Significance
The evolution of land plants from algae is an age-old question in biology. The entire terrestrial flora stems from a grade of algae, the streptophyte algae. Recent phylogenomic studies have pinpointed the Zygnematophyceae as the modern-day streptophyte algal lineage that is most closely related to the algal land plant ancestor. Here, we provide insight into the biology of this ancestor that might have aided in its conquest of land. Specifically, we uncover the existence of stress-signaling pathways and the potential for intimate plastid-nucleus communication. Plastids act as environmental sensors in land plants; our data suggest that this feature was present in a common ancestor they shared with streptophyte algae.
Keywords: plant terrestrialization, abscisic acid, plastid-nucleus communication, charophyte algae, stress physiology
Abstract
Streptophytes are unique among photosynthetic eukaryotes in having conquered land. As the ancestors of land plants, streptophyte algae are hypothesized to have possessed exaptations to the environmental stressors encountered during the transition to terrestrial life. Many of these stressors, including high irradiance and drought, are linked to plastid biology. We have investigated global gene expression patterns across all six major streptophyte algal lineages, analyzing a total of around 46,000 genes assembled from a little more than 1.64 billion sequence reads from six organisms under three growth conditions. Our results show that streptophyte algae respond to cold and high light stress via expression of hallmark genes used by land plants (embryophytes) during stress–response signaling and downstream responses. Among the strongest differentially regulated genes were those associated with plastid biology. We observed that among streptophyte algae, those most closely related to land plants, especially Zygnema, invest the largest fraction of their transcriptional budget in plastid-targeted proteins and possess an array of land plant-type plastid-nucleus communication genes. Streptophyte algae more closely related to land plants also appear most similar to land plants in their capacity to respond to plastid stressors. Support for this notion comes from the detection of a canonical abscisic acid receptor of the PYRABACTIN RESISTANCE (PYR/PYL/RCAR) family in Zygnema, the first found outside the land plant lineage. We conclude that a fine-tuned response toward terrestrial plastid stressors was among the exaptations that allowed streptophytes to colonize the terrestrial habitat on a global scale.
Land plants are the most species-rich group of photosynthetic eukaryotes. The entire lineage traces back to a common ancestor shared with higher branching streptophyte algae; it was a member of this algal grade that successfully rose above its substrate, conquered land, and gave rise to the diversity of land plants we observe today (1). Many aspects of land plant evolution remain enigmatic. A long-standing assumption is that the streptophyte algal progenitors of land plants must have possessed exaptations that allowed them to deal with the stresses of a dry environment (1, 2). Given that the terrestrial stress response in plants is intimately linked to plastid biology, these exaptations might also have been plastid-associated (3). Indeed, the coding capacity of plastid genomes (3, 4) reveals a division between the three lower branching [Mesostigmatophyceae, Chlorokybophyceae, and Klebsormidiophyceae (KCM)] and the three higher branching streptophyte algae [Charophyceae, Coleochaetophyceae, and Zygnematophyeae (ZCC)] (Fig. 1 A and B). Land plants are also unique in that their plastids can differentiate into a broad range of plastid subtypes (5), a process mediated by nuclear control over plastid function.
Fig. 1.
Streptophyte algae used in this study. (A) Nomarski and dissection micrographs of the three lower-branching (KCM) and three higher-branching (ZCC) streptophyte algae cultured for transcriptome profiling. Colored text next to the strain numbers indicates the habitat (freshwater or soil) from which the strains were originally isolated, and colored backgrounds indicate the substrate the strains have been grown on in the laboratory (light purple, liquid medium; light tan, agar plates). A, antheridium; O, oogonium. (B) Cladogram (based on ref. 42) showing the relationship of the streptophytes (black), consisting of land plants and streptophyte algae (KCM and ZCC), to the chlorophyte Chlamydomonas (gray). (C) Steady-state light curves and calculated rETRs of the streptophyte algae analyzed in this study. Control conditions (20 °C) and cold conditions (4 °C) were set to 50 μmol of photons m−2⋅s−1, whereas HL (20 °C) conditions were set to ∼600 μmol of photons m−2⋅s−1. Error bars indicate SD between the biological triplicates. Cha., Chara; Chl., Chlorokybus; Col., Coleochaete; Kle., Klebsormidium; Mes., Mesostigma; Zyg., Zygnema.
Land plants exhibit remarkable control over their plastids derived from endosymbiosis (3). The process of plastid differentiation (5) is intimately linked to control over organellar transcription (6). Angiosperms are unique in having both a nuclear-encoded and plastid-encoded plastid RNA polymerase [NEP and PEP, respectively (7)], whereas most algae rely solely on PEP. In land plants, the activities of PEP-associated proteins (PAPs) add additional layers of control over gene expression (8). The bulk of plastid transcriptional activity is devoted to perpetuation of its biochemistry, foremost photosynthesis (9). Due to endosymbiotic gene transfer (10), more than 90% of the plastid proteome is the product of genes residing in the nucleus (11). Efficient and accurate plastid-nucleus communication is thus an essential part of algae and plant biology.
Retrograde-signaling pathways transmit information on the plastid’s condition to the nucleus. This involves biogenic signaling during plastid development, which is tied to NEP/PEP and PAPs (7, 8), and operational signaling (12), which is mainly associated with the control of photosynthesis. In Arabidopsis, an array of proteins has been characterized that sense the plastid’s status, from which the nucleus then alters its plastid-associated nuclear gene expression accordingly (12–14). In land plants, recent data show that (i) retrograde signaling is woven into the broader cellular-signaling network on many levels and (ii) carotenoid-derived metabolites appear to be key in plastid-nucleus communication (15–17). Indeed, the production of carotenoid metabolites is a reoccurring theme in plant abiotic stress signaling (16, 18). The carotenoid-derived signal abscisic acid (ABA) has been known for decades as one of the major stress-responsive phytohormones.
Stress physiology has been studied in detail in the model land plant Arabidopsis. Stress is perceived by the organism’s so-called “perceptron,” a complex regulatory network that receives input from phytohormones (19), such as jasmonic and salicylic acid (20), as well as ABA (21). ABA biosynthesis commences in the plastid using xanthophylls as a source, and only the last two steps occur in the cytosol (22). Regarding ABA perception, several proteins have been proposed to be involved, including G proteins (23). However, the canonical ABA-signaling pathway (reviewed in ref. 21) consists of the cytosolic PYR/PYL/RCAR receptor (24), which negatively regulates PROTEIN PHOSPHATASE 2C proteins (PP2Cs), such as ABSCISIC ACID INSENSITIVE [ABI (25)]. In turn, inhibition of PP2Cs lifts their negative regulation of SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASEs (SnRKs), such as OPEN STOMATA 1 (OST1), that activate ABA-responsive element binding factors [e.g., AREB1(26)], triggering the ABA-induced (stress) response (27). The ABA-signaling pathway appears conserved across land plant diversity (28), but parts of it (e.g., PYR/PYL/RCAR receptors) have not been found outside of land plants (29–31). The same is true for parts of the ABA biosynthesis pathway (30). That said, ABA has been detected in various algae (32), including streptophyte algae (33–35), suggesting that they utilize alternative subsets of enzymes for synthesizing ABA.
New technologies have accelerated the pace at which molecular links between land plants and their closest algal relatives have been discovered (36–38). Using comparative RNA sequencing (RNA-seq) on stress-treated representatives of each of the six major streptophyte algal classes, we show that streptophyte algae possess previously undetected components of the general embryophytic stress response and ABA-signaling pathway. Moreover, the ZCC-grade streptophyte algae appear to have a particularly tight genetic interaction between the nucleus and plastid, suggesting that, as in land plants (18), their plastid is an integral part of the cell-signaling network.
Results and Discussion
Transcriptome Profiling of the Stress Response Across Streptophyte Algal Diversity.
We cultivated one representative from each of the six major streptophyte algal classes (Mesostigma viride, Chlorokybus atmophyticus, Klebsormidium flaccidum, Chara globularis, Coleochaete scutata, and Zygnema circumcarinatum; Fig. 1 A and B), under equal growth conditions (Materials and Methods). Using pulse amplitude modulation fluorometry, we generated light curves and determined that all six species have a similar relative electron transport rate (rETR) at 50 μmol of photons m−2⋅s−1 and 20 °C, which served as the control condition (Fig. 1C). At ∼600 μmol of quanta m−2⋅s−1, the rETR of all species was saturated. This condition was hence selected as the high light (HL) treatment, and it was to this treatment that we exposed the algae for 1 h in subsequent experiments. Cold is a major terrestrial stressor and is known to induce plastid-derived reactive oxygen species (ROS), among other effects triggering plastid retrograde signaling (12, 39); we applied cold stress by subjecting the algae to 4 °C (at 50 μmol of quanta m−2⋅s−1) for 24 h.
