Significance
Discovering where a gene acts, when it is required, and the consequence of its perturbation are essential for understanding gene function. These tasks require genetic tools that allow precise control of gene expression at will. In this paper, we combine a split intein strategy and a bipartite expression system to develop a refined method of controlling transgene expression in Caenorhabditis elegans. The new system specifies transgene expression only in cells where two distinct promoters are active, allowing precise spatiotemporal control. Our work provides the C. elegans community with a new genetic tool for precise control of transgene expression. We speculate that a similar split intein strategy could also be applied to other bipartite expression systems in other genetic model organisms.
Keywords: split cGAL, intein, GAL4-UAS, Caenorhabditis elegans, PKA
Abstract
Bipartite expression systems, such as the GAL4-UAS system, allow fine manipulation of gene expression and are powerful tools for interrogating gene function. Recently, we established cGAL, a GAL4-based bipartite expression system for transgene control in Caenorhabditis elegans, where a single promoter dictates the expression pattern of a cGAL driver, which then binds target upstream activation sequences to drive expression of a downstream effector gene. Here, we report a split strategy for cGAL using the split intein gp41-1 for intersectional control of transgene expression. Split inteins are protein domains that associate, self-excise, and covalently ligate their flanking peptides together. We split the DNA binding domain and transcriptional activation domain of cGAL and fused them to the N terminal of gp41-1-N-intein and the C terminal of gp41-1-C-intein, respectively. In cells where both halves of cGAL are expressed, a functional cGAL driver is reconstituted via intein-mediated protein splicing. This reconstitution allows expression of the driver to be dictated by two promoters for refined spatial control or spatiotemporal control of transgene expression. We apply the split cGAL system to genetically access the single pair of MC neurons (previously inaccessible with a single promoter), and reveal an important role of protein kinase A in rhythmic pharyngeal pumping in C. elegans. Thus, the split cGAL system gives researchers a greater degree of spatiotemporal control over transgene expression, and will be a valuable genetic tool in C. elegans for dissecting gene function with finer cell-specific resolution.
A fundamental goal of biology is to understand how an organism uses its full complement of genes to determine its development, morphology, cellular and tissue functions, and behaviors. Each gene may act in different cells and at different times for various biological processes. Thus, genetic tools that enable precise control of gene expression both spatially and temporally are extremely valuable for dissecting gene function. With its powerful genetics and small size, Caenorhabditis elegans is an important genetic model for studying various biological processes and has contributed to the understanding of fundamental mechanisms underlying biology (1). While a variety of tissue- and cell-specific promoters have long been available to the C. elegans community, genetic access for each individual cell type, especially each anatomical neuron type, has not yet been achieved. Providing this type of access would allow much finer resolution of genetic analysis, accelerating full dissection of gene function and understanding of the biology of the worm.
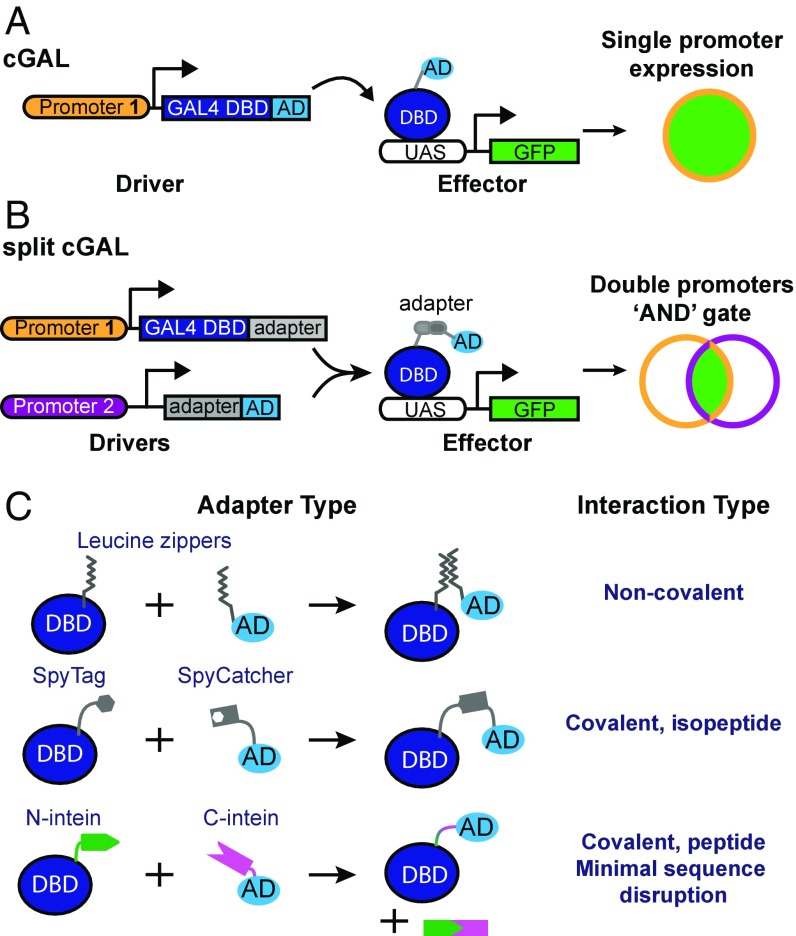
We recently developed cGAL, a GAL4-based bipartite expression system, for controlling transgene expression in C. elegans (2). As with other bipartite expression systems in which a driver specifies the expression pattern of the transgene and an effector dictates the nature of the transgenic perturbation, the cGAL system uses the DNA binding domain (DBD) from a cryophilic yeast strain Saccharomyces kudriavzevii and the synthetic VP64 activation domain (AD) for the driver. The cGAL driver triggers expression of the effector gene by binding upstream activation sequence (UAS) sites only in cells in which the promoter used in the driver construct is active (Fig. 1A).
