Significance
The major human pathogen, Staphylococcus aureus, produces a broad-spectrum metallophore, staphylopine (StP), to acquire transition metals under metal-limited conditions. The first step of the metal acquisition process is StP/metal recognition by the extracytoplasmic solute-binding protein CntA from the StP/metal transportation system. We determined the crystal structures of CntA/StP/metal (Ni2+, Co2+, and Zn2+) complexes and apo CntA, deciphering a sophisticated StP/metal-recognition mechanism. Moreover, we uncovered that StP/metal recognition is indispensable for StP-mediated metal acquisition. Because the StP-like biosynthetic and trafficking pathways are present in various pathogens, these discoveries provide critical insights into the investigations of metal-acquisition mechanisms in microbes.
Keywords: staphylopine, metal acquisition, CntA, CRISPR/Cas9, genome editing
Abstract
Metal acquisition is vital to pathogens for successful infection within hosts. Staphylopine (StP), a broad-spectrum metallophore biosynthesized by the major human pathogen, Staphylococcus aureus, plays a central role in transition-metal acquisition and bacterial virulence. The StP-like biosynthesis loci are present in various pathogens, and the proteins responsible for StP/metal transportation have been determined. However, the molecular mechanisms of how StP/metal complexes are recognized and transported remain unknown. We report multiple structures of the extracytoplasmic solute-binding protein CntA from the StP/metal transportation system in apo form and in complex with StP and three different metals. We elucidated a sophisticated metal-bound StP recognition mechanism and determined that StP/metal binding triggers a notable interdomain conformational change in CntA. Furthermore, CRISPR/Cas9-mediated single-base substitution mutations and biochemical analysis highlight the importance of StP/metal recognition for StP/metal acquisition. These discoveries provide critical insights into the study of novel metal-acquisition mechanisms in microbes.
Transition metals, such as iron, copper, zinc, nickel, and cobalt, play vital roles in numerous biological processes. Because of these metals’ unique redox and inorganic chemical properties, they serve as catalytic centers for enzymatic reactions, contribute to overall protein stability, or serve as signaling agents. To thrive, all organisms have to maintain adequate levels of transition metals for their cellular metalloproteins, which account for more than 30% of all proteins in biological systems (1, 2).
In the context of microbial infections, because transition metals are essential for microbial survival and pathogenesis, host cells produce high concentrations of metal-chelating proteins, such as transferrin (3), lactoferrin (4), and calprotectin (5), to restrict metal availability. In response, pathogens have evolved diverse strategies to subvert metal sequestration. The battle between pathogens and hosts for transition metals constitutes an important and constant component for the pathogenesis of microbial infections (6).
Staphylococcus aureus, a major human pathogen, is the leading cause of hospital- and community-acquired infections. This microbe can cause a wide variety of infections, ranging from minor skin infections to life-threatening diseases (7, 8). The pathogen is equipped with sophisticated virulence-regulatory mechanisms (9–11) and diverse transition-metal acquisition systems (12–15), enabling its success in infections. In addition to its iron and manganese acquisition systems, S. aureus possesses several other transition-metal transportation systems, such as Nik, NixA, Adc, and Cnt (15–18). The Nik system belongs to the PepT family of ABC transporters and is important for bacterial nickel acquisition. It is composed of an extracytoplasmic solute-binding protein (SBP) NikA and four other proteins NikBCDE to form an integral transportation channel. Through the utilization of small-molecule chelators, such as l-histidine, the Nik system effectively transports nickel, determines the urease activity, and plays a critical role in bacterial colonization of mouse urinary tract (17, 19, 20). NixA is an additional nickel acquisition system in S. aureus. It is a high-affinity metal transporter and belongs to the NiCoT family of secondary transporters. The NixA system is also important for urease activity and plays a synergistic role in kidney colonization in addition to the Nik system (17). Adc is a recently discovered zinc-acquisition system in S. aureus. It is similar to Zn-specific ABC permeases in other Gram-positive bacteria and is comprised of a metal ion-recruiting component AdcA and an ABC transporter AdcBC (18). Cnt was originally discovered as a cobalt and nickel-acquisition system in S. aureus (15). Later, it was shown to transport multiple metals, including nickel, cobalt, zinc, iron, copper, and manganese, depending on the culture conditions (21). Recent studies showed that it functioned as a zinc-acquisition system under zinc-limited conditions, such as during infection and in the presence of calprotectin (18). The Adc system is the first system utilized by S. aureus to import zinc, and the Cnt system will be induced when the cellular zinc demand cannot be met by the Adc system (18).
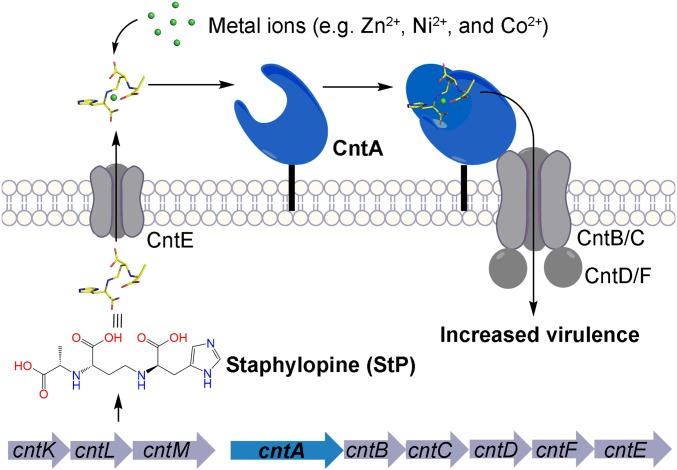
One striking feature of the Cnt system is that it utilizes a broad-spectrum nicotianamine-like metallophore staphylopine (StP) biosynthesized by the first three genes cntKLM in the pathway to facilitate metal acquisition (15–18, 21) (Fig. 1). CntK is a histidine racemase, CntL is a nicotianamine-synthase-like enzyme, and CntM is an octopine dehydrogenase that adds a pyruvate moiety to the StP precursor. A wide range of bacteria possess the StP-like biosynthesis locus. However, they differ in both the number and the function of the biosynthetic proteins, indicating that different StP-like chemicals are present in different microbes (18, 21). Recent investigations in Pseudomonas aeruginosa uncovered a new nicotianamine-like metallophore pseudopaline that also mediates bacterial zinc acquisition under zinc-poor conditions (22–24). In contrast to the biosynthetic pathway of StP, the pseudopaline biosynthetic pathway lacks the histidine racemase and incorporates l-histidine and α-ketoglutarate instead of d-histidine and pyruvate into the scaffold of the metallophore (22–24).