For each algal species and condition (except M. viride, which did not yield usable RNA after cold treatment), we sequenced 150-bp paired-end reads of mRNA libraries derived from three independent biological replicates on the Illumina HiSeq platform, generating 1,640,342,307 reads and 492.1 Gbp of sequence data (Fig. S1 and Table S1). Using the TRINITY pipeline, we de novo assembled the raw data for each species (reads from all libraries/conditions were combined) and retrieved contigs to which RSEM (RNA-Seq by Expectation Maximization) mapped reads at a density of greater than one transcript per million [TPM; trimmed mean of M values (TMM)-normalized]. Using DIAMOND and reciprocal blastx/tblastn, we filtered for contigs that had their best hit against Chloroplastida or transcripts encoding putative orthologs to Klebsormidium nitens (33) proteins. We retained between 4,729 and 13,939 contigs for the six species (Fig. S1 and Dataset S1) that were used for downstream analyses.
To evaluate the global gene expression response to the HL and cold treatments in streptophyte algae, we mapped all transcript data onto Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways occurring in plants (Fig. 2 and Fig. S2). We detected between 1,496 and 2,619 unique KEGG orthologs in the six species examined, of which 7.3–25.3% showed a differential response [|log2(fold change)| > 1] upon HL and 33.4–78.1% upon cold treatment. Cold treatment had a particularly dramatic effect on gene expression in the KCM-grade algae Klebsormidium and Chlorokybus, with 78.1% and 65.0%, respectively, of the genes responding. In the case of Klebsormidium, 41.1% of the KEGG orthologs responded by greater than eightfold down-regulation (in contrast to a mere 9.5% in Chlorokybus); hence, Klebsormidum’s responses in specific pathways and genes must be interpreted with caution, as various pleiotropic effects might have occurred. Nevertheless, as is to be expected from cold and HL treatment, we observed a consistently strong effect on plastid biology. All species showed enrichment for gene ontology (GO) terms with a photosynthesis (biological processes) or plastid (cellular compartment) affiliation (Fig. S3). For photosynthesis-related genes, between 10.0% and 63.9% of KEGG orthologs responded to HL, while 65.2–83.3% were responsive to cold treatment (Fig. 2B). This is consistent with the significantly enriched GO terms found among differentially responding [|log2(fold change)| > 1] genes. In summary, our HL and cold treatments triggered a plastid-associated differential gene expression response.
Fig. 2.
Differential gene expression in streptophyte algae upon cold and HL cues. Treatments were as follows: cold (turquoise) = 24 h at 4 °C, HL (yellow) = 1 h at ∼600 μmol of photons m−2⋅s−1. For each species, TMM-normalized gene expression data of genes with the same KEGG ortholog assignment were summed and used to calculate log2(fold changes) relative to control conditions (50 μmol of photons m−2⋅s−1, 20 °C). For each K-number, a colored tick indicates its expression relative to the control. The color of the ticks corresponds to a gradient (Scale) from down-regulation in red [log2(fold change) ≤ −3], to no change in blue [log2(fold change) = 0], to up-regulation in green [log2(fold change) ≥ 3]. (A) Overview across all KEGG orthologs, arranged hierarchically by KEGG pathways (small boxes in light gray shading) and higher order BRITE (a KEGG database containing functional hierarchies of biological entities) categories (two types of darker gray with white labels). Within each KEGG pathway category, data have been hierarchically sorted by the breadth of detection of specific KEGG orthology groups within the RNA-seq data. ENVIRON. INFO, environmental information. (B) Close-up view of plastid-associated KEGG pathways. Next to the KEGG pathway photosynthesis, the relative amounts of up- and down-regulated K-numbers are shown (normalized for each species independently). Z, zeatin biosynthesis. Note that no gene expression data were obtained for cold-treated Mesostigma. C.a., C. atmophyticus; C.g., C. globularis; C.s., C. scutata; K.f., K. flaccidum; M.v., M. viride; Z.c., Z. circumcarinatum.
Z. circumcarinatum Devotes a Larger Transcriptional Budget to Plastid-Targeted Proteins than Other Streptophyte Algae.
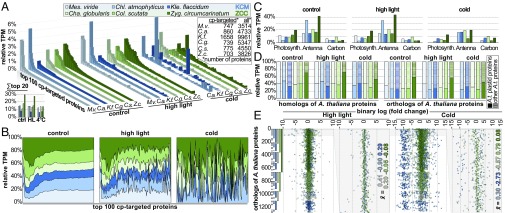
Stress impacts the expression of important plastid-associated genes [e.g., photosynthesis- and plastid redox-associated nuclear gene expression (12)]; plastid-nucleus communication thus alters the composition of the plastid proteome and relative abundance of individual plastid-targeted proteins under stress conditions. Since we observed that streptophyte algal plastid biology was strongly influenced by our treatments, we first sought to assess the nuclear contribution to the plastid proteomes of streptophyte algae. For this, we used three subcellular localization prediction tools on all ORFs with full-length amino (N) termini and calculated the relative expression level distribution among those ORFs predicted to encode plastid-targeted proteins. Considering the top 100 most highly expressed plastid-targeted proteins in each organism, the ZCC-grade streptophyte algae C. scutata and Z. circumcarinatum were found to invest most heavily (Fig. 3 A and B). This is pronounced in the latter species, for which a higher “shoulder” on the expression level distribution is apparent (Fig. 3A). In Zygnema, the top 20 most highly expressed plastid-targeted proteins sum up to about 30% of the entire transcript budget under control conditions, compared with less than 20% in all other species (Fig. 3A). The KCM-grade streptophyte algae were characterized by having relatively few very-high-abundance transcripts. However, it should be noted that important genes, such as that encoding the plastid stroma protein RIBULOSE BISPHOSPHATE CARBOXYLASE SMALL CHAIN (KfRBCS), were not among those ORFs with analyzable N termini in the Klebsormidium data, while the transcript for CaRBCS was the second most highly expressed gene in Chlorokybus in this analysis. We thus also performed additional analyses independent of our subcellular predictions.
Fig. 3.
Transcript abundance for plastid-targeted proteins increases along the trajectory of streptophyte algal evolution. (A) Transcripts were in silico-translated, and those with intact N termini were extracted. Subcellular localizations of these proteins were predicted using three tools (TargetP, LOCALIZER, and PredAlgo) and considered targeted to the plastid if indicated by at least two programs. The top 100 highest expressed transcripts (based on TMM-normalized TPM expression) coding for plastid-targeted proteins were extracted, and their relative expression was plotted (TPMTMM-normalized of plastid-targeted proteins/all TPMTMM-normalized of transcripts with intact N termini). The table in the top right corner shows the number of proteins used for the analysis. (Inset) Bar graph shows the sum of the relative expression of the top 20 highest expressed plastid-targeted proteins. (B) Pooled relative expression data—of all species—on the top 100 highest expressed plastid-targeted proteins shown in A. (C) TPMTMM-normalized expression levels of all transcripts assigned to the KEGG pathway photosynthesis (Photosynth.), antenna proteins, and carbon fixation for all six species, normalized against all TPMTMM-normalized expression levels for which a K-number could be assigned (i.e., mapped onto KEGG). (D) Relative expression levels of transcripts coding for homologs (Left) and (based on a reciprocal best blastp approach) predicted orthologs (Right) of A. thaliana proteins that are listed in the curated subcellular localization Plant Proteome Database as plastid-targeted proteins. Filled bars show homologs/orthologs of Arabidopsis plastid-targeted proteins, and striped bars show other (i.e., nonplastid localized) homologs/orthologs of Arabidopsis proteins (a depiction of the data in relation to all Chloroplastida sequences is shown in Fig. S4A). (E) Dot plot of the relative [log2(fold change)] expression changes in orthologs of Arabidopsis plastid-targeted proteins upon HL and cold treatment versus control. Each position on the y axis represents an ortholog found in at least one of the species. Orthologs (total = 1,271) were sorted hierarchically (Z-C-C-K-C-M, with Zyg. given the highest priority) according to the number of species in which they were detected. The sorting is depicted as a vertical bar graph (far left): Orthologs detected in (first) Zyg. make up one contiguous dark-green block, (second) Col. make up two grass-green blocks, (third) Cha. make up four light-green blocks, and so on (2n−1). Next to each dot plot is a zoom-in into the 95 orthologs detected among all species; numbers indicate the average (x̄) expression changes relative to control among these 95 shared orthologs. The legend for the color code in A applies to all displayed items. No gene expression data were obtained for cold-treated Mes. A.t., A. thaliana; C.a., C. atmophyticus; C.g., C. globularis; Cha., Chara; Chl., Chlorokybus; Col., Coleochaete; C.s., C. scutata; K.f., K. flaccidum; Kle., Klebsormidium; Mes., Mesostigma; M.v., M. viride; Z.c., Z. circumcarinatum; Zyg., Zygnema.
