Significance
This work resolves a long-standing controversy regarding the activation mechanism and photocycle of cryptochromes (CRYs), a broadly represented group of photosensors. We demonstrate a high degree of correspondence between the capability of the tryptophan (Trp) chain to photoreduce the flavin cofactor and biological activity. These results establish that the evolutionary pressure to conserve the Trp chain derives from maintaining a flavin photocycle that, in this case, entrains the fly circadian clock. Furthermore, we reengineer CRY to be more biologically photoresponsive by manipulating photoreduction by the Trp chain. As CRYs are important signaling molecules in animals, plants, and bacteria, and are increasingly used as optogenetic tools, the defined mechanism is relevant to many signaling processes and will enable novel applications.
Keywords: light sensor, redox potential, tryptophan, electron transfer, signal transduction
Abstract
Cryptochromes (CRYs) entrain the circadian clocks of plants and animals to light. Irradiation of the Drosophila cryptochrome (dCRY) causes reduction of an oxidized flavin cofactor by a chain of conserved tryptophan (Trp) residues. However, it is unclear how redox chemistry within the Trp chain couples to dCRY-mediated signaling. Here, we show that substitutions of four key Trp residues to redox-active tyrosine and redox-inactive phenylalanine tune the light sensitivity of dCRY photoreduction, conformational activation, cellular stability, and targeted degradation of the clock protein timeless (TIM). An essential surface Trp gates electron flow into the flavin cofactor, but can be relocated for enhanced photoactivation. Differential effects of Trp-mediated flavin photoreduction on cellular turnover of TIM and dCRY indicate that these activities are separated in time and space. Overall, the dCRY Trp chain has evolutionary importance for light sensing, and its manipulation has implications for optogenetic applications of CRYs.
Cryptochromes (CRYs) are flavin-containing blue-light sensors that play key roles in the circadian clocks of animals and plants (1–3). In CRYs, as well as the homologous photolyase (PL) DNA repair enzymes, the photoinduced electron transfer (ET) process along a chain of conserved tryptophan (Trp) residues reduces the excited state of the FAD cofactor (1, 4, 5). The type 1 insect CRYs are well-studied in this respect, but the importance of Trp-mediated ET in light sensing is a complex and open question (3, 6). Drosophila CRY (dCRY) functions as a primary light-sensing protein for the entrainment of the fly circadian clock (2). The C-terminal extension of dCRY comprises a short helical C-terminal tail (CTT) that binds alongside the FAD cofactor analogous to how substrate DNA lesions bind to PLs (7–9) (Fig. 1). Light-induced release of the CTT gates engagement of cellular targets important for entraining the circadian clock, such as timeless (TIM), a key component of the circadian gene repressor complex (10–12), and jetlag (JET), an E3 ubiquitin ligase responsible for TIM ubiquitination (12, 13). Photoexcitation of dCRY also stimulates self-degradation through ubiquitin-mediated proteolysis (14). In addition to its influence on the core clock components, dCRY affects neuronal firing by signaling to redox-sensitive ion channels (15, 16).
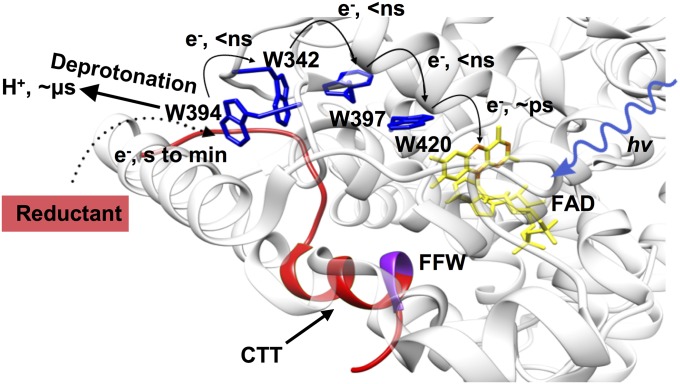
Fig. 1.
Electron conduit for photoreduction of dCRY. The Trp-tetrad (W420, W397F, W342, and W394; blue) reductively quenches the FAD-excited state (yellow flavin) of dCRY (Protein Data Bank ID code 4GU5). The displaceable CTT (red) associates closely with the protein core via the Phe-Phe-Trp (FFW) motif (purple). Photoexcited FAD oxidizes the most proximal Trp residue (in picoseconds), and the electron hole then propagates to a surface residue that can be reduced by external reductants. e−, electron; hv, photon.
Many factors may contribute to dCRY photosensing via the Trp chain, including radical stability, accessibility of oxidized sites, temperature, and redox environment. Light photoreduces the dCRY FAD cofactor to the anionic semiquinone (ASQ) state (17–20). The ASQ state promotes active-site histidine protonation that destabilizes the CTT against the FAD pocket, thereby linking flavin photochemistry to protein conformational changes known to be important for downstream signaling (21). In the CRY/PL family, FAD photoreduction is facilitated by three invariant Trp residues (the so-called Trp “triad”) that reductively quench the FAD-excited singlet state through a multistep ET process (5, 22–24). Recently, a fourth surface Trp residue conserved among PLs and CRYs has also been shown to participate in flavin photoreduction (4, 25), thereby expanding the functional redox chain to a Trp “tetrad” (Fig. 1). In dCRY, the photoexcited oxidized flavin cofactor (FADox) is reduced by the most proximal Trp, W420, in picoseconds (5). In nanoseconds, the W420 cation radical (W420•+) then gains an electron from Trp residues along the chain (W397, W342). This migration of the electron hole to the protein surface stabilizes charge separation, and hence FAD photoreduction. However, rapid recombination reactions within the internal chain limit radical propagation. Net FAD•− formation then depends on rates of reductive quenching at the protein surface.