Fig. 1.
Scheme of the proposed StP-medicated metal acquisition process. StP is biosynthesized by CntKLM and exported out of cells by CntE. After capturing transition metals, StP/metal complexes are recognized by CntA and imported into cells by CntABCDF.
In addition to the StP biosynthesis genes, the cnt operon possesses an StP exporter encoded by the cntE gene and an StP/metal importer encoded by the five genes cntABCDF (15, 18, 21) (Fig. 1). The CntA protein of the importer complex is an extracytoplasmic SBP and belongs to the NikA/Opp family of proteins that recognize and transport diverse solutes, such as oligopeptides and metals (17, 25). CntA plays a primary role in StP/metal recognition and importation initiation (15, 18, 21). CntBC are two membrane proteins that form an integral channel and may play a role in StP/metal transportation (15, 18, 21). CntDF are two membrane-associated ATP-binding proteins that provide energy for the transportation (15, 18, 21). Previous independent genetic studies targeting cntA, cntKLM, and cntE have demonstrated the importance of the genes for bacterial virulence, implicating the critical role of this pathway in bacterial fitness and virulence (15, 18, 26). Despite the established biosynthetic pathway and the essential role of this pathway in pathogenesis, the molecular mechanisms of how StP/metal complexes are recognized and transported are currently unknown.
In this study, we report crystal structures of the StP/metal recognition complexes CntA/StP/metal (Co2+, Ni2+, and Zn2+), elucidating the detailed StP/metal recognition mechanism. By comparing the crystal structure of ligand-free CntA, also obtained in this study, we unveiled the ligand-capture mechanism that StP/metal binding triggers a drastic interdomain conformational change in CntA. Through structure-guided mutagenesis, we discovered the key StP/metal-binding residues and demonstrated the critical role of StP/metal recognition in StP/metal transportation and acquisition. These findings shed light on the exploration of metal-acquisition mechanisms in other microbes.
Results
Overall 3D Structure of apo-CntA.
The amino acid-sequence similarity of CntA with well-characterized Ni-binding proteins [e.g., Brucella suis NikA (36% identities), Escherichia coli NikA (34% identities), and Campylobacter jejuni NikZ (27% identities)] indicates the role of CntA in metal recognition (19). In addition, previous studies have demonstrated a role of CntA in metal binding and uptake (18, 21). However, the lack of tertiary structure information of CntA prevents a deeper understanding of the detailed metal-recognition mechanism. Thus, we first sought to determine the crystal structure of CntA to probe its function biochemically.
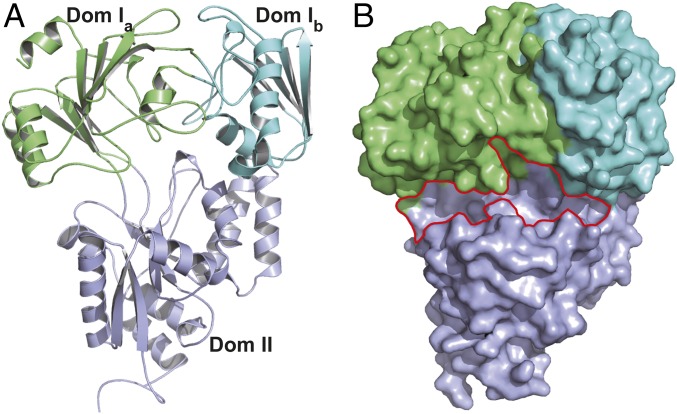
To facilitate the crystallization of the CntA protein, we cloned, expressed, and purified different truncations of the protein and screened 500 crystallization conditions for each truncation. One construct harboring the amino acids from G26 to K532 (the residues were renumbered as G1 to K507 for future analysis) was successfully crystallized. The protein was crystallized in the space group P61, and the crystal was diffracted to 2.90-Å resolution (SI Appendix, Table S1). The asymmetric unit contains a single protein monomer that is composed of three domains (Fig. 2A): two top domains (Ia and Ib) and one bottom domain (II). Residues G1-Y38, K161-Q250, and S478-Y506 belong to domain Ia, residues E39-K160 belong to domain Ib, and residues P251-G477 belong to domain II (SI Appendix, Fig. S1). The overall protein structure features an α/β-fold, common to extracytoplasmic SBPs (27). In total, the protein contains 17 α-helices and 16 β-sheets. The domain Ia contains five α-helices and seven β-sheets, the domain Ib contains three α-helices and four antiparallel β-sheets, and the domain II comprises nine α-helices and five β-sheets (Fig. 2B and SI Appendix, Fig. S2). The structure exhibits an open conformation with two exposed ligand-binding sites surrounded by domains Ia and II as well as Ia, Ib, and II, respectively (Fig. 2).
Fig. 2.