Reliable plastid-targeting prediction can be challenging for nonmodel organisms (e.g., 40). To confirm our observations, we used two additional approaches. First, for each species, we considered the size of the transcript budget invested in plastid biology-associated KEGG categories. Expression of photosynthesis-associated and antenna-associated proteins made up 19.8% and 41.6%, respectively, of all Zygnema gene transcript data for which KEGG orthologs were identified (Fig. 3C). Chlorokybus also invests a remarkable 35.3% into antenna proteins. Second, we utilized the well-curated Plant Proteome Database of Arabidopsis (41) and screened our streptophyte algal RNA-seq data for transcripts homologous and orthologous to Arabidopsis plastid proteins. In both the homology and orthology datasets, Zygnema again invested the largest fraction of its transcriptional budget into plastid-targeted proteins (Fig. 3D and Fig. S4A). In summary, the streptophyte algal lineage that is phylogenetically closest to the land plants (according to ref. 42) invests more heavily in transcription of plastid-targeted proteins than do the KCM algae. This is in line with our proposal that along the trajectory of streptophyte algal and land plant evolution, plastids became more dependent on the nucleus (3). Increases in the nuclear contribution to the plastid proteome might be a consequence of higher nuclear dependency. In the case of Zygnema, the fact that the organism is biplastidic must also be taken into account (43).
What about the influence of stress on the nuclear contribution to the plastid proteome? Upon stress treatment, the aforementioned transcript budget pattern generally held. On average, HL treatment had little influence on overall expression levels of plastid-targeted proteins (average changes between 1.5% and 19.5% were observed among the top 100 most highly expressed genes based on prediction tool data and 0.8–3.7% based on Arabidopsis thaliana orthology data). Cold treatment led to the most pronounced response (Fig. 3B), reducing expression of the top 20 plastid-targeted proteins by 15.8% and 22.0% in Chlorokybus and Klebsormidium, respectively, and more than 50% in Coleochaete and Zygnema. Not so in Chara, where expression was, in fact, increased by 20%. Across all streptophyte algae, we detected 1,271 orthologs of the A. thaliana plastid proteome, of which 95 were found in all six species (Fig. 3E). Among these 95 shared protein orthologs, the average expression changes were less than twofold upon all expression treatments, except for Klebsormidium, with an average 6.6-fold down-regulation upon cold. Zygnema responded to both treatments with a mere 1.06-fold change in expression. The proteins that were, on average, influenced the most by HL treatment were photosystem I components [potentially due to the Mehler reaction (44, 45)]. In stark contrast, a whole variety of factors responded to cold treatment, but were nevertheless dominated by photosystem II and light-harvesting components (Fig. S4B). In summary, the nuclear-encoded plastid proteome of ZCC-grade streptophyte algae responded, on average, less in terms of fold change but more in terms of overall transcript budget (especially toward cold) than the KCM-grade streptophyte algae.
ZCC-Grade Streptophyte Algae Possess a Plant-Like Stress Response.
Land plants have a fine-tuned stress–response system built upon an array of receptors and a cascade of downstream signal transduction pathways; together, they have been described as the plant perceptron (19). To better understand stress–response signaling in streptophyte algae, we screened our data for orthologs of the well-curated Arabidopsis protein data. Across all streptophyte algae, we detected 8,585 transcripts encoding orthologs of Arabidopsis proteins, of which 266 were detected in all six species. To calculate whether these 266 shared genes followed the same expression trend, we categorized them as (i) up-regulated [log2(fold change) ≥ 1], (ii) nonregulated [|log2(fold change)| ≯ 1], and (iii) down-regulated [log2(fold change) ≤ −1] (Fig. 4). Upon HL treatment, there were no genes coregulated between all six species; within-grade comparisons revealed coregulation of 50.4% between KCM, 52.6% between all ZCC, and 75.2% between Zygnema and Coleochaete. Upon cold treatment, 3.4% of these 266 genes were coregulated between the five species with RNA-seq data; within-grade comparisons revealed 35.7% coregulation (6.0% nonregulation) between KC, 27.1% between ZCC, and 50.0% between Zygnema and Coleochaete. When comparing within-species coregulation upon the two treatments, we observed a clear difference between the coregulation pattern of KCM and ZCC algae: Chlorokybus and Klebsormidium coregulated a mere 29.3% and 30.1% of their genes, respectively, while Chara, Coleochaete, and Zygnema coregulated 66.9%, 62.4%, and 68.0%, respectively. At the same time, the KCM-grade algae had the most differentially responding [|log2(fold change)| > 1] genes (Fig. 4). It should be noted that the species investigated here were isolated from a broad range of soil and freshwater habitats (Fig. 1A). While some strains and species are limited to a certain habitat, others are not and can, under laboratory conditions, be cultivated using various types of media. Future ecophysiological studies utilizing different strains and habitat combinations will be essential for testing the robustness of the patterns observed here. At the present time, our data are most consistent with the idea that ZCC-grade streptophyte algae have a more fine-tuned stress response, especially toward cold.
Fig. 4.
Coregulation of gene expression changes of Arabidopsis orthologs is higher in the ZCC-grade than in KCM-grade streptophyte algae. Using a reciprocal blastp approach of in silico-translated proteins, orthologs of Arabidopsis proteins were detected in all six streptophyte algal datasets. Orthologs shared by all six species, only the ZCC-grade species, only Zygnema and Coleochaete (ZC) species, or only the KCM-grade species were plotted; their relative expression changes [log2(fold changetreatment/control)] are depicted as heat map gradients from down-regulation, red [log2(fold change) ≤ −3], to no change, blue [log2(fold change) = 0], to up-regulation, green [log2(fold change) ≥ 3]. Plots at the top show comparisons of gene expression responses toward the two treatments between species, sorted by Zygnema or Mesostigma/Chlorokybus (in the case of orthologs restricted to KCM). Plots at the bottom show within-species comparisons of gene expression response, sorted by cold treatment. (Right) Corresponding relative amount of coregulation [distinguishing between the categories (i) up, defined by log2(fold change) ≥ 1; (ii) no change; or (iii) down, defined as log2(fold change) ≤ −1] of the 266 orthologs shared by all species is plotted. Note that no gene expression data were obtained for cold-treated Mesostigma. C.a., C. atmophyticus; C.g., C. globularis; C.s., C. scutata; KC, Klebsormidium and Chlorokybus; K.f., K. flaccidum; M.v., M. viride; Z.c., Z. circumcarinatum.
To elucidate the molecular chassis underpinning the stress response in streptophyte algae, we analyzed the 50 most cold- and HL-responsive transcripts [i.e., transcripts with the highest and lowest log2(fold change)] coding for Arabidopsis protein orthologs (Fig. 5 and Fig. S5). Regardless of the species and treatment, many photosystem, light-harvesting, and photooxidative stress-related proteins were identified, such as catalase (e.g., 29- and 15-fold cold-induced CAT orthologs in Zygnema and Coleochaete, respectively) and an ortholog of PEROXIREDOXIN Q that was up-regulated 64.1-fold upon cold (P < 0.05) and is known to ward off ROS stress in plants (46). We also detected various stress-responsive orthologs of plastid lipid-associated proteins that are known to be involved in thylakoid maintenance (47) (e.g., fibrillin; FIB1b orthologs were up-regulated fourfold and more upon HL in Klebsormidium, Chara, and Zygnema). Early light-induced proteins (ELIPs) are light-harvesting complex-related proteins that dampen (cold-induced) photooxidative damage (48). ELIPs are a classical stress-responding gene family in land plants (e.g., ref. 49) and, recently, a cold-responsive ELIP has been characterized in the streptophyte alga Spirogyra (50). We found an ortholog of ELIP (AT3G22840) in all streptophyte algae. This gene was especially cold-responsive, being up-regulated between >18-fold and >76-fold in Chlorokybus and Chara, respectively (significant at P < 0.01). In contrast, orthologs of the CHAOS-encoding gene that regulates light-harvesting complex protein targeting to the thylakoids (51), including ELIP (48), were strongly down-regulated in Zygnema (17-fold), Chara (55.8-fold), and Klebsormidium (675.8-fold).
