverse regulation. (B) FPKM values reveal the expression of Sprr2b in CFs following 4 or 10 days of a swim and 3 or 10 days after TAC relative to controls. (C) qRT-PCR confirms changes in Sprr2b expression in CFs isolated by differential plating. Sprr2b is not detected in CMs. (D) qRT-PCR for Sprr2b expression in cardiac endothelial cells (CD31+), circulating cells (CD45+), and mesenchymal/fibroblasts (CD31−/CD45−) isolated by FACS. qRT-PCR data normalized to Gapdh. All data represent mean ± SEM. *P < 0.05; ****P < 0.0001, one-way ANOVA compared with control. n = 3 (swim and TAC), n = 4 (control). ND, not detected. (E) Immunostaining of sections from mouse hearts of the indicated treatment for SPRR2B (red), cardiac troponin T (blue), WGA (green), and nuclei (DAPI, blue). (Scale bar: 25 μm.)
TGF-β1 and H2O2 Synergistically Induce Sprr2b Expression, Which Is Required for CF Proliferation.
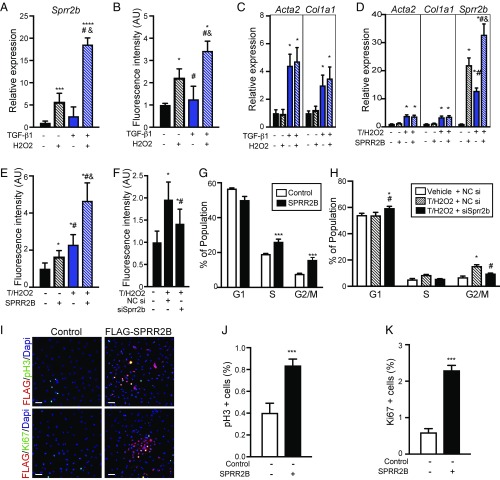
Since HF is associated with TGF-β signaling and increased ROS burden, we evaluated the potential contribution of SPRR2B to TGF-β and ROS-induced changes in fibroblast plasticity. Treatment of CFs with TGF-β1 (10 ng/mL) and H2O2 (50 μM) was found to maximize fibroblast activation and proliferation while minimizing cell death (Fig. S2). TGF-β1 and H2O2 synergistically stimulated the expression of Sprr2b (Fig. 2A) and also led to a synergistic increase in CF proliferation (Fig. 2B). In contrast, myofibroblast genes were not synergistically induced by TGF-β1 and H2O2 (Fig. 2C). To determine whether SPRR2B impacts fibroblast activation and/or proliferation, we transfected vehicle or TGF-β1/H2O2–treated CFs with a plasmid driving the expression of human SPRR2B. SPRR2B did not have an effect on Acta2 or Col1a1 expression (Fig. 2D), but significantly increased proliferation in both control and TGF-β1/H2O2–treated CFs (Fig. 2E). In contrast, knockdown of endogenous Sprr2b expression using two distinct siRNA oligos (Fig. S3 A and B) suppressed TGF-β1/H2O2–induced CF proliferation (Fig. 2F and Fig. S3C). These results reveal that induction of Sprr2b expression in CFs by TGF-β1/H2O2 at least partially underlies the ability of this treatment to induce cell proliferation.
Fig. 2.
Synergistic activation of Sprr2b by TGF-β1 and H2O2 is required for CF proliferation. (A–C) Primary neonatal CFs were treated with combinations of TGF-β1 (10 ng/mL) and H2O2 (50 μM) for 24 h. (A) qRT-PCR was performed to evaluate Sprr2b expression (n = 6). One-way ANOVA. (B) CyQuant dye DNA incorporation was performed to evaluate proliferation (n = 8). One-way ANOVA. (C) qRT-PCR for Acta2 and Col1a1 was performed to evaluate myofibroblast activation (n = 6). Two-way ANOVA with Bonferroni post hoc. (D) SPRR2B transfection of CFs followed by qRT-PCR for Acta2 and Col1a1 (n = 6). Two-way ANOVA with Bonferroni post hoc. (E) CyQuant assay was used to evaluate the impact of SPRR2B overexpression on CF proliferation (n = 8). One-way ANOVA. (F) CyQuant assay was used to evaluate the effect of siRNA-mediated Sprr2b knockdown on TGF-β1/H2O2–induced CF proliferation (n = 8). One-way ANOVA. NC, no-target control siRNA. (G) Flow cytometry shows G2/M and S populations increase in response to SPRR2B overexpression in CFs relative to control (n = 3). One-way ANOVA. (H) Flow cytometry reveals siRNA-mediated knockdown of Sprr2b in CFs blocks the increase in G2/M and S populations by TGF-β1/H2O2 treatment, relative to control (n = 3). One-way ANOVA. (I) Primary CFs were transfected with control or Flag-SPRR2B expression vectors (transfection efficiency was ∼36%). Immunofluorescent detection of FLAG and phosphohistone H3 or Ki67 reveals increased mitosis and proliferation in SPRR2B-transfected CFs. (Scale bar: 50 μm.) (J) Quantification of pH3+ cells from I. (K) Quantification of Ki67+ cells from I. n = 6 wells with five nonoverlapping fields of view each. Unpaired Student’s t test. qRT-PCR data normalized to Gapdh. All data represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 compared with control. #P < 0.05 compared with second condition; &P < 0.05 compared with third condition.