Fig. 1.
Schematic of cGAL and split cGAL strategies. (A) The original cGAL bipartite system, in which a single promoter governs expression of the cGAL driver (2). The driver is composed of the DBD from S. kudriavzevii Gal4p, which recognizes UAS, and a transcriptional AD, which recruits transcriptional machinery. The cGAL driver then specifies expression of the effector gene (i.e., GFP), under the control of UAS, in cells where the promoter is active. (B) Using split strategies, two promoters can be used, providing an “AND” gate to achieve intersectional control of transgene expression. (C) Splitting the driver components requires a way to reconstitute the split components in cells expressing both. Leucine zippers allow for noncovalent reconstitution of the DBD and AD. The SpyTag/SpyCatcher domains reconstitute via covalent formation of an isopeptide bond. Split intein domains recognize one another and associate, after which they covalently ligate the flanking sequences and self-excise.
However, the extent of transgene control with cGAL in C. elegans is limited by available promoters, because the expression pattern is dictated by the single promoter used in each driver. In particular, the majority of neurons in C. elegans are not genetically accessible with single promoters (3), which hinders our understanding of the functional importance of different genes and neurons for different behaviors. Furthermore, it is generally impossible to achieve spatial and temporal regulation of transgene expression at the same time using a single promoter in cGAL drivers. Here, we addressed these limitations of the original cGAL system by designing an intersectional split cGAL strategy that provides an “AND” gate for refined transgene control using two distinct promoters (Fig. 1B).
Deletion mutant studies in yeast analysis showed that the original Gal4p protein from Saccharomyces cerevisiae has two functional modules: the DBD and transcriptional AD (4, 5). Independently, neither is sufficient to drive the expression of the effector gene downstream of UAS sites. Luan et al. (6) took advantage of this modular independence and designed a split GAL4 system for the Drosophila community by fusing the DBD and AD to one half of an antiparallel leucine zipper adapter pair, and putting the fusions under the control of two different promoters. With this design, only in cells where both promoters are active would both components be expressed, and the two antiparallel leucine zipper adapters allow the DBD and AD to associate via noncovalent interactions to reconstitute a functional GAL4 driver (6) (Fig. 1C). This “split” system gives spatially restricted expression of GAL4 in cells at the intersection of two promoters, and has made split GAL4 a powerful tool for Drosophila researchers to precisely control transgene expression, particularly in the nervous system (6, 7).
Since the introduction of the split GAL4 system in Drosophila, new adapter protein domains that mediate covalent interactions have been discovered. One example is the SpyTag/SpyCatcher system, an adapter system engineered from Streptococcus pyogenes (8). When proteins are tagged with this adapter pair, SpyTag/SpyCatcher will associate and form a covalent isopeptide bond, uniting their protein partners (8).
Another example is a class of protein domains called inteins. The first intein was characterized in the yeast VMA1 gene as an internal portion of the protein (the “intein”) that was capable of simultaneously self-excising and mediating intramolecular ligation of the two flanking sequences (termed “exteins”) in cis (9–11). Later, sequence analysis in cyanobacteria revealed the presence of split inteins, which could mediate protein splicing in trans (12, 13). Here, two separate genes encode two peptide products, each having one extein and one-half of the split intein. The two peptides associate via their split intein domains and undergo intermolecular protein splicing, excising the two split intein domains and fusing the two exteins via a peptide bond (9). The split inteins gp41-1 and Npu DnaE are among the most robust and fastest described in the literature (14, 15). However, neither SpyTag/SpyCatcher nor these split inteins have been tested in assembling a functional GAL4 driver from the DBD and AD.
With the sole exception of split intein gp41-1, the other three protein adapters described above have been reported to successfully associate proteins in C. elegans (3, 16–19). To establish a robust split cGAL system, we systematically compared the efficiency of all four adapters in reassociating the DBD and AD to reconstitute a functional cGAL driver (Fig. 1 B and C). We determined that the gp41-1 split intein is the best adapter for our split cGAL system. We also show that split cGAL allows simultaneous spatiotemporal control or refined spatial control of transgene expression in C. elegans with two different promoters. Finally, we applied our split cGAL system to reveal a critical role of protein kinase A (PKA) in the single pair of cholinergic MC pharyngeal neurons in the feeding behavior of C. elegans.