The role of flavin reduction in CRY signal transduction is debatable. Some CRY variants that have Phe substitutions in the Trp tetrad are reportedly unable to photoreduce but still remain biologically active in several assays (19, 25, 26). However, FAD photoreduction correlates well with conformational activation of CRY (17, 20, 21, 27), and the Trp triad greatly facilitates this process. Adding to the conundrum, the dCRY flavin has been shown to have an unusually high redox potential, which would imply that the ASQ is the most stable form in the dark (28). Nonetheless, a reduced ground state is not consistent with the action spectrum for phase shifting of the fly clock, which instead matches well to that of FADox (10, 29).
Herein, we explore the dCRY photoreduction process by substituting residues of the electron-donating Trp tetrad to both redox-active and -inactive residues and then characterizing their properties and biological activities. We correlate the photoreduction properties of the dCRY variants with their capacity to catalyze both TIM and dCRY proteolysis in insect cells. Differences in these two activities among the variants unambiguously define the role of flavin photoreduction in dCRY function and have important implications for how protein turnover is timed in the circadian oscillator.
Results
Light Intensity and Reducing Environment Affect the Extent of dCRY Flavin Photoreduction.
Upon illumination with blue light (440 nm), the dCRY FADox readily reduces to the ASQ (FAD•−; Fig. 2A). This reaction depends upon ET from the Trp tetrad to the photoexcited FAD singlet state (FAD*) (5). To assess the role of the tetrad residues (W420, W397, W342, and W394) in photoreduction, we substituted each to both Phe and Tyr. Tyr residues have a standard redox potential at pH 7 (Eo′) in the range of Trp (∼1.0–1.4 V) (30, 31), whereas the Phe redox potential is substantially higher (>2.2 V) and may exceed that of the peptide backbone. Although the FADox/FAD•− couple can range greatly depending on the protein (6, 32), FAD* is high enough in energy (∼2.5 V) (33) to readily oxidize Trp and Tyr, but not Phe.
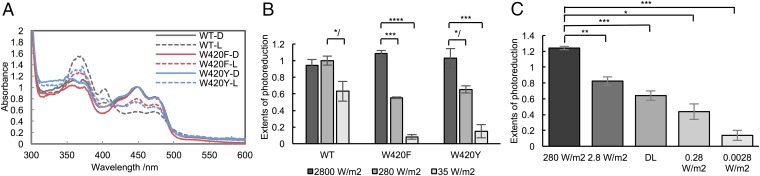
Fig. 2.
Light intensity affects the extent of flavin photoreduction. (A) Photoreduction of WT, W420F, and W420Y, followed by UV–visible absorption spectroscopy. WT and W420 variants photoreduce to different extents at 280 W/m2 after 15 min with 2 mM Tris(2-carboxyethyl)phosphine (TCEP) as a reductant. D, dark sample, solid line; L, light-exposed sample, dashed line. (B) Extent of photoreduction is affected by light intensity (440 nm; 2,800, 280, or 35 W/m2). Values are shown relative to WT at moderate light intensity (280 W/m2), which is normalized to 1.0. WT reduction extents appear smaller than those of variants, because WT is slightly reduced by solvent reductants in the dark state. (C) Extent of photoreduction under different light intensities for WT dCRY with 2 mM GSH. A deuterium lamp (DL) indicates the reduction by the light source used for monitoring UV–visible absorbance. Error bars reflect the SEM for a sample size of at least three. */P < 0.1, *P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001; two-tailed, unpaired t test.
To distinguish the reactivity of the Trp-tetrad variants, many of which were quite unreactive to photoreduction, responses were measured at light intensities that differed by five orders of magnitude (2.8 kW/m2 to 0.0028 W/m2). Bright sunlight has an irradiance of ∼250 W/m2 in the visible-light absorption of FADox (400–520 nm), whereas dawn and dusk conditions are in the range of 2–5 W/m2. Biochemical and physiological responses of dCRY require only ∼10 W/m2; however, the rhythmic responses of flies can be entrained by values as low as 0.2 W/m2 (34). Considering the flavin-proximal W420 residue first, its Phe and Tyr variants reduce similar to WT under high light intensity (2.8 kW/m2); however, at lower light intensities, the rates and extents of reduction decrease dramatically (Fig. 2B and Fig. S1). WT dCRY produces a significant amount of the ASQ at light intensities as low as 0.0028 W/m2 (Fig. 2C), whereas the W420Y and W420F reduce similar to WT at 2.8 kW/m2, but are equally insensitive to lower light intensity (35 W/m2) (Fig. 2B). This behavior is consistent with CRY Phe substitution variants being reported as inactive at exposures of ∼10–50 W/m2 (9, 19, 35). The extent of photoreduction reflects a photochemical equilibrium between forward photoreduction and reoxidation of the flavin, primarily by oxygen (21) (SI Notes). Reoxidation rates are small and similar for the W420 variants; under anaerobic conditions, even relatively unreactive variants fully reduce at moderate light intensity (Fig. S1). Furthermore, solution reductants influence the photoreduction rate constants (Fig. S1), thereby indicating that reductive quenching of an accessible site is the rate-limiting step in sustained photoreduction. For example, glutathione (GSH), the primary cellular reduced thiol, effectively stabilizes the dCRY ASQ (Fig. S1). Unreactive Phe variants of the Trp chain may still accumulate the ASQ in the reducing environment of the cell, allowing for integration of the light response over many minutes (34).
Residue Substitutions Within Trp Tetrad Differentially Affect Photoreduction.
The photoreduction properties of several Trp-tetrad substitutions in dCRY [W420F, W397F, and W342F (4, 9, 19, 35)] have considerably lessened photosensitivity relative to WT (Fig. 3 A and B). W420F is the most defective, whereas W397F shows some activity at moderate light intensity. Nevertheless, all Phe replacements within the canonical Trp triad (W420, W397, and W342) still retain partial light sensitivity. In contrast, Phe substitution of the surface Trp, W394F, nearly abolishes sustained flavin photoreduction. In general, Tyr substitutions are more active at forming the ASQ than Phe substitutions, with the difference in efficacy more pronounced as the distance from the flavin increases (Fig. 3 A and B). W394Y is quite active for photoreduction, followed by W397Y and W342Y, which are both more active than highly inactive W420Y. Despite their lower rate constants for photoreduction than WT, W397Y and W342Y reduce to nearly the same extent as WT after 10 min of light exposure at 280 W/m2.
