Significance
How protein aggregation leads to neurodegenerative disorders such as Huntington’s disease remains poorly understood. Here we show that polyglutamine (polyQ) aggregation in yeast results in the formation of amorphous inclusions and less frequent fibrils. This is accompanied by significant changes in proteome composition as well as distortions in mitochondria and lipid droplet morphology that do not arise from direct interactions of these organelles with polyQ inclusions or fibrils. This contrasts with recent observations in mammalian cells, where the same polyQ proteins formed amyloid-like fibrils that distort endoplasmic reticulum membranes. These results demonstrate that the same polyQ expansion protein can adopt different nonnative conformations that utilize distinct mechanisms to target a variety of cellular structures.
Keywords: neurodegeneration, protein aggregation, cryo-electron microscopy, cryo-focused ion beam milling, label-free mass spectrometry
Abstract
Huntington’s disease is caused by the expansion of a polyglutamine (polyQ) tract in the N-terminal exon of huntingtin (HttEx1), but the cellular mechanisms leading to neurodegeneration remain poorly understood. Here we present in situ structural studies by cryo-electron tomography of an established yeast model system of polyQ toxicity. We find that expression of polyQ-expanded HttEx1 results in the formation of unstructured inclusion bodies and in some cases fibrillar aggregates. This contrasts with recent findings in mammalian cells, where polyQ inclusions were exclusively fibrillar. In yeast, polyQ toxicity correlates with alterations in mitochondrial and lipid droplet morphology, which do not arise from physical interactions with inclusions or fibrils. Quantitative proteomic analysis shows that polyQ aggregates sequester numerous cellular proteins and cause a major change in proteome composition, most significantly in proteins related to energy metabolism. Thus, our data point to a multifaceted toxic gain-of-function of polyQ aggregates, driven by sequestration of endogenous proteins and mitochondrial and lipid droplet dysfunction.
Expansion of a polyglutamine (polyQ) tract in otherwise unrelated proteins is the cause of several inherited neurodegenerative diseases, including ataxias and Huntington’s disease (HD) (1). However, the exact length of polyQ tract that can be tolerated, the affected cell types, and the clinical phenotypes observed for the different mutant proteins vary widely, consistent with an important role of the sequences flanking the polyQ tract and the cellular environment in modulating pathology (2–9).
HD is caused by the expansion of a polyQ repeat in the N-terminal exon of huntingtin (HttEx1), which leads to protein aggregation and cell death (2, 10, 11). However, the link between huntingtin aggregation and neurodegeneration remains poorly understood, partly because toxic loss- and gain-of-function effects of mutant huntingtin likely coexist in HD. As no huntingtin homologs exist in yeast, Saccharomyces cerevisiae can be utilized to study cellular toxic gain-of-function effects caused by mutant HttEx1 in isolation (12, 13).
Here we explore the mechanisms of HttEx1 aggregation and toxicity in yeast by combining MS-based proteome and interactome analyses with in situ structural studies by cryo-electron tomography (cryo-ET). While quantitative MS allows the global and comprehensive analysis of cellular phenomena (14), cryo-ET provides 3D images of fully hydrated cells in a close-to-native state and at molecular resolution (15). Our data show that polyQ-expanded proteins in yeast form unstructured inclusions that coexist with less frequent fibrils. Potentially harmful interactions of the aggregates with cellular membranes were not detected. This contrasts with our recent study of polyQ inclusions in mammalian cells, where inclusions consist of fibrils that interact extensively with cellular endomembranes (16). These findings suggest that differences in the protein homeostasis network (17) are critical in shaping aggregate morphology and toxicity. Furthermore, expression of polyQ expansion proteins in yeast resulted in morphological alterations of mitochondria and lipid droplets (LDs) and in reduced levels of proteins related to energy metabolism, some of which were sequestered by polyQ aggregates.
Results
PolyQ Aggregation in Yeast.
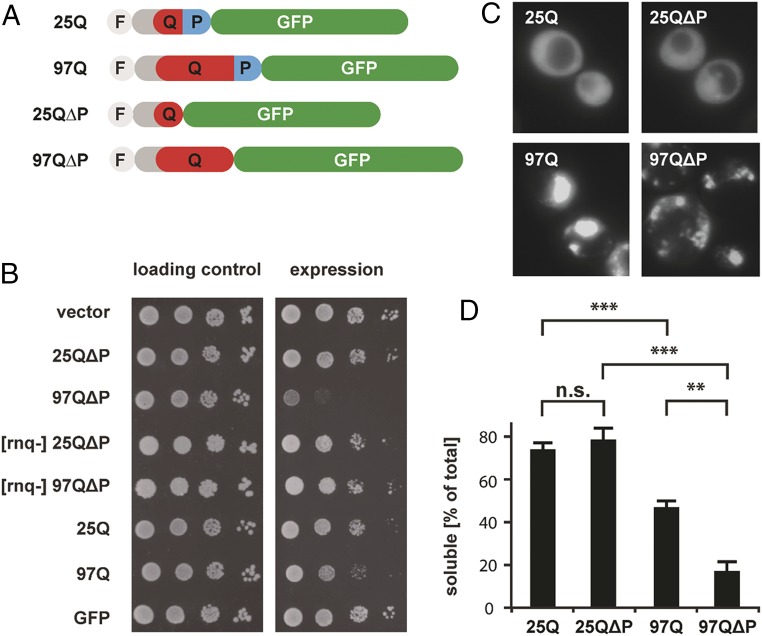
PolyQ-length–dependent toxicity can be reproduced in yeast cells (12, 13). Expression of HttEx1 with an expanded polyQ stretch and a C-terminal GFP tag (97Q) resulted in a growth defect, whereas expression of a construct with a short polyQ stretch (25Q) or GFP alone did not cause any observable toxicity (Fig. 1 A and B). As shown previously (4, 9), deletion of the proline-rich region located at the C terminus of the polyQ expansion (97QΔP) enhanced polyQ-length–dependent toxicity (Fig. 1 A and B) and caused an increase in the amount of detergent-insoluble polyQ aggregates (Fig. 1D and Fig. S1 A and C). Deletion of the proline-rich region in a construct with short polyQ length (25QΔP) did not result in toxicity (Fig. 1 A and B). As expected, the constructs with expanded polyQ tracts were more aggregation-prone than those with short polyQ lengths (Fig. 1D and Fig. S1 B and C) and were localized in fluorescent inclusions visible by light microscopy (Fig. 1C). Inclusions of 97QΔP were smaller and more numerous than those formed by 97Q containing the proline-rich flanking region (Fig. 1C). Thus, 97QΔP likely presents an overall larger aggregate surface than 97Q.