Structural characterizations of apo-CntA. (A) The overall structure of apo-CntA. Domains Ia, Ib, and II are colored lime, aquamarine, and light blue, respectively. (B) The surface structure of apo-CntA. The potential ligand binding pockets are highlighted.
Structural Characterizations of CntA/StP/Metal Complexes.
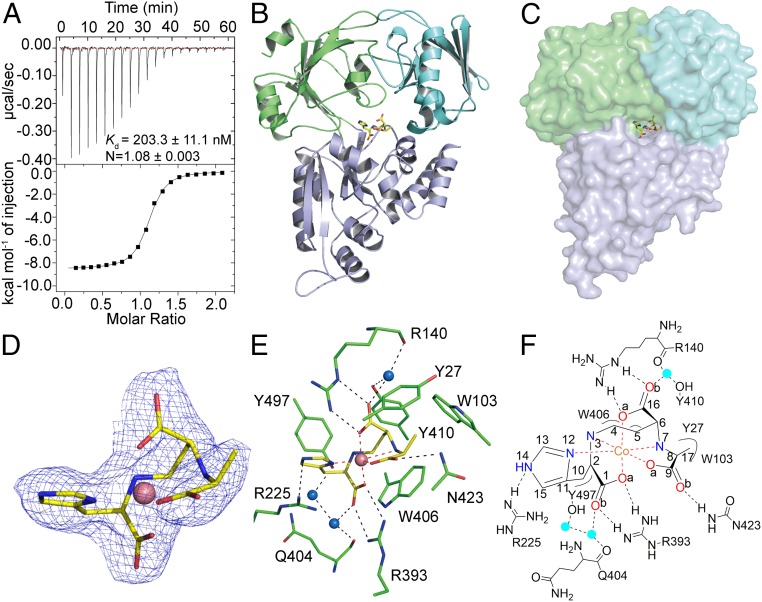
We employed isothermal titration calorimetry (ITC) to examine the interactions between StP/metal and CntA before the crystallization of CntA/StP/metal tertiary complexes. The titration of CntA with a solution containing StP/metal produced a significant amount of heat. The measured dissociation constant Kd values for the interactions between CntA and StP/Co2+, StP/Ni2+, and StP/Zn2+, were 203.3 nM (Fig. 3A), 167.8 nM (SI Appendix, Fig. S3A), and 952.3 nM (SI Appendix, Fig. S3B), respectively. In all cases, the stoichiometry is 1:1, indicating that one CntA molecule binds one molecule of StP/metal. However, the titration of CntA with solutions containing StP or metals alone only produced basal levels of heat (SI Appendix, Fig. S3 C and D), implicating no enthalpy-driven binding between CntA and StP or metals. The direct binding to metal-bound metallophore produced from the same operon was also observed in other SBPs. For instance, MbnE, an SBP from methanotrophic bacteria, directly binds copper-bound methanobactin, a peptide-derived natural product produced from the same operon of MbnE (28).
Fig. 3.
Structural characterizations of the CntA/StP/Co2+ complex. (A) ITC assay for the binding between CntA and StP/Co2+. Kd, the dissociation constant; N, the number of binding sites per CntA. (B) The overall structure of the CntA/StP/Co2+ complex. StP is colored yellow. (C) The surface structure of the CntA/StP/Co2+ complex. (D) Detailed view of the electron density of StP/Co2+. 2Fo − Fc map (1.0 σ) is shown as a blue mesh. (E and F) Detailed interactions between CntA and StP/Co2+. Dashed lines represent hydrogen bonds or coordination bonds. Water molecules are blue spheres.
To further probe the binding affinity differences between CntA and StP/metals, we performed a competitive binding assay in the presence of StP and all three metals (Ni2+, Co2+, and Zn2+). Seventy micromolar 6XHis-tag cleaved CntA protein was incubated with 560 μM StP and a metal mixture containing 140 μM Zn2+, 140 μM Ni2+, and 140 μM Co2+. StP was in excess to ensure all of the metals were chelated by StP. The mixture was subjected to a desalting separation to remove unbounded StP/metals. We then performed the inductively coupled plasma-atomic emission spectrometry (ICP-AES) analysis to determine the metal content in the protein. The content of Zn2+ was lower than that of Co2+ or Ni2+ in the protein (SI Appendix, Fig. S4). The result indicated that CntA bound StP/Co2+ and StP/Ni2+ tighter than that of StP/Zn2+, consistent with the results of the ITC assay.
To investigate the detailed CntA-mediated metal recognition mechanism, we cocrystallized CntA, StP, and three different metals (Co2+, Ni2+, and Zn2+). The CntA/StP/Co2+, CntA/StP/Ni2+, and CntA/StP/Zn2+ complexes were crystallized in the space groups P1, P22121, and P21212, respectively, and the crystals were diffracted to 2.47, 2.12, and 2.03-Å resolution, respectively (SI Appendix, Table S1). The asymmetric units of CntA/StP/Co2+, CntA/StP/Ni2+, and CntA/StP/Zn2+ complexes contain two, one, and one monomer(s), respectively. In contrast to the open conformation observed in the ligand-free CntA structure, all CntA/StP/metal complexes exhibit closed compact conformations (Fig. 3 B and C and SI Appendix, Fig. S5). Each monomer in all three complex structures contains a well-defined StP/metal molecule (Fig. 3D and SI Appendix, Fig. S6) residing in the central pocket surrounded by all three domains (Ia, Ib, and II) (Fig. 3C and SI Appendix, Fig. S5 B and D).