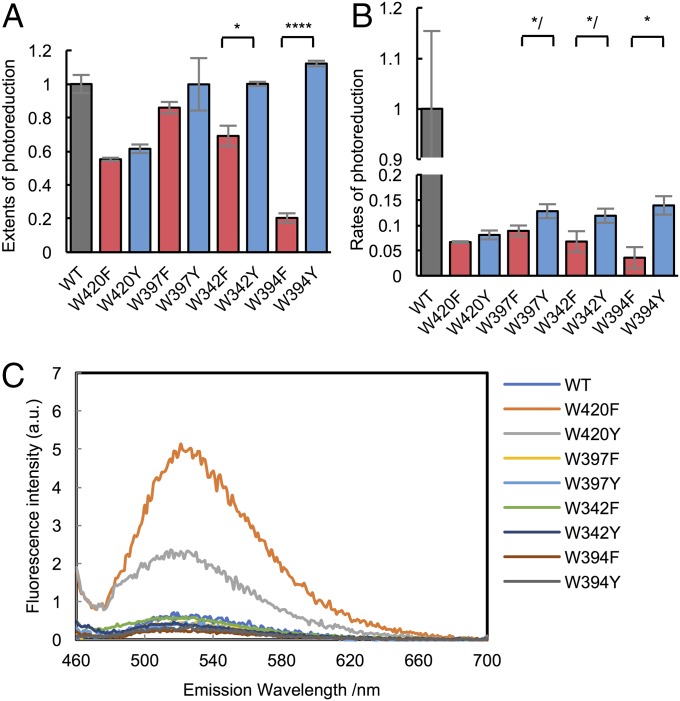
Fig. 3.
Photoreduction of the Trp-tetrad Phe and Tyr variants. (A and B) Extents and rates of photoreduction upon Trp substitution vary considerably for different positions (WT, gray; Phe, red; Tyr, blue) at moderate light intensity (440 nm, 280 W/m2, 15 min). Normalization as in Fig. 2B. Error bars reflect the SEM for n ≥ 3. P values are shown for differences between F and Y variants. */P < 0.1, *P < 0.05, ****P < 0.001; two-tailed, unpaired t test. (C) Fluorescence emission spectra of each variant, excitation (λex = 450 nm). Only Trp420 substitutions affect fluorescence emission.
Trp420 is the closest tetrad residue to the flavin, and thus dominates picosecond ET quenching of the FAD-excited state (5). Residue substitutions that cause slower single-step ET reactions to FAD* should increase the excited-state lifetime, the fluorescence quantum yield, and the intensity of the emission spectrum. Only two Trp-tetrad variants affect the FAD fluorescence emission intensity: W420F and W420Y (Fig. 3C). The emission intensity increases substantially for W420F, indicating that the Trp residue indeed rapidly reduces FAD*. In contrast, W420Y has a fluorescence intensity intermediate to that of WT and W420F, which is consistent with slower ET from Tyr than Trp to FAD*. All other variants have highly quenched FAD fluorescence because initial ET from neighboring W420 is maintained. Interestingly, W420Y has a low extent of net flavin photoreduction, similar to that of W420F, despite showing enhanced flavin fluorescence quenching. A slowed ET rate to FAD* from Y420 may indicate that Tyr is more difficult to oxidize than Trp, and hence more efficient recombination of Y420•+ with the reduced flavin. The other Tyr substitutions indicate that this curtailment of net reduction mitigates as the site of recombination resides further from the flavin (Fig. 3C).
FAD Photoreduction Correlates with dCRY Conformational Changes in Trp-Tetrad Variants.
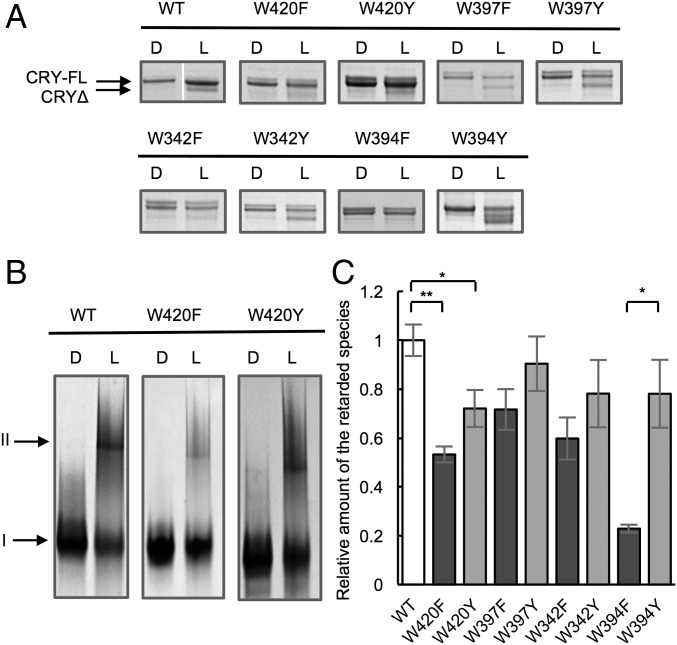
Photoreduction of dCRY causes displacement of the CTT from the protein core and thereby increases its sensitivity to proteolytic cleavage (18, 20). As such, band shifts on SDS/PAGE after limited trypsinolysis reflect light activation (Fig. 4A; note the shift from 62 to 50 kDa). In addition, migration of dCRY on a native PAGE gel is sensitive to photoactivation, with the light-excited state showing an expanded and/or aggregated conformation that is retarded during electrophoresis (Fig. 4B). Formation of this slower migrating species is largely irreversible and includes only a small fraction of the total protein; however, the amount formed correlates with the degree of photoreduction (Fig. 4C). Both the native gel and proteolytic sensitivity assay indicate that the light-induced conformational changes in Trp-tetrad variants correlate well with their photosensitivity for forming the ASQ. Those variants that are largely insensitive to light (W420F and W394F) show little conformational activation, whereas the W342Y, W397Y, and W394Y variants again behave similar to WT (Fig. 3). In line with W397F,Y producing similar amounts of ASQ as WT, W397F,Y also undergoes commensurate degrees of conformational change.