Fig. 5.
Top 50 most stress-responsive Arabidopsis orthologs detected among ZCC-grade streptophyte algae. Based on edgeR, relative gene expression changes and statistical support for these changes were calculated. The top 50 Arabidopsis orthologs with the strongest up- and down-regulation were extracted and plotted as word clouds. The word size corresponds to log2 changes in transcript abundance [log2(fold changetreatment/control)], and the font color indicates changes in a factor upon cold (blue) and HL (yellow) treatment; significant changes (Benjamini–Hochberg adjusted P < 0.05) are indicated by dark blue and orange, respectively [for each species, the arrowheads in the key indicate the word size of the (first) highest and 50th highest depicted changes]. A comparison of all species is shown in Fig. S5, and a table format is shown in Dataset S2.
Arabidopsis phytochromes sense light and temperature (52). The transcription factor REVEILLE (RVE) was recently found to be important for integrating both cues (53). In C. scutata, we found that both the phytochromobilin-associated gene GENOMES UNCOUPLED 3 [GUN3 (13, 54)] and RVE1 were up-regulated (greater than eightfold and >19-fold, respectively; P < 0.01) upon cold treatment and that the phytochrome B gene LONG HYPOCOTYL 3 (HY3) was greater than twofold down-regulated upon cold treatment. This suggests that there are regulatory modules that integrate temperature sensing and plastid biology [through GUN3 (13)] conserved between Arabidopsis and ZCC-grade streptophyte algae, such as Coleochaete. Phytochrome and RVE1 have been shown to act in concert in influencing seed dormancy (53), which is a classical ABA-regulated process. This warrants attention as upstream of many stress–response factors, we find the canonical ABA-signaling cascade (reviewed in ref. 21).
There are recurring themes in land plant stress response. Due to the connectivity of the perceptron of plants (19), multiple cues are transduced via similar circuits. Hence, there are certain hallmark proteins and signaling factors of stress response. The late embryogenesis abundant (LEA) proteins were first found in seeds and tied to dormancy and ABA (55). LEAs are now considered classical factors induced by abiotic stresses, such as cold (56), and are known to increase stress tolerance when overexpressed [e.g., in cold tolerance in tobacco (57)]. In land plants, promoters of LEA genes often contain ABA-response elements (58). We found between two- and 32-fold induction of LEAs in all cold-treated streptophyte algae, except for Chara (and Mesostigma, for which we were not able to generate such data).
Coleochaete and Zygnema are noteworthy in terms of their differential expression of proteins that are known to be ABA-responsive in Arabidopsis. For example, Coleochaete was found to up- and down-regulate various cell wall-associated enzymes, which is commonly observed in land plants under stress (59); in a recent study, we found that Coleochaete stood out in terms of its suite of potential cell wall modification proteins (38). Most interesting, however, are CsATHB33 and SLAC1 HOMOLOGUE 1 (SLAH1). ATHB33 (HOMEOBOX PROTEIN 33) is one of three transcription factors differentially expressed in the HL-sensitive and ELIP-misregulating cryptochrome mutants (60). Indeed, we found the CsCRY3 orthologs of Coleochaete to be greater than fourfold and >13-fold up-regulated (P < 0.01) upon HL and cold treatment, respectively. Hence, the up-regulation of CsATHB33 suggests that there is a conserved regulatory module that involves ATHB33, ELIP, and cryptochrome. SLAH1 is closely related to SLOW ANION CHANNEL-ASSOCIATED 1 (SLAC1), which has been detected in K. nitens genome data (61). We found a SLAH1 ortholog in Zygnema whose expression was almost 25-fold up-regulated upon cold treatment. SLAC1 is known to be an important player in canonical ABA-mediated stomatal closure (62); SLAH1 also responds to ABA, having been found down-regulated upon ABA treatment in Arabidopsis roots (63). Recently, it was shown that retrograde signals converge in the regulation of SLAC1, tying it to the canonical ABA-mediated drought response (64).
Stomata responses toward ABA across land plant evolution are currently debated (65, 66), and Lind et al. (61) showed that ABA-signaling components can only physically interact with SLAC1 ion channels in stomata-bearing streptophytes. It is hence instructive that some of the responsiveness of this signaling module was already present in the closest streptophyte algal ancestors of land plants, which obviously had no stomata but very likely ABA signaling. Similarly, upon cold stress, among the top 50 down-regulated genes in Zygnema was a gene coding for the canonical ABA receptor protein PYR1-LIKE 8 [PYL8 (24)] (discussed below). This is noteworthy, given that the PYR/PYL ABA receptors are suggested to have evolved in land plants after they diverged from their closest streptophyte relatives (29, 30).
Land Plant-Like Plastid-Nucleus Communication in Streptophyte Algae.
Plastids are increasingly recognized as central players in plant stress–response signaling (12, 18). Retrograde signaling translates signals from the plastid into a nuclear response. At the same time, the nucleus exercises direct control over the plastid through, for example, the plastid-targeted PAPs. We utilized Arabidopsis orthology data to better understand the evolutionary origin of retrograde-signaling genes known from land plants in their algal progenitors.
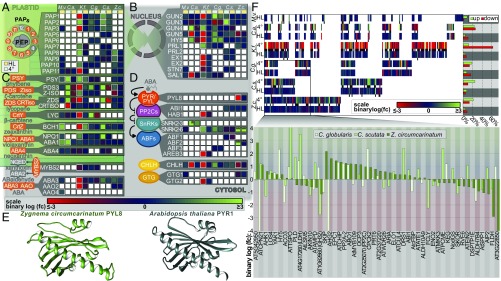
The Arabidopsis nuclear genome encodes 12 PAPs (8). We detected a total of 10 PAPs across the six streptophyte algae examined (Fig. 6A). If found to be differentially expressed, these PAPs were usually up-regulated and more responsive to cold [34.7% log2(fold change) ≥ 1 up-regulation and 8.6% log2(fold change) ≤ −1 down-regulation] than to HL [4.8% log2(fold change) ≥ 1 up-regulation and no log2(fold change) ≤ −1 down-regulation]; the only down-regulation of PAPs was observed in the cold-hypersensitive Klebsormidium. Interestingly, in Coleochaete, PAP1, PAP2, and PAP3 [pTAC3, pTAC2, and pTAC10, respectively (8)] were up-regulated in response to both HL and cold. Our data suggest that streptophyte algae have a battery of PAPs that modulate the in organello plastid response to stresses.
Fig. 6.
Retrograde and ABA signaling in streptophyte algae upon cold and HL treatment. Changes in levels of transcripts coding for proteins (based on a reciprocal best blastp approach) predicted to be orthologous to A. thaliana plastid-targeted PAPs (A) and canonical retrograde-signaling factors (B) are shown (additional putative factors that did not meet the expression criterion cutoff are shown in Fig. S6). Note also that since the role of PTM in retrograde signaling has recently been challenged (104), it is only shown in the supporting information (Fig. S6). Changes in levels of transcripts coding for orthologs of A. thaliana ABA biosynthesis (C) and ABA-signaling factors (D) are shown. A white outline in A–D indicates statistically significant differential regulation (Benjamini–Hochberg adjusted P < 0.05). One asterisk indicates that calculations were done based on TPM expression values, not edgeR (which relies on read counts); two asterisks (ZcSNRK2-6 and ZcABF2) additionally indicate expression of <1 TPM. (E) Best tertiary protein structure for the PYL8 ortholog detected in Z. circumcarinatum (computed by I-TASSER) and its closest structural analog (template modeling score = 0.895), AtPYR1 (Protein Data Bank ID code 3K3K). (F) Changes in levels of transcripts coding for orthologs of A. thaliana proteins that are known to respond to ABA. (Upper Left) Below each colored tick in the heat map (representing the relative expression), a black tick indicates statistically significant differential regulation (Benjamini–Hochberg adjusted P < 0.05). (Upper Right) Bar graph depicts the relative amount (normalized by the number of orthologs detected for each species) of these ABA-responsive orthologs. (Bottom) Close-up view of the cold-induced expression changes, plotted as a bar graph, of orthologs shared by at least Zygnema and another ZCC-grade streptophyte algae. Relative expression data in A–D and F are depicted as a gradient from down-regulation, red [log2(fold change [fc]) ≤ −3], to no change, blue [log2(fold change) = 0], to up-regulation, green [log2(fold change) ≥ 3]. No gene expression data were obtained for cold-treated Mesostigma. Treatments were as follows: cold (turquoise in A–D) = 24 h at 4 °C, HL (yellow in A–D) = 1 h at ∼600 μmol of photons s−1⋅m−2. C.a., C. atmophyticus; C.g., C. globularis; C.s., C. scutata; K.f., K. flaccidum; M.v., M. viride; Z.c., Z. circumcarinatum.
