Significance
Mice that have homozygous deletion of the p53 tumor suppressor protein universally die of malignancy, generally before 6 months of age. We show that hemizygous deficiency of RALBP1 (RLIP76 or Rlip) confers a degree of protection from spontaneous malignancy that has never previously been observed. This discovery introduces a paradigm for p53 function, in which Rlip plays a central role as an effector that appears necessary for the cancer susceptibility of p53 null mice. Because p53 loss has a powerful effect on genomic instability that contributes to the initiation and promotion of cancers and to drug and radiation resistance in humans, our findings provide a method for prevention and therapy of p53-deficient cancer.
Keywords: RALBP1, TP53, cancer prevention, cancer signalling, cytokine
Abstract
TP53 (p53) is a tumor suppressor whose functions are lost or altered in most malignancies. p53 homozygous knockout (p53−/−) mice uniformly die of spontaneous malignancy, typically T-cell lymphoma. RALBP1 (RLIP76, Rlip) is a stress-protective, mercapturic acid pathway transporter protein that also functions as a Ral effector involved in clathrin-dependent endocytosis. In stark contrast to p53−/− mice, Rlip−/− mice are highly resistant to carcinogenesis. We report here that partial Rlip deficiency induced by weekly administration of an Rlip-specific phosphorothioate antisense oligonucleotide, R508, strongly inhibited spontaneous as well as benzo(a)pyrene-induced carcinogenesis in p53−/− mice. This treatment effectively prevented large-scale methylomic and transcriptomic abnormalities suggestive of inflammation found in cancer-bearing p53−/− mice. The remarkable efficiency with which Rlip deficiency suppresses spontaneous malignancy in p53−/− mice has not been observed with any previously reported pharmacologic or genetic intervention. These findings are supported by cross-breeding experiments demonstrating that hemizygous Rlip deficiency also reduces the spontaneous malignancy phenotype of p53+/− mice. Rlip is found on the cell surface, and antibodies directed against Rlip were found to inhibit growth and promote apoptosis of cell lines as effectively as Rlip siRNA. The work presented here investigates several features, including oxidative DNA damage of the Rlip–p53 association in malignant transformation, and offers a paradigm for the mechanisms of tumor suppression by p53 and the prospects of suppressing spontaneous malignancy in hereditary cancer syndromes such as Li-Fraumeni.
The p53 gene (human TP53) is a 53-kDa stress-responsive, genome-protective, tumor suppressor protein that loses its normal function as a result of genetic alterations found in most cancers (1–3). The powerful tumor suppressor function of p53 is evident from the universal susceptibility of p53 knockout mice to spontaneous cancer, usually T-cell lymphoma, within 6 mo of age (4), and from the inability to abrogate this phenotype with any previous genetic manipulation (Dataset S1). The pathogenic role of loss of p53 function in spontaneous neoplasia in humans is evident in Li-Fraumeni syndrome, an autosomal dominant genetic disorder of p53 deficiency that carries a lifetime cancer risk of 70% in men, and approaching 100% in women. Surveillance is the only effective means to prevent death from malignancy (5, 6). Deficiency of p53 function in sporadic cancers has been associated with treatment failure resulting from dysregulated expression of p53-linked stress response genes that protect cancer cells from apoptosis and provide metabolic defenses against oxidative stress and xenobiotic toxins such as chemotherapy drugs (7–9).
Rlip (encoded by RALBP1 [18p11.22]) is a stress-responsive ATPase enzyme of the mercapturic acid pathway that catalyzes the transmembrane efflux of glutathione (GSH)-electrophile thioether conjugates (GS-E) formed through glutathione S-transferase-catalyzed conjugation of GSH with exogenous and endogenous electrophilic toxins (9–30). Because Rlip-catalyzed efflux of GS-E prevents product/feedback inhibition of several mercapturic acid pathway enzymes, its loss promotes apoptosis exerted by xenobiotics and GS-E, derived from oxidative degradation of ω-6 fatty acids (31). Its ATPase activity is coupled with clathrin-dependent endocytosis (CDE) (26), the RAL-regulated first step in the internalization and trafficking of membrane vesicles containing receptor-bound cancer-promoting growth hormones (24, 32, 33). CDE regulates signaling downstream of receptors for insulin, EGF, TNFα, FGF1, and many other peptide hormones (34–37); Rlip, a key component of CDE, links RAL, RAS, RHO, and RAC signaling (38–45). CDE and GS-E transport are severely deficient in Rlip−/− mice (15, 20, 26).
Oxidative metabolism of ω-6 polyunsaturated fatty acids in response to radiant (X-ray, UV light, heat) or oxidative stress yields lipid hydroperoxides, which degrade to toxic lipid alkenals; principally, 4-hydroxynonenal (4HNE). 4HNE is metabolized primarily to a glutathione conjugate (GS-HNE) that is removed from cells by Rlip (11, 17, 19, 20, 29, 30). Recombinant Rlip protein is the most potent biological agent for defending cells and animals from the toxicity of stressors that generate massive amounts of 4HNE: ionizing radiation and chemical warfare agents (46). Interestingly, the apoptotic activity of 4HNE is directed selectively toward malignant cells, as evident from apoptosis of cancer cells and dramatic regression of melanoma, neuroblastoma, and cancers of the lung, colon, kidney, pancreas, and prostate by Rlip-depletion/inhibition in mouse models (16, 18, 22, 24, 25). An existential need of cancer cells for Rlip is underscored by resistance to chemical carcinogenesis in Rlip−/− mice to a degree exceeding that for any other previously reported genetic intervention (26). The diametrically opposite cancer susceptibility of Rlip−/− and p53−/− mice led us to hypothesize a mutually inhibitory and functionally opposed relationship between Rlip and p53 in carcinogenesis.
Results
Rlip Deficiency Suppresses Malignancy in p53−/− Mice.