Fig. 4.
dCRY conformational changes correlate with flavin photoreduction in each Trp-tetrad variant. (A) Release of CTT, monitored by trypsin digestion, SDS/PAGE, and Coomassie staining. There is a small shift in band position from full-length CRY (CRY-FL) to CRY with CTT cleaved by trypsin (CRYΔ) (D, dark sample; L, light-exposed sample; 440 nm, 280 W/m2, 15 min). For WT, D and L samples were run on the same gel, but in nonadjacent lanes. (B) Native PAGE of Coomassie-stained proteins shows the appearance of a retarded species that correlates with the degree of photoreduction (440 nm, 280 W/m2, 15 min). Band I represents the native conformation. Band II represents the expanded and/or aggregated conformation. (C) Quantification of the relative amount of the retarded species. Values are relative to that of WT. Error bars reflect the SEM for n = 3. *P < 0.05, **P < 0.01; two-tailed, unpaired t test.
Relocation of the Surface Trp Tunes dCRY Light Sensitivity.
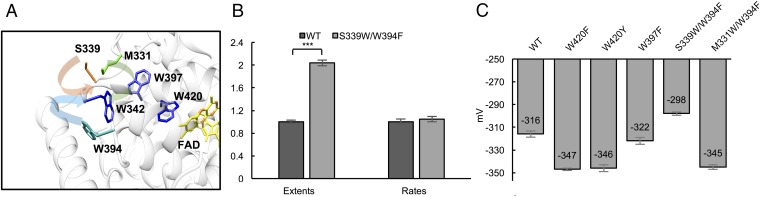
Flavin photoreduction produces a relatively stable radial at W394 on the protein surface (4); reductive quenching of this radical is required to stabilize the ASQ. We reasoned that altering the positioning and solvent exposure of the fourth Trp may then affect the extent of photoreduction. Of residues within ∼7.0 Å from Trp342, M331 and S339 are solvent-exposed (34.4 Å2 and 85.7 Å2 of surface area, respectively) and appear to be unimportant for stabilizing local conformation (Fig. 5A). Both the M331W/W394F and S339W/W394F dCRY variants express well, and the S339W/W394F variant purifies in a partially reduced state under ambient room light (Fig. S2). In contrast, the WT protein (and other variants) purify mainly with FADox. Upon reoxidation in the dark, S339W/W394F again partially reduces under ambient light. When tested for photoreduction at moderate light intensity, Trp substitutions at both positions 331 and 339 recover light sensitivity of the W394F variant, with the S339W having an activity comparable to that of WT (Fig. S2). The CTT of the S339W/W394F variant also partially releases from the protein core with room light (Fig. S2). Thus, the precise location of the terminal Trp can be altered without disrupting efficient and sustained photoreduction. Under low light intensity (between 0.28 and 2.8 W/m2), which reduces photoreduction to a level where WT and S339W/W394F can be compared, the extent of photoreduction for S339W/W394F exceeds that of WT (Fig. 5B). Thus, relocation of the surface Trp residue improves dCRY light sensitivity.
Fig. 5.
Relocation of surface Trp tunes the light sensitivity of dCRY and alters the flavin redox potential. (A) Position of dCRY surface Trp394 relative to other surface sites with similar proximity to Trp342 (Protein Data Bank ID code 4GU5). Compared with Trp394 (solvent accessibility = 85.7 Å2, blue) Ser-339 (solvent accessibility = 85.7 Å2, orange) and Met-331 (solvent accessibility = 34.4 Å2, green) are more surface-exposed. (B) S339W/W394F improves light sensitivity compared with WT at low light intensity [∼ <1 W/m2, with Tris(2-carboxyethyl)phosphine (TCEP) as the reductant]. Normalization as in Fig. 2B. ***P < 0.001. (C) dCRY variants have altered FADox/ASQ redox potentials (E0′) relative to WT. Notably, S339W/W394F has a higher redox potential than WT. Error bars reflect the SEM for n = 3.
A Low Flavin Redox Potential Ensures That the dCRY Reduction Is Accessed by Light.
For signaling to depend upon photoreduction, dCRY FADox must be stable in cells. Typically, reduced/oxidized GSH ratios set eukaryotic cellular redox potentials in the range of −260 to −300 mV (36). The tendency of S339W/W394F to purify as partially reduced and its enhanced photoreduction rates suggested an altered flavin redox potential for this variant.
To measure the dCRY FADox/ASQ redox couple, we employed a spectroscopic method of equilibrating dCRY with a reference dye of similar redox potential that also gives an absorbance change distinguishable from that of the flavin (details are provided in SI Materials and Methods). When dCRY was reduced with the dye safranin T [Eo′ = −289 V vs. the standard hydrogen electrode (SHE)] (Fig. S3), the plot of log ([oxidized]/[reduced]) dye vs. log ([oxidized]/[reduced]) dCRY produced a slope of 0.87 (Fig. S3), which is close to the expected value of 1 for a one-electron reaction. The intercept indicates a redox potential for dCRY of −316 mV vs. SHE (Fig. 5C and Fig. S3). Testing of several Trp-tetrad variants revealed that their flavin redox potentials were altered relative to WT. For example, W420F and W420Y have potentials ∼30 mV lower than WT (Fig. 5C), perhaps owing to their close proximity to the flavin. In contrast, substitution at the next most remote Trp, W397F, has a potential similar to that of WT. Surprisingly, the surface Trp substitution S339W increases the flavin redox potential by ∼20 mV relative to WT, which may partly explain why FAD in W394F/S339W is partially reduced upon purification and in weak room light (Fig. 5C). In contrast, the M331W/W394F variant is ∼30 mV lower than WT, which is consistent with its diminished photoreduction properties. Notably, changes to surface residues 20 Å from the flavin cofactor impact its redox potential.