Results
Comparing Protein Adapter Domains for Reconstitution of the Split cGAL Driver.
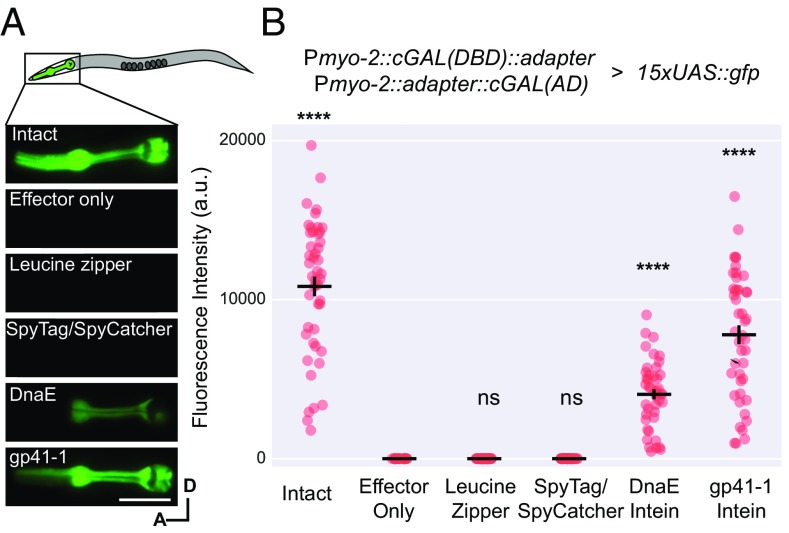
To construct our split system, we first wished to test the ability of different adapters to reconstitute a functional cGAL driver from its two modular halves (DBD and AD). We separated our cGAL driver into its modular components and appended one of four different adapters (antiparallel leucine zipper, SpyTag/SpyCatcher, Npu DnaE split intein, and gp41-1 split intein) (Fig. 1C). One half, the cGAL(DBD)-adapter, contained the DBD from S. kudriavzevii and an adapter domain; the other half, adapter-cGAL(AD), contained the cognate adapter domain and the VP64 activation domain (Fig. 1B). After placing each gene fusion under the control of a pharyngeal muscle-specific promoter (myo-2 promoter), we then injected each pair of split cGAL-adapter constructs together at equal concentrations into a transgenic strain with an integrated 15xUAS::gfp effector [unc-119 (ed3); syIs300] and performed quantitative fluorescence imaging to assess GFP levels in pharyngeal muscles (Fig. 2).
Fig. 2.
The gp41-1 intein is most efficient in reconstituting split cGAL components. (A) Diagram of the pharynx region, representative fluorescence images, and (B) quantification of animals with intact cGAL driver, GFP effector only, or the indicated split cGAL driver pairs. The intact cGAL and effector only serve as positive and negative controls. Two independent extrachromosomal transgenic lines were assayed for all groups except the effector alone control, which had only one. a.u., artificial units. Bars are mean ± SEM. From left to right, n = 47, 23, 41, 42, 44, 45. ****P < 0.0001, one-way ANOVA and Dunnett’s multiple comparisons test to compare the means to the mean of the effector alone. A, anterior; D, dorsal. ns, not significant. (Scale bar, 20 μm.)
To our surprise, we found that neither the antiparallel leucine zipper nor the SpyTag/SpyCatcher could reconstitute the split cGAL(DBD) and cGAL(AD) to drive expression of GFP in pharyngeal muscles (Fig. 2) (not statistically significant compared with effector only, one-way ANOVA with Tukey’s correction, P > 0.9999), although both adapters have been shown to bring together other proteins successfully in C. elegans (3, 16, 17, 20). In contrast, both intein adapters restored the transcriptional activity of split cGAL (Fig. 2) (statistically significant compared with effector only, one-way ANOVA with Tukey’s correction, P < 0.0001). Split cGAL with the DnaE and gp41-1 adapters achieved 37% and 72% of transcriptional activator activity of the intact cGAL, respectively (Fig. 2B). The gp41-1 intein in particular showed the brightest and most robust expression.
To rule out the possibility that the high level of GFP expression observed with gp41-1–mediated split cGAL is due to recombination of the injected DNA constructs that might have generated an intact cGAL driver fragment in the extrachromosomal array, we injected and integrated each split cGAL driver transgene separately. When individually crossed to the GFP effector (syIs300), neither half produced fluorescence (Fig. S1). We only observed GFP fluorescence in the cross progeny containing both cGAL halves and the GFP effector transgene, demonstrating that split drivers are essential for driving the expression of the effector (Fig. S1).