Fig. 1.
Toxicity and aggregation of HttEx1 constructs are influenced by polyQ repeat length and flanking regions. (A) Schematic representation of the HttEx1 constructs used in this study. Light gray indicates the N-terminal Flag-tag (F), dark gray the 17 N-terminal amino acids of HttEx1, red the polyQ stretch (Q), blue the proline-rich region (P), and green the GFP tag. (B) Growth test of cells expressing different HttEx1 constructs. Serial dilutions of cells were spotted on plates containing galactose to express the indicated constructs (Right) or on glucose plates as loading control (Left). Expression of 97Q and 97Q∆P causes mild and strong toxicity, respectively, that is dependent on the presence of the [RNQ+] prion (52). (C) Light microscopy imaging of cells expressing the indicated HttEx1 constructs. (D) Percentage of total HttEx1 in the soluble fraction after differential centrifugation (see also Fig. S1C). **P < 0.01, *** P < 0.001 by t test; n.s., not significant.
Proteomic Profiling of PolyQ Expression.
To investigate the cellular response to different polyQ aggregates, we performed global proteome analyses using a quantitative label-free MS workflow (18). We detected an extensive remodeling of the cellular proteome in the presence of polyQ-expanded constructs (Fig. 2A and Dataset S1). Cytosolic chaperones and other components of the cellular stress response, including Hsp26, Hsp104, or SSA1/2, were increased in abundance by 1.5-fold to fourfold (Dataset S1). Gene Ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis demonstrated that proteins with functions in transcription regulation and translation were also increased. In contrast, the abundance of proteins related to energy metabolism, especially the main mitochondrial metabolic pathways such as the tricarboxylic acid (TCA) cycle or oxidative phosphorylation, was significantly reduced upon expression of expanded polyQ.
Fig. 2.
PolyQ aggregation causes proteome alterations and coaggregation of endogenous proteins. Complete proteome (A), proteome of a LD-enriched light fraction obtained by fractionation of cell lysates in a sucrose gradient (B), and interactome (C) analysis of polyQ-expressing cells. (A and B) Heat maps show unsupervised hierarchical clustering of the median MaxLFQ intensities (log2, z-scored) of significantly increased or reduced proteins [ANOVA, 5% false-discovery rate (FDR)]. Enriched GO-annotations and KEGG pathways for the indicated clusters are shown (Fisher’s test, 5% FDR). Note that mitochondrial proteins are strongly reduced in the global and LD proteomes upon polyQ expansion. (C, Upper) Volcano plots show pairwise comparisons of the interactomes of the indicated HttEx1 constructs. Significantly changed proteins are indicated in red (Welch’s t test, S0: 1, 5% FDR). (Lower) The histograms (log counts) show the shift of selected annotations (green: prions according to ref. 53; blue: KEGG TCA cycle) compared with all proteins (gray).
Nonproductive interactions of aggregates with endogenous proteins may contribute to toxicity by the sequestration and functional impairment of the interacting proteins (19–21). Therefore, we analyzed the interactions of cellular proteins with the different HttEx1 constructs. Compared with control proteins with short polyQ stretches, the polyQ-expanded constructs showed a dramatic increase in the number of interacting proteins (Fig. 2C). This likely reflects the cellular interactions of different polyQ-expanded protein species, ranging from large inclusions to smaller oligomeric species and monomers. In agreement with previous work (19, 22, 23), proteins involved in a wide variety of cellular pathways, including transcription, protein synthesis, protein quality control and metabolism, as well as prion-like and mitochondrial proteins, were enriched in the 97Q and 97QΔP interactomes (Fig. 2C and Dataset S2). Some of the proteins enriched in these interactomes, e.g., those related to transcription/translation, were also increased in the total proteome (Fig. 2A), presumably reflecting a cellular attempt to compensate for their sequestration. In contrast, mitochondrial proteins were both globally reduced (Fig. 2A) and preferentially sequestered (Fig. 2C and Dataset S2) by the polyQ aggregates, consistent with reports of mitochondrial dysfunction upon expanded polyQ expression from yeast to HD patients (24, 25). Interactors included mitochondrially translated proteins, suggesting that at least some mitochondria–polyQ interactions occur upon mitochondrial disruption.
Both polyQ-expanded constructs had very similar interaction profiles, and the same was true for both control proteins with short polyQ length (Fig. 2C and Dataset S2), indicating that the major driver of protein interactions is the expansion of the polyQ stretch and the concomitant increase in aggregation propensity. Together, these data suggest that even though 97QΔP expression leads to higher toxicity than 97Q, both constructs interfere with similar cellular pathways and therefore may cause toxicity by similar mechanisms but with different propensities.