Because the structures of three CntA/StP/metal complexes possess almost identical overall conformation [0.21-Å root-mean-square deviation (rmsd) between CntA/StP/Co2+ and CntA/StP/Ni2+ and 0.25-Å rmsd between CntA/StP/Co2+ and CntA/StP/Zn2+] and detailed StP-recognition modes (SI Appendix, Fig. S7), we used the structure of CntA/StP/Co2+ for subsequent analysis. The close inspection of the ligand-binding pocket revealed extensive interactions between StP and CntA. In total, 10 residues (Y27, W103, R140, R225, R393, Q404, W406, Y410, N423, and Y497) engage in 14 direct or water-mediated contacts with metal-bound StP (Fig. 3 E and F and SI Appendix, Table S2). In particular, three arginine residues (R140, R225, and R393) and one asparagine residue (N423) play a primary role in metal-bound StP recognition. R140 and R393 form two salt bridges with the side-chain carboxyl groups of StP. R225 forms a direct hydrogen bond with the side-chain imidazole nitrogen atom of StP, while N423 forms a direct hydrogen bond with the side-chain carboxyl oxygen atom of StP. In addition, four residues (R140, Q404, Y410, and Y497) provide four additional water-mediated hydrogen bonds with the side-chain carboxyl oxygen atoms of StP. Moreover, five aromatic residues (Y27, W103, W406, Y410, and Y497) directly contact two main-chain carbon atoms (C4 and C5) and two side-chain carbon atoms (C10 and C17) of StP through van der Waals interactions (Fig. 3 E and F and SI Appendix, Table S2). Together, the sophisticated metal-bound StP recognition network likely ensures the high selectivity of StP/metal binding. To experimentally test the selectivity of StP/metal recognition by CntA, we titrated CntA with two well-studied but structurally distinct metallophore/metal complexes (EDTA/Co2+ and nicotianamine/Co2+) (29) using ITC. Neither of the titrations produced significant amounts of heat (SI Appendix, Fig. S8), suggesting no enthalpy-driven binding between CntA and EDTA/Co2+ or nicotianamine/Co2+.
In all cases, no direct interaction was observed between the central metal and the protein. The central metal adopts an octahedral coordination geometry regardless of the nature of metals. This octahedral coordination geometry is also observed in other Ni-binding proteins (e.g., B. suis NikA, C. jejuni NikZ, and Yersinia pestis YntA) (19) despite the difference in metal-binding small molecules.
STP/Metal Recognition Triggers a Dramatic Interdomain Conformational Change.
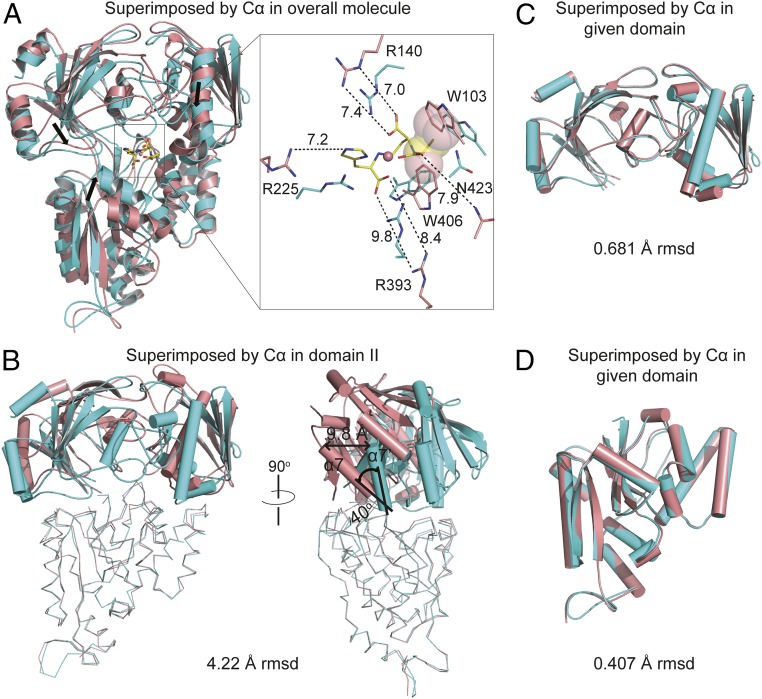
To examine the dynamics of the StP/metal-recognition process, we superposed the structure of apo-CntA with the structure of StP/Co2+-bound CntA, revealing a drastic overall conformational change with 4.22-Å rmsd over 503 residues (Fig. 4A). The key residues (R140, R225, R393, and N423) that directly contact the side-chain carboxyl oxygen and imidazole nitrogen atoms of StP in the CntA/StP/Co2+ structure are all distant from the metal-bound StP in the apo-CntA structure (>7-Å distance, Fig. 4A). In addition, the aromatic rings of two aromatic residues (W103 and W406) that participate in van der Waals interactions with StP in the CntA/StP/Co2+ structure have strong clashes with carbons 8 and 17 of StP in the apo-CntA structure (Fig. 4A). All of these structural differences likely drive the conformational change of apo-CntA to effectively capture the metal-bound StP.
Fig. 4.
StP/metal binding triggers a drastic conformational change in CntA. (A) Structural comparison of the CntA/StP/Co2+ complex and apo-CntA by superimposing Cα atoms in the overall molecule. The proteins of the CntA/StP/Co2+ complex and apo-CntA are colored cyan and salmon, respectively. The black arrows represent the directions that each domain moves to capture StP/metal. Distances between the side-chain atoms of StP and the amino acids of apo-CntA are indicated. The clashes between StP and W103 and W406 of apo-CntA are viewed as semitransparent spheres. (B) Structural comparison of the CntA/StP/Co2+ complex and apo-CntA by superimposing Cα atoms in domain II. The structural changes between helices α7 of the CntA/StP/Co2+ complex and apo-CntA are indicated. The domain II is viewed as ribbon, and the domains Ia and Ib are viewed as cartoon. (C) Overlay of Cα atoms in domains Ia and Ib reveals the structural similarity of the CntA/StP/Co2+ complex and apo-CntA subunits. (D) Overlay of Cα atoms in the domain II reveals the structural similarity of the CntA/StP/Co2+ complex and apo-CntA subunits.