To test whether successful reconstitution of gp41-1–mediated split cGAL is dependent on protein splicing, we mutated the first cysteine of gp41-1 N-intein to alanine [referred to as cGAL(DBD)-gp41-1-N-intein (C1A)]. Because this cysteine is essential for jointing two exteins (14), we predicted that cGAL(DBD)-gp41-1-N-intein (C1A) would not be able to join cGAL(DBD) and cGAL(AD) together. Indeed, we found that unlike wild-type Pmyo-2::cGAL(DBD)-gp41-1-N-intein (mean 15,218.0 ± SEM 722.3), the mutated Pmyo-2::cGAL(DBD)-gp41-1-N-intein (C1A) almost completely abolished GFP expression in pharyngeal muscles (mean 97.3 ± SEM 13.6 vs. negative control 12.0 ± 1.0) when injected into a strain with both Pmyo-2:: gp41-1-C-intein-cGAL(AD) and 15xUAS::gfp (Fig. S2). The C1A mutant driver produced GFP levels that were barely visible under standard dissecting fluorescence microscopy. Based on these results, we concluded that, among the adapters that we tested, the split cGAL system using gp41-1–mediated protein splicing is most effective for reconstituting a functional cGAL in C. elegans. From this point onwards, we will refer to cGAL(DBD)-gp41-1-N-intein as cGAL-N, and to gp41-1-C-intein-cGAL(AD) as cGAL-C, unless stated otherwise.
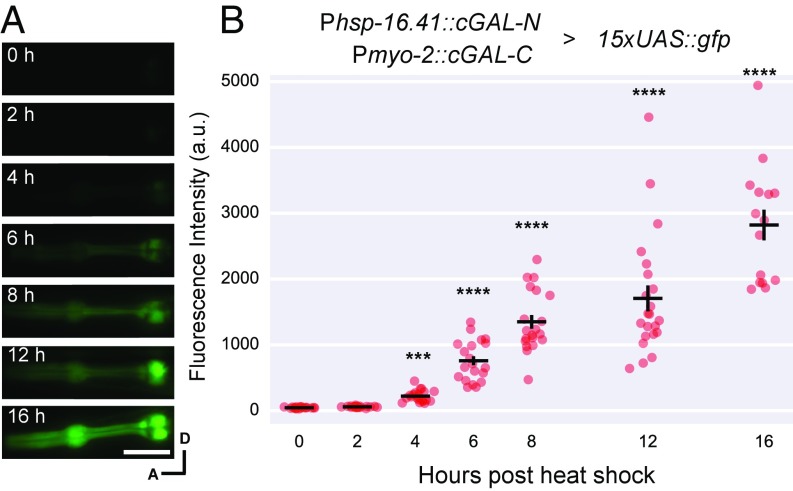
Spatial and Temporal Control with Split cGAL.
During development, genes are turned on at different times to perform their functions. The determination of critical time windows for such genes requires genetic tools that provide temporal control of transgene expression. The use of heat-shock promoters is a common way to impart temporal control but it sacrifices spatial control; conversely, tissue-specific promoters in transgenes generally cannot provide temporal control at the same time. We explored the possibility that the split cGAL system could simultaneously achieve spatial and temporal control of transgene expression. As we reported previously, cGAL driver constructs in the Fire vector pPD117.01 containing the let-858 3′UTR are more robustly expressed than in the Fire vector pPD49.26 with the unc-54 3′UTR (2). Thus, we built new split cGAL drivers in the backbone with the let-858 3′UTR. We used a heat-shock promoter (hsp-16.41 promoter) to drive the expression of cGAL-N, and the constitutive pharyngeal muscle promoter (myo-2 promoter) to drive the expression of cGAL-C. In cross progeny that were triple heterozygotes for Phsp-16.41::cGAL-N, Pmyo-2::cGAL-C, and 15xUAS::gfp, no GFP expression in pharyngeal muscles was observed without heat shock (Fig. 3). Starting 4 h after heat-shock treatment (33 °C for 1 h), we observed a steady increase of GFP expression in pharyngeal muscles all of the way up to 16 h after heat shock, the last time point that we assayed (Fig. 3). The induction of the GFP effector seemed to be slow, relative to that from a direct heat-shock promoter::GFP fusion (Discussion). Furthermore, we also showed that the conditional expression of the GFP effector in pharyngeal muscles after heat shock required both Phsp-16.41 and Pmyo-2 split cGAL drivers (Fig. S2).
Fig. 3.
Using split cGAL for spatiotemporal control of gene expression. (A) Representative fluorescence images and (B) quantification of fluorescence in the pharynx of animals that were triple heterozygotes for a conditional cGAL-N driver with the heat shock promoter (syIs435), a tissue-specific cGAL-C driver with the myo-2 promoter (syIs433) and a 15xUAS::GFP effector (syIs300). a.u., artificial units. Bars are mean ± SEM. Each column is a separate group of animals that were imaged at the indicated time point after heat shock, n = 21, 20, 21, 20, 20, 22, and 15, from left to right. ***P < 0.001 and ****P < 0.0001, one-way ANOVA and Dunnett’s multiple comparisons test to compare the means to the mean of no heat-shock control. cGAL-N and cGAL-C represent the two halves of the gp-41-1–mediated split cGAL driver. A, anterior; D, dorsal. (Scale bar, 20 μm.)