Structure of PolyQ Aggregates.
We employed cryo-ET to investigate the effects of expanded polyQ constructs on cellular ultrastructure. Cells expressing the HttEx1 proteins were vitrified and thinned using cryo-focused ion beam (FIB) milling (26), resulting in 150- to 250-nm-thick lamellas that were examined by cryo-ET (Fig. S2).
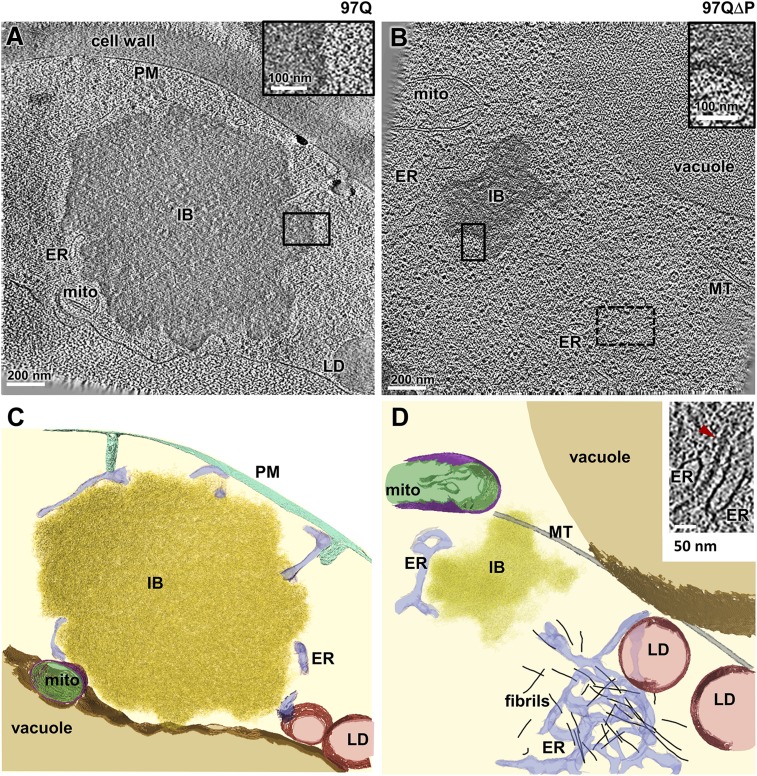
Large, globular electron-dense structures were found in cells expressing high levels of 97Q and 97QΔP (Fig. 3 and Movie S1) but not in those expressing control proteins. Correlative cryo-light microscopy revealed that the electron-dense structures appeared in cells with 97Q or 97QΔP foci (Fig. S3), indicating that they represent inclusions of polyQ-expanded proteins. The inclusions in 97Q-expressing cells were larger than those in 97QΔP-expressing cells (700 ± 300 nm vs. 420 ± 170 nm in diameter; mean ± SD; P < 0.05 by unpaired t test), consistent with our observations by light microscopy (Fig. 1C) and previous reports (4). PolyQ inclusions were typically found in close proximity to mitochondria and the endoplasmic reticulum (ER) and often at the sites in which these organelles engaged in membrane contacts, as reported for stress-induced aggregates (27). Except for their difference in size, the 97Q and 97QΔP inclusions were morphologically indistinguishable, as they both consisted of an apparently amorphous density delimited by an irregular boundary (Fig. 3 and Movie S1).
Fig. 3.
97Q and 97QΔP inclusion bodies are morphologically similar. (A and B) Tomographic slices of yeast cells expressing the 97Q (A) or 97QΔP (B) constructs. ER, endoplasmic reticulum; IB, inclusion body; LD, lipid droplet; mito, mitochondria; PM, plasma membrane. (Insets) Higher magnifications of the boxed areas. (C and D) Corresponding 3D renderings. Color coding: cytosol, light yellow; endoplasmic reticulum, blue; fibrils, black; inclusion bodies, gold; lipid droplets, red; mitochondria inner membrane and matrix, green; mitochondria outer membrane, purple; plasma membrane, turquoise; vacuole, brown. Note that, other than their difference in size, 97Q and 97Q∆P inclusions are morphologically indistinguishable. (Inset) Higher magnification of a different tomographic slice of the area marked by the dotted rectangle in B showing a fibril (red arrowhead) in the vicinity of ER membranes. Tomographic slices are 2.1 nm thick.
Amyloid-like fibrils were also observed in the cytoplasm of some cells expressing the polyQ-expanded HttEx1 constructs (Fig. 3D, Inset and Movie S1) but at a lower frequency than amorphous inclusions (one of seven and 6 of 14 aggregates analyzed were fibrillar in 97Q- and 97QΔP-expressing cells, respectively). The fibrils were 340 ± 110 nm (mean ± SD) in length and 16 ± 3 nm in diameter, considerably thicker than actin, intermediate filaments, or prion fibers. Also, unlike prions (28, 29), the fibrils appeared in clusters without apparent internal organization, in most cases close to the ER. These data suggest that the fibrils correspond to another, amyloid-like form of polyQ aggregate. Despite their close proximity to various organelles, no aberrant interactions of either fibrils or unstructured polyQ inclusions with cellular membranes were detectable, in contrast to our recent observations in mammalian cells (16). We note, however, that such interactions may be transient, escaping detection.
Morphological Alterations in Cells Containing PolyQ Aggregates.