SPRR2B Accelerates Cell Cycle Progression in CFs.
To further evaluate the role of SPRR2B in CF proliferation, we assessed cell cycle progression by flow cytometry and immunostaining. TGF-β1/H2O2 treatment led to a significant increase of primary CFs in the S and G2/M phases, and decreased the proportion of cells in G1 (Fig. S3D). Importantly, forced expression of SPRR2B further stimulated CF proliferation in response to TGF-β1/H2O2 treatment (Fig. 2G). In contrast, siRNA-mediated knockdown of endogenous Sprr2b blocked the increase in cell cycle progression observed upon TGF-β1/H2O2 treatment (Fig. 2H). Immunostaining for FLAG-tagged SPRR2B and phospho-histone H3 (pH3) or Ki67 to identify SPRR2B-expressing CFs in M or S phases of the cell cycle confirmed the positive influence of SPRR2B on cell cycle progression (Fig. 2 I–K).
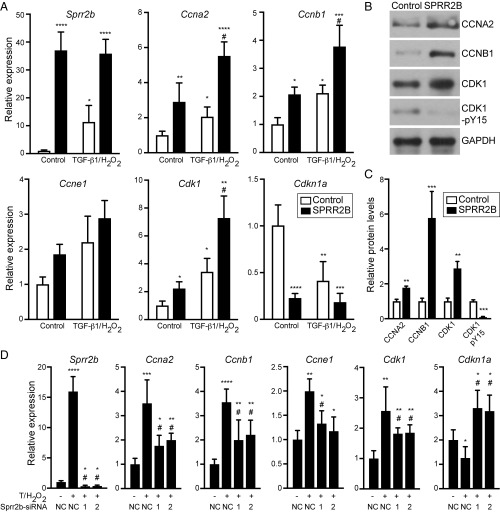
Since Sprr2b induction by TGF-β1 and ROS drives CF proliferation in vitro, we evaluated our RNA-seq dataset for genes associated with mitosis and cell cycle that cluster with Sprr2b expression changes. Several promitotic cyclins and cyclin-dependent kinases are up-regulated in TAC relative to swim, while important cyclin-dependent kinase inhibitors (CDKNs) are suppressed (Table S1), consistent with the dysregulation of p53-dependent gene programs identified by Ingenuity Pathway Analysis (Table S2). Interestingly, the expression of the mitotic cyclins Ccna2 and Ccnb1, and Cdk1 were synergistically induced in CFs by TGF-β1/H2O2 and SPRR2B (Fig. 3A). In contrast, expression of Cdkn1a, a CDK1 inhibitor, was silenced by SPRR2B overexpression (Fig. 3A). Western blot for the protein products of relevant mitotic genes confirmed the alterations upon SPRR2B overexpression (Fig. 3 B and C). In addition to alterations in CDK1 protein levels, SPRR2B also led to a reduction in the phosphorylation of CDK1 at Y15—a cell cycle-inhibitory phosphorylation conferred by Wee1/Myt1 kinases (Fig. 3 B and C) (15). To determine whether SPRR2B is essential for the alterations in mitotic regulators by TGF-β1/H2O2, we again used siRNA knockdown of endogenous Sprr2b in CFs, which significantly attenuated the alterations in mitotic regulators (Fig. 3D). These data indicate SPRR2B is necessary and sufficient for the acceleration of cell cycle in TGF-β1/H2O2–treated CFs.
Fig. 3.
Sprr2b is necessary and sufficient for alterations in gene expression consistent with accelerated cell cycle. (A) The expression of promitotic cyclins and cyclin-dependent kinases (Ccna2, Ccnb1, Ccne1, Cdk1) and Cyclin-dependent kinase 1 inhibitor (Cdkn1a/p21) were examined by qRT-PCR in primary CFs overexpressing SPRR2B with or without TGF-β1/H2O2 and analyzed by two-way ANOVA with Bonferroni post hoc (n = 6). (B) Western blot evaluates alterations in CCNA2, CCNB1, CDK1 protein levels and an inhibitory phosphorylation of CDK1 (pY15). GAPDH is included as a loading control. (C) Quantification of B. n = 3 experiments each, analyzed by unpaired Student’s t test. (D) TGF-β1/H2O2–dependent alterations in the expression of mitotic genes were evaluated by qRT-PCR upon knockdown of Sprr2b with two independent siRNAs (1 and 2). n = 3 experiments each, analyzed by two-way ANOVA with Bonferroni post hoc. qRT-PCR data normalized to Gapdh. All data represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. #P < 0.05 compared with second condition. NC, nontargeting control siRNA.
SPRR2B Binds USP7, Stabilizing USP7–MDM2 Interaction.
To gain insight into the potential mechanism whereby SPRR2B drives CF cell cycle progression, we examined the expression of genes encoding proteins known to interact with SPRR2B. The best characterized role of SPRR2B is as part of the keratinocyte cornified envelope (CEnv), following cross-linking by TGM1 and TGM3 (16–21). Since the CEnv program is induced by oxidative stress in the skin, we began by evaluating the expression of CEnv program genes (Lor, Ivl, Flg, Tgm1, and Tgm3) in either CFs or primary mouse dermal fibroblasts (DFs) subjected to optimal doses of TGF-β1 and H2O2 (Figs. S2 and S4). Although the entire CEnv program was present in DFs at baseline and was highly enriched in response to TGF-β1/H2O2, only the expression of Sprr2b was stimulated by TGF-β1/H2O2 in CFs (Fig. 4A). This finding suggests that SPRR2B may play a noncanonical role in CFs due to the absence of typical binding partners.