After heat shock, we observed noticeable background GFP expression in the excretory cell in animals containing Phsp-16.41::cGAL-N and 15xUAS::gfp, but not in those containing Pmyo-2::cGAL-C and 15xUAS::gfp (Fig. S3A). In the presence of the 15xUAS::gfp effector, this ectopic expression of GFP in the excretory canal cell was also observed in worms carrying the cGAL-N split driver under control of the ubiquitous eft-3 promoter but not those with myo-2 promoter (pharyngeal muscle promoter), rab-3 promoter (pan-neuronal promoter), or unc-17 promoter (cholinergic neurons) (Fig. S3B), suggesting that the cGAL-N split driver may interact with an unknown transcriptional activator that is specifically expressed in the excretory cell and thus can nonspecifically drive the effector gene in this cell. We believe this effect is limited to only the cGAL-N half, as a strain carrying Peft-3::cGAL-C; 15xUAS::gfp does not display ectopic GFP expression in the excretory cell (five independent lines). Thus, if the ectopic expression in excretory cell of the promoter in the cGAL-N driver is a concern, the promoter can be swapped to drive cGAL-C instead.
Refined Spatial Control with Split cGAL.
C. elegans has 302 neurons in the adult hermaphrodite and 385 neurons in the adult male (21, 22). Despite the relative simplicity of these nervous systems, however, many anatomical neuron types cannot be genetically approached using single promoters. As the expression pattern of many C. elegans genes are well-characterized, it has been suggested that most anatomical neuron types in C. elegans can be genetically accessed with the intersection of two different promoters (3).
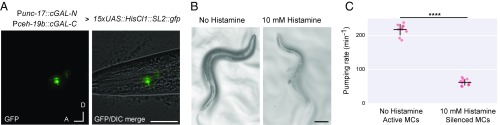
We wanted to determine if gp41-1–mediated split cGAL could be used as an “AND” gate to spatially restrict transgene expression with two overlapping promoters (Fig. 1B). As a test case, we chose to examine the regulation of pharyngeal pumping by the paired MC pharyngeal neurons for several reasons. Previous work with laser ablation showed that MC neurons are the major excitatory neurons for fast pumping, and their ablation resulted in significantly reduced pumping and growth rate (23, 24). However, there were no previously described single promoters that gave specific access to this neuron type. Thus, we chose to design split cGAL drivers to genetically access the MC pharyngeal neurons. Moreover, we could validate our MC driver by using our existing neuronal effector strain kit (2) to silence MC neurons and look for phenocopying of the MC-ablated animals.
The unc-17 and ceh-19b promoters are proposed to specifically overlap in the MC neurons (www.wormweb.org/neuralnet). We made two split cGAL constructs, Punc-17::cGAL-N and Pceh-19b::cGAL-C, and injected both together in an integrated HisCl1 effector line (syIs371, 15xUAS::HisCl1::SL2::gfp). HisCl1 encodes a histamine-gated chloride channel, capable of silencing neurons when histamine is applied (25). As expected, we observed bright green fluorescence in the pair of MC neurons (Fig. 4A), suggesting the two split cGAL components successfully reconstituted and drove the expression of both HisCl1 and GFP in MC neurons. However, we also found an additional pair of neurons with weaker GFP fluorescence, likely to be the sensory ADF neurons, as both unc-17 and ceh-19b were also reported to be expressed in ADF (26, 27) (Fig. S4).
Fig. 4.
Using split cGAL for cell-specific expression and perturbation of MC neurons. (A) Representative images showing specific GFP labeling of bilateral MC motor neurons with the combination of two split cGAL driver constructs using unc-17 and ceh-19b promoters. As indicated, each promoter drives one of the split cGAL components. Coinjection and integration of the components (syIs483) is capable of specifically driving a 15xUAS::HisCl1::SL2::gfp effector (syIs371) in MC neurons. A, anterior; D, dorsal. (Scale bar, 20 μm.) (B) Light microscopy of syIs483; syIs371 animals. In the absence of histamine, MC neurons retain their activity and produce pigmented, healthy adults (Left). Raising animals on 10 mM histamine (Right) activates the syIs371 effector to chronically silence the MC neurons, reduces pumping, and produces unhealthy animals with decreased size and pigmentation. (Scale bar, 100 μm.) (C) Quantification of pumping rate of syIs483; syIs371 animals with or without histamine. Each column represents a separate group of animals with indicated treatments. Bars are mean ± SEM n = 10, 11 from left to right. ****P < 0.0001, unpaired Student’s t test. cGAL-N and cGAL-C represent the two halves of the gp-41-1–mediated split cGAL driver.
To functionally validate the MC split cGAL driver, we silenced MC neurons expressing the HisCl1 channel by exposing animals to 10 mM histamine and quantified pharyngeal pumping rate. Animals with both the MC split cGAL driver and the HisCl1 effector raised on 10 mM histamine from hatching grew up to be thinner and less pigmented than counterparts raised in the absence of histamine (Fig. 4B). Those animals treated with histamine also pumped much slower (62.3 ± 2.8 pumps/min, mean ± SEM, n = 11), compared with the worms of the same genotype but not treated with histamine (217.2 ± 5.3 pumps/min, mean ± SEM, n = 10) (Fig. 4C). We also found that neither half of the split MC driver alone was sufficient to inhibit pumping in the presence of the HisCl1 effector and histamine (Fig. S6). This result is consistent with previous observations in worms with laser-ablated MC neurons (23, 24).