Cryo-FIB–milled lamellas of cells expressing 97QΔP often contained dead cells in which neither intact organelles nor macromolecules were visible. Instead, a characteristic electron-dense multilayered structure lined the cell interior (Fig. S4), surrounding vesicles reminiscent of those observed in the vacuolar lumen of live cells and occasionally containing remnants of organelles such as mitochondria (Fig. S4B). Consistent with toxicity assays (Fig. 1B), dead cells were found less often upon expression of 97Q (P < 0.05 by χ2 test) (Fig. S4C) and were essentially absent in preparations of control cells (P < 0.05 by χ2 test) (Fig. S4C).
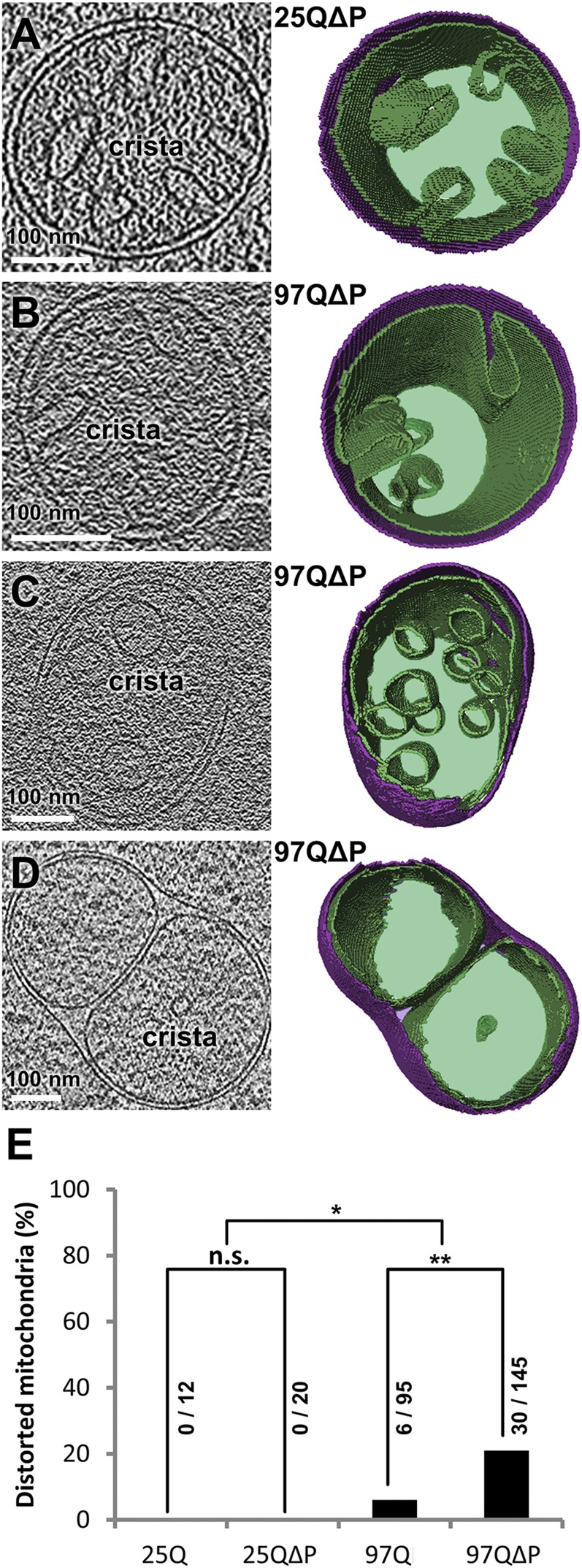
Interestingly, mitochondrial morphology was altered in otherwise healthy-looking cells expressing 97Q or 97QΔP to an extent correlating with the relative toxicity of these constructs. Inner membrane cristae were distorted or even absent in ∼20% of mitochondria in 97QΔP-expressing cells, significantly more than in 97Q cells (P < 0.01 by χ2 test) (Fig. 4), whereas no distorted mitochondria were observed in cells expressing the control proteins (P < 0.05 by χ2 test) (Fig. 4). Therefore, in line with our proteomics results (Fig. 2), the cryo-ET data suggest that mitochondrial dysfunction plays an important role in polyQ-induced toxicity in yeast.
Fig. 4.
Alterations in mitochondria morphology in cells expressing polyQ-expanded HttEx1. (A–D, Left) Tomographic slices of mitochondria from cells expressing 25QΔP (A) or 97QΔP (B–D). Tomographic slices are 2.1 nm (A and D) or 1.4 nm (B and C) thick. (Right) Corresponding 3D renderings. The outer mitochondrial membrane is shown in purple, and the inner mitochondrial membrane and matrix are shown in green. B shows a mitochondrion with normal cristae morphology, C shows a mitochondrion with globular cristae, and D shows a mitochondrion with almost no visible cristae. (E) Quantification of the percentage of distorted mitochondria (all mitochondria distortions pooled). The numbers above the bars indicate the number of distorted mitochondria and the total number of mitochondria observed in the tomograms taken for each condition. Note that the frequency of mitochondrial distortions correlates with the toxicity of the HttEx1 constructs. *P < 0.05, **P < 0.01 by χ2 test; n.s., not significant.