Next, when we overlaid the bottom domains (domain II) or the top domains (domain Ia and Ib) of apo-CntA and ligand-bound CntA, a dramatic conformational change was observed in the other domains of the two protein structures, including a 40-degree rigid-body rotation and a 9.8-Å translation (using α7 and α7′ when the bottom domains were superimposed for illustration, Fig. 4B and SI Appendix, Fig. S9). However, little structural change was observed within the given domains with 0.681-Å rmsd of the top domain (Fig. 4C) and 0.407-Å rmsd of the bottom domain (Fig. 4D) compared with 4.22-Å rmsd between overall structures, thus revealing an interdomain conformational-change mechanism for StP/metal recognition by CntA. This movement effectively closed up the large ligand-binding cavities observed in the apo-CntA structure (Fig. 2B) and ensured tight binding between CntA and metal-bound StP.
The interdomain conformational-change mechanism was also observed in other SBPs. As shown in SI Appendix, Figs. S10–S12, ligand binding induced a significant domain movement in all of the SBPs analyzed. However, different degrees of the structural change were observed in different SBPs. Binding of EDTA/Fe3+ to B. suis NikA triggered a drastic conformational change (3.37-Å rmsd between overall structures), whereas binding of l-histidine/Ni2+ to Y. pestis YntA only induced a slight structural change (1.58-Å rmsd between overall structures) (SI Appendix, Fig. S10) (19). Binding of a nonapeptide to Lactococcus lactis OppA triggered a dramatic conformational change (4.49-Å rmsd between overall structures), while binding of a dipeptide to Salmonella typhimurium OppA induced a modest structural change (2.83-Å rmsd between overall structures) (SI Appendix, Fig. S11) (30–32). In addition, binding of Cd2+ to Streptococcus pneumoniae PsaA (PsaA belongs to class A SBPs and the four aforementioned proteins belong to class C SBPs; refs. 27 and 33) induced a conformational change with 1.22-Å rmsd between overall structures (SI Appendix, Fig. S12) (34). However, little structural change was observed within the given domains in all of the SBPs analyzed (SI Appendix, Figs. S10–S12). These analyses implicated that the interdomain conformational-change mechanism was ubiquitous in SBPs and some factors, such as the ligand-recognition mode of the SBP and the size of the ligand, could affect the degree of the structural change.
CntA Possesses a Unique Ligand-Recognition Mode.
CntA has been suggested to employ a distinct metal-recognition mechanism compared with other proteins in the Ni-binding protein family (19). We superposed the structure of ligand-bound CntA with the structures of S. aureus NikA/l-His/Ni2+ (20) and B. suis NikA/EDTA/Fe3+ (19) to probe the differences in metal-recognition mode. As shown in SI Appendix, Fig. S13A, the metal-binding site in the structure of SaNikA/l-His/Ni2+ is away from that of the structure of CntA/StP/Co2+. In addition, the ligand-recognition residues of SaNikA locate in the left region of the pocket, whereas the ligand-recognition residues of CntA distribute across the entire pocket. The metal-binding sites in the structures of CntA/StP/Co2+ and BsNikA/EDTA/Fe3+ are adjacent (SI Appendix, Fig. S13B). However, some of the key StP-binding residues, including Y497, R225, and N423, are missing in the iron-binding protein (SI Appendix, Fig. S13B), thus revealing the unique StP/metal-recognition mode in CntA.
StP/Metal Recognition Is Essential for Metal Acquisition.
The structural analysis of the CntA/StP/metal complexes has unveiled the StP–protein interaction in detail. To further assess the roles of the residues in StP recognition, we mutated the StP-recognition residues Y27, W103, R140, R225, R393, W406, Y410, N423, and Y497 to alanine, respectively. We purified the proteins and analyzed the proteins using both size-exclusion chromatography and polyacrylamide gel electrophoresis. All of the proteins shared similar elution volumes and had similar purities (SI Appendix, Fig. S14), implicating that the single amino acid mutation did not affect protein expression or stability. We next systematically examined the abilities of the mutant proteins in StP/metal binding by ITC. As shown in Fig. 5A and SI Appendix, Fig. S15, different StP/metal-binding properties for the mutant proteins were observed. The mutation of N423 to alanine slightly increased the dissociation constant (Kd) by ∼2.5-fold. Mutation of Y27, W103, or Y410 to alanine dramatically increased the dissociation constant by ∼109-, 123-, and 65-fold, respectively. No detectable binding was observed when R140, R225, R393, W406, or Y497 was mutated to alanine.
Fig. 5.
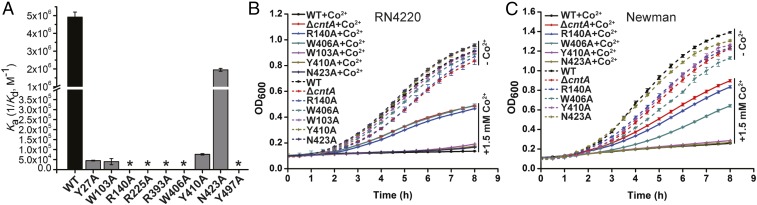
Metal-bound STP recognition by CntA is essential for metal acquisition. (A) Single amino acid mutation of the StP-binding residues differentially reduces the binding affinity between the protein and metal-bound StP. The binding affinity Ka values were calculated from ITC assays. *, no detectable binding by ITC. (B and C) Growth curve measurements of various RN4220 (B) and Newman strains (C) in the presence or absence of 1.5 mM Co2+. The assays were performed in the chemically defined medium. Dashed lines, in the absence of 1.5 mM Co2+; solid lines, in the presence of 1.5 mM Co2+.