Fig. 4.
SPRR2B is a binding partner for the MDM2 deubiquitinase USP7 in CFs. (A) qRT-PCR for Sprr2b and genes encoding keratinocyte proteins in primary CFs and DFs with or without TGF-β1/H2O2. (B) Schematic of human SPRR2B protein, highlighting predicted protein–protein interactions related to cell cycle control. The predicted USP7 interaction domain contains a canonical tyrosine phosphorylation site at Y67. (C) Fluorescent detection of phalloidin (green) SPRR2B (red) and nuclei (blue) by confocal microscopy reveals both cytoplasmic and nuclear localization of SPRR2B in CFs. (Scale bar: 10 μm.) (D) NIH 3T3 cells were cotransfected with HA-tagged USP7 with or without Flag-tagged SPRR2B. Immunoprecipitation of HA-USP7, followed by immunoblotting for Flag-SPRR2B or endogenous MDM2 revealed complex formation particularly in the presence of TGF-β1/H2O2. (E) Quantification of D, analyzed by two-way ANOVA with Bonferroni post hoc (n = 3). qRT-PCR data normalized to Gapdh. All data represent mean ± SEM. *P < 0.05, ***P < 0.001 compared with control. #P < 0.05 compared with second condition.
We therefore evaluated predicted interacting motifs in the SPRR2B protein. Eukaryotic linear motif prediction identified four motifs potentially related to cell cycle control, including a C-terminal interacting module for USP7, an MDM2 and p53 deubiquitinase (Fig. 4B) (22). Consistent with our observation of endogenous SPRR2B protein localization in hearts of mice subjected to TAC surgery, immunofluorescent detection of Flag epitope-tagged SPRR2B in NIH 3T3 cells revealed localization both in the cytoplasm and nucleus, where relevant interactions with USP7/MDM2 would likely occur (Fig. 4C). To test the predicted interaction between SPRR2B and USP7, we overexpressed an HA-tagged USP7 construct alone or in combination with Flag-SPRR2B in NIH 3T3 cells. SPRR2B coimmunoprecipitated with USP7, and formation of this unique protein complex was stimulated by TGF-β1/H2O2 treatment (Fig. 4 D and E). The presence of SPRR2B also facilitated an interaction between USP7 and endogenous MDM2 and the accumulation of MDM2 protein, indicating SPRR2B is a component of the p53–E3 ubiquitin ligase protein complex in CFs.
Tyrosine Phosphorylation of SPRR2B Promotes USP7 Binding and CF Proliferation.
SPRR2B was predicted to be a phosphoprotein in a high-throughput mass spectrometry screen of non-small cell lung cancer cells (23). We find that Flag epitope-tagged SPRR2B is a substrate for tyrosine (Y) phosphorylation in NIH 3T3 cells and primary mouse CFs (Fig. 5A and Fig. S5). Phosphorylation of SPRR2B in both the nuclear and cytoplasmic compartments is strongly stimulated by TGF-β1/H2O2. SPRR2s have been shown to interact with the Src family of nonreceptor tyrosine kinases (NRTKs) via SH3 domain repeats, but the consequence of these interactions is unknown (24, 25). Inhibition of NRTK activity with saracatinib (50 nM) attenuated TGF-β1/H2O2–dependent USP7-SPRR2B binding (Fig. 5 B and C). Since human SPRR2B has only two tyrosine residues (Y3 and Y67), and Y67 is in the USP7-interacting region, we tested whether USP7 binding is dependent upon SPRR2B-pY67. Although a SPRR2B-Y67F phospho-mutant does not completely abolish the USP7 interaction, the Y67F mutation attenuates the stimulation of SPRR2B–USP7 interaction and accumulation of MDM2 protein upon TGF-β1/H2O2 treatment (Fig. 5 D and E). Importantly, SPRR2B-Y67F overexpression fails to stimulate the TGF-β1/H2O2–dependent changes in cell cycle-related genes and induction of CF proliferation that are observed upon wild-type SPRR2B overexpression (Fig. 5 F and G). These data suggest that the phosphorylation of SPRR2B by Src family NRTKs at Y67 facilitates USP7 interaction, MDM2 accumulation, and cell cycle progression.
Fig. 5.