Regulation of Pharyngeal Pumping by PKA in C. elegans.
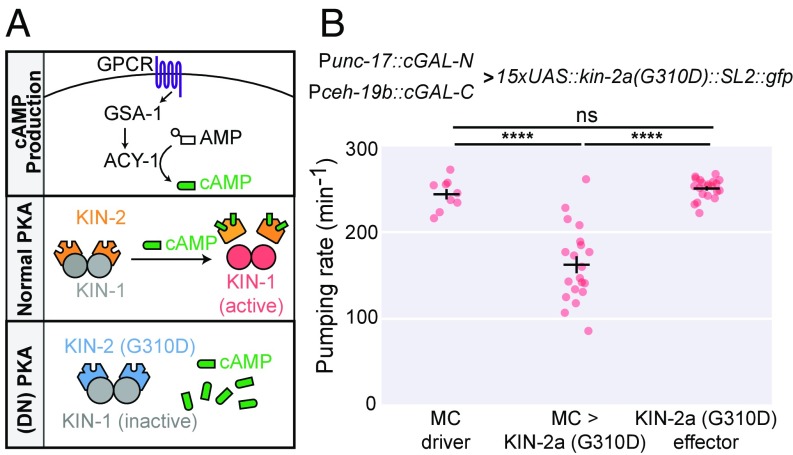
PKA is one of the major targets of the second messenger cyclic adenosine monophosphate (cAMP) (28). The PKA holoenzyme is a tetramer, consisting of two catalytic subunits and two regulatory subunits. In the absence of cAMP, the kinase activity of the catalytic subunits is inhibited by the regulatory subunits. When cAMP levels increase, cAMP binds to the regulatory subunit, leading to its dissociation from the catalytic subunit and subsequent disinhibition of PKA (28) (Fig. 5A). In C. elegans, the catalytic and regulatory subunits of PKA are encoded by kin-1 and kin-2, respectively. Null mutants for both kin-1 and kin-2 are lethal (29, 30), preventing detailed genetic analysis of PKA signaling in C. elegans.
Fig. 5.
Dominant-negative inhibition of PKA signaling in MC neurons reduces pharyngeal pumping rate. (A) Diagram of PKA signaling. Ligand binding to a G protein-coupled receptor (GPCR) activates the Gαs subunit GSA-1. GSA-1 goes on to activate adenylyl cyclases (i.e., ACY-1), causing conversion of adenosine monophosphate (AMP) to cAMP (Top). In wild-type signaling, cAMP dissociates the inhibitory KIN-2 subunits from the catalytic KIN-1 subunits, leading to the activation of PKA (Middle). The G310D mutation of KIN-2 is essentially insensitive to cAMP and remains associated with KIN-1, causing dominant-negative PKA [(DN) PKA] (Bottom). (B) Quantification of pumping rate in animals expressing the dominant-negative KIN-2a(G310D) in the MC neurons. Here, the 15xUAS::kin-2a(G310D)::SL2::gfp effector is an extrachromosomal array, and two independent lines were used (syEx1596 and syEx1597). The MC driver is an integrated transgene (syIs483) with both Punc-17::cGAL-N and Pceh-19b::cGAL-C. Bars are mean ± SEM n = 9, 20, and 21 from left to right. ****P < 0.0001, one-way ANOVA and Tukey’s multiple comparisons test to compare all three means. ns, not significant. cGAL-N and cGAL-C represent the two halves of the gp-41-1–mediated split cGAL driver. ns, not significant.
Genetic studies using partial loss-of-function kin-2 mutants revealed that PKA signaling in the nervous system is involved in the regulation of pharyngeal pumping in C. elegans (30, 31). Because the MC neurons are the major excitatory motor neurons for pharyngeal pumping (23, 24), we hypothesized that normal PKA activity in the MC neurons is necessary for rapid pumping. To test this hypothesis, we used the split cGAL system to block PKA activity specifically in MC neurons. We first created an effector strain with an extrachromosomal array of 15xUAS::kin-2a(G310D)::SL2:::gfp. The G310D mutation in isoform a of the regulatory subunit KIN-2 prevents its binding with cAMP, thereby maintaining its inhibitory interaction with the catalytic subunit KIN-1 even when cAMP is elevated, and produces a dominant negative form of PKA (32, 33) (Fig. 5A). Neither this effector strain nor the split cGAL driver strain for MC neurons showed any defect in pharyngeal pumping rate. However, cross progeny from these two parent strains displayed a 33% decrease in pharyngeal pumping rate (162.3 ± 9.8 pumps/min vs. 243.6 ± 5.9 and 250.3 ± 2.5 pumps/min for driver and effector alone, respectively; mean ± SEM) (Fig. 5B). These results support the conclusion that PKA signaling in MC neurons is essential for normal fast pharyngeal pumping in C. elegans.