We next sought to examine the role of StP/metal recognition in StP/metal transportation and acquisition by determining intracellular metal accumulation. We mutated the selected StP/metal-recognition residues N423, W103, Y410, R140, and W406 to alanine, respectively, in the S. aureus genome by using the recently developed CRISPR/Cas9-based genome editing tool pCasSA (SI Appendix, Fig. S16) (35). All of the mutants except W103A were created in both the RN4220 and Newman strains, whereas the W103A mutant was only constructed in the RN4220 strain. Because the overloading of transition metals is toxic to bacteria and high concentrations of transition metals have a greater repression on the growth of the wild-type S. aureus strain than that of the StP importer complex (cntABCDF) deletion mutant (21), we subjected the strains to growth curve analysis in chemically defined medium (CDM). All of the strains exhibited similar growth without the supplementation of a high concentration of cobalt (dashed lines in Fig. 5 B and C and SI Appendix, Fig. S17). However, distinct growth behaviors were observed when the cells were cultured under the condition containing 1.5 mM cobalt (solid lines in Fig. 5 B and C and SI Appendix, Fig. S17). A high concentration of cobalt drastically repressed the growth of the wild-type strain, whereas the deletion of cntA significantly increased bacterial growth. The growth rate of the cntA deletion mutant was similar to that of the importer complex cntABCDF deletion mutant in the presence of a high concentration of cobalt (21), implicating that the disruption of the StP/metal-recognition protein is sufficient to terminate StP/metal transportation. A single amino acid mutation of the indispensable StP/metal binding residues (R140 and W406) dramatically relieved the growth repression, whereas a single amino acid mutation of the weak (N423) or strong (W103 and Y410) StP/metal binding residues affected growth repression only slightly.
Next, we compared the intracellular metal contents of various strains using ICP-AES. We cultured the strains in CDM in the presence of 1 μM Co2+. In agreement with the growth experiments, the complete deletion of cntA drastically impaired metal accumulation (SI Appendix, Fig. S18). The single amino acid mutation of the indispensable StP/metal binding residues (R140 and W406) significantly reduced metal accumulation (SI Appendix, Fig. S18). However, the single amino acid mutations of the weak (N423) or strong (Y410) StP/metal binding residues (SI Appendix, Fig. S18A) reduced metal accumulation only slightly. The phenotypes of the mutants observed in both the growth curve assay and ICP-AES experiment can be complemented by introducing a single-copy wild-type cntA gene back into the mutants (SI Appendix, Figs. S17 and S18B). Taken together, these results demonstrate that StP/metal recognition is essential for StP/metal acquisition and that weak binding of StP/metal by CntA is sufficient for StP/metal acquisition under the conditions tested.
Discussion
The tug-of-war between host and bacterial pathogens for nutrient transition metals is a critical factor in microbial infections (2, 36–41). The discovery of the metallophore, StP (20, 21), provides insights into the mechanisms of metal acquisition. Our work unveils the detailed StP-recognition and transportation-initiation mechanisms found through the structural characterizations of the CntA/StP/metal complexes as well as ligand-free CntA. Structure-guided mutagenesis further underscores the roles of StP/metal recognition in metal transportation, demonstrating that targeting StP/metal recognition could be a feasible strategy to counter bacterial infections.
Previous studies showed that the presence of zinc attenuated cnt expression and, thereby, inhibited nickel and cobalt transportation (15). Later, it was shown that the cnt system could transport multiple metals, and the metals it transported depended on the culture conditions (21). Recent investigations revealed that the cnt system functioned as a zinc acquisition system under zinc-limited conditions, such as during infection and in the presence of calprotectin (18). Our biochemical data (crystal structures, ITC assay, and the competition assay) showed that CntA could recognize all three StP/metal (StP/Ni2+, StP/Co2+, and StP/Zn2+) complexes and CntA bound StP/Ni2+ and StP/Co2+ tighter than StP/Zn2+. The kind of metal that the cnt system recognizes and transports likely depends on the growth conditions of the bacteria.
The StP-like synthesis loci are present in a wide variety of bacteria, such as Bacillus hemicellulosilyticus, Brevibacillus brevis, Staphylococcus epidermidis, Streptococcus pneumonia, S. aureus, Yersinia pseudotuberculocis, and P. aeruginosa (18, 21). The StP-like synthesis loci can broadly be divided into two groups based on whether they contain the major facilitator superfamily (MFS) or the EamA family of transporters (18). Interestingly, the CntABCDF importer homologs are present in the MFS-containing loci, whereas a variety of importers are associated with the EamA-containing loci (18). To get insights into the types of the ligands recognized by the CntA homologs in other bacteria, we performed a sequence alignment of CntA with its homologs from four other bacteria (Staphylococcus epidermidis, Streptococcus pneumonia, Bacillus hemicellulosilyticus, and Brevibacillus brevis) containing the MFS-type StP-like synthesis loci. The results showed that most of the StP-recognition residues of CntA except N423 and Y497 were highly conserved in the CntA homologous proteins (SI Appendix, Fig. S19). N423 was mutated to Thr or Ser in Staphylococcus epidermidis, Streptococcus pneumonia, and Brevibacillus brevis, and Y497 was mutated to Phe in Bacillus hemicellulosilyticus and Brevibacillus brevis. Because mutation of N423 to Ala only slightly reduced the binding affinity and Y497 participated in the binding majorly by van der Waals interactions (Figs. 3F and 5A), the mutations of N423 to Thr or Ser and Y497 to Phe probably cannot affect StP recognition by CntA homologs, indicating that these bacteria can recognize StP/metal. The native metallophores utilized by these bacteria need to be further explored.