USP7 binding and cell cycle progression is facilitated by NRTK-dependent tyrosine phosphorylation of SPRR2B. (A) Fractionation of Flag-SPRR2B–transfected NIH 3T3 cells and immunoprecipitation of Flag followed by immunoblot for phosphotyrosine. Representative of three independent experiments. Loading controls are HDAC1 (nucleus) and SOD1 (cytoplasm). (B) NIH 3T3 cells transfected with HA-USP7 and Flag-SPRR2B were treated with saracatinib (50 nM) with or without TGF-β1/H2O2. Coimmunoprecipitation of HA reveals stimulation of Flag-SPRR2B interaction by TGF-β1/H2O2 is blocked by saracatinib treatment. TCL, total cell lysate. (C) Quantification of B. n = 3. (D) Coimmunoprecipitation of HA-USP7/Flag-SPRR2B (wild type or Y67F) reveals USP7 association and MDM2 accumulation is dependent upon SPRR2B-Y67 phosphorylation. (E) Quantification of D. n = 3. (F) Expression of genes encoding cell cycle regulators was evaluated by qRT-PCR in CFs transfected with SPRR2B or SPRR2B-Y67F with or without TGF-β1/H2O2. n = 6. qRT-PCR data normalized to Gapdh and analyzed by one-way ANOVA. (G) CyQuant assay was used to evaluate proliferation of CFs in response to SPRR2B or SPRR2B-Y67F transfection with or without TGF-β1/H2O2. n = 8. Analyzed by two-way ANOVA with Bonferroni post hoc. All data represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; #P < 0.05 compared with second condition.
SPRR2B Destabilizes the Nuclear p53 Pool.
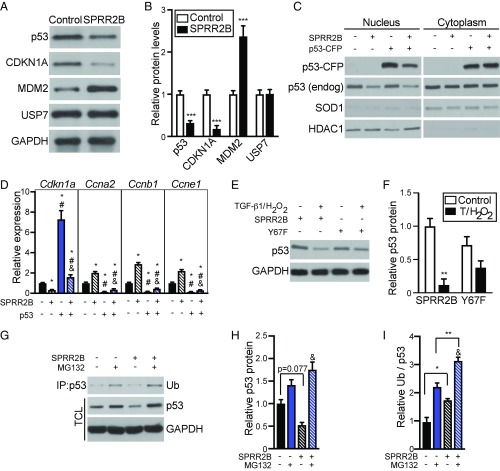
Since SPRR2B promotes cell cycle progression and the accumulation of MDM2, we hypothesized that SPRR2B may influence p53 protein levels in CFs. TGF-β1/H2O2–treated CFs overexpressing SPRR2B displayed a significant decrease in p53 protein relative to control cultures (Fig. 6 A and B). The reduction in p53 was reflected in significant attenuation of the p53 target gene and cell cycle inhibitor, CDKN1A/p21. We also made use of a Zn2+-inducible CFP-tagged p53 overexpression construct to test the effect of SPRR2B on p53 protein levels, localization, and target gene expression in NIH 3T3 cells. SPRR2B led to reduced levels of both p53-CFP and endogenous p53, specifically in the nucleus, where ubiquitin-mediated degradation of p53 occurs (Fig. 6C). Importantly, p53-CFP overexpression induced the expression of Cdkn1a and inhibited the promitotic cyclins, an effect that was reversed upon SPRR2B overexpression (Fig. 6D). In contrast, SPRR2B-Y67F failed to facilitate p53 degradation in response to TGF-β1/H2O2 treatment, confirming the importance of SPRR2B phosphorylation (Fig. 6 E and F). MG132 blocked the SPRR2B-dependent degradation of p53, revealing SPRR2B stimulates ubiquitin-mediated p53 degradation (Fig. 6 G and H). Indeed, SPRR2B overexpression stimulated the ubiquitination of p53 as revealed by coimmunoprecipitation (Fig. 6 G and I). These data link SPRR2B stimulation by TGF-β1/H2O2 to MDM2-dependent p53 degradation and CF proliferation.
Fig. 6.
SPRR2B facilitates ubiquitin-mediated proteasome degradation of p53. (A) Primary CFs were transfected with Flag-SPRR2B or control and analyzed by Western blot. (B) Quantification of A. n = 3. Analyzed by Student’s two-tailed unpaired t test. (C) Nuclear and cytoplasmic fractions were isolated from NIH 3T3 cells that were transfected with indicated combination of SPRR2B and p53-CFP. Western blot was performed to detect exogenous or endogenous p53. Loading controls are HDAC1 (nucleus) and SOD1 (cytoplasm). (D) qRT-PCR was used to evaluate the expression of genes encoding cell cycle regulators in NIH 3T3 cells that were transfected with p53-CFP with or without SPRR2B (n = 6). qRT-PCR data normalized to Gapdh and analyzed by one-way ANOVA. (E) NIH 3T3 cells were transfected with SPRR2B or SPRR2B-Y67F with or without TGF-β1/H2O2, followed by Western blot for endogenous p53. (F) Quantification of E analyzed by two-way ANOVA with Bonferroni post hoc (n = 3). (G–I) Primary CFs were treated with ZnSO4 (50 μM) and transfected with Flag-SPRR2B with or without MG132 (10 μM). (G) Coimmunoprecipitation of Flag was followed by immunoblot for monoubiquitin (Ub). Western blot was also performed to detect endogenous p53 in total cell lysate (TCL). (H) Quantification of total p53 levels in G relative to GAPDH loading controls. (I) Quantification of immunoprecipitated ubiquitin in G relative to total GAPDH-normalized p53 levels. Analyzed by two-way ANOVA with Bonferroni post hoc (n = 3). All data represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001; #P < 0.05 compared with second condition; &P < 0.05 compared with third condition.
SPRR2B Expression Correlates with Fibroblast Proliferation in Human HF.