Discussion
In this study, we describe our development of a split cGAL system using the intein gp41-1 to mediate protein splicing and produce a transcriptionally competent cGAL driver from its split components (DBD and AD). We demonstrate that split cGAL can achieve refined spatial and spatiotemporal control of transgene expression in C. elegans using two separate promoters. We also build a cell-type–specific split cGAL driver to specifically manipulate PKA activity in the MC pharyngeal neurons, and discover that inhibiting the PKA pathway in MC neurons results in a decrease in pumping rate.
To engineer the split cGAL system, we experimented with four methods of reconstituting cGAL DBD and AD, and determined that among the four adapters tested, gp41-1 is the most effective, recapitulating over 70% of the intact cGAL driver’s performance (Fig. 2B). Several reasons may explain this. First, gp41-1 brings the DBD and AD together with a canonical peptide bond. Second, the kinetics of gp41-1–mediated protein splicing are fast, about 10 times faster than Npu DnaE (14). This may explain why gp41-1 outperformed DnaE (Fig. 2B). Third, similar to DnaE, gp41-1 excises itself and leaves a minimal peptide sequence between DBD and AD of the reconstituted cGAL after protein splicing. Reconstitution of cGAL using the leucine zipper or SpyTag/SpyCatcher results in larger extraneous protein domains between the DBD and AD, likely leading to spatial and steric constraints with negative functional consequences. Compared with the intact cGAL driver, our split constructs with the DnaE and gp41-1 inteins have additional 13 and 14 amino acids between the cGAL DBD and AD, in contrast to 80 and 126 amino acids for the leucine zipper and SpyTag/SpyCatcher. This, together with other factors, such as weak association and poor kinetics, may account for the failure of leucine zipper and SpyTag/SpyCatcher to successfully reconstitute split cGAL. In support of this explanation, Luan et al. (6) observed a 48% reduction in function of the S. cerevisiae GAL4 transcriptional activator when split with the leucine zipper in Drosophila (6). Our results suggest that the gp41-1 intein would be an excellent tool for other aspects of protein engineering for C. elegans requiring protein splicing. For example, a similar split Q bipartite system using the same leucine zipper pair as we tested was also described in C. elegans (20). It would be interesting to determine if the gp41-1 intein can boost the performance of the split Q system.
Although gp41-1–mediated protein splicing is fast (14), expressing a gene using split cGAL is likely to introduce a temporal delay, compared with a direct heat-shock promoter::gene fusion. The heat-shock promoter must first drive expression of one half of the cGAL driver, which then must undergo protein splicing with the other cGAL driver half before driving the expression of the effector gene. However, this delay comes at the benefit of adding spatial control of the heat-shock promoter, should the experimenter need conditional expression in a restricted subset of cells where the hsp-16.41 promoter is active. The gp41-1–mediated protein splicing rate in vivo is likely to be dependent on the concentration and stoichiometry of both split cGAL components within the cell, as well as temperature. This complexity has unique implications for using the split cGAL system to achieve spatiotemporal control of gene expression. For example, the temperature and the duration of the heat-shock protocol to induce transgene expression will influence the timing and the expression level of the split cGAL component under the control of the heat-shock promoter. The cellular environment of different cell types may also influence the time scale of the intein-mediated protein splicing. Thus, in studies where timing of gene expression is a critical factor, we recommend characterizing the temporal dynamics of a split driver combination with GFP or any other fluorescently tagged effector.
Our study on PKA in MC neurons highlights the importance of genetic tools that allow highly refined spatial control of gene activity, which ultimately will help in understanding the cell-specific roles of genes. PKA has not been reported in forward genetic screens for mutants that are defective in pumping, likely due to the fact that the null alleles of kin-1 and kin-2 are lethal. When studying lethal or toxic alleles, expression often must be limited to a small subset of cells. Precise and systemic expression of these alleles is most efficiently achieved with bipartite systems. We used a split cGAL driver and a dominant-negative PKA effector and showed that PKA activity in MC neurons is necessary for normal pumping. This finding is in line with a previous observation that serotonin potentiates pumping rate by activating the G protein-coupled receptor SER-7 and Gαs signaling to increase cholinergic transmission from MC neurons (30). PKA may regulate pumping rate by modulating the firing rate of MC neurons or controlling the release of acetylcholine from MC neurons.
With its precision of transgene control, we expect that split cGAL will be particularly useful in providing genetic access to cell types that could not be accessed before, such as many of the individual anatomical neuron types in C. elegans. Besides providing highly specific genetic targeting, split cGAL can be used to perturb gene activity and cellular processes with great efficiency, because split cGAL drivers can be reused in combination with UAS effectors by crossing. For example, with our recently published UAS effector toolkit that contains effector strains to manipulate and record neuronal activity (2), any new split cGAL drivers for a neuron type can be crossed with these effector strains to interrogate the function of the neuron in a relevant behavior. Similarly, if new UAS effector strains are generated (i.e., new reporters or interesting alleles of native C. elegans genes), they can be crossed to currently available split cGAL drivers to test gene function in cells of interest. As more strains are built, documented, and described, they will contribute to a growing repository of tried and true reagents available to the community at large for extensive, rigorous, and rapid analysis of neural circuits and gene function in C. elegans. Furthermore, split cGAL can also be combined with several other systems that have been developed for spatiotemporal control of transgene expression in C. elegans (20, 34–40) to further improve the precision of transgene expression or achieve orthogonal control of different transgenes to interrogate gene function.