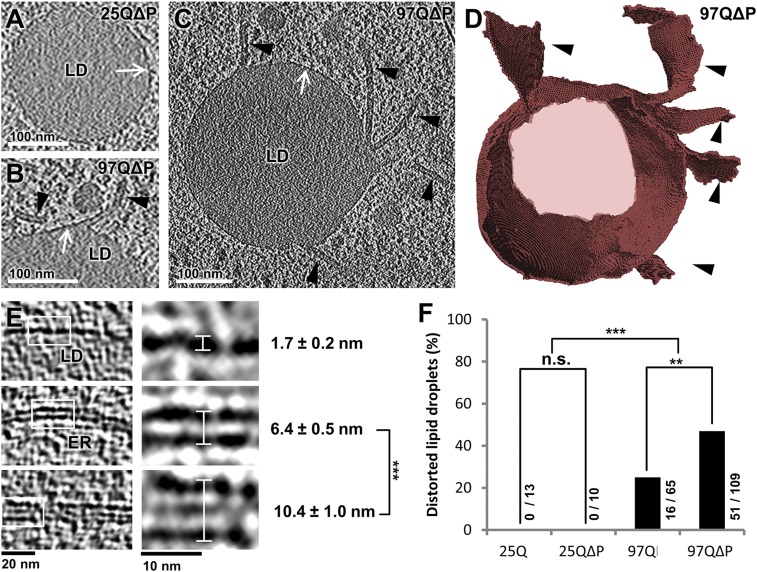
Another striking feature of cells expressing polyQ-expanded constructs was a dramatic distortion in the morphology of LDs. LDs are the main cellular lipid storage depots and play important roles in the regulation of lipid metabolism. LD alterations have been linked to HD (30, 31), but the detailed study of LD structure in HD has been hindered by the sensitivity of LD morphology to the techniques of sample preparation used in conventional EM (32). Our cryo-ET data show that, unlike the smooth boundary of LDs in control cells, large planar protrusions often emanated from the surface of LDs in cells expressing polyQ-expanded constructs (Fig. 5 A–D). A detailed analysis demonstrated that the protrusion boundary was continuous with the LD phospholipid monolayer (Fig. 5B). The protrusions were significantly thicker than phospholipid bilayers such as the ER membrane (10.4 ± 1.0 nm vs. 6.4 ± 0.5 nm, mean ± SD, P < 0.001 by unpaired t test) (Fig. 5E), and an intermediate density was often observed between the delimiting monolayers (Fig. 5E, Bottom). The frequency of these distortions correlated with the toxicity of the different polyQ constructs, as they were found to be nearly twice as frequent in LDs of 97QΔP-expressing cells than in cells expressing 97Q (P < 0.01 by χ2 test) (Fig. 5F) and never occurred in cells expressing the control polyQ proteins (P < 0.001 by χ2 test) (Fig. 5F). Neither mitochondrial nor LD distortions seemed to be caused by direct physical contact with inclusions or fibrils.
Fig. 5.
Expanded polyQ-induced distortions in LDs. (A–C) Tomographic slices of LDs from cells expressing 25QΔP (A) or 97QΔP (B and C). White arrows point to the LD phospholipid monolayer, and black arrowheads mark planar protrusions. (D) 3D rendering of C. (E) Tomographic slices at lower (Left) and higher (Right) magnification of a LD phospholipid monolayer (Top), a phospholipid bilayer (ER membrane; Middle), and a LD planar protrusion (Bottom). The measured thickness of each of these structures is shown (white lines). Tomographic slices are 2.1 nm (A) or 1.4 nm (B–E) thick. ***P < 0.001 by t test. (F) Quantification of the percentage of distorted LDs. The numbers beside or above the bars indicate the number of distorted LDs and the total number of LDs observed in the tomograms taken for each condition. Note that the frequency of LD distortions correlates with the toxicity of the HttEx1 constructs. **P < 0.01, ***P < 0.001 by χ2 test; n.s., not significant.
Since the proteome data hinted at metabolic alterations (Fig. 2) that may be related to such changes in LD morphology, we analyzed by MS the composition of an LD-enriched light fraction obtained by fractionation of cell lysates in a sucrose gradient. We did not detect any changes in the lipid composition of whole cells or of the light fraction that were indicative of polyQ-length–dependent effects on the lipidome of LDs (Fig. S5). Also, no significant changes were found in the abundance of LD proteins such as FAT1, TGL1, or DGA1. However, there was a strong decrease in the levels of mitochondrial proteins in the LD-enriched light fraction of lysates from cells expressing expanded polyQ constructs (Fig. 2B). To investigate this phenomenon, we performed MS-based protein correlation profiling (PCP) (33), which allows the generation of characteristic profiles reflecting the subcellular localization of proteins along the complete sucrose gradient (Fig. S6A and Dataset S3). Whereas in control lysates LD proteins were almost exclusively found in the light fractions of the gradient, mitochondrial proteins showed a bimodal distribution. The majority of mitochondrial proteins migrated to higher densities, but a second peak was detected in the light fraction in agreement with previous findings (33). Notably, the content of mitochondrial proteins in the LD-enriched light fraction was strongly reduced in cells expressing polyQ-expanded constructs (Fig. S6B), indicating that the colocalization of LD and mitochondrial proteins reflects a bona fide interaction between these organelles, which may be disrupted upon expression of proteins with expanded polyQ tracts. Thus, the morphological alterations observed in mitochondria and LDs of cells expressing polyQ-expanded proteins may be linked with a set of mitochondrial proteins whose distribution between these two compartments is disturbed.
Together, our cryo-ET and proteomic analyses strongly suggest that mitochondrial and LD dysfunctions contribute to polyQ toxicity in yeast, with 97QΔP causing more severe impairment than 97Q.