We next evaluated the expression of SPRR2B in cardiac tissue isolated from HF patients undergoing ventricular assist device implantation, compared with normal heart tissue. SPRR2B was undetectable in 9 of 16 normal heart samples and significantly enriched in human HF (Fig. 7A). Immunostaining for SPRR2B in tissue derived from HF patients (n = 3) supported these data, as SPRR2B expression correlated well with vimentin staining—suggesting a nonmyocyte localization similar to that seen in the TAC mouse model (Fig. 7B). Importantly, healthy hearts (n = 3) were largely devoid of SPRR2B expression (Fig. 7B). We therefore examined the relationship between SPRR2B expression and markers of myofibroblast activation in cultures of primary human CFs (hCFs) isolated from three independent HF patients, compared with one hCF isolate obtained from healthy cardiac tissue. SPRR2B expression was significantly increased in HF hCFs compared with healthy hCFs, positively correlating with the expression of POSTN, ACTA2, COL1A1, and COL3A1 (Fig. 7C).
Fig. 7.
SPRR2B is significantly enriched in the failing human heart and positively correlates with promitotic factors in human HF fibroblasts. (A) qRT-PCR was used to evaluate SPRR2B expression in left ventricle tissue isolated HF patients or healthy hearts. n = 16 (Healthy), 15 (HF). ND, not detected. (B) Immunohistochemistry of representative sections from failing or healthy hearts costained for WGA (white), SPRR2B (green), and VIMENTIN (VIM, red). DAPI (blue) stains nuclei. SPRR2B is associated with vimentin-positive cells in the failing heart, indicated by white arrows. (Scale bar: 25 μm.) (C) qRT-PCR was used to evaluate the expression of SPRR2B and genes encoding myofibroblast markers in human CFs isolated from healthy or failing (HF) hearts (n = 3 HF and one healthy isolate). (D) qRT-PCR was used to evaluate the expression of SPRR2B and myofibroblast markers in healthy human CFs treated with indicated combinations of TGF-β1 (10 ng/mL), IGF1 (100 ng/mL), and H2O2 (25 μM) (n = 3). (E) qRT-PCR was used to evaluate the expression of SPRR2B and mitotic regulators in human HF CFs treated with vehicle or IGF1/H2O2. (F) CyQuant dye incorporation to measure proliferation of human HF CFs treated with vehicle or IGF1/H2O2. All qRT-PCR data normalized to Gapdh and analyzed by one-way ANOVA (n = 3, unless indicated). CyQuant data analyzed by unpaired Student’s t test (n= 3). All data represent mean ± SEM. *P < 0.05; **P < 0.01. (G) Model highlighting the role of SPRR2B in regulating cell cycle progression in CFs. Heart disease (in vivo) or TGF-β1/ROS (in vitro) induce, while exercise (in vivo) or IGF1 (in vitro) repress SPRR2B gene expression in CFs. Phosphorylation of SPRR2B facilitates interaction with USP7/MDM2, leading to MDM2 accumulation and degradation of the nuclear p53 pool, relieving constraints on cell cycle progression and promoting fibrosis in heart disease.
To further evaluate the regulation and function of SPRR2B, we cultured normal healthy hCFs in a combination of H2O2 (25 μM) and TGF-β1 (10 ng/mL) or insulin-like growth factor 1 (IGF1) (100 ng/mL) to mimic pathological and physiological conditions, respectively; IGF1 stimulates physiological hypertrophy in skeletal myoblasts and CMs (12). We found that IGF1/H2O2 significantly reduced SPRR2B expression in healthy hCFs, whereas TGF-β1/H2O2 elevated SPRR2B expression relative to control treatment, again positively correlating with markers of myofibroblast activation (Fig. 7D). We therefore asked whether IGF1/H2O2 treatment of hCFs from HF patients might attenuate SPRR2B expression and normalize the expression of cell cycle regulators. While all three HF hCF isolates proliferated to a similar extent in response to varying concentrations of H2O2 (Fig. S6A), none of the HF hCF isolates displayed a significant increase in myofibroblast markers upon TGF-β1/H2O2 treatment, further indicating a high level of basal activation (Fig. S6B). IGF1/H2O2 treatment of HF hCFs decreased SPRR2B expression, and this reduction correlated with an increase in CDKN1A as well as a decrease in CDK1 expression (Fig. 7E). Importantly, IGF1/H2O2 treatment also significantly reduced proliferation of primary HF hCFs (Fig. 7F). We conclude that SPRR2B dysregulation is a conserved aspect of mouse and human HF that controls cell cycle regulators and CF proliferation in disease.
Discussion
This study reveals a unique and conserved molecular mechanism that stimulates CF accumulation during pathological cardiac remodeling. Expression of the SPRR2B gene is elevated in human HF and in a mouse model of pressure overload-induced ventricular hypertrophy and lost during exercise. We demonstrate that, within the heart, SPRR2B is restricted to CFs, where it functions as a regulatory subunit of the USP7/MDM2-containing p53 E3 ubiquitination ligase complex, facilitating MDM2 accumulation and p53 destruction (Fig. 7G). Thus, SPRR2B relieves a major constraint on CF cell cycle progression in response to pathological signals and may contribute to the development of pathological cardiac fibrosis.