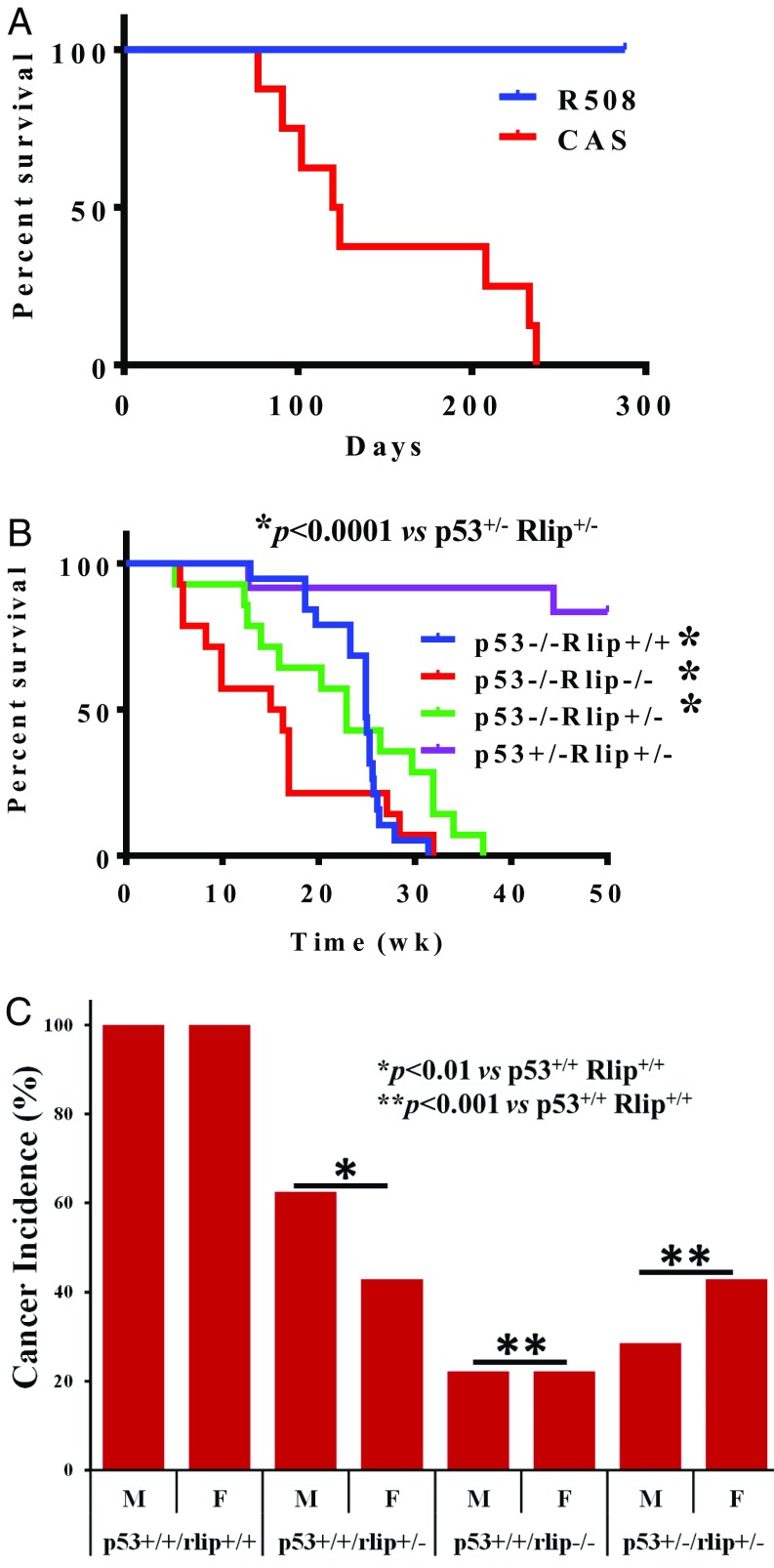
Previous studies demonstrated that antisense oligonucleotides exert potent antineoplastic effects and that the phosphorothioate oligonucleotide R508 is the most potent (16, 18, 22, 24, 25). We report here that a single 200 µg i.p. dose of R508 given to wild-type (wt) mice reduced Rlip protein to 56 ± 12% of control in the liver, and to a lesser extent in all other tissues, at 24 h (P < 0.001), with gradual recovery by day 7 (SI Appendix, Fig. S1). Reduction of blood glucose by 26%, triglycerides by 32%, and cholesterol by 48% on R508 treatment showed expected pharmacodynamic effects, given that these metabolic alterations are characteristic of Rlip knockout mice (16, 23, 47). We then treated C57BL/6 p53−/− mice with weekly i.p. injections of 200 µg R508 or control scrambled antisense (CAS) beginning at age 8 wk and reduced Rlip protein to 47 ± 7% (P < 0.0001) and Rlip mRNA to 49 ± 13% (P < 0.001). Two sequential independent experiments were performed, each giving the same dramatic results: prevention of malignancy in 100% of R508-treated p53−/− mice, whereas all control mice died of T-cell lymphomas by age 34 wk, with median survival of 122 d (Fig. 1A and SI Appendix, Figs. S2–S5). CAS- or PBS-treated mice had extensive tissue injury in multiple organs, whereas R508-treated p53−/− mice had no histological evidence of tissue injury or any type of malignancy. We next crossbred P53+/− with Rlip−/− mice and found that offspring with partial or complete Rlip deficiency failed to develop tumors, independent of p53 status. All p53+/−Rlip+/− offspring were healthy, and 11/13 mice alive at 48 wk of age, as well as two killed at earlier ages because of malocclusion, were cancer-free (Fig. 1B). Median survival of p53−/−Rlip+/+; p53−/−Rlip−/− and p53−/−Rlip+/− mice was 24.9, 15.65, and 22.9, wk respectively. Comparing p53+/−Rlip+/− with p53−/−Rlip+/+; p53−/−Rlip−/− and p53−/−Rlip+/− via Log-rank (Mantel-Cox) and Gehan-Breslow-Wilcoxon test (GraphPad Prizm version 6) disclosed a significant difference in survival rates of mice (P < 0.0001). Only male p53−/−/Rlip−/− and female p53−/−Rlip+/− were viable, but they developed inanition resulting from malocclusion or hydrocephalus at a median age of 12 and 23 wk, respectively. However, these mice were also all free of malignancy at necropsy. In a chemical carcinogenesis model, 75% of male and 60% of female p53+/−Rlip+/− mice (P < 0.001) treated with B[a]P were free of any malignancy at 32 wk age, whereas all wild-type (p53+/+Rlip+/+) mice developed stomach or lung adenocarcinoma (Fig. 1C). Sixty percent of p53+/+Rlip+/− males and 40% of females (P < 0.01) developed adenocarcinoma; this rate was intermediate between wild-type (100%) and p53+/+Rlip−/− (20%; P < 0.001) previously reported by us (26). These results clearly indicate that wild-type (p53+/+Rlip+/+) mice had significantly higher (P < 0.001) incident of chemically induced cancer than any other genotype (Fig. 1C). Statistical significance was determined by Fisher’s exact test (two-tailed), using GraphPad software. Thus, hemizygous Rlip deficiency also exerted a strong dominant negative effect on the spontaneous malignancy phenotype of p53−/− mice.
Fig. 1.
Rlip deficiency prevents carcinogenesis. (A) Tumor-free survival curve is shown for C57BL/6J p53−/− mice treated weekly with i.p. injection of 0.2 mL PBS containing CAS or R508 (0.2 mg each) for up to 24 wk. All surviving animals were killed at 32 wk of age in the first and 36 wk of age in the second experiment. (B) Overall survival curves for double-knockout mice of the indicated genotype are shown. No treatment was given, and the mice were killed if there was the appearance of morbidity. (C) Mice of the indicated genotypes were administered 3 mg benzo(a)pyrene in corn oil by gavage at age 8 wk and 12 wk. All mice killed during the study because of the appearance of morbidity as well as those killed at the end of the study at 24 wk underwent necropsy to determine the incidence of cancer (lung + gastric).