Retrograde signaling transmits plastid-derived signals to the nucleus. Transduction of these signals rests upon plastid-localized transmitters and nucleocytoplasmic receivers (12). We screened our streptophyte algal RNA-seq data for the presence of 18 retrograde-signaling components known from land plants. In total, 12 were detected in our streptophyte algal RNA-seq datasets (Fig. 6B). However, there are likely more, including, for example, GUN1 in Zygnema and the key light-signaling mediators GOLDEN2-LIKE 1/2 (GLK1/2) (67), which were identified by also considering transcripts below the chosen cutoff (Fig. S6). These results also align well with a reciprocal blastp screening of the retrograde-signaling repertoire encoded in the K. nitens genome (33), in which 11 components were detected (Fig. S6). Again, cold induced the strongest response, triggering, on average, 28.1% up-regulation and 26.8% down-regulation; HL induced 4.8% up-regulation and 7.1% down-regulation [up = log2(fold change) ≥ 1 and down = log2(fold change) ≤ −1]. Some retrograde-signaling factors showed consistent responses toward both HL and cold, especially within the ZCC-grade streptophyte algae.
Various abiotic stresses funnel into plastid retrograde signaling (68). Recently, there has been growing appreciation of the involvement of retrograde signaling in drought stress and its overlap with HL stress (69, 70). Furthermore, recent data highlight the impact of drought (desiccation) on photosynthesis-related gene expression in Zygnema (71). The same seems to apply to the basal-branching land plant Physcomitrella, where a major share of stress-induced gene expression changes are plastid-associated (72). The conditions discussed here (HL, cold, and drought) are similar in that they are intertwined with ROS (73) and retrograde signaling (17, 74). Tolerance toward terrestrial stressors during plant terrestrialization might thus have included components of the retrograde-signaling pathway. The presence of these retrograde-signaling pathways is underpinned by our data, although their physiological impact remains to be determined.
Carotenoid-derived metabolites are often involved in retrograde signaling (18). We detected most components of the carotenoid pathway in all streptophyte algae and, with the exception of Klebsormidium, cold treatment induced their expression (Fig. 6C). Carotenoid cleavage products, such as β-cyclocitral, have been shown to be major players in retrograde signaling (12, 16). Cold stress-induced regulation of carotenogenesis speaks to an involvement of carotenoid-mediated stress signaling in streptophyte algae.
Zygnema Possesses Genes for ABA Signaling.
The stress hormone ABA is a carotenoid-derived metabolite. In our transcriptomic survey of streptophyte algae, we did not detect all components of the ABA biosynthesis pathway downstream of zeaxanthin, but ABA itself has been detected in the streptophyte algae (e.g., K. nitens and Chara australis, refs. 33, 35). Furthermore, desiccation stress in Klebsormidium crenulatum was shown to induce components of the ABA-signaling pathway (36). The detection of canonical (PYR/PYLs|PP2Cs|SnRKs) and controversial (G protein-based or CHLH/GUN5) ABA-signaling components (21) was patchy (Fig. 6D). We found orthologs of the PP2C protein ABI1 in Coleochaete and Zygnema and an ortholog of HAB1 (HYPERSENSITIVE TO ABA1) in K. flaccidum (our data); the latter is consistent with the presence of PP2C genes in the genome of K. nitens (33). SnRKs were detected in all streptophyte algae except Zygnema, but SnRK2.6 has been detected in Spirogyra (75). Indeed, when we surveyed our raw RNA-seq data, we detected an ortholog of SnRK2.6/OST1 [Expect (E) values of 0 in tblastn and blastx searches] that was expressed at a very low level, but was induced by HL and cold (average TPMTMM-normalized: control = 0, cold = 0.07, and HL = 0.2). In a complementation experiment, K. nitens SnRK2.6 (OST1) was shown to be capable of replacing AtSnRK2.6 in terms of interacting with SLAC1, as well as being suppressed by the PP2C ABI1 (61). Since we detected an additional component of ABA signaling that is thought to have evolved at the base of land plants (i.e., a PYR/PYL receptor) (29, 30), this suggests that the genetic toolkit for the ABA-signaling pathway was present already during the earliest steps of land plant evolution.
The PYR/PYL/RCAR proteins are the receptors in the canonical ABA-signaling pathway (21, 24), and they are believed to have evolved at the base of the embryophyte lineage (i.e., after the origin of land plants) (29). However, we detected a gene coding for an ortholog of PYL8 in Zygnema, ZcPYL8, which is down-regulated in response to cold stress more than 15-fold, potentially indicating a feedback loop due to stress-induced elevation of ABA levels. ZcPYL8 is a 186-aa protein with a 143-aa PYR/PYL/RCAR domain. It contains 14 of the 27 residues that are, according to Yin et al. (76), relevant for binding ABA or interacting with ABI. I-TASSER (iterative threading assembly refinement) modeling of ZcPYL8 recovered AtPYR1 (Protein Data Bank ID code 3K3K) as its closest structure (template modeling score = 0.895; Fig. 6E). With ZcPYL8, Zygnema has a putative functional ortholog of the canonical PYR/PYL/RCAR ABA receptors. The detection (and regulation) of a PYL8 ortholog in Zygnema not only underpins the phylogenomic placement of the Zygnematophyceae as the closest algal relatives of land plants (42), but adds a key factor to the list of exaptations that might have enabled plant terrestrialization.
ABA signaling induces an array of downstream targets. We screened our streptophyte RNA-seq data for orthologs of all 593 Arabidopsis proteins listed by The Arabidopsis Information Resource (TAIR) as responsive to ABA. Across all streptophyte algae, we detected 230 orthologs of these proteins, of which 9.7–23.5% and 32.1–75.4% were regulated upon HL and cold treatment, respectively. These orthologs tended to be especially responsive to cold (Fig. 6F), showing between 2.5-fold (Chara) and sixfold (Zygnema) more up-regulation and 1.6-fold (Zygnema) and 9.4-fold (Klebsormidium) more down-regulation compared with HL treatment. The only exception was Chara, which responded with 1.2-fold less down-regulation. Interestingly, orthologs of ABA-responsive proteins in ZCC tended to be more induced (1.4-, 2.5-, and 1.6-fold more up-regulation). Among the downstream factors responding were well-known ABA- and stress-associated factors, such as the drought responsive DRS1 (77); the ABA-induced negative regulator of ABA signaling, PP2A-2 (78); and the positive regulator of ABA-mediated stress signaling, HISTIDINE KINASE 1 (AHK1) (79). In land plants, there is an AHK1-associated interplay between three players: cytokinin, ABA, and stress (79). The detection and regulation of AHK1 hint that this interplay might be similarly wired in streptophyte algae. Indeed, in Zygnema, we detected cold-induced greater than threefold up-regulation of orthologs of CYTOKININ RESPONSE FACTOR 1 or RESPONSE REGULATOR 2. Further connections to other stress hormones might include ethylene, which was recently shown to be an active phytohormone in the Zygnematophyceae Spirogyra (80). Accordingly, among the top 50 cold-responsive genes in Coleochaete is an ortholog of the ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 4 (ATERF-4) that is involved in both ethylene and ABA response (81).
In summary, we detected both ABA-associated signaling and general (land plant) stress–response-signaling networks in streptophyte algae. The extent to which ABA-mediated signaling in these algae mirrors that in land plants remains obscure. On balance, however, our data suggest that the signaling circuits that unite stress, ABA, and plastid-nucleus communication in land plants are present and responsive to similar cues in the algal ancestors of plants.