SPRR2B function has previously been studied almost exclusively in the context of keratinocyte differentiation, where it is cross-linked into the cell membrane to form the CEnv during the process of epidermal barrier formation and programmed cell death (26). Although SPRR2B has not been directly implicated in cell death mechanistically, its incorporation into the CEnv is an essential step leading to cornification death. Interestingly, the USP7 interaction motif within the C terminus of SPRR2B identified in our study is cross-linked by TGM1 and TGM3 in keratinocytes (21). Thus, an interaction between SPRR2B and USP7 that may suppress p53 activity and block death signaling is unlikely in keratinocytes or DFs. In contrast, CFs do not express SPRR2B cross-linking enzymes or any of the CEnv components other than SPRR2B. The only transglutaminase we found to be significantly expressed in CFs is tissue transglutaminase (TGM2), which has a significantly lower affinity for SPRRs (21, 27, 28). Our study reveals that, in the absence of CEnv partners, SPRR2B plays a noncanonical role in the nucleus, where it contributes to cell cycle acceleration via p53 degradation. More broadly, our study expands on a paradigm whereby fibroblasts express gene programs that reflect their developmental origin and may influence their function (29).
The unique interaction between SPRR2B and USP7 that leads to MDM2 accumulation and p53 degradation suggests a more complex regulation of cell cycle control in CFs. USP7 is a context-dependent deubiquitinase that stabilizes either MDM2 or p53, accelerating or blocking cell cycle progression, respectively (9). Thus, regulation of MDM2–USP7 interaction is a primary determinant of p53 stability and cell cycle control. MDM2 interacts with USP7 via an N-terminal MATH domain (competitive interaction with p53) and at a C-terminal Ubl domain (responsible for p53 deubiquitination) (9, 30). Motifs present in SPRR2B predict the interaction with USP7 is through the C-terminal Ubl domain, and not through the MATH domain that might preclude MDM2-dependent p53 degradation (22). We also found that NRTK-dependent phosphorylation of SPRR2B at Y67 facilitated complex formation with USP7 and p53 degradation. We therefore anticipate SPRR2B may be the target of signaling pathways and additional binding partners that coordinate the regulation of cell cycle progression. Eukaryotic linear motif analysis predicted interaction sites for CDKs and CKS1, which suggest that cyclin–CDK–CKS1 complexes may directly interact with SPRR2B once in the nucleus (22). A proteomics approach might identify additional binding partners for SPRR2B and provide further insight into the regulation of the SPRR2B–USP7–MDM2–p53 complex. Taken together, our findings suggest that SPRR2B controls whether USP7/MDM2 is self-destructive or acts as the p53 E3 ubiquitin ligase, relieving constraints on cell cycle in CFs. Possible explanations for these findings include reduced deubiquitination of p53 by USP7, increased deubiquitination of MDM2 by USP7, or increased MDM2 activity, which may arise either via steric hindrance or alterations in the stoichiometry of complex formation by SPRR2B.
The development of cardiac fibrosis is associated with proliferation of Tcf21+ resident CFs and the transformation of quiescent CFs into Postn+ activated fibroblasts (31). Activated fibroblasts, called myofibroblasts, are characterized by the abundant expression of contractile proteins and secretion of ECM (3, 4). Although it is currently not clear whether fibroblast proliferation and myofibroblast activation are coincident or are separable cellular processes, cell proliferation and differentiation are mutually exclusive in many contexts, including progenitor cell self-renewal versus differentiation and smooth muscle cell phenotypic modulation (32–34). Even though SPRR2B gene expression positively correlates with both myofibroblast differentiation markers and promitotic factors, SPRR2B only functionally impacts CF proliferation. It remains possible that the heart harbors a heterogeneous population of CFs, some of which are proliferative SPRR2B+ cells and some of which are SPRR2B− myofibroblasts. Further studies using single-cell transcriptomics are required to discern the differences between individual CFs in health and disease that might differentially influence CF proliferation and ECM secretion.
RNA-seq revealed inverse regulation of Sprr2b expression by exercise and disease specifically in CFs. We were able to recapitulate this dichotomy in CFs derived from HF patients using in vitro models of physiological (IGF1/H2O2) and pathologic (TGF-β1/H2O2) signaling. These data suggest that regulation of Sprr2b expression represents a conserved transcriptional response to extrinsic signals in CFs. The Sprr2b promoter is complex, with several evolutionarily conserved transcription factor-binding sites upstream of the transcriptional start site and within the first intron that are relevant to cardiac stress signaling (e.g., Smad, NRF2, AHR, GABP, and SP1). Further study of this promoter region in CFs may reveal a paradigm of differential transcriptional regulation by pathological and physiological signaling.
In conclusion, this study defines an evolutionarily conserved mechanism controlling p53 stability that regulates resident CF proliferation, but not their differentiation into myofibroblasts. Scar formation by myofibroblasts is an essential aspect of cardiac repair that provides structural support in pressure overload and prevents ventricle rupture after myocardial infarction (35, 36). However, accumulation of fibroblasts in the injured heart ultimately leads to pathological fibrosis and deterioration of cardiac function. Our findings indicate a provocative therapeutic strategy may be blocking excessive CF proliferation and accumulation while allowing preexisting resident CFs to differentiate into ECM-producing cells. Chemotherapeutic agents typically target replication checkpoint controls, a general approach that would not be amenable to cardiac therapeutics. Targeting CF-enriched and stress-responsive promitotic factors such as SPRR2B may prevent pathological cardiac remodeling while limiting off-target effects.