Methods
Bacterial Strains, Primers, Plasmids, and Growth Conditions.
Bacterial strains and plasmids used in this study are listed in SI Appendix, Table S3. Primers used in this study are listed in SI Appendix, Table S4. E. coli strains were grown in Luria–Bertani (LB) broth with shaking at 220 rpm. S. aureus strains were cultured in Tryptic Soy Broth with shaking at 250 rpm. Growth and metal analysis experiments were performed in CDM (21). Antibiotics were used at the following concentrations: kanamycin (50 μg/mL for E. coli), carbenicillin (50 μg/mL for E. coli), and chloramphenicol (10 μg/mL for the S. aureus Newman strain and 5 μg/mL for the S. aureus RN4220 strain).
Other Procedures.
Detailed procedures are available in SI Appendix, SI Procedures.
Supplementary Material
Acknowledgments
We thank staffs from BL18U1 and BL19U1 beamlines of National Facility for Protein Science Shanghai at Shanghai Synchrotron Radiation Facility for assistance during data collection and Prof. Taeok Bae for reading the manuscript and giving critical comments. This work was financially supported by National Key R&D Program of China Grant 2017YFA0506800; National Natural Science Foundation of China Grants 91753127 and 31700123; Shanghai Committee of Science and Technology, China Grant 17ZR1449200; and the “Young 1000 Talents” program (to Q.J.) and China Postdoctoral Science Foundation Grant 2017M620178 (to W.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystal structures of apo-CntA, CntA/StP/Co2+, CntA/StP/Ni2+, and CntA/StP/Zn2+ have been deposited in the RCSB Protein Data Bank, www.wwpdb.org (PDB ID codes: 5YH5, 5YHE, 5YH8, and 5YHG, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718382115/-/DCSupplemental.
References
- 1.Kim BE, Nevitt T, Thiele DJ. Mechanisms for copper acquisition, distribution and regulation. Nat Chem Biol. 2008;4:176–185. doi: 10.1038/nchembio.72. [DOI] [PubMed] [Google Scholar]
- 2.Palmer LD, Skaar EP. Transition metals and virulence in bacteria. Annu Rev Genet. 2016;50:67–91. doi: 10.1146/annurev-genet-120215-035146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schade AL, Caroline L. Raw hen egg white and the role of iron in growth inhibition of Shigella dysenteriae, Staphylococcus aureus, Escherichia coli and Saccharomyces cerevisiae. Science. 1944;100:14–15. doi: 10.1126/science.100.2584.14. [DOI] [PubMed] [Google Scholar]
- 4.Oram JD, Reiter B. Inhibition of bacteria by lactoferrin and other iron-chelating agents. Biochim Biophys Acta. 1968;170:351–365. doi: 10.1016/0304-4165(68)90015-9. [DOI] [PubMed] [Google Scholar]
- 5.Damo SM, et al. Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc Natl Acad Sci USA. 2013;110:3841–3846. doi: 10.1073/pnas.1220341110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nolan EM. BIOCHEMISTRY. A metal shuttle keeps pathogens well fed. Science. 2016;352:1055–1056. doi: 10.1126/science.aaf8755. [DOI] [PubMed] [Google Scholar]
- 7.Archer GL. Staphylococcus aureus: A well-armed pathogen. Clin Infect Dis. 1998;26:1179–1181. doi: 10.1086/520289. [DOI] [PubMed] [Google Scholar]
- 8.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 9.Cheung AL, Ying P. Regulation of alpha- and beta-hemolysins by the sar locus of Staphylococcus aureus. J Bacteriol. 1994;176:580–585. doi: 10.1128/jb.176.3.580-585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 11.Sun F, et al. In the Staphylococcus aureus two-component system sae, the response regulator SaeR binds to a direct repeat sequence and DNA binding requires phosphorylation by the sensor kinase SaeS. J Bacteriol. 2010;192:2111–2127. doi: 10.1128/JB.01524-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meiwes J, et al. Isolation and characterization of staphyloferrin A, a compound with siderophore activity from Staphylococcus hyicus DSM 20459. FEMS Microbiol Lett. 1990;55:201–205. doi: 10.1111/j.1574-6968.1990.tb13863.x. [DOI] [PubMed] [Google Scholar]
- 13.Haag H, et al. Isolation and biological characterization of staphyloferrin B, a compound with siderophore activity from staphylococci. FEMS Microbiol Lett. 1994;115:125–130. doi: 10.1111/j.1574-6968.1994.tb06626.x. [DOI] [PubMed] [Google Scholar]
- 14.Kehl-Fie TE, et al. MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect Immun. 2013;81:3395–3405. doi: 10.1128/IAI.00420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Remy L, et al. The Staphylococcus aureus Opp1 ABC transporter imports nickel and cobalt in zinc-depleted conditions and contributes to virulence. Mol Microbiol. 2013;87:730–743. doi: 10.1111/mmi.12126. [DOI] [PubMed] [Google Scholar]
- 16.Eitinger T, Suhr J, Moore L, Smith JA. Secondary transporters for nickel and cobalt ions: Theme and variations. Biometals. 2005;18:399–405. doi: 10.1007/s10534-005-3714-x. [DOI] [PubMed] [Google Scholar]
- 17.Hiron A, et al. A nickel ABC-transporter of Staphylococcus aureus is involved in urinary tract infection. Mol Microbiol. 2010;77:1246–1260. doi: 10.1111/j.1365-2958.2010.07287.x. [DOI] [PubMed] [Google Scholar]