The Hepatic Transcriptome and Methylome of R508-Treated p53−/− Mice Are Near Normal Even in Aged Animals.
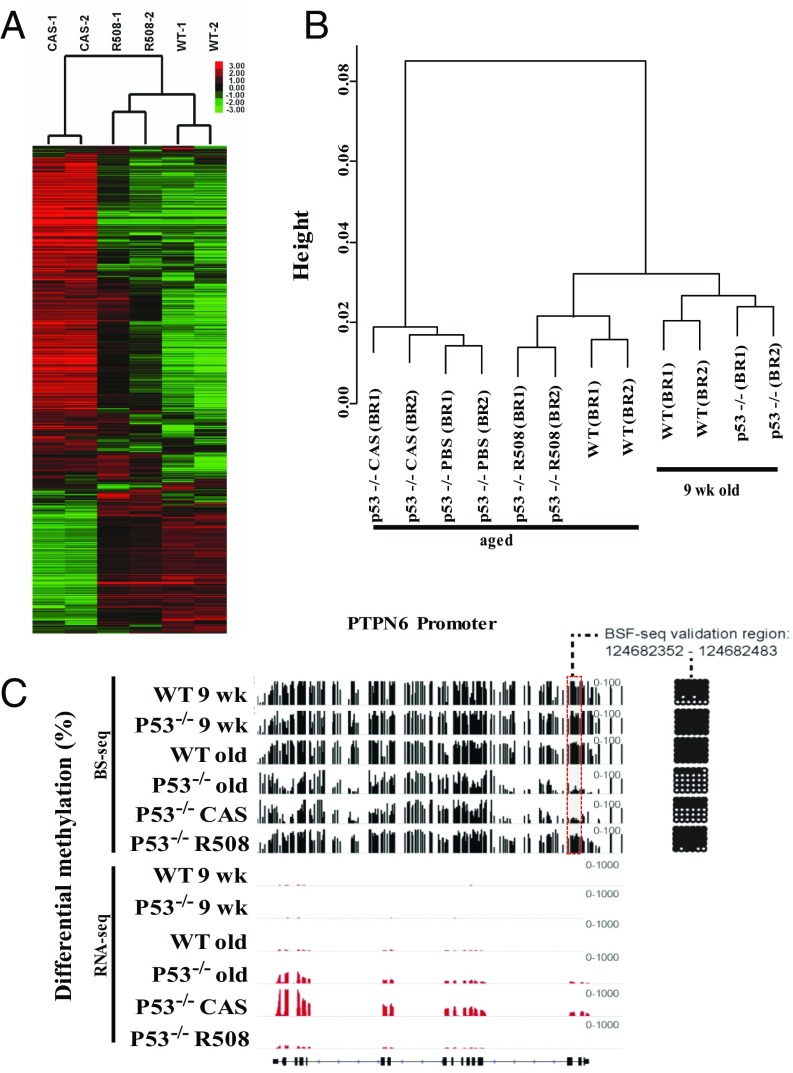
RNA was prepared from livers at the time of autopsy (18–24 wk), and the transcriptomes of R508- and CAS-treated p53−/− mice were compared with wt, using RNA-Seq. Unsupervised hierarchical clustering analyses of RNA-Seq results on hepatic tissues revealed that the transcriptomes of R508-treated p53−/− mice were very similar to wt and quite different from p53−/− (Fig. 2A). Gene ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways terms showed differential expression of cell cycling, T-cell immunity, inflammation, cancer, and developmental and stem cell processes in untreated control p53−/− compared with R508-treated mice (Dataset S2) that was consistent with previous reports for RNA changes in the livers of cancer-bearing as well as cancer-free p53−/− mice (48, 49). The transcriptomes of young (9 wk) p53−/− or wt mice were quantitatively similar to those of aged (32 wk) wt mice and distinct from cancer-bearing p53−/− mice at older ages, indicating that the abnormal transcriptome of p53−/− mice was not congenital but acquired, either as a result of aging or as a consequence of lymphoma-induced cytokine storm (Dataset S2).
Fig. 2.
Rlip deficiency reverts transcriptomic and methylomic abnormalities in p53 knockout mice. (A) Hierarchical clustering of transcriptional changes. RPKM values were log2 transformed, and the genes were filtered based on i) at least one sample has log2 RPKM ≥ 2 and ii) standard deviation across all six samples ≥1. This filtering step resulted in 5,024 genes. The log2 RPKM data of filtered genes were subject to hierarchical clustering with correlation dissimilarity as distance and average linkage using Cluster 3.0. The clustering result was visualized and exported using Java Treeview. (B) Clustering of promotor DNA methylation. WGBS (BS-Seq) was performed on liver tissues. Young (9-wk) wt and p53−/− mice were aging controls. The aged (32-wk) wt mice were cancer-free controls for the R508-treated 53−/− mice and the aged (18- to 24-wk) PBS- or CAS-treated p53−/− mice were controls for R508-treatement. For clustering displayed, promoters were defined using RefSeq (±1000 bp of transcription start site) and were selected if average CpG site methylation level was >50% in at least one sample and methylation range was >25%. Hierarchical clustering was performed using average linkage algorithm and visualized by Java Treeview. (C) Visualization and bisulfite sequencing validation of WGBS results. WGBS results (black bars) were aligned with RNA-Seq (transcriptomic) data (red bars), shown with exons and introns (bottom line). Bisulfite sequencing validation on the promoter region (gray box) are shown in the grid on the right, with black dots indicating methylated and white dots indicating unmethylated sites within the PTPN6 gene promoter. The height of the bars represent degree of methylation and height of the red bars is proportional to mRNA expression.