Conclusion
The rise of land plants depended in great measure on their ability to sense and respond to the terrestrial environment. Their developmental plasticity bears the signatures of selection for adaptability that likely go back to the very origin of land plants (recently discussed in ref. 82). We have shown that Zygnema and, to some degree, Coleochaete, the algae phylogenetically closest to land plants (42), have many of the stress-signaling factors until now only known from land plants. At the same time, these algae possess sophisticated anterograde plastid-nucleus communication and many of the components known to mediate retrograde signaling in land plants. The latter is particularly relevant in light of recent hypotheses putting the plastid at the center of land plant cell signaling. Taken together, our data suggest that embryophyte stress-signaling circuits were already present at the time that streptophytes began their conquest of land more than 450 million y ago.
Materials and Methods
Culture Handling.
Four algae were obtained from the culture collections Sammlung von Algenkulturen (SAG) Göttingen and cultured in Erddekokt and Salze (ES) or ES plus peptone (ESP) with 1% agar: C. atmophyticus SAG34.98 in ES, K. flaccidum SAG2307 in ESP, C. scutata SAG50.90 in ES, and Z. circumcarinatum SAG698-1a in ESP. M. viride CCAC 1140 was obtained from the Culture Collection of Algae at the University of Cologne (CCAC) and cultivated in liquid ESP with 1% saturated CaSO4 and 5 × 10−6 g/L vitamin B12. C. globularis was obtained from the pond nursery Wasserpflanzen Rehberg and grown in a Weck jar with ES and heat-sterilized aquaria sand. All algae were cultivated (climate-controlled) at 20 °C and 12-h/12-h light (L)/dark (D) conditions (L = 50 μmol of photons m−2⋅s−1 from an LED light source). Cold stress treatment was applied by shifting the algae for 24 h to 4 °C at 50 μmol of quanta m−2⋅s−1. HL treatment was applied by subjecting the algae (at 20 °C) for 1 h to ∼600 μmol of photons m−2⋅s−1.
Light Curves.
Steady-state light curves were generated using a FluorCam FC 800MF (Photon Systems Instruments). Modulated blue light (0, 6, 30, 56, 83, 109, 134, 184, 231, 348, 457, and 838 μmol of photons m−2⋅s−1; emission maximum at 440 nm) served as a source for the actinic light. A red measuring light (625 nm) was used.
RNA Extraction and Sequencing.
RNA was extracted from growing (∼0.3 × 104 cells per milliliter per day) Mesostigma cultures (6.6 ± 0.6 × 104 cells per milliliter), 39-d-old Chlorokybus, 13-d-old Klebsormidium, 21-d-old Chara and Zygnema, and 12-wk-old Coleochaete. All RNA was extracted from at least three biological replicates (per species, per treatment) 6 h after the light went on (i.e., cold-treated algae were subjected to 6 h L, 12 h D, and 6 h L at 4 °C; HL-treated algae were subjected to 5 h L and 1 h HL). RNA extraction was performed using TRIzol reagent (Invitrogen, Thermo Fisher Scientific) and an additional DNase I (Thermo Fisher Scientific) treatment, both according to the manufacturer’s instructions. The RNA quantity and quality were assessed using a NanoDrop (Thermo Fisher Scientific) spectrophotometer and a formamide gel, and then shipped to Genome Québec or BGI Tech Solutions on dry ice, where it was assessed again using a Bioanalyzer (Agilent Technologies). No usable RNA was obtained from six replicates of cold-treated Mesostigma. Libraries were prepared in biological triplicates using TruSeq RNA Library Preparation Kits (Illumina), and all replicates (only duplicates for HL-treated Zygnema) were sequenced on an Illumina HiSeq 4000 platform, yielding a total of 492.1 Gbp (1,640,342,307 150-bp paired-end reads) of data (Table S1).
Data Processing.
The raw Illumina data were inspected using FastQC version 0.11.5 (83), trimmed using Trimmomatic version 0.36 (settings: ILLUMINACLIP:TruSeq3-PE-2.fa:2:30:10:2:TRUE HEADCROP:10 TRAILING:3 SLIDINGWINDOW:4:20 MINLEN:36) (84) and assessed again via FastQC. For assembly, all samples (i.e., replicates, treatments) from a species were pooled and de novo-assembled using Trinity version 2.4.0 (85). Using the Trinity pipeline, abundance estimates were calculated using RSEM version 1.2.5 (86) and Bowtie version 1 (87). For expression data, we used the Trinity pipeline to calculate TMM-normalized TPM (88) and significant gene expression changes via edgeR (89) and Benjamini–Hochberg false discovery rate correction. For each gene, a representative isoform was selected based on the highest expression; only genes with an average (over all samples) TPMTMM-normalized ≥ 1 were used for downstream analysis. All transcripts with TPMTMM-normalized ≥ 1 were used as a query for a DIAMOND blastx (90) search against refseq_protein (version: September 2016) and a reciprocal blastx/tblastn search against the protein data of A. thaliana [TAIR10 (91)] and K. nitens v1.1 (33). Contamination was omitted by keeping only those transcripts that retrieved (i) as the best DIAMOND hit (E value cutoff of 10−5) a Viridiplantae (Chloroplastida) sequence or (ii) a reciprocal best blastx/tblastn hit (E value cutoff of 10−5) against K. nitens. All sequencing data (reads and assembly) have been deposited under the National Center for Biotechnology Information Bioproject ID PRJNA399177.
Analysis.
We used a Perl script to extract the best in silico-translated protein based on EMBOSS (European Molecular Biology Open Software Suite) getorf (92) and blastx alignments from their best (Chloroplastida) hits in the refseq sequences or a database consisting of protein data from the genomes of Micromonas pusilla CCMP1545 v3 (93), Chlamydomonas reinhardtii v5.5 (94), K. nitens v1.1 (33), Physcomitrella patens v3.3 (95), Selaginella moellendorffii v1.0 (96), and A. thaliana [TAIR10 (91)]. These in silico proteins were used for all downstream analyses, including orthology assignment-based reciprocal blastp against A. thaliana TAIR10 and K. nitens, HMMER-based eggNOG-mapper runs (97) for orthologous group assignment based on eggNOG 4.5.1 (98), and GhostKOALA runs versus KEGG (99). For localization predictions, we used a Perl script to select those transcripts whose ORFs included the N-terminal region and predicted the subcellular localization of the proteins encoded by these ORFs using TargetP (100), LOCALIZER (101), and PredAlgo (102). We considered a protein to be plastid localized if two of these three programs predicted a plastid (chloroplast) localization. Word clouds of any kind were generated using wordle.net in advanced settings. GO term enrichment was done based on eggNOG-mapper–assigned GO terms and BiNGO (103) with hypergeometric tests, Benjamini–Hochberg false discovery rate correction, and a custom reference annotation for each organism (based on all GO terms assigned to the reference transcripts); transcripts with a |log2(fold change)| ≥ 1 were considered differentially up-regulated or down-regulated.
Supplementary Material
Acknowledgments
We thank Dr. Gregor Christa (University of Aveiro) for help with pulse amplitude modulation fluorometry. J.d.V. acknowledges funding through a German Research Foundation Research Fellowship (VR 132/1-1). J.M.A. acknowledges funding from the Natural Sciences and Engineering Research Council of Canada. J.M.A. is a Senior Fellow of the Canadian Institute for Advanced Research, Program in Integrated Microbial Biodiversity.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The Transcriptome Shotgun Assembly projects have been deposited at DNA Data Bank of Japan/European Molecular Biology Laboratory (European Nucleotide Archive)/GenBank under accession nos. GFXX00000000, GFZF00000000, GFXY00000000, GFZG00000000, GFXZ00000000, and GFYA00000000. Single-read data have been deposited at the National Center for Biotechnology Information (NCBI) Sequence Read Archive under accession nos. SRR5948987–SRR5949030 and SRR5990072–SRR5990080. The NCBI Bioproject ID is PRJNA399177.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719230115/-/DCSupplemental.