- 18.Grim KP, et al. The metallophore staphylopine enables Staphylococcus aureus to compete with the host for zinc and overcome nutritional immunity. MBio. 2017;8:e01281-17. doi: 10.1128/mBio.01281-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lebrette H, et al. Promiscuous nickel import in human pathogens: Structure, thermodynamics, and evolution of extracytoplasmic nickel-binding proteins. Structure. 2014;22:1421–1432. doi: 10.1016/j.str.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 20.Lebrette H, et al. Novel insights into nickel import in Staphylococcus aureus: The positive role of free histidine and structural characterization of a new thiazolidine-type nickel chelator. Metallomics. 2015;7:613–621. doi: 10.1039/c4mt00295d. [DOI] [PubMed] [Google Scholar]
- 21.Ghssein G, et al. Biosynthesis of a broad-spectrum nicotianamine-like metallophore in Staphylococcus aureus. Science. 2016;352:1105–1109. doi: 10.1126/science.aaf1018. [DOI] [PubMed] [Google Scholar]
- 22.Lhospice S, et al. Pseudomonas aeruginosa zinc uptake in chelating environment is primarily mediated by the metallophore pseudopaline. Sci Rep. 2017;7:17132. doi: 10.1038/s41598-017-16765-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mastropasqua MC, et al. Growth of Pseudomonas aeruginosa in zinc poor environments is promoted by a nicotianamine-related metallophore. Mol Microbiol. 2017;106:543–561. doi: 10.1111/mmi.13834. [DOI] [PubMed] [Google Scholar]
- 24.McFarlane JS, Lamb AL. Biosynthesis of an opine metallophore by Pseudomonas aeruginosa. Biochemistry. 2017;56:5967–5971. doi: 10.1021/acs.biochem.7b00804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tame JR, Dodson EJ, Murshudov G, Higgins CF, Wilkinson AJ. The crystal structures of the oligopeptide-binding protein OppA complexed with tripeptide and tetrapeptide ligands. Structure. 1995;3:1395–1406. doi: 10.1016/s0969-2126(01)00276-3. [DOI] [PubMed] [Google Scholar]
- 26.Ding Y, Fu Y, Lee JC, Hooper DC. Staphylococcus aureus NorD, a putative efflux pump coregulated with the Opp1 oligopeptide permease, contributes selectively to fitness in vivo. J Bacteriol. 2012;194:6586–6593. doi: 10.1128/JB.01414-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berntsson RP, Smits SH, Schmitt L, Slotboom DJ, Poolman B. A structural classification of substrate-binding proteins. FEBS Lett. 2010;584:2606–2617. doi: 10.1016/j.febslet.2010.04.043. [DOI] [PubMed] [Google Scholar]
- 28.Dassama LM, Kenney GE, Ro SY, Zielazinski EL, Rosenzweig AC. Methanobactin transport machinery. Proc Natl Acad Sci USA. 2016;113:13027–13032. doi: 10.1073/pnas.1603578113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker EL, Waters BM. The role of transition metal homeostasis in plant seed development. Curr Opin Plant Biol. 2011;14:318–324. doi: 10.1016/j.pbi.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 30.Sleigh SH, Tame JR, Dodson EJ, Wilkinson AJ. Peptide binding in OppA, the crystal structures of the periplasmic oligopeptide binding protein in the unliganded form and in complex with lysyllysine. Biochemistry. 1997;36:9747–9758. doi: 10.1021/bi970457u. [DOI] [PubMed] [Google Scholar]
- 31.Berntsson RP, et al. Selenomethionine incorporation in proteins expressed in Lactococcus lactis. Protein Sci. 2009;18:1121–1127. doi: 10.1002/pro.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berntsson RP, Thunnissen AM, Poolman B, Slotboom DJ. Importance of a hydrophobic pocket for peptide binding in lactococcal OppA. J Bacteriol. 2011;193:4254–4256. doi: 10.1128/JB.00447-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheepers GH, Lycklama A Nijeholt JA, Poolman B. An updated structural classification of substrate-binding proteins. FEBS Lett. 2016;590:4393–4401. doi: 10.1002/1873-3468.12445. [DOI] [PubMed] [Google Scholar]
- 34.Begg SL, et al. Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae. Nat Commun. 2015;6:6418. doi: 10.1038/ncomms7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen W, Zhang Y, Yeo WS, Bae T, Ji Q. Rapid and efficient genome editing in Staphylococcus aureus by using an engineered CRISPR/Cas9 system. J Am Chem Soc. 2017;139:3790–3795. doi: 10.1021/jacs.6b13317. [DOI] [PubMed] [Google Scholar]
- 36.Weinberg ED. Nutritional immunity. Host’s attempt to withold iron from microbial invaders. JAMA. 1975;231:39–41. doi: 10.1001/jama.231.1.39. [DOI] [PubMed] [Google Scholar]
- 37.Cassat JE, Skaar EP. Metal ion acquisition in Staphylococcus aureus: Overcoming nutritional immunity. Semin Immunopathol. 2012;34:215–235. doi: 10.1007/s00281-011-0294-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cassat JE, Skaar EP. Iron in infection and immunity. Cell Host Microbe. 2013;13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma Z, Jacobsen FE, Giedroc DP. Coordination chemistry of bacterial metal transport and sensing. Chem Rev. 2009;109:4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischbach MA, Lin H, Liu DR, Walsh CT. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat Chem Biol. 2006;2:132–138. doi: 10.1038/nchembio771. [DOI] [PubMed] [Google Scholar]
- 41.Neumann W, Gulati A, Nolan EM. Metal homeostasis in infectious disease: Recent advances in bacterial metallophores and the human metal-withholding response. Curr Opin Chem Biol. 2017;37:10–18. doi: 10.1016/j.cbpa.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.