Epigenetic dysregulation including changes in global DNA methylation patterns are hallmarks of cancer (50–52). Therefore, we conducted whole-genome bisulfite sequencing (WGBS), with confirmation of selected differentially methylated regions (DMRs) between cancer-free and cancer-presenting mice by conventional bisulfite sequencing. DMR validation at PTPN6, a tyrosine phosphatase that regulates hematopoietic cell signaling and growth, is shown as an example (Fig. 2C). DMR validation is also displayed for HOXA5 (Fig. S6). The TP53 deletion was confirmed by the absence of DNA sequence alignment to the region of targeted deletion (SI Appendix, Figs. S6 and S7). Hierarchical clustering analyses revealed that the methylome of cancer-bearing PBS or CAS-treated p53−/− mice was clearly distinct from the R508-treated p53−/− mice, which closely resembled young or aged wt mice and the young p53−/− mice (Fig. 2C and Dataset S3). Whereas 14,367 DMRs were found between p53−/− and wt mice, only 869 DMRs were found between R508-treated p53−/− and wt mice. Thus, ∼95% of all methylation abnormalities were prevented by R508 treatment. CpG island hypermethylation, repetitive element (SINE/LINE) hypomethylation, and gains or losses of DNA methylation in promoters and gene bodies of immune modulatory, cancer developmental genes, and cell cycle genes in p53−/− were prevented by R508 (SI Appendix, Figs. S8 and S9). Promoter hypomethylation of critical immune system genes involved in leukocyte activation, T-cell proliferation, and cytokine production found in control p53−/− mice was essentially absent in R508-treated p53−/− mice (Fig. 2B). For example, PTPN6 promoter hypomethylation was well correlated with gene activation in cancer-bearing mice, with promoter-mediated gene silencing in wt and R508-treated mice (Fig. 2C). Compared with 31 hypermethylated DMRs in promoters of genes within the ontology term embryonic morphogenesis [including homeobox genes A-D (HOXA-D) clusters], only three were hypermethylated in R508-treated p53−/− mice (SI Appendix, Fig. S9). Ontology terms related to Rlip function enriched in R508-treated p53−/− mice included GSH metabolism, xenobiotic metabolism, oxidative stress, transport, endocytosis, vesicular transport, mitochondrial abnormalities, cell cycling, mitosis, PI3K and MAPK signaling, angiogenesis, and cancer pathways (Fig. 2C and Dataset S2).
Rlip Regulates the Activity of Cancer-Signaling and Cytokine Pathways.
Results obtained here suggest that Rlip depletion may reduce a response to inflammatory signals, especially as Rlip is a rate determinant of CDE, a process that broadly regulates peptide hormone signaling (33, 53). In support of this hypothesis, we observed that activation or protein content of PI3K, AKT, mTOR, p70S6K, RB, and BCL2 was reduced, and that of HSF1, JNK, p27 (CDK1B), and BCLXL was increased in hepatic tissues of R508-treated mice (SI Appendix, Fig. S10). That inflammatory signaling is affected by Rlip is supported by our studies using shRNA to reduce Rlip in p53−/− mouse embryonic fibroblasts (MEFs), where we see effects similar to those seen in liver of Rlip depletion on AKT, PI3K, CDK4, JNK, BCL2, BAX, mTOR, P70S6K, RB, and p27. Through a series of studies to examine the role of Rlip in CDE, we confirmed its rate determining role in benign as well as malignant cells and showed that STAT3 signaling, downstream of FGF, is regulated by Rlip (SI Appendix, Figs. S11–S13).
Lymphoma Prevention by R508 Treatment Is Independent of Effects on Oxidative DNA Damage.
Because oxidative stress promotes carcinogenesis and can regulate DNA methylation (54, 55), we considered the possibility that reduced oxidative DNA damage underlies cancer prevention. 4HNE levels, a surrogate for overall oxidative stress (56), were approximately eightfold higher in control p53−/− mice than in R508-treated mice, largely reflecting the presence of tissue damage. 4HNE levels in R508-treated p53−/− mice were significantly lower than in control p53−/− mice, similar to those in Rlip−/− mice, but still twofold higher than wt (SI Appendix, Fig. 14A). Surprisingly, despite much lower oxidative stress, the levels of 8-OHdG (a measure of oxidative DNA damage) (57) in R508-treated p53−/− mice were not significantly different from in the control p53−/− groups (SI Appendix, Fig. S14B). Potent prevention of malignancy without affecting oxidative DNA damage supports an existential requirement of Rlip for malignant transformation of genetically damaged cells.
Rlip, p53, and HSF1 Interactions.
The dramatic change in cancer susceptibility phenotype of p53−/− mice on Rlip depletion to only a hemizygous level is consistent with previous work demonstrating that p53+/+Rlip+/− mice are highly resistant to chemical carcinogenesis (15, 26) and suggests a haploinsufficiency phenomenon, in which deficiency of a protein with multiple binding partners simultaneously alters the quantity and relative ratios of multiple heterodimers, resulting in disproportionate phenotypic effects. HSF1 was an obvious candidate in haploinsufficiency interactions because HSF1 binds p53 during stress-induced nuclear translocation, its nuclear translocation for regulation of chaperone expression is inhibited by Rlip, and its deficiency converts the histological profile of malignancy in p53−/− mice from lymphoma to adenocarcinoma (58–60). Because binding of p53 with HSF1 (58) or with Rlip (21) has been reported ex vivo and in vitro but not in vivo, we examined this possibility in present studies. Proximity ligation assay demonstrated cytosolic and nuclear interactions of p53 with Rlip in MEF (SI Appendix, Fig. 14C). Furthermore, proximity ligation assay also demonstrated p53-Rlip and HSF1-p53 interactions in mouse liver (SI Appendix, Fig. 14 D–F). These interactions between HSF1, p53, and Rlip suggest haploinsufficiency interactions between the three proteins.
Antibodies Targeting Rlip.
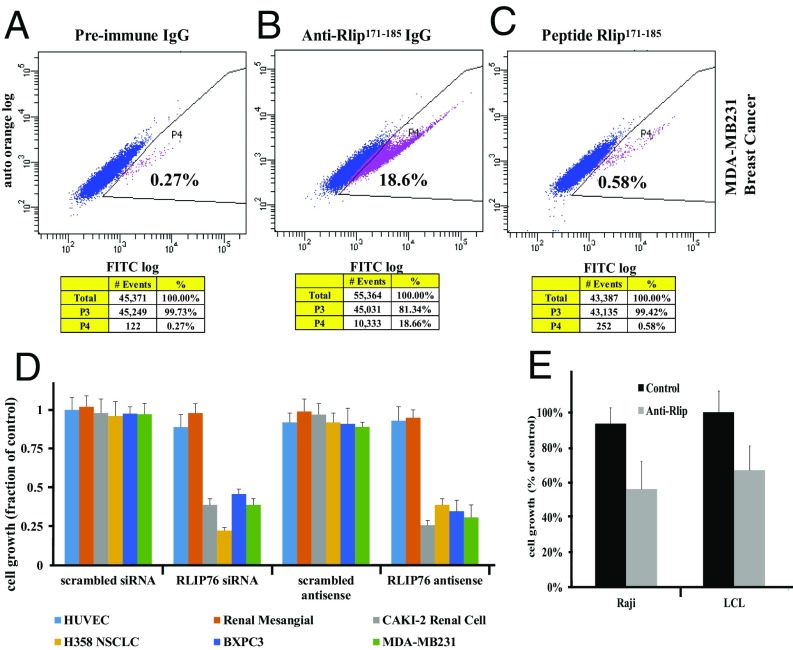
Humanized monoclonal antibodies are likely to be more pharmacologically suitable than antisense for the treatment of p53-deficient malignancy or cancer prevention in Li-Fraumeni syndrome. Using polyclonal antibodies specific for the N171–185 peptide and a cleavable biotinylation reagent (Sulfo-NHS-SS-Biotin), we observed the presence of the Rlip171–185 epitope on the cell surface (SI Appendix, Fig. S15). This was confirmed by flow cytometry on MDA-MB231 breast cancer cells, and the specificity of the cell surface detection was confirmed using competitive inhibition by the N171–185 peptide (Fig. 3 A–C). These antibodies inhibited the growth of multiple histologies of cancer cell lines including melanoma (independent of B-Raf, CDKN2A, p53, or MAPK3 status), estrogen receptor positive (ER+) breast cancer, small cell lung cancer, and both squamous and adenocarcinoma histology of nonsmall cell lung cancer (SI Appendix, Fig. S16). These antibodies inhibited p53-deficient nonsmall cell lung cancer, VHL-deficient renal cell cancer, triple-negative breast cancer, and p53/p16/SMAD-4 deficient pancreatic cancer cell lines as effectively as antisense or siRNA (Fig. 3D). Nonmalignant HUVEC and renal mesangial cells were unaffected. Importantly, these antibodies also inhibited growth (Fig. 3E) and induced apoptosis (SI Appendix, Fig. S17) in mouse and human lymphoma cell lines.