References
- 1.Delwiche CF, Cooper ED. The evolutionary origin of a terrestrial flora. Curr Biol. 2015;25:R899–R910. doi: 10.1016/j.cub.2015.08.029. [DOI] [PubMed] [Google Scholar]
- 2.Becker B, Marin B. Streptophyte algae and the origin of embryophytes. Ann Bot. 2009;103:999–1004. doi: 10.1093/aob/mcp044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Vries J, Stanton A, Archibald JM, Gould SB. Streptophyte terrestrialization in light of plastid evolution. Trends Plant Sci. 2016;21:467–476. doi: 10.1016/j.tplants.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 4.de Vries J, Archibald JM, Gould SB. The carboxy terminus of YCF1 contains a motif conserved throughout >500 Myr of streptophyte evolution. Genome Biol Evol. 2017;9:473–479. doi: 10.1093/gbe/evx013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jarvis P, López-Juez E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol. 2013;14:787–802. doi: 10.1038/nrm3702. [DOI] [PubMed] [Google Scholar]
- 6.Liebers M, et al. Regulatory shifts in plastid transcription play a key role in morphological conversions of plastids during plant development. Front Plant Sci. 2017;8:23. doi: 10.3389/fpls.2017.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Börner T, Aleynikova AY, Zubo YO, Kusnetsov VV. Chloroplast RNA polymerases: Role in chloroplast biogenesis. Biochim Biophys Acta. 2015;1847:761–769. doi: 10.1016/j.bbabio.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Pfannschmidt T, et al. Plastid RNA polymerases: Orchestration of enzymes with different evolutionary origins controls chloroplast biogenesis during the plant life cycle. J Exp Bot. 2015;66:6957–6973. doi: 10.1093/jxb/erv415. [DOI] [PubMed] [Google Scholar]
- 9.Klimmek F, Sjödin A, Noutsos C, Leister D, Jansson S. Abundantly and rarely expressed Lhc protein genes exhibit distinct regulation patterns in plants. Plant Physiol. 2006;140:793–804. doi: 10.1104/pp.105.073304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- 11.Martin W, Herrmann RG. Gene transfer from organelles to the nucleus: How much, what happens, and why? Plant Physiol. 1998;118:9–17. doi: 10.1104/pp.118.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ. Learning the language of the chloroplast: Retrograde signaling and beyond. Annu Rev Plant Biol. 2016;67:25–53. doi: 10.1146/annurev-arplant-043015-111854. [DOI] [PubMed] [Google Scholar]
- 13.Susek RE, Ausubel FM, Chory J. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell. 1993;74:787–799. doi: 10.1016/0092-8674(93)90459-4. [DOI] [PubMed] [Google Scholar]
- 14.Lee J, et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Avendaño-Vázquez A-O, et al. An uncharacterized apocarotenoid-derived signal generated in ζ-carotene desaturase mutants regulates leaf development and the expression of chloroplast and nuclear genes in Arabidopsis. Plant Cell. 2014;26:2524–2537. doi: 10.1105/tpc.114.123349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramel F, et al. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA. 2012;109:5535–5540. doi: 10.1073/pnas.1115982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gläßer C, et al. Meta-analysis of retrograde signaling in Arabidopsis thaliana reveals a core module of genes embedded in complex cellular signaling networks. Mol Plant. 2014;7:1167–1190. doi: 10.1093/mp/ssu042. [DOI] [PubMed] [Google Scholar]
- 18.Kleine T, Leister D. Retrograde signaling: Organelles go networking. Biochim Biophys Acta. 2016;1857:1313–1325. doi: 10.1016/j.bbabio.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 19.Scheres B, van der Putten WH. The plant perceptron connects environment to development. Nature. 2017;543:337–345. doi: 10.1038/nature22010. [DOI] [PubMed] [Google Scholar]
- 20.Turner JG, Ellis C, Devoto A. The jasmonate signal pathway. Plant Cell. 2002;14:S153–S164. doi: 10.1105/tpc.000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: Emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 22.Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- 23.Pandey S, Nelson DC, Assmann SM. Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell. 2009;136:136–148. doi: 10.1016/j.cell.2008.12.026. [DOI] [PubMed] [Google Scholar]
- 24.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 25.Nishimura N, et al. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 2010;61:290–299. doi: 10.1111/j.1365-313X.2009.04054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furihata T, et al. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshida T, et al. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 2015;38:35–49. doi: 10.1111/pce.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komatsu K, et al. Functional analyses of the ABI1-related protein phosphatase type 2C reveal evolutionarily conserved regulation of abscisic acid signaling between Arabidopsis and the moss Physcomitrella patens. Plant Mol Biol. 2009;70:327–340. doi: 10.1007/s11103-009-9476-z. [DOI] [PubMed] [Google Scholar]
- 29.Umezawa T, et al. Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 2010;51:1821–1839. doi: 10.1093/pcp/pcq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hauser F, Waadt R, Schroeder JI. Evolution of abscisic acid synthesis and signaling mechanisms. Curr Biol. 2011;21:R346–R355. doi: 10.1016/j.cub.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C, Liu Y, Li S-S, Han G-Z. Insights into the origin and evolution of the plant hormone signaling machinery. Plant Physiol. 2015;167:872–886. doi: 10.1104/pp.114.247403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tietz A, Ruttkowski U, Köhler R, Kasprik W. Further investigations on the occurrence and the effects of abscisic acid in algae. Biochem Physiol Pflanz. 1989;184:259–266. [Google Scholar]
- 33.Hori K, et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun. 2014;5:3978. doi: 10.1038/ncomms4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hackenberg D, Pandey S. Heteromeric G proteins in green algae: An early innovation in the evolution of the plant lineage. Plant Signal Behav. 2014;9:e28457. doi: 10.4161/psb.28457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beilby MJ, Turi CE, Baker TC, Tymm FJM, Murch SJ. Circadian changes in endogenous concentrations of indole-3-acetic acid, melatonin, serotonin, abscisic acid and jasmonic acid in Characeae (Chara australis Brown) Plant Signal Behav. 2015;10:e1082697. doi: 10.1080/15592324.2015.1082697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holzinger A, Becker B. Desiccation tolerance in the streptophyte green alga Klebsormidium: The role of phytohormones. Commun Integr Biol. 2015;8:e1059978. doi: 10.1080/19420889.2015.1059978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delaux P-M, et al. Algal ancestor of land plants was preadapted for symbiosis. Proc Natl Acad Sci USA. 2015;112:13390–13395. doi: 10.1073/pnas.1515426112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Vries J, de Vries S, Slamovits CH, Rose LE, Archibald JM. How embryophytic is the biosynthesis of phenylpropanoids and their derivatives in streptophyte algae? Plant Cell Physiol. 2017;58:934–945. doi: 10.1093/pcp/pcx037. [DOI] [PubMed] [Google Scholar]
- 39.Maruta T, et al. H2O2-triggered retrograde signaling from chloroplasts to nucleus plays specific role in response to stress. J Biol Chem. 2012;287:11717–11729. doi: 10.1074/jbc.M111.292847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fuss J, Liegmann O, Krause K, Rensing SA. Green targeting predictor and ambiguous targeting predictor 2: The pitfalls of plant protein targeting prediction and of transient protein expression in heterologous systems. New Phytol. 2013;200:1022–1033. doi: 10.1111/nph.12433. [DOI] [PubMed] [Google Scholar]
- 41.Sun Q, et al. PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res. 2009;37:D969–D974. doi: 10.1093/nar/gkn654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wickett NJ, et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci USA. 2014;111:E4859–E4868. doi: 10.1073/pnas.1323926111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Vries J, Gould SB. The monoplastidic bottleneck in algae and plant evolution. J Cell Sci. 2017;131:jcs203414. doi: 10.1242/jcs.203414. [DOI] [PubMed] [Google Scholar]
- 44.Mehler AH. Studies on reactions of illuminated chloroplasts. I. Mechanism of the reduction of oxygen and other Hill reagents. Arch Biochem Biophys. 1951;33:65–77. doi: 10.1016/0003-9861(51)90082-3. [DOI] [PubMed] [Google Scholar]
- 45.Takagi D, Takumi S, Hashiguchi M, Sejima T, Miyake C. Superoxide and singlet oxygen produced within the thylakoid membranes both cause photosystem I photoinhibition. Plant Physiol. 2016;171:1626–1634. doi: 10.1104/pp.16.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dietz KJ. Peroxiredoxins in plants and cyanobacteria. Antioxid Redox Signal. 2011;15:1129–1159. doi: 10.1089/ars.2010.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh DK, McNellis TW. Fibrillin protein function: The tip of the iceberg? Trends Plant Sci. 2011;16:432–441. doi: 10.1016/j.tplants.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 48.Hutin C, et al. Early light-induced proteins protect Arabidopsis from photooxidative stress. Proc Natl Acad Sci USA. 2003;100:4921–4926. doi: 10.1073/pnas.0736939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adamska I, Ohad I, Kloppstech K. Synthesis of the early light-inducible protein is controlled by blue light and related to light stress. Proc Natl Acad Sci USA. 1992;89:2610–2613. doi: 10.1073/pnas.89.7.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han JW, Kim GH. An ELIP-like gene in the freshwater green alga, Spirogyra varians (Zygnematales), is regulated by cold stress and CO2 influx. J Appl Phycol. 2013;25:1297–1307. [Google Scholar]