Discussion
In HD and related neurodegenerative pathologies, the expansion of a polyQ tract in specific disease proteins leads to protein aggregation and neuronal death (1). How the aggregation process and cellular toxicity are related is not well understood. Here we combined quantitative proteomic analyses by MS with high-resolution cryo-ET to provide insights into the cellular mechanisms of polyQ toxicity. The well-documented influence of flanking sequences on the aggregation propensity and toxicity of expanded polyQ tracts in yeast (4, 9) allowed us to study the effects of aggregating proteins with an identical number of polyQ repeats but different levels of toxicity. Although the toxicity of polyQ-expanded HttEx1 constructs was exacerbated by deletion of the C-terminal proline-rich sequence, our proteomic and cryo-ET analysis revealed surprisingly similar interaction profiles and morphological properties of 97Q and 97QΔP aggregates.
Both polyQ-expanded constructs engaged in numerous aberrant interactions with endogenous proteins and caused a major remodeling of the cellular proteome. The pathways affected were similar for both constructs. This suggests that these cellular interactions are mediated mainly by the polyQ region, whereas the C-terminal proline-rich sequence mainly modulates polyQ aggregation propensity, in agreement with previous reports (5–8, 34). The pathways identified here overlap partially with a recent study of the polyQ interactome in neuroblastoma cells (35), indicating that some, but not all, of the cellular effects of expanded polyQ expression are common to both systems.
We applied a FIB milling/cryo-ET workflow to produce 3D images of HttEx1 polyQ aggregates within their fully hydrated, unstained cellular environment. In these close-to-native conditions, polyQ inclusions appeared unstructured, with indistinguishable morphologies for 97Q and 97QΔP, although 97Q inclusions were generally larger. In addition to the amorphous aggregates, we also observed cytoplasmic fibrils at a lower frequency. These fibrils most likely represent amyloid-like aggregates of polyQ-expanded HttEx1 similar to those observed in vitro (11, 12) and in situ within mammalian cells (16). The fibrils were sometimes found close to amorphous inclusions, indicating that both aggregated species coexist, as recently shown in mammalian cells (36). It is possible that the balance between these types of aggregates is influenced by other proteins with prion-like domains, chaperones, the length of the polyQ tract, or the expression level of the HttEx1 proteins. Despite their proximity to and even direct contact with membranes, polyQ inclusions and fibrils did not cause membrane deformations or any other visible deleterious effects.
The properties of polyQ aggregates in yeast contrast with our recent observations of polyQ inclusions in primary murine neurons and human cells (16), where 97Q and 64Q HttEx1 constructs give rise to exclusively fibrillar inclusions. In these cells, polyQ fibrils strongly interact with and deform cellular membranes, suggesting that fibrils may be disease-relevant and not simply inert end-stage products of aggregation (37). Therefore, the morphology and cellular interactions of similar HttEx1 polyQ constructs vary under different cellular environments. In this respect, notable differences between the chaperone machineries of yeast and mammals may be particularly relevant. For example, while Hsp104 is a designated disaggregase in yeast that can remodel polyQ aggregates (13), metazoans have lost this enzyme and instead employ a trimeric chaperone complex that acts on fibrillar forms of HttEx1 (38).
We have recently shown that heat shock-induced aggregates of endogenous proteins in yeast also adopt an amorphous morphology (39). These aggregates appeared similar to but less compact than polyQ inclusions, consistent with the notion that coaggregation with endogenous proteins may result in the unstructured morphology of polyQ inclusions. Heat shock-induced aggregates also associated with the membranes of the ER without causing visible deformations. Therefore, the interaction of ER membranes with cytoplasmic aggregates may be a general phenomenon (27, 40).
We found morphological distortions in the cristae of mitochondria in cells expressing 97Q and, to a larger extent, upon expression of 97QΔP. Furthermore, the total levels of proteins related to mitochondrial function were strongly reduced upon expression of the polyQ-expanded constructs, and mitochondrial proteins were substantially sequestered by polyQ aggregates. Thus, our data suggest that the previously reported polyQ-induced mitochondrial dysfunction documented in HD patients (24, 25) and model systems such as yeast (41, 42) is caused by aberrant interactions between polyQ aggregates and mitochondrial proteins that lead to a global dysregulation of mitochondrial proteins. While the exact nature of the toxic interactions remains to be elucidated, polyQ oligomers (35) small enough to escape detection by EM may play an important role. Given that 97QΔP is less soluble than 97Q but forms smaller inclusions, the concentration of polyQ oligomers is probably higher in 97QΔP-expressing cells, leading to more severe toxicity. Thus, the size of the aggregating species may play an important role in its toxicity (35, 37). Additionally, we cannot rule out contributions from upstream toxic events.
Proteome analysis revealed further metabolic alterations that could underlie our observation of structural distortions in LDs of cells expressing polyQ-expanded proteins. Similar to our observations in mitochondria, the degree of LD distortion correlated with toxicity and was most prominent in 97QΔP-expressing cells. LDs often showed planar protrusions that seemed to be delimited by monolayers but were substantially thicker than bona fide bilayers, constituting a distinct type of membrane structure. This morphology is reminiscent of defatted LDs (43) and conditions in which LD fatty acid mobilization to peroxisomes may become rate-limiting (44, 45), although the polyQ-length–dependent changes in LD lipid composition observed here were minimal. Mitochondria might also be implicated in this interface, as the most dramatic change in the proteome of an LD-enriched fraction was a strong reduction of mitochondrial proteins.
Interestingly, an increasing number of studies have linked alterations in LD function with neurodegeneration (46). Furthermore, huntingtin is widely expressed in different organs, suggesting that HD be may not only a neurological syndrome but also a systemic disease (47). HD patients often suffer from metabolic disorders that may be related to the defects in triglyceride storage observed in HD mouse models (48, 49), which may arise from LD abnormalities in the adipocytes of these mice. Further work will be necessary to address whether the 97Q-induced alterations of LD morphology observed here in yeast are relevant to understanding adipocyte dysfunction in HD patients.