CRY Activity in Insect Cell Culture.
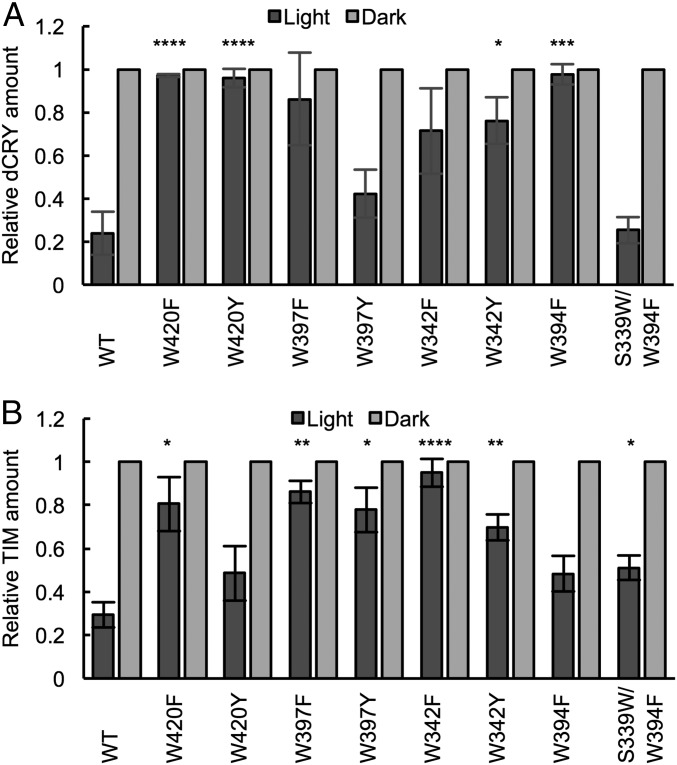
We evaluated the ability of the CRY variants to undergo light-induced proteolysis and catalyze light-induced TIM proteolysis in insect cell culture. Embryonic Drosophila S2 cells were transiently transfected with constructs expressing epitope-tagged CRY, JET, and TIM at 1:1:2 ratios. Cells were either left under red light (“dark”) or exposed to 600 lx (∼6 W/m2) of light for 1 h, at which time dCRY and TIM levels were evaluated in cell lysates by Western blots. For WT dCRY, both dCRY and TIM are stable in the dark and degraded to ∼20% of their dark level after light exposure (Fig. 6A and Fig. S4). In the case of the dCRY variants, the Phe and Tyr residue substitutions give increased TIM and dCRY light stabilities that track remarkably well with their photoreduction sensitivities (Fig. 3). For light-induced dCRY degradation, W420F and W420Y show nearly no activity and W342F, W397Y, and W342F are also highly impaired. In contrast, W397Y degrades in light consistent with its ability to achieve levels of sustained reduction and conformational change comparable to WT. Importantly, the surface substitution W394F shows no light-dependent degradation, yet nearly WT activity is recovered in S339W/W394F. Thus, a redox-active surface residue is critical to stabilize charge separation in the Trp chain, but its location only need be in proximity to W342. For TIM degradation, the W420F, W342F, and W397F variants all show severely reduced TIM degradation, whereas the Tyr variants of the same residues are more active, consistent with the greater redox capability of Tyr (Fig. 6B). Notably, W397F and particularly W397Y show substantial activity toward TIM, despite their low expression levels (Fig. S4). An interesting outlier is the W394F variant, which while displaying nearly no sustained photoreduction, little conformational activation, and no CRY turnover in light, does induce substantial photoinduced TIM proteolysis. However, it should be noted that the W394F variant is very stable and has a high level of expression in both light and dark (Fig. S4). Thus, even a small amount of photoreduced protein, which can be produced with GSH under anaerobic conditions (Fig. S5), may contribute to the observed TIM degradation.
Fig. 6.
Effect of Trp-tetrad substitutions on dCRY stability and dCRY-mediated TIM degradation in S2 insect cell culture when proteins are expressed with JET under light or dark conditions. (A and B) Quantification of dCRY and TIM levels in light, relative to dark. Black column, light-exposed sample; gray column, dark sample. All light values are shown relative to dark (normalized to 1). Error bars reflect the SEM for n = 4. *P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001; two-tailed, unpaired t test of differences in light-state levels of the variant compared with WT.
Discussion
Here, we show that (i) dCRY has a low enough FADox/ASQ redox couple to limit formation of the ASQ in the dark, (ii) dCRY photoreduction is sensitive enough to be biologically relevant, (iii) substitutions in the Trp tetrad alter photoreduction to varying extents, (iv) dCRY and TIM turnover in insect cells correlates well with the photoreduction properties of a wide range of dCRY variants, and (v) dCRY degradation requires more sustained FAD photoreduction than TIM degradation.