Fig. 3.
Anticancer effects of targeting Rlip by antibodies. Presence of the Rlip171-185 peptide epitope on the surface of the MDA-MB231 cells was determined by flow cytometry, using (A) preimmune antibody and (B) anti-Rlip171-185 antibody and (C) by including the Rlip171-185 peptide with anti-Rlip171-185 antibody to competitively inhibit specific binding. The stained cells were analyzed using the Beckman Coulter Cytomics FC500, Flow Cytometry Analyzer. Results were processed using CXP2.2 analysis software from Beckman Coulter. The percentage of cells displaying cell surface fluorescence is given in each panel. (D) Effect of Rlip depletion by R508 or Rlip-siRNA on cell growth inhibition was determined by MTT assay at 48 h after transfection with lipofectamine. CAS and control scrambled siRNA with the same sequence served as controls. The nonmalignant transformed cell lines were human umbilical vein endothelial cell (HUVEC) and renal mesangial. The malignant cell lines were CAKI-2 renal cell carcinoma, H358 human lung bronchioalveolar nonsmall cell carcinoma, BXPC3 pancreatic carcinoma, and MDA-MB-231 ER/PR/Her2-negative breast carcinoma. (E) Growth inhibition of Raji and LCL lymphoma cell lines by anti-Rlip171-185 antibody (20 µg/mL) was measured using an MTT assay. The control consisted of antibody heat inactivated by incubation for 2 min at 70 °C.
Discussion
Rlip−/− and p53−/− mice are polar opposites in the spectrum of cancer susceptibility (4, 26). Present studies have shown that pharmacologically mediated partial suppression of Rlip protein prevented the appearance of lymphoma or any other malignancy in p53−/− mice. Normalization of the methylome and transcriptome of R508-treated mice strongly supported the results of gross, histological, and immunohistochemical findings of the absence of any malignancy at necropsy. In vivo studies showing diminished activation of key cell cycling, cell growth, replication checkpoint, and cytokine signaling pathways at the protein level was also consistent with the absence of malignancy in R508-treated mice. R508-induced hypoglycemia and hypolipidemia confirm target specificity, showing predicted pharmacodynamic endpoints that reflect Rlip deficiency (16, 23, 47). These surprising results are confirmed by our findings that p53−/−Rlip−/+ heterozygous mice are resistant to both spontaneous and chemical carcinogenesis, supporting a haploinsufficiency mechanism.
Rlip functions intersect with those of p53 at multiple levels including stress response, drug/xenobiotic/radiation resistance, transcriptional control of chaperones, apoptosis, and cell cycling. Rlip is an ATPase that catalyzes the efflux of proapoptotic electrophilic metabolites arising from lipid peroxidation and xenobiotic exposure (10–17, 19, 29, 30). Potent antiapoptotic function of Rlip has been established through the development of recombinant Rlip protein as the most effective drug for the treatment and prevention of radiation and chemical weapon poisoning (15, 20, 46). Thus, an existential requirement of its antiapoptotic effect after malignant transformation could potentially explain the lack of lymphoma in Rlip-deficient mice. Because Rlip regulates the rate of CDE, and consequently the intensity of cancer-promoting signals downstream of cell growth hormones and cytokines (16, 26, 40, 44, 47, 61–63), its deficiency could delay the appearance of lymphoma by slowing growth after malignant transformation. Regulation of CDE by p53 (64, 65) suggests another possible site of direct interaction.
Narrowing the key cancer-promoting processes or the subset of genes whose expression is critical for lymphoma formation would have been possible by subtraction of expression profiles of R508-treated p53−/− mice with malignancy from aged p53−/− mice with altered transcriptomes but no malignancy; unfortunately, we observe no such cases. For the same reasons, our results cannot establish a cause–effect relationship between methylomic aberrations and lymphomagenesis in p53−/− mice or between correction of methylomic aberrations and lymphoma suppression by R508 treatment. However, a nearly normal methylome in cancer-free R508-treated p53−/− mice despite persistent extensive oxidative DNA damage clearly shows that methylomic aberrations are not an obligatory consequence of either p53 deficiency or oxidative DNA damage in the setting of partial Rlip deficiency. The absence of Rlip–p53 heterodimers in both Rlip−/− and the p53−/− mice also rules out any direct role of such a heterodimer in mechanisms of DNA methylation or the opposite cancer susceptibility phenotype of Rlip−/− and p53−/− mice.
The abrupt change in the cancer susceptibility phenotype of p53−/− mice caused by hemizygous loss of Rlip supports a haploinsufficiency interaction. HSF1 is a reasonable candidate in such a mechanism because it interacts with both p53 and Rlip. The HSF1–p53 complex translocates to the nucleus in response to stress (58). HSF1 loss in p53−/− mice switches the cancer phenotype, from primarily lymphoma to primarily adenocarcinoma or sarcoma, without altering the overall incidence of malignancy (60). Rlip inhibits chaperone transcription by sequestering HSF1 in a tubulin-bound complex that dissociates on stress exposure, releasing HSF1 for nuclear translocation (59). Consistent with this, numerous chaperones are up-regulated in Rlip−/− mice (15). A mutually inhibitory relationship is evident from inhibition of transport and antiapoptotic activities of Rlip by HSF1 (27). Rlip–p53 interactions were indicated by inhibition of the transport activity of Rlip by p53 and reduced binding of Rlip to mutant p53 in neuroblastoma cells (21). Present studies show an Rlip–p53 complex exists in vitro and in vivo, and indicate that this complex may also be present in the nucleus. We propose a model in which the three proteins coordinately regulate transcriptional responses to stress, while Rlip serves as an antiapoptotic effector and feedback inhibitor of p53 and HSF1. Only the p53-HSF1 dimer can exist in the absence of Rlip, and only the Rlip-HSF1 can exist in the absence of p53; conceptually, the former would play a role in cancer prevention, and the latter in promotion. There are numerous other examples of haploinsufficiency interactions that cause abrupt changes in cancer susceptibility of p53−/− mice (Dataset S1), but none of these result in the dramatic phenotypic switch from universal cancer susceptibility to nearly complete resistance.