Materials and Methods
All animal work was approved by the University Committee on Animal Research at the University of Rochester. Human tissue samples were collected following informed consent, and all experiments involving human subjects were performed under the auspices of protocols approved by University of Rochester or Cleveland Clinic Research Subjects Review Board.
Animal Models.
TAC and swim training models were performed as described in SI Materials and Methods.
Primary Cell Isolations.
Isolation of adult mouse ventricular fibroblasts, neonatal mouse ventricular and dermal fibroblasts, and human CFs is described in SI Materials and Methods. CFs were used between passages 1 and 7 for experiments.
Constructs and Mutagenesis.
Constructs and their sources may be found in Tables S3 and S4.
RNA Isolation and Analyses.
RNA-seq and qRT-PCR analysis are described in SI Materials and Methods.
Proliferation Assays.
Proliferation was assessed by CyQuant assay (Invitrogen) using manufacturer instructions using a multiwell plate reader (BMG FluoStar Optima).
Flow Cytometry.
Cells were trypsinized with 2.5% trypsin–EDTA, resuspended in PBS with 0.2% BSA, and then fixed with cold 70% ethanol. Cell suspensions were then stained with propidium iodide containing RNase A (BD Pharmingen). Flow cytometry was accomplished using a three-laser, 12-color BD LSR-II with excitation at 610/20 nm (BD Biosciences). Data were analyzed using FlowJo.
Cellular Fractionation.
Separation of the nuclear and nonnuclear cellular fractions was performed as described in Materials and Methods.
Western Blotting and Immunoprecipitation.
Western blotting and immunoprecipitation were performed as described in SI Materials and Methods. Antibodies used are listed in Table S5.
Immunofluorescence.
Immunofluorescence was performed as described in SI Materials and Methods. Primary antibody dilutions may be found in Table S5.
Image Analysis.
Masson Trichrome staining was quantified with the use of the K-means clustering algorithm in NIH ImageJ. Nuclear counting was performed by particle analysis with NIH ImageJ. Western blot densitometry was performed in NIH ImageJ using the Gels plugin.
Statistical Analysis.
Data are presented as the mean ± SEM. Statistical differences between two groups were determined using Student’s unpaired t test with Welch’s correction. For comparisons between more than two groups, one-way or two-way ANOVA was used, as indicated in figure legend. Significance was considered as P < 0.05.
Supplementary Material
Acknowledgments
This work was supported by NIH/National Heart, Lung, and Blood Institute Grants R01HL133761 and R01HL120919 (to E.M.S.), T32HL007937 and F32HL136066 (to R.M.B.), T32HL066988 (to J.K.L.), and T32HL066988 and F32HL134206 (to P.Q.); American Heart Association Grant 15POST25550114 (to J.K.L.); and a Pilot Study Grant from the Aab Cardiovascular Research Institute at the University of Rochester School of Medicine and Dentistry. E.M.S. was supported in part by a grant from Novartis Pharmaceuticals.
Footnotes
Conflict of interest statement: E.M.S. is the recipient of a research grant from Novartis Pharmaceuticals.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE89885).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717423115/-/DCSupplemental.
References
- 1.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 2.Mozaffarian D, et al. Writing Group Members; American Heart Association Statistics Committee; Stroke Statistics Subcommittee Heart disease and stroke statistics—2016 update: A report from the American Heart Association. Circulation. 2016;133:e38–e360. doi: 10.1161/CIR.0000000000000350. [DOI] [PubMed] [Google Scholar]
- 3.Hinz B, et al. The myofibroblast: One function, multiple origins. Am J Pathol. 2007;170:1807–1816. doi: 10.2353/ajpath.2007.070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat Rev Mol Cell Biol. 2002;3:349–363. doi: 10.1038/nrm809. [DOI] [PubMed] [Google Scholar]
- 5.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 6.Ubil E, et al. Mesenchymal-endothelial transition contributes to cardiac neovascularization. Nature. 2014;514:585–590. doi: 10.1038/nature13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamaki Y, et al. Metastasis-associated protein, S100A4 mediates cardiac fibrosis potentially through the modulation of p53 in cardiac fibroblasts. J Mol Cell Cardiol. 2013;57:72–81. doi: 10.1016/j.yjmcc.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Vousden KH, Lu X. Live or let die: The cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 9.Faesen AC, Luna-Vargas MP, Sixma TK. The role of UBL domains in ubiquitin-specific proteases. Biochem Soc Trans. 2012;40:539–545. doi: 10.1042/BST20120004. [DOI] [PubMed] [Google Scholar]
- 10.Kaplan ML, et al. Cardiac adaptations to chronic exercise in mice. Am J Physiol. 1994;267:H1167–H1173. doi: 10.1152/ajpheart.1994.267.3.H1167. [DOI] [PubMed] [Google Scholar]