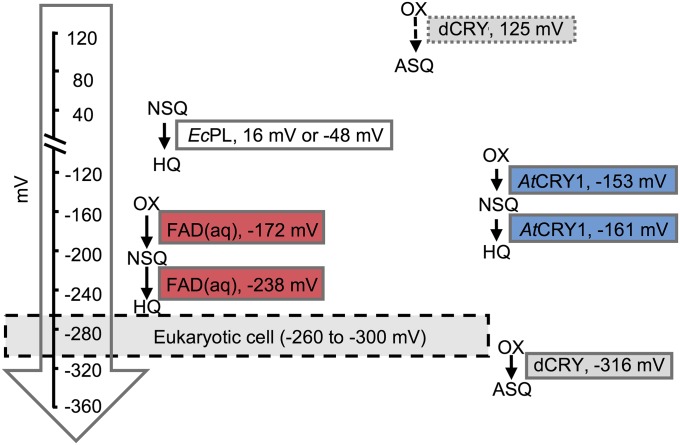
The −316-mV redox potential of the FADox/ASQ couple indicates that the protein will be primarily oxidized within the cell and susceptible to photoactivation. The FADox/FADH• couple of Arabidopsis thaliana CRY [−153 mV (32)] is higher than the dCRY FADox/FAD•− couple, as would be expected for formation of a neutral rather than anionic product. A much higher potential of 125 mV has been previously measured for dCRY by a different method (28) (Fig. 7). In our hands, dCRY can only be reduced to the ASQ by low-potential reductants such as dithionite and Cr(II)-EDTA, which is inconsistent with a positive potential. It is also telling that the WT protein purifies from both insect cells and Escherichia coli in the oxidized state, but the S339W/W394F variant purifies partially reduced and has a potential upshifted into the range of an E. coli cell [−280 mV (37)]. Importantly, dCRY can be photoreduced in ambient oxygen to an appreciable extent with very low levels of light, albeit after many minutes (Fig. 2). The extent of photoreduction will increase in low-potential environments and with the availability of suitable cellular reductants, as has been demonstrated for plant CRY2 (38). Thus, slow but kinetically stable photoreduction may explain how dCRY can integrate very weak light signals over long times (e.g., >1/2 h) (34).
Fig. 7.
Redox potentials of flavins in CRYs and PLs. Comparison of the flavin redox potentials (vs. SHE) in PL, CRY, and free flavin in aqueous (aq) solution (6, 32, 44). The dashed arrow indicates a previously measured redox potential of dCRY WT (28). AtCRY1, Arabidopsis thalania CRY1; EcPL, E. coli PL; HQ, hydroquinone; NSQ, neutral semiquinone; OX, oxidized flavin.
It has been reported that dCRY proteins with substitutions in the Trp triad are insensitive to sustained photoreduction but retain light-induced TIM degradation and CRY degradation activity (19, 35). In contrast, we find substantial defects in both activities for Trp-tetrad variants that are photoreduction-impaired, whereas photoreduction-enabled variants retain activity (Fig. 6). Overall, dCRY photosensitivity correlates well with dCRY light stability in insect cell culture. For example, the W394F variant, which is completely inactive for sustained photoreduction and dCRY degradation, reactivates upon replacement of a Trp or Tyr residue at another surface location. Interestingly, light-induced, ubiquitin-mediated degradation of plant CRY2 was also curtailed in Trp-triad variants (26), but this was attributed to structural defects, not loss of photoreduction activity. The rescue of dCRY W394F light-induced degradation by the photoactive S394W/W394F variant rules against a structural explanation for loss of activity. Light-induced TIM degradation also depends on photoreduction. In general, W-to-F substitutions curtail TIM-directed activity, with the exception of W394F. Interestingly, W394F still destabilizes TIM in light but has high light stability and low photoreduction. However, for complete dCRY proteolysis, each molecule must undergo photoactivation, whereas only a few active dCRY molecules may turn over multiple TIM proteins. Furthermore, TIM degradation rates also reflect the amount of dCRY, and light-stable molecules will produce higher steady-state levels. In the case of W394F, even a small active dCRY fraction may be enough to degrade TIM. Bolstering the view that the requirements for TIM- and self-degradation differ, a similar separation in these functions was also observed in dCRY variants of a conserved His residue that participates in CTT displacement (21).
The ability to undergo sustained versus transient photoreduction may distinguish the activities of the variants. Only changes at the 420 position affect initial reduction of the flavin-excited state. However, the resulting “hole” must migrate to and be quenched at the protein surface for sustained photoreduction. Variants of 394 that prevent stable photoreduction may still allow ASQ formation and recombination on a fast time scale, and thus may form some ASQ under constant illumination. Transient EPR and optical spectroscopy support this assertion (4): Phe substitutions of the first three flavin proximal Trp residues in CRY proteins produce no transient radicals upon blue-light laser excitation, whereas the W394F variant of dCRY produces a short-lived radical. If TIM degradation is rapid compared with dCRY degradation, the W394F variant may be capable of activating enough JET to degrade TIM. Indeed, the W420Y, which also does not undergo sustained photoreduction and does not destabilize dCRY in the light, does catalyze substantial TIM degradation and likely forms a transient radical, as reflected in its reduced FAD* fluorescence compared with W420F (Figs. 3 and 6). Furthermore, W420Y undergoes more irreversible conformational activation than W420F (Fig. 4B), which may belie a short-lived reduced state.
Given the apparent importance of dCRY photoreduction, why then have others found in both flies and plants that substitutions of the Trp-tetrad residues produce proteins whose photoreduction properties do not correlate well with their biological activities (19, 26, 35, 39)? One explanation is that stable flavin photoreduction does not require all of the Trp-triad residues. Although the photoreduction rates drop considerably with single substitutions, many of the variants still accumulate a substantial amount of the ASQ, especially in a reducing environment. Indeed, several cellular components have been shown to facilitate reduction of FAD* (40), and illumination of S2 cells expressing dCRY results in flavin radicals (27, 41). Thus, higher light levels, longer exposures, and efficient reductants may compensate for Trp-triad defects (3). Second, JET levels also affect the ability of dCRY to degrade TIM in light (12). In transfection experiments, large amounts of JET may compliment low levels of photoreduced CRY to degrade TIM. Furthermore, increasing JET levels in pacemaker neurons augments TIM degradation in response to a light pulse, similar to dCRY overexpression (42). Light conditions also differ among studies. One study showing that the dCRY W397F substitution did not affect TIM and CRY degradation used 366-nm UVA instead of blue light (19), which photoreduces FADox less efficiently but likely promotes greater radical generation and redox imbalance in cells. With blue light, W397F accumulates a substantial amount of the ASQ. In plants, CRY1 and CRY2 have also been tested for photosensitivity after substitution of Trp triad residues to Ala and Phe (26, 39). As has been discussed elsewhere (3), interpretations of the plant-CRY physiological studies are not straightforward. For both CRY1 and CRY2, several of the Trp substitutions are constitutively active in the dark (26, 39). Furthermore, the more flavin-proximal Trp substitutions produce proteins that do alter physiological light responses (figures 2 and 4 of ref. 26 and figure 3 of ref. 39), particularly at lower light levels (figure S11 of ref. 39). Thus, CRY signaling in these experiments is not inconsistent with a role for flavin photoreduction (3).