Materials and Methods
MS.
Samples were prepared as described (23, 50) for proteome and interactome analyses. Protein mixtures were desalted, separated by liquid chromatography, and subsequently loaded in a quadrupole spectrometer via a nano-electrospray source for analysis. The data were processed using the MaxQuant package (51).
Cryo-ET.
Cells were vitrified by plunge freezing and loaded into a cryo-focus ion beam instrument to produce 150- to 250-nm-thick lamellas (26). Low-dose tomographic tilt series of lamellas were recorded in transmission electron microscopes operated at 300 kV and equipped with a direct electron detector (Figs. 3 and 5 and Fig. S2) or a CCD camera (Figs. S3 and S4). Upon tomographic reconstruction, the final pixel size was 2.08 or 1.40 nm (direct detector) or 2.28 nm (CCD).
Supplementary Material
Acknowledgments
We thank Felix Bäuerlein, Jürgen Cox, Felix Meissner, Günter Pfeifer, Jürgen Plitzko, Jan Rieckmann, Eri Sakata, Tillman Schäfer, and Martin Spitaler for helpful discussions and experimental help. This work was funded by European Commission 7th Research Framework Program Grant GA ERC-2012-SyG_318987–ToPAG and the Erasmus+ Program; by German Research Foundation Grants EXC 1010 SyNergy, Munich Cluster for Systems Neurology, EXC 81, Cellular Networks: From Molecular Mechanisms to Quantitative Understanding of Complex Functions Cluster GSC-1006, Graduate School of Quantitative Biosciences Munich, and SFB/TRR 83, Molecular Architecture and Cellular Functions of Lipid/Protein Assemblies Collaborative Research Centre; and by the International Max Planck Research School Graduate School.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717978115/-/DCSupplemental.
References
- 1.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 2.Arrasate M, Finkbeiner S. Protein aggregates in Huntington’s disease. Exp Neurol. 2012;238:1–11. doi: 10.1016/j.expneurol.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arndt JR, Chaibva M, Legleiter J. The emerging role of the first 17 amino acids of huntingtin in Huntington’s disease. Biomol Concepts. 2015;6:33–46. doi: 10.1515/bmc-2015-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci USA. 2006;103:11045–11050. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crick SL, Ruff KM, Garai K, Frieden C, Pappu RV. Unmasking the roles of N- and C-terminal flanking sequences from exon 1 of huntingtin as modulators of polyglutamine aggregation. Proc Natl Acad Sci USA. 2013;110:20075–20080. doi: 10.1073/pnas.1320626110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen K, et al. Control of the structural landscape and neuronal proteotoxicity of mutant Huntingtin by domains flanking the polyQ tract. eLife. 2016;5:e18065. doi: 10.7554/eLife.18065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darnell G, Orgel JP, Pahl R, Meredith SC. Flanking polyproline sequences inhibit beta-sheet structure in polyglutamine segments by inducing PPII-like helix structure. J Mol Biol. 2007;374:688–704. doi: 10.1016/j.jmb.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 8.Lakhani VV, Ding F, Dokholyan NV. Polyglutamine induced misfolding of huntingtin exon1 is modulated by the flanking sequences. PLOS Comput Biol. 2010;6:e1000772. doi: 10.1371/journal.pcbi.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dehay B, Bertolotti A. Critical role of the proline-rich region in Huntingtin for aggregation and cytotoxicity in yeast. J Biol Chem. 2006;281:35608–35615. doi: 10.1074/jbc.M605558200. [DOI] [PubMed] [Google Scholar]
- 10.Macdonald M. The Huntington’s Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 11.Scherzinger E, et al. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 12.Muchowski PJ, et al. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci USA. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krobitsch S, Lindquist S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc Natl Acad Sci USA. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 15.Beck M, Baumeister W. Cryo-electron tomography: Can it reveal the molecular sociology of cells in atomic detail? Trends Cell Biol. 2016;26:825–837. doi: 10.1016/j.tcb.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 16.Bauerlein FJB, et al. In situ architecture and cellular interactions of PolyQ inclusions. Cell. 2017;171:179–187. doi: 10.1016/j.cell.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 17.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 18.Cox J, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park SH, et al. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell. 2013;154:134–145. doi: 10.1016/j.cell.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 20.Olzscha H, et al. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144:67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- 21.Hipp MS, Park SH, Hartl FU. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 2014;24:506–514. doi: 10.1016/j.tcb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Schaffar G, et al. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol Cell. 2004;15:95–105. doi: 10.1016/j.molcel.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 23.Ripaud L, et al. Overexpression of Q-rich prion-like proteins suppresses polyQ cytotoxicity and alters the polyQ interactome. Proc Natl Acad Sci USA. 2014;111:18219–18224. doi: 10.1073/pnas.1421313111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Damiano M, Galvan L, Déglon N, Brouillet E. Mitochondria in Huntington’s disease. Biochim Biophys Acta. 2010;1802:52–61. doi: 10.1016/j.bbadis.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 25.Schon EA, Przedborski S. Mitochondria: the next (neurode)generation. Neuron. 