In conclusion, our studies establish a strong dominant negative effect of Rlip deficiency on the cancer susceptibility phenotype conferred by Rlip loss and support an existential need of Rlip for cancer formation in the mouse model. Our findings are significant because they create a paradigm for defining the role of p53 in carcinogenesis. A very broad impact in cancer prevention as well as therapy is expected because of abnormal p53 function in most human cancers. Creating a chronic partial Rlip deficiency could be a method for prevention of malignancy in individuals with Li-Fraumeni syndrome. Because targeted depletion of Rlip causes regression of malignancy in preclinical models regardless of the p53 status of the neoplasm, Rlip inhibitors may prove to exert a very broad-spectrum therapeutic effect. The potent antiapoptotic and stress-defense functions of Rlip indicate potential application in drug- or radiation-resistant malignancy. Because only a hemizygous state of Rlip deficiency would be needed, the likelihood of adverse effects would be mitigated. Indeed, collateral health benefit could also be realized through reduction of blood glucose, insulin resistance, and hyperlipidemia (16, 23, 47).
Methods
To study suppression of spontaneous carcinogenesis by R508 (phosphorothioate antisense oligonucleotide; 508GGCTCCTGAATTGGCTTTTTC529) or CAS (control scrambled phosphorothioate oligonucleotide, CATCGAAATCGTTGCAGTTAC), p53−/− mice were treated with 200 µg R508 or CAS by i.p. injection starting at 8 wk of age until killing. To study suppression of spontaneous carcinogenesis cross-bred mice, p53−/−Rlip+/+, p53−/−Rlip+/−, p53−/−Rlip−/−, and p53+/−Rlip+/− mice were monitored three times per week for distress or overt malignancy, and all surviving mice were killed at age 48 wk. Chemical carcinogenesis was studied in 10–15 mice of each genotype per treatment group. Mice were administered 3 mg B[a]P in 0.1 mL corn oil or corn oil alone by gavage at the age of 8 and 12 wk. Raji and human LCL lymphoma cell lines were grown in RPMI1640 medium and MEFs in DMEM containing 10% FBS and 1% penicillin/streptomycin at 37 °C in 5% CO2. Cytotoxicity, signaling, and endocytosis studies were performed as described in SI Appendix, Methods. RNA-Seq studies, whole-genome bisulfite sequencing, and validation studies were conducted in triplicate for each biological replicate in p53−/− CAS or R508-treated mice liver. The RNA-sequencing run was performed in a Illumina HiSEq. 2500 platform with HiSeq SBS V4 Kits, and reads were aligned using Tophat v2.0 to mouse reference genome mm9 (66, 67). To validate RNA-Seq results, qRT-PCR was conducted with primers, prevalidated from BioRad (PrimePCR, cat. 10025636), for the following genes: Cib3, Dlk1, Cyp4a32, Fzd10, Gpr3, Tff1, and Six3. For the whole-genome bisulfite sequencing, 200 ng DNA was sonicated to get 200-bp-size DNA (68), and library templates were prepared for sequencing. Reads were aligned to in silico bisulfite-converted mm9 genome using Bismark aligner (69), using the default settings. Quantitative validation was carried out for the PTPN6 and HOXA5 gene promoters by conventional bisulfite sequencing (70).
Statistical methods used for RNA-Seq and WGBS analyses are given in SI Appendix. The statistical significance of differences between control and treatment groups was determined by ANOVA followed by Bonferroni correction and Benjamini-Hochberg procedure with a false-discovery rate <0.05. The heat map of the P values of top differentially expressed genes by Euclidean distance and an average linkage strategy for the four groups (wt, PBS-p53−/−, CAS-p53−/−, and R508-p53−/−) are presented. Changes in tumor size and body weight during the experiments were visualized by scatter plot.
Animal experiments were approved by the Institutional Animal Care and Use Committee at City of Hope and performed under approved protocol no. 11016.
Supplementary Material
Acknowledgments
We thank the personnel in the Animal Research Center, Pathology, Genomics, and Bioinformatics Cores of the City of Hope for their invaluable assistance. The authors are grateful to Dr. Brian Armstrong (Microscope Core Lab, City of Hope) and Lucy Brown (Analytical Cytometry Core, City of Hope) for technical assistance in the fluorescence microscope and flow cytometry analyses, respectively. A.D.R. holds the Samuel Rahbar Chair in Diabetes and Drug Discovery. This work was supported by NIH-R01-CA77495 (to S.A.), NIH/National Cancer Institute Grant CA129383 (to S.K.S.), the City of Hope Cancer Center Support Grant P30 CA33572, the Perricone Foundation, Texas Tech University Health Sciences Center Ethel S. Neely & Emma S. Treadwell Endowment (S.A.), and the University Medical Center Chair of Excellence. Funding from the Department of Defense Grant W81XWH-16-1-0641 (to S.S.S.) is also acknowledged.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719586115/-/DCSupplemental.