- 51.Klimyuk VI, et al. A chromodomain protein encoded by the arabidopsis CAO gene is a plant-specific component of the chloroplast signal recognition particle pathway that is involved in LHCP targeting. Plant Cell. 1999;11:87–99. doi: 10.1105/tpc.11.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Legris M, et al. Phytochrome B integrates light and temperature signals in Arabidopsis. Science. 2016;354:897–900. doi: 10.1126/science.aaf5656. [DOI] [PubMed] [Google Scholar]
- 53.Jiang Z, Xu G, Jing Y, Tang W, Lin R. Phytochrome B and REVEILLE1/2-mediated signalling controls seed dormancy and germination in Arabidopsis. Nat Commun. 2016;7:12377. doi: 10.1038/ncomms12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kohchi T, et al. The Arabidopsis HY2 gene encodes phytochromobilin synthase, a ferredoxin-dependent biliverdin reductase. Plant Cell. 2001;13:425–436. doi: 10.1105/tpc.13.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dure L, 3rd, Greenway SC, Galau GA. Developmental biochemistry of cottonseed embryogenesis and germination: Changing messenger ribonucleic acid populations as shown by in vitro and in vivo protein synthesis. Biochemistry. 1981;20:4162–4168. doi: 10.1021/bi00517a033. [DOI] [PubMed] [Google Scholar]
- 56.Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- 57.Hara M, Terashima S, Fukaya T, Kuboi T. Enhancement of cold tolerance and inhibition of lipid peroxidation by citrus dehydrin in transgenic tobacco. Planta. 2003;217:290–298. doi: 10.1007/s00425-003-0986-7. [DOI] [PubMed] [Google Scholar]
- 58.Hundertmark M, Hincha DK. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics. 2008;9:118. doi: 10.1186/1471-2164-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tenhaken R. Cell wall remodeling under abiotic stress. Front Plant Sci. 2015;5:771. doi: 10.3389/fpls.2014.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kleine T, Kindgren P, Benedict C, Hendrickson L, Strand A. Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol. 2007;144:1391–1406. doi: 10.1104/pp.107.098293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lind C, et al. Stomatal guard cells co-opted an ancient ABA-dependent desiccation survival system to regulate stomatal closure. Curr Biol. 2015;25:928–935. doi: 10.1016/j.cub.2015.01.067. [DOI] [PubMed] [Google Scholar]
- 62.Vahisalu T, et al. SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature. 2008;452:487–491. doi: 10.1038/nature06608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qiu J, Henderson SW, Tester M, Roy SJ, Gilliham M. SLAH1, a homologue of the slow type anion channel SLAC1, modulates shoot Cl- accumulation and salt tolerance in Arabidopsis thaliana. J Exp Bot. 2016;67:4495–4505. doi: 10.1093/jxb/erw237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pornsiriwong W, et al. A chloroplast retrograde signal, 3′-phosphoadenosine 5′-phosphate, acts as a secondary messenger in abscisic acid signaling in stomatal closure and germination. eLife. 2017;6:e23361. doi: 10.7554/eLife.23361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McAdam SA, et al. Abscisic acid controlled sex before transpiration in vascular plants. Proc Natl Acad Sci USA. 2016;113:12862–12867. doi: 10.1073/pnas.1606614113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hõrak H, Kollist H, Merilo E. Fern stomatal responses to ABA and CO2 depend on species and growth conditions. Plant Physiol. 2017;174:672–679. doi: 10.1104/pp.17.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martín G, et al. Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat Commun. 2016;7:11431. doi: 10.1038/ncomms11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fernández AP, Strand A. Retrograde signaling and plant stress: Plastid signals initiate cellular stress responses. Curr Opin Plant Biol. 2008;11:509–513. doi: 10.1016/j.pbi.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 69.Chan KX, Crisp PA, Estavillo GM, Pogson BJ. Chloroplast-to-nucleus communication: Current knowledge, experimental strategies and relationship to drought stress signaling. Plant Signal Behav. 2010;5:1575–1582. doi: 10.4161/psb.5.12.13758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Estavillo GM, et al. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell. 2011;23:3992–4012. doi: 10.1105/tpc.111.091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rippin M, Becker B, Holzinger A. Enhanced desiccation tolerance in mature cultures of the streptophytic green alga Zygnema circumcarinatum revealed by transcriptomics. Plant Cell Physiol. 2017;58:2067–2084. doi: 10.1093/pcp/pcx136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cuming AC, Cho SH, Kamisugi Y, Graham H, Quatrano RS. Microarray analysis of transcriptional responses to abscisic acid and osmotic, salt, and drought stress in the moss, Physcomitrella patens. New Phytol. 2007;176:275–287. doi: 10.1111/j.1469-8137.2007.02187.x. [DOI] [PubMed] [Google Scholar]
- 73.Choudhury FK, Rivero RM, Blumwald E, Mittler R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017;90:856–867. doi: 10.1111/tpj.13299. [DOI] [PubMed] [Google Scholar]
- 74.Dietz K-J, Turkan I, Krieger-Liszkay A. Redox- and reactive oxygen species-dependent signaling into and out of the photosynthesizing chloroplast. Plant Physiol. 2016;171:1541–1550. doi: 10.1104/pp.16.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ju C, et al. Conservation of ethylene as a plant hormone over 450 million years of evolution. Nat Plants. 2015;1:14004. doi: 10.1038/nplants.2014.4. [DOI] [PubMed] [Google Scholar]
- 76.Yin P, et al. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol. 2009;16:1230–1236. doi: 10.1038/nsmb.1730. [DOI] [PubMed] [Google Scholar]
- 77.Lee I, Ambaru B, Thakkar P, Marcotte EM, Rhee SY. Rational association of genes with traits using a genome-scale gene network for Arabidopsis thaliana. Nat Biotechnol. 2010;28:149–156. doi: 10.1038/nbt.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pernas M, García-Casado G, Rojo E, Solano R, Sánchez-Serrano JJ. A protein phosphatase 2A catalytic subunit is a negative regulator of abscisic acid signalling. Plant J. 2007;51:763–778. doi: 10.1111/j.1365-313X.2007.03179.x. [DOI] [PubMed] [Google Scholar]
- 79.Tran L-S, et al. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:20623–20628. doi: 10.1073/pnas.0706547105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van de Poel B, Cooper ED, Van Der Straeten D, Chang C, Delwiche CF. Transcriptome profiling of the green alga Spirogyra pratensis (Charophyta) suggests an ancestral role for ethylene in cell wall metabolism, photosynthesis, and abiotic stress responses. Plant Physiol. 2016;172:533–545. doi: 10.1104/pp.16.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol Biol. 2005;58:585–596. doi: 10.1007/s11103-005-7294-5. [DOI] [PubMed] [Google Scholar]
- 82.de Vries J, Archibald JM. Plant evolution: Landmarks on the path to terrestrial life. New Phytol. 2018;217:1428–1434. doi: 10.1111/nph.14975. [DOI] [PubMed] [Google Scholar]
- 83.FASTQC 2016 A quality control tool for high throughput sequence data. Available at www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed February 18, 2017.
- 84.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 91.Lamesch P, et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 93.Worden AZ, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- 94.Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lang D, et al. The Physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution. Plant J. 2008;93:515–533. doi: 10.1111/tpj.13801. [DOI] [PubMed] [Google Scholar]
- 96.Banks JA, et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 2011;332:960–963. doi: 10.1126/science.1203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huerta-Cepas J, et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol. 2017;34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huerta-Cepas J, et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44:D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 100.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 101.Sperschneider J, et al. LOCALIZER: Subcellular localization prediction of both plant and effector proteins in the plant cell. Sci Rep. 2017;7:44598. doi: 10.1038/srep44598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tardif M, et al. PredAlgo: A new subcellular localization prediction tool dedicated to green algae. Mol Biol Evol. 2012;29:3625–3639. doi: 10.1093/molbev/mss178. [DOI] [PubMed] [Google Scholar]
- 103.Maere S, Heymans K, Kuiper M. BiNGO: A cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 104.Page MT, et al. Seedlings lacking the PTM protein do not show a genomes uncoupled (gun) mutant phenotype. Plant Physiol. 2017;174:21–26. doi: 10.1104/pp.16.01930. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.