2011;70:1033–1053. doi: 10.1016/j.neuron.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rigort A, et al. Focused ion beam micromachining of eukaryotic cells for cryoelectron tomography. Proc Natl Acad Sci USA. 2012;109:4449–4454. doi: 10.1073/pnas.1201333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou C, et al. Organelle-based aggregation and retention of damaged proteins in asymmetrically dividing cells. Cell. 2014;159:530–542. doi: 10.1016/j.cell.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawai-Noma S, et al. In vivo evidence for the fibrillar structures of Sup35 prions in yeast cells. J Cell Biol. 2010;190:223–231. doi: 10.1083/jcb.201002149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saibil HR, et al. Heritable yeast prions have a highly organized three-dimensional architecture with interfiber structures. Proc Natl Acad Sci USA. 2012;109:14906–14911. doi: 10.1073/pnas.1211976109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez-Vicente M, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aditi K, Shakarad MN, Agrawal N. Altered lipid metabolism in Drosophila model of Huntington’s disease. Sci Rep. 2016;6:31411. doi: 10.1038/srep31411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujimoto T, Ohsaki Y, Suzuki M, Cheng J. Imaging lipid droplets by electron microscopy. Methods Cell Biol. 2013;116:227–251. doi: 10.1016/B978-0-12-408051-5.00012-7. [DOI] [PubMed] [Google Scholar]
- 33.Krahmer N, et al. Protein correlation profiles identify lipid droplet proteins with high confidence. Mol Cell Proteomics. 2013;12:1115–1126. doi: 10.1074/mcp.M112.020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhattacharyya A, et al. Oligoproline effects on polyglutamine conformation and aggregation. J Mol Biol. 2006;355:524–535. doi: 10.1016/j.jmb.2005.10.053. [DOI] [PubMed] [Google Scholar]
- 35.Kim YE, et al. Soluble oligomers of PolyQ-expanded huntingtin target a multiplicity of key cellular factors. Mol Cell. 2016;63:951–964. doi: 10.1016/j.molcel.2016.07.022. [DOI] [PubMed] [Google Scholar]
- 36.Caron NS, Hung CL, Atwal RS, Truant R. Live cell imaging and biophotonic methods reveal two types of mutant huntingtin inclusions. Hum Mol Genet. 2014;23:2324–2338. doi: 10.1093/hmg/ddt625. [DOI] [PubMed] [Google Scholar]
- 37.Tipping KW, van Oosten-Hawle P, Hewitt EW, Radford SE. Amyloid fibres: Inert end-stage aggregates or key players in disease? Trends Biochem Sci. 2015;40:719–727. doi: 10.1016/j.tibs.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scior A, et al. Complete suppression of Htt fibrilization and disaggregation of Htt fibrils by a trimeric chaperone complex. EMBO J. 2018;37:282–299. doi: 10.15252/embj.201797212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner J, Schaffer M, Fernández-Busnadiego R. Cryo-electron tomography-the cell biology that came in from the cold. FEBS Lett. 2017;591:2520–2533. doi: 10.1002/1873-3468.12757. [DOI] [PubMed] [Google Scholar]
- 40.Escusa-Toret S, Vonk WI, Frydman J. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat Cell Biol. 2013;15:1231–1243. doi: 10.1038/ncb2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solans A, Zambrano A, Rodríguez M, Barrientos A. Cytotoxicity of a mutant huntingtin fragment in yeast involves early alterations in mitochondrial OXPHOS complexes II and III. Hum Mol Genet. 2006;15:3063–3081. doi: 10.1093/hmg/ddl248. [DOI] [PubMed] [Google Scholar]
- 42.Ocampo A, Zambrano A, Barrientos A. Suppression of polyglutamine-induced cytotoxicity in Saccharomyces cerevisiae by enhancement of mitochondrial biogenesis. FASEB J. 2010;24:1431–1441. doi: 10.1096/fj.09-148601. [DOI] [PubMed] [Google Scholar]
- 43.Yatsu LY, Jacks TJ. Spherosome membranes: half unit-membranes. Plant Physiol. 1972;49:937–943. doi: 10.1104/pp.49.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wanner G, Theimer RR. Membranous appendices of spherosomes (oleosomes) : Possible role in fat utilization in germinating oil seeds. Planta. 1978;140:163–169. doi: 10.1007/BF00384916. [DOI] [PubMed] [Google Scholar]
- 45.Binns D, et al. An intimate collaboration between peroxisomes and lipid bodies. J Cell Biol. 2006;173:719–731. doi: 10.1083/jcb.200511125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Welte MA. Expanding roles for lipid droplets. Curr Biol. 2015;25:R470–R481. doi: 10.1016/j.cub.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Burg JM, Björkqvist M, Brundin P. Beyond the brain: widespread pathology in Huntington’s disease. Lancet Neurol. 2009;8:765–774. doi: 10.1016/S1474-4422(09)70178-4. [DOI] [PubMed] [Google Scholar]
- 48.Fain JN, Del Mar NA, Meade CA, Reiner A, Goldowitz D. Abnormalities in the functioning of adipocytes from R6/2 mice that are transgenic for the Huntington’s disease mutation. Hum Mol Genet. 2001;10:145–152. doi: 10.1093/hmg/10.2.145. [DOI] [PubMed] [Google Scholar]
- 49.Phan J, Hickey MA, Zhang P, Chesselet MF, Reue K. Adipose tissue dysfunction tracks disease progression in two Huntington’s disease mouse models. Hum Mol Genet. 2009;18:1006–1016. doi: 10.1093/hmg/ddn428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hornburg D, et al. Deep proteomic evaluation of primary and cell line motoneuron disease models delineates major differences in neuronal characteristics. Mol Cell Proteomics. 2014;13:3410–3420. doi: 10.1074/mcp.M113.037291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 52.Meriin AB, et al. Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J Cell Biol. 2002;157:997–1004. doi: 10.1083/jcb.200112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.