References
- 1.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 2.Levine AJ, Oren M. The first 30 years of p53: Growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zilfou JT, Lowe SW. Tumor suppressive functions of p53. Cold Spring Harb Perspect Biol. 2009;1:a001883. doi: 10.1101/cshperspect.a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donehower LA, Lozano G. 20 years studying p53 functions in genetically engineered mice. Nat Rev Cancer. 2009;9:831–841. doi: 10.1038/nrc2731. [DOI] [PubMed] [Google Scholar]
- 5.Ariffin H, et al. Whole-genome sequencing analysis of phenotypic heterogeneity and anticipation in Li-Fraumeni cancer predisposition syndrome. Proc Natl Acad Sci USA. 2014;111:15497–15501. doi: 10.1073/pnas.1417322111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villani A, et al. Biochemical and imaging surveillance in germline TP53 mutation carriers with Li-Fraumeni syndrome: A prospective observational study. Lancet Oncol. 2011;12:559–567. doi: 10.1016/S1470-2045(11)70119-X. [DOI] [PubMed] [Google Scholar]
- 7.Nantasanti S, Toussaint MJ, Youssef SA, Tooten PC, de Bruin A. Rb and p53 liver functions are essential for xenobiotic metabolism and tumor suppression. PLoS One. 2016;11:e0150064. doi: 10.1371/journal.pone.0150064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sablina AA, et al. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shetzer Y, et al. The paradigm of mutant p53-expressing cancer stem cells and drug resistance. Carcinogenesis. 2014;35:1196–1208. doi: 10.1093/carcin/bgu073. [DOI] [PubMed] [Google Scholar]
- 10.Awasthi S, et al. Novel function of human RLIP76: ATP-dependent transport of glutathione conjugates and doxorubicin. Biochemistry. 2000;39:9327–9334. doi: 10.1021/bi992964c. [DOI] [PubMed] [Google Scholar]
- 11.Awasthi S, et al. Functional reassembly of ATP-dependent xenobiotic transport by the N- and C-terminal domains of RLIP76 and identification of ATP binding sequences. Biochemistry. 2001;40:4159–4168. doi: 10.1021/bi002182f. [DOI] [PubMed] [Google Scholar]
- 12.Awasthi S, et al. ATP-dependent human erythrocyte glutathione-conjugate transporter. II. Functional reconstitution of transport activity. Biochemistry. 1998;37:5239–5248. doi: 10.1021/bi972131r. [DOI] [PubMed] [Google Scholar]
- 13.Awasthi S, et al. ATP-dependent human erythrocyte glutathione-conjugate transporter. I. Purification, photoaffinity labeling, and kinetic characteristics of ATPase activity. Biochemistry. 1998;37:5231–5238. doi: 10.1021/bi972130z. [DOI] [PubMed] [Google Scholar]
- 14.Awasthi S, et al. Adenosine triphosphate-dependent transport of doxorubicin, daunomycin, and vinblastine in human tissues by a mechanism distinct from the P-glycoprotein. J Clin Invest. 1994;93:958–965. doi: 10.1172/JCI117102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Awasthi S, et al. RLIP76 is a major determinant of radiation sensitivity. Cancer Res. 2005;65:6022–6028. doi: 10.1158/0008-5472.CAN-05-0968. [DOI] [PubMed] [Google Scholar]
- 16.Awasthi S, et al. A central role of RLIP76 in regulation of glycemic control. Diabetes. 2010;59:714–725. doi: 10.2337/db09-0911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng JZ, et al. Accelerated metabolism and exclusion of 4-hydroxynonenal through induction of RLIP76 and hGST5.8 is an early adaptive response of cells to heat and oxidative stress. J Biol Chem. 2001;276:41213–41223. doi: 10.1074/jbc.M106838200. [DOI] [PubMed] [Google Scholar]
- 18.Leake K, Singhal J, Nagaprashantha LD, Awasthi S, Singhal SS. RLIP76 regulates PI3K/Akt signaling and chemo-radiotherapy resistance in pancreatic cancer. PLoS One. 2012;7:e34582. doi: 10.1371/journal.pone.0034582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma R, Singhal SS, Wickramarachchi D, Awasthi YC, Awasthi S. RLIP76 (RALBP1)-mediated transport of leukotriene C4 (LTC4) in cancer cells: Implications in drug resistance. Int J Cancer. 2004;112:934–942. doi: 10.1002/ijc.20516. [DOI] [PubMed] [Google Scholar]
- 20.Awasthi S, et al. RLIP76 and cancer. Clin Cancer Res. 2008;14:4372–4377. doi: 10.1158/1078-0432.CCR-08-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagaprashantha L, Vartak N, Awasthi S, Awasthi S, Singhal SS. Novel anti-cancer compounds for developing combinatorial therapies to target anoikis-resistant tumors. Pharm Res. 2012;29:621–636. doi: 10.1007/s11095-011-0645-9. [DOI] [PubMed] [Google Scholar]
- 22.Drake KJ, et al. RALBP1/RLIP76 mediates multidrug resistance. Int J Oncol. 2007;30:139–144. [PubMed] [Google Scholar]
- 23.Figarola JL, et al. COH-SR4 reduces body weight, improves glycemic control and prevents hepatic steatosis in high fat diet-induced obese mice. PLoS One. 2013;8:e83801. doi: 10.1371/journal.pone.0083801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singhal SS, et al. Regression of lung and colon cancer xenografts by depleting or inhibiting RLIP76 (Ral-binding protein 1) Cancer Res. 2007;67:4382–4389. doi: 10.1158/0008-5472.CAN-06-4124. [DOI] [PubMed] [Google Scholar]
- 25.Singhal SS, et al. RLIP76: A target for kidney cancer therapy. Cancer Res. 2009;69:4244–4251. doi: 10.1158/0008-5472.CAN-08-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singhal SS, et al. Glutathione-conjugate transport by RLIP76 is required for clathrin-dependent endocytosis and chemical carcinogenesis. Mol Cancer Ther. 2011;10:16–28. doi: 10.1158/1535-7163.MCT-10-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singhal SS, Yadav S, Drake K, Singhal J, Awasthi S. Hsf-1 and POB1 induce drug sensitivity and apoptosis by inhibiting Ralbp1. J Biol Chem. 2008;283:19714–19729. doi: 10.1074/jbc.M708703200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stuckler D, et al. RLIP76 transports vinorelbine and mediates drug resistance in non-small cell lung cancer. Cancer Res. 2005;65:991–998. [PubMed] [Google Scholar]
- 29.Yadav S, et al. Identification of membrane-anchoring domains of RLIP76 using deletion mutant analyses. Biochemistry. 2004;43:16243–16253. doi: 10.1021/bi0482811. [DOI] [PubMed] [Google Scholar]
- 30.Yang Y, et al. Cells preconditioned with mild, transient UVA irradiation acquire resistance to oxidative stress and UVA-induced apoptosis: Role of 4-hydroxynonenal in UVA-mediated signaling for apoptosis. J Biol Chem. 2003;278:41380–41388. doi: 10.1074/jbc.M305766200. [DOI] [PubMed] [Google Scholar]
- 31.Burg D, Mulder GJ. Glutathione conjugates and their synthetic derivatives as inhibitors of glutathione-dependent enzymes involved in cancer and drug resistance. Drug Metab Rev. 2002;34:821–863. doi: 10.1081/dmr-120015695. [DOI] [PubMed] [Google Scholar]