- 11.Braitsch CM, Kanisicak O, van Berlo JH, Molkentin JD, Yutzey KE. Differential expression of embryonic epicardial progenitor markers and localization of cardiac fibrosis in adult ischemic injury and hypertensive heart disease. J Mol Cell Cardiol. 2013;65:108–119. doi: 10.1016/j.yjmcc.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McMullen JR, et al. Protective effects of exercise and phosphoinositide 3-kinase(p110alpha) signaling in dilated and hypertrophic cardiomyopathy. Proc Natl Acad Sci USA. 2007;104:612–617. doi: 10.1073/pnas.0606663104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McMullen JR, et al. The insulin-like growth factor 1 receptor induces physiological heart growth via the phosphoinositide 3-kinase(p110alpha) pathway. J Biol Chem. 2004;279:4782–4793. doi: 10.1074/jbc.M310405200. [DOI] [PubMed] [Google Scholar]
- 14.McMullen JR, et al. Phosphoinositide 3-kinase(p110alpha) plays a critical role for the induction of physiological, but not pathological, cardiac hypertrophy. Proc Natl Acad Sci USA. 2003;100:12355–12360. doi: 10.1073/pnas.1934654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayeni JO, et al. Dual phosphorylation of Cdk1 coordinates cell proliferation with key developmental processes in Drosophila. Genetics. 2014;196:197–210. doi: 10.1534/genetics.113.156281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinert PM, Marekov LN. Initiation of assembly of the cell envelope barrier structure of stratified squamous epithelia. Mol Biol Cell. 1999;10:4247–4261. doi: 10.1091/mbc.10.12.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steinert PM, et al. Transglutaminase crosslinking and structural studies of the human small proline rich 3 protein. Cell Death Differ. 1999;6:916–930. doi: 10.1038/sj.cdd.4400568. [DOI] [PubMed] [Google Scholar]
- 18.Candi E, et al. Acquisition of ordered conformation by the N-terminal domain of the human small proline rich 2 protein. Biochem Biophys Res Commun. 1999;262:395–400. doi: 10.1006/bbrc.1999.1215. [DOI] [PubMed] [Google Scholar]
- 19.Candi E, et al. Transglutaminase cross-linking properties of the small proline-rich 1 family of cornified cell envelope proteins. Integration with loricrin. J Biol Chem. 1999;274:7226–7237. doi: 10.1074/jbc.274.11.7226. [DOI] [PubMed] [Google Scholar]
- 20.Steinert PM, Candi E, Kartasova T, Marekov L. Small proline-rich proteins are cross-bridging proteins in the cornified cell envelopes of stratified squamous epithelia. J Struct Biol. 1998;122:76–85. doi: 10.1006/jsbi.1998.3957. [DOI] [PubMed] [Google Scholar]
- 21.Tarcsa E, et al. Structural and transglutaminase substrate properties of the small proline-rich 2 family of cornified cell envelope proteins. J Biol Chem. 1998;273:23297–23303. doi: 10.1074/jbc.273.36.23297. [DOI] [PubMed] [Google Scholar]
- 22.Dinkel H, et al. ELM 2016—data update and new functionality of the eukaryotic linear motif resource. Nucleic Acids Res. 2016;44:D294–D300. doi: 10.1093/nar/gkv1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornbeck PV, et al. PhosphoSitePlus: A comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizuguchi Y, et al. SPRR2A enhances p53 deacetylation through HDAC1 and down regulates p21 promoter activity. BMC Mol Biol. 2012;13:20. doi: 10.1186/1471-2199-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demetris AJ, et al. Small proline-rich proteins (SPRR) function as SH3 domain ligands, increase resistance to injury and are associated with epithelial-mesenchymal transition (EMT) in cholangiocytes. J Hepatol. 2008;48:276–288. doi: 10.1016/j.jhep.2007.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eckhart L, Lippens S, Tschachler E, Declercq W. Cell death by cornification. Biochim Biophys Acta. 2013;1833:3471–3480. doi: 10.1016/j.bbamcr.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 27.Martinet N, et al. Epidermal and hair follicle transglutaminases. Partial characterization of soluble enzymes in newborn mouse skin. J Biol Chem. 1988;263:4236–4241. [PubMed] [Google Scholar]
- 28.Candi E, et al. Biochemical, structural, and transglutaminase substrate properties of human loricrin, the major epidermal cornified cell envelope protein. J Biol Chem. 1995;270:26382–26390. doi: 10.1074/jbc.270.44.26382. [DOI] [PubMed] [Google Scholar]
- 29.Furtado MB, et al. Cardiogenic genes expressed in cardiac fibroblasts contribute to heart development and repair. Circ Res. 2014;114:1422–1434. doi: 10.1161/CIRCRESAHA.114.302530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Epping MT, et al. TSPYL5 suppresses p53 levels and function by physical interaction with USP7. Nat Cell Biol. 2011;13:102–108. doi: 10.1038/ncb2142. [DOI] [PubMed] [Google Scholar]
- 31.Kanisicak O, et al. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nat Commun. 2016;7:12260. doi: 10.1038/ncomms12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das A, et al. Stromal-epithelial crosstalk regulates kidney progenitor cell differentiation. Nat Cell Biol. 2013;15:1035–1044. doi: 10.1038/ncb2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, et al. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen AT, et al. Smooth muscle cell plasticity: Fact or fiction? Circ Res. 2013;112:17–22. doi: 10.1161/CIRCRESAHA.112.281048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeda N, et al. Cardiac fibroblasts are essential for the adaptive response of the murine heart to pressure overload. J Clin Invest. 2010;120:254–265. doi: 10.1172/JCI40295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frangogiannis NG. The inflammatory response in myocardial injury, repair, and remodelling. Nat Rev Cardiol. 2014;11:255–265. doi: 10.1038/nrcardio.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.