The contrasting properties of the W394F variant of dCRY with respect to light stability (dCRY degradation) and TIM degradation may relate to different requirements of these processes on dCRY activation. In the fly brain, the role of dCRY depends on the neuronal cluster in which it operates (42, 43). The action of dCRY on TIM for light resetting appears to be most important in the so-called “evening” cells. In the so-called “morning” cells, dCRY contributes to light-dependent phase shifting but may be unlinked to TIM degradation (42, 43). Hence, dCRY self-degradation or some other light-dependent function (15, 16) may constitute the key entrainment event in these cells. Our data support an uncoupling of dCRY-mediated TIM proteolysis and light-dependent dCRY proteolysis. Based on the behavior of the W394F variant, the former occurs quickly and requires only transient reduction of the flavin cofactor, and not conversion of the entire dCRY pool to the ASQ. In contrast, light-dependent dCRY instability requires sustained photoreduction over longer times. Sustained photoreduction alters dCRY conformational stability (20, 21) and likely promotes ubiquitination, followed by proteosomal degradation. Thus, dCRY photoreduction may send different signals depending on the time scale involved and the cellular environment.
Supplementary Material
Acknowledgments
We thank Jenna Chong for help with protein purification. This work was supported by grants from the NIH: R01GM054339 (to M.W.Y.) and R35GM122535 (to B.R.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719376115/-/DCSupplemental.
References
- 1.Chaves I, et al. The cryptochromes: Blue light photoreceptors in plants and animals. Annu Rev Plant Biol. 2011;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- 2.Conrad KS, Manahan CC, Crane BR. Photochemistry of flavoprotein light sensors. Nat Chem Biol. 2014;10:801–809. doi: 10.1038/nchembio.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmad M. Photocycle and signaling mechanisms of plant cryptochromes. Curr Opin Plant Biol. 2016;33:108–115. doi: 10.1016/j.pbi.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 4.Nohr D, et al. Extended electron-transfer in animal cryptochromes mediated by a tetrad of aromatic amino acids. Biophys J. 2016;111:301–311. doi: 10.1016/j.bpj.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kao Y-T, et al. Ultrafast dynamics and anionic active states of the flavin cofactor in cryptochrome and photolyase. J Am Chem Soc. 2008;130:7695–7701. doi: 10.1021/ja801152h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu B, Liu H, Zhong D, Lin C. Searching for a photocycle of the cryptochrome photoreceptors. Curr Opin Plant Biol. 2010;13:578–586. doi: 10.1016/j.pbi.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy C, et al. Updated structure of Drosophila cryptochrome. Nature. 2013;495:E3–E4. doi: 10.1038/nature11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zoltowski BD, et al. Structure of full-length Drosophila cryptochrome. Nature. 2011;480:396–399. doi: 10.1038/nature10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Czarna A, et al. Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function. Cell. 2013;153:1394–1405. doi: 10.1016/j.cell.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 10.Busza A, Emery-Le M, Rosbash M, Emery P. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science. 2004;304:1503–1506. doi: 10.1126/science.1096973. [DOI] [PubMed] [Google Scholar]
- 11.Dissel S, et al. A constitutively active cryptochrome in Drosophila melanogaster. Nat Neurosci. 2004;7:834–840. doi: 10.1038/nn1285. [DOI] [PubMed] [Google Scholar]
- 12.Peschel N, Chen KF, Szabo G, Stanewsky R. Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Curr Biol. 2009;19:241–247. doi: 10.1016/j.cub.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 13.Koh K, Zheng X, Sehgal A. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science. 2006;312:1809–1812. doi: 10.1126/science.1124951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozturk N, VanVickle-Chavez SJ, Akileswaran L, Van Gelder RN, Sancar A. Ramshackle (Brwd3) promotes light-induced ubiquitylation of Drosophila cryptochrome by DDB1-CUL4-ROC1 E3 ligase complex. Proc Natl Acad Sci USA. 2013;110:4980–4985. doi: 10.1073/pnas.1303234110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fogle KJ, Parson KG, Dahm NA, Holmes TC. CRYPTOCHROME is a blue-light sensor that regulates neuronal firing rate. Science. 2011;331:1409–1413. doi: 10.1126/science.1199702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fogle KJ, et al. CRYPTOCHROME-mediated phototransduction by modulation of the potassium ion channel β-subunit redox sensor. Proc Natl Acad Sci USA. 2015;112:2245–2250. doi: 10.1073/pnas.1416586112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berndt A, et al. A novel photoreaction mechanism for the circadian blue light photoreceptor Drosophila cryptochrome. J Biol Chem. 2007;282:13011–13021. doi: 10.1074/jbc.M608872200. [DOI] [PubMed] [Google Scholar]