- 32.Brown MS, Goldstein JL. Receptor-mediated endocytosis: Insights from the lipoprotein receptor system. Proc Natl Acad Sci USA. 1979;76:3330–3337. doi: 10.1073/pnas.76.7.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sorkin A, von Zastrow M. Endocytosis and signalling: Intertwining molecular networks. Nat Rev Mol Cell Biol. 2009;10:609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jean S, et al. Extended-synaptotagmin-2 mediates FGF receptor endocytosis and ERK activation in vivo. Dev Cell. 2010;19:426–439. doi: 10.1016/j.devcel.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 35.Pinilla-Macua I, Sorkin A. Methods to study endocytic trafficking of the EGF receptor. Methods Cell Biol. 2015;130:347–367. doi: 10.1016/bs.mcb.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singhal SS, et al. Rlip76 transports sunitinib and sorafenib and mediates drug resistance in kidney cancer. Int J Cancer. 2010;126:1327–1338. doi: 10.1002/ijc.24767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsujimoto M, Yip YK, Vilcek J. Tumor necrosis factor: Specific binding and internalization in sensitive and resistant cells. Proc Natl Acad Sci USA. 1985;82:7626–7630. doi: 10.1073/pnas.82.22.7626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fillatre J, et al. Dynamics of the subcellular localization of RalBP1/RLIP through the cell cycle: The role of targeting signals and of protein-protein interactions. FASEB J. 2012;26:2164–2174. doi: 10.1096/fj.11-196451. [DOI] [PubMed] [Google Scholar]
- 39.Hinoi T, et al. Post-translational modifications of Ras and Ral are important for the action of Ral GDP dissociation stimulator. J Biol Chem. 1996;271:19710–19716. doi: 10.1074/jbc.271.33.19710. [DOI] [PubMed] [Google Scholar]
- 40.Jullien-Flores V, et al. RLIP76, an effector of the GTPase Ral, interacts with the AP2 complex: Involvement of the Ral pathway in receptor endocytosis. J Cell Sci. 2000;113:2837–2844. doi: 10.1242/jcs.113.16.2837. [DOI] [PubMed] [Google Scholar]
- 41.Kashatus DF, et al. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat Cell Biol. 2011;13:1108–1115. doi: 10.1038/ncb2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee S, Goldfinger LE. RLIP76 regulates HIF-1 activity, VEGF expression and secretion in tumor cells, and secretome transactivation of endothelial cells. FASEB J. 2014;28:4158–4168. doi: 10.1096/fj.14-255711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee S, Wurtzel JG, Singhal SS, Awasthi S, Goldfinger LE. RALBP1/RLIP76 depletion in mice suppresses tumor growth by inhibiting tumor neovascularization. Cancer Res. 2012;72:5165–5173. doi: 10.1158/0008-5472.CAN-12-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossé C, L’Hoste S, Offner N, Picard A, Camonis J. RLIP, an effector of the Ral GTPases, is a platform for Cdk1 to phosphorylate epsin during the switch off of endocytosis in mitosis. J Biol Chem. 2003;278:30597–30604. doi: 10.1074/jbc.M302191200. [DOI] [PubMed] [Google Scholar]
- 45.Wurtzel JG, et al. RLIP76 regulates Arf6-dependent cell spreading and migration by linking ARNO with activated R-Ras at recycling endosomes. Biochem Biophys Res Commun. 2015;467:785–791. doi: 10.1016/j.bbrc.2015.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prasanna PG, et al. Radioprotectors and radiomitigators for improving radiation therapy: The small business innovation research (SBIR) gateway for accelerating clinical translation. Radiat Res. 2015;184:235–248. doi: 10.1667/RR14186.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singhal J, Nagaprashantha L, Vatsyayan R, Awasthi S, Singhal SS. RLIP76, a glutathione-conjugate transporter, plays a major role in the pathogenesis of metabolic syndrome. PLoS One. 2011;6:e24688. doi: 10.1371/journal.pone.0024688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morris SM, et al. Effect of p53 genotype on gene expression profiles in murine liver. Mutat Res. 2008;640:54–73. doi: 10.1016/j.mrfmmm.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 49.Uren AG, et al. Large-scale mutagenesis in p19(ARF)- and p53-deficient mice identifies cancer genes and their collaborative networks. Cell. 2008;133:727–741. doi: 10.1016/j.cell.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goding CR, Pei D, Lu X. Cancer: Pathological nuclear reprogramming? Nat Rev Cancer. 2014;14:568–573. doi: 10.1038/nrc3781. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki H, Yamamoto E, Maruyama R, Niinuma T, Kai M. Biological significance of the CpG island methylator phenotype. Biochem Biophys Res Commun. 2014;455:35–42. doi: 10.1016/j.bbrc.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 52.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 53.Singhal J, et al. 2′-Hydroxyflavanone: A novel strategy for targeting breast cancer. Oncotarget. 2017;8:75025–75037. doi: 10.18632/oncotarget.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ames BN, Shigenaga MK, Gold LS. DNA lesions, inducible DNA repair, and cell division: Three key factors in mutagenesis and carcinogenesis. Environ Health Perspect. 1993;101:35–44. doi: 10.1289/ehp.93101s535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deferme L, et al. Dynamic interplay between the transcriptome and methylome in response to oxidative and alkylating stress. Chem Res Toxicol. 2016;29:1428–1438. doi: 10.1021/acs.chemrestox.6b00090. [DOI] [PubMed] [Google Scholar]
- 56.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 57.Shigenaga MK, Gimeno CJ, Ames BN. Urinary 8-hydroxy-2′-deoxyguanosine as a biological marker of in vivo oxidative DNA damage. Proc Natl Acad Sci USA. 1989;86:9697–9701. doi: 10.1073/pnas.86.24.9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Q, Martinez JD. P53 is transported into the nucleus via an Hsf1-dependent nuclear localization mechanism. Mol Carcinog. 2011;50:143–152. doi: 10.1002/mc.20713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu Y, Mivechi NF. HSF-1 interacts with Ral-binding protein 1 in a stress-responsive, multiprotein complex with HSP90 in vivo. J Biol Chem. 2003;278:17299–17306. doi: 10.1074/jbc.M300788200. [DOI] [PubMed] [Google Scholar]
- 60.Min JN, Huang L, Zimonjic DB, Moskophidis D, Mivechi NF. Selective suppression of lymphomas by functional loss of Hsf1 in a p53-deficient mouse model for spontaneous tumors. Oncogene. 2007;26:5086–5097. doi: 10.1038/sj.onc.1210317. [DOI] [PubMed] [Google Scholar]
- 61.Morinaka K, et al. Epsin binds to the EH domain of POB1 and regulates receptor-mediated endocytosis. Oncogene. 1999;18:5915–5922. doi: 10.1038/sj.onc.1202974. [DOI] [PubMed] [Google Scholar]
- 62.Nakashima S, et al. Small G protein Ral and its downstream molecules regulate endocytosis of EGF and insulin receptors. EMBO J. 1999;18:3629–3642. doi: 10.1093/emboj/18.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamaguchi A, Urano T, Goi T, Feig LA. An Eps homology (EH) domain protein that binds to the Ral-GTPase target, RalBP1. J Biol Chem. 1997;272:31230–31234. doi: 10.1074/jbc.272.50.31230. [DOI] [PubMed] [Google Scholar]
- 64.Enari M, Ohmori K, Kitabayashi I, Taya Y. Requirement of clathrin heavy chain for p53-mediated transcription. Genes Dev. 2006;20:1087–1099. doi: 10.1101/gad.1381906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Endo Y, et al. Regulation of clathrin-mediated endocytosis by p53. Genes Cells. 2008;13:375–386. doi: 10.1111/j.1365-2443.2008.01172.x. [DOI] [PubMed] [Google Scholar]
- 66.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li R, Li Y, Kristiansen K, Wang J. SOAP: Short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 68.Hahn MA, Li AX, Wu X, Pfeifer GP. Cancer Epigenetics. Methods Mol Biol. 2015;1238:273–287. doi: 10.1007/978-1-4939-1804-1_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krueger F, Andrews SR. Bismark: A flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li LC, Dahiya R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.