- 18.Ozturk N, Selby CP, Annayev Y, Zhong D, Sancar A. Reaction mechanism of Drosophila cryptochrome. Proc Natl Acad Sci USA. 2011;108:516–521. doi: 10.1073/pnas.1017093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozturk N, Selby CP, Zhong D, Sancar A. Mechanism of photosignaling by Drosophila cryptochrome: Role of the redox status of the flavin chromophore. J Biol Chem. 2014;289:4634–4642. doi: 10.1074/jbc.M113.542498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaidya AT, et al. Flavin reduction activates Drosophila cryptochrome. Proc Natl Acad Sci USA. 2013;110:20455–20460. doi: 10.1073/pnas.1313336110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganguly A, et al. Changes in active site histidine hydrogen bonding trigger cryptochrome activation. Proc Natl Acad Sci USA. 2016;113:10073–10078. doi: 10.1073/pnas.1606610113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z, et al. Determining complete electron flow in the cofactor photoreduction of oxidized photolyase. Proc Natl Acad Sci USA. 2013;110:12966–12971. doi: 10.1073/pnas.1311073110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeugner A, et al. Light-induced electron transfer in Arabidopsis cryptochrome-1 correlates with in vivo function. J Biol Chem. 2005;280:19437–19440. doi: 10.1074/jbc.C500077200. [DOI] [PubMed] [Google Scholar]
- 24.Biskup T, et al. Variable electron transfer pathways in an amphibian cryptochrome: Tryptophan versus tyrosine-based radical pairs. J Biol Chem. 2013;288:9249–9260. doi: 10.1074/jbc.M112.417725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Müller P, Yamamoto J, Martin R, Iwai S, Brettel K. Discovery and functional analysis of a 4th electron-transferring tryptophan conserved exclusively in animal cryptochromes and (6-4) photolyases. Chem Commun (Camb) 2015;51:15502–15505. doi: 10.1039/c5cc06276d. [DOI] [PubMed] [Google Scholar]
- 26.Li X, et al. Arabidopsis cryptochrome 2 (CRY2) functions by the photoactivation mechanism distinct from the tryptophan (trp) triad-dependent photoreduction. Proc Natl Acad Sci USA. 2011;108:20844–20849. doi: 10.1073/pnas.1114579108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoang N, et al. Human and Drosophila cryptochromes are light activated by flavin photoreduction in living cells. PLoS Biol. 2008;6:e160. doi: 10.1371/journal.pbio.0060160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paulus B, et al. Spectroscopic characterization of radicals and radical pairs in fruit fly cryptochrome–Protonated and nonprotonated flavin radical-states. FEBS J. 2015;282:3175–3189. doi: 10.1111/febs.13299. [DOI] [PubMed] [Google Scholar]
- 29.Suri V, Qian Z, Hall JC, Rosbash M. Evidence that the TIM light response is relevant to light-induced phase shifts in Drosophila melanogaster. Neuron. 1998;21:225–234. doi: 10.1016/s0896-6273(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 30.Warren JJ, Winkler JR, Gray HB. Redox properties of tyrosine and related molecules. FEBS Lett. 2012;586:596–602. doi: 10.1016/j.febslet.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shih C, et al. Tryptophan-accelerated electron flow through proteins. Science. 2008;320:1760–1762. doi: 10.1126/science.1158241. [DOI] [PubMed] [Google Scholar]
- 32.Balland V, Byrdin M, Eker APM, Ahmad M, Brettel K. What makes the difference between a cryptochrome and DNA photolyase? A spectroelectrochemical comparison of the flavin redox transitions. J Am Chem Soc. 2009;131:426–427. doi: 10.1021/ja806540j. [DOI] [PubMed] [Google Scholar]
- 33.He TF, et al. Femtosecond dynamics of short-range protein electron transfer in flavodoxin. Biochemistry. 2013;52:9120–9128. doi: 10.1021/bi401137u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vinayak P, et al. Exquisite light sensitivity of Drosophila melanogaster cryptochrome. PLoS Genet. 2013;9:e1003615. doi: 10.1371/journal.pgen.1003615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oztürk N, Song SH, Selby CP, Sancar A. Animal type 1 cryptochromes. Analysis of the redox state of the flavin cofactor by site-directed mutagenesis. J Biol Chem. 2008;283:3256–3263. doi: 10.1074/jbc.M708612200. [DOI] [PubMed] [Google Scholar]
- 36.Gutscher M, et al. Real-time imaging of the intracellular glutathione redox potential. Nat Methods. 2008;5:553–559. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- 37.Ritz D, Beckwith J. Roles of thiol-redox pathways in bacteria. Annu Rev Microbiol. 2001;55:21–48. doi: 10.1146/annurev.micro.55.1.21. [DOI] [PubMed] [Google Scholar]
- 38.Engelhard C, et al. Cellular metabolites enhance the light sensitivity of Arabidopsis cryptochrome through alternate electron transfer pathways. Plant Cell. 2014;26:4519–4531. doi: 10.1105/tpc.114.129809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao J, et al. Trp triad-dependent rapid photoreduction is not required for the function of Arabidopsis CRY1. Proc Natl Acad Sci USA. 2015;112:9135–9140. doi: 10.1073/pnas.1504404112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El-Esawi M, et al. Cellular metabolites modulate in vivo signaling of Arabidopsis cryptochrome-1. Plant Signal Behav. 2015;10:e1063758. doi: 10.1080/15592324.2015.1063758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herbel V, et al. Lifetimes of Arabidopsis cryptochrome signaling states in vivo. Plant J. 2013;74:583–592. doi: 10.1111/tpj.12144. [DOI] [PubMed] [Google Scholar]
- 42.Tang CH, Hinteregger E, Shang Y, Rosbash M. Light-mediated TIM degradation within Drosophila pacemaker neurons (s-LNvs) is neither necessary nor sufficient for delay zone phase shifts. Neuron. 2010;66:378–385. doi: 10.1016/j.neuron.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshii T, et al. Cryptochrome-dependent and -independent circadian entrainment circuits in Drosophila. J Neurosci. 2015;35:6131–6141. doi: 10.1523/JNEUROSCI.0070-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buckel W, Thauer RK. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na(+) translocating ferredoxin oxidation. Biochim Biophys Acta. 2013;1827:94–113. doi: 10.1016/j.bbabio.2012.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.