Materials and Methods
Strains.
The C. elegans strains were maintained at 20 °C, as previously described (41). All of the strains used in this study are described in detail in SI Appendix.
Molecular Biology.
All plasmids were constructed in the worm expression vectors pPD49.26 or pPD117.01 from the Fire kit (Addgene). All constructs were generated by standard molecular cloning procedures with restriction digest, PCR, and Gibson assembly or T4 ligation. The coding sequences in the constructs were verified by Sanger sequencing. The complete list of the plasmids and oligos used in this study are listed in Tables S1–S3.
Transgenic Animals.
The standard microinjection procedure for C. elegans was used to generate transgenic worms with extrachromosomal arrays, some of which were then integrated into the genome using X-ray treatment (42). The concentrations and compositions of DNA constructs in the injection mixtures of the transgenic worms are described in SI Appendix.
Fluorescence Imaging.
Worms were paralyzed in M9 buffer supplemented with 30 mg/mL of 2,3-Butanedione monoxime (Sigma). All fluorescent images for quantification of GFP fluorescence in the pharynx were taken with a Leica DMI6000 inverted microscope equipped with a 40× oil objective and an Andor iXon Ultra 897 EMCCD camera, using Metamorph software (Molecular Devices). A region of interest outlining the entire pharynx, as well as a background region of interest, was selected for each image. The background-subtracted mean fluorescence intensity was used to quantify the GFP fluorescence in the pharyngeal muscles of each worm.
Pumping Analysis.
For the histamine experiments, animals were raised from eggs on regular NGM plates or NGM plates with10 mM histamine dihydrochloride (Sigma), seeded with 150 μL of OP50 bacteria. Gravid animals were bleached and their eggs were transferred to corresponding plates. Animals were assayed 72 h later.
For the dominant-negative kin-2 experiments, L4 animals were picked on regular NGM plates seeded with OP50 bacteria. The next day, each adult was transferred to a new NGM plate seeded with OP50 and allowed to acclimate for 10 min before assaying.
For both experiments, each worm was recorded for 1 min under a Wild Makroskop M420 dissecting microscope equipped with a Unibrain 501b camera. The pumping rate for each worm was determined as total pumping events over the 1-min recording.
Heat-Shock Treatment.
L4 cross progeny were picked 1 d before being placed onto new NGM plates. The next day, plates were sealed with Parafilm and put in a 33 °C water bath for 1 h with the agar side down. After heat shock, worms were recovered at room temperature and imaged at different times after heat shock.
Statistical Analysis.
All of the quantification plots were made using custom written Python scripts in Jupyter Notebook (43). Unpaired Student’s t test and one-way ANOVA with Tukey’s or Dunnett’s tests (GraphPad Prism) were applied when appropriate, as indicated in the figure legends.
Supplementary Material
Acknowledgments
We thank D. Tirrell and H. Schwartz for insightful discussion; S. J. Walton and S. Gharib for technical help; members of the P.W.S. laboratory and H. Chiu for editorial comments on the manuscript; and V. Gradinaru and D. Sieburth for sharing reagents. This work was supported by National Institute of Mental Health Grant R21MH115454 (to P.W.S.); National Institute of General Medical Sciences Grant K99GM126137 (to H.W.); a Della Martin Fellowship in Mental Illness (to H.W.); and the Howard Hughes Medical Institute, with which P.W.S. was an investigator. J.L. and A.J.H. were supported by National Institute of General Medical Sciences training Grant T32GM007616, and K.P.Y. was supported by U.S. Army Research Office, Institute for Collaborative Biotechnologies Grant W911NF-09-0001.
Footnotes
The authors declare no conflict of interest.
Data deposition: Some of the plasmids used in this study have been deposited in Addgene (catalog nos. 107130–107137, and 107139, available at https://www.addgene.org/Paul_Sternberg/).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720063115/-/DCSupplemental.
References
- 1.Corsi AK, Wightman B, Chalfie M. A transparent window into biology: A primer on Caenorhabditis elegans. Genetics. 2015;200:387–407. doi: 10.1534/genetics.115.176099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang H, et al. cGAL, a temperature-robust GAL4-UAS system for Caenorhabditis elegans. Nat Methods. 2017;14:145–148. doi: 10.1038/nmeth.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chelur DS, Chalfie M. Targeted cell killing by reconstituted caspases. Proc Natl Acad Sci USA. 2007;104:2283–2288. doi: 10.1073/pnas.0610877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keegan L, Gill G, Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science. 1986;231:699–704. doi: 10.1126/science.3080805. [DOI] [PubMed] [Google Scholar]
- 5.Ma J, Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987;48:847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- 6.Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006;52:425–436. doi: 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venken KJT, Simpson JH, Bellen HJ. Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron. 2011;72:202–230. doi: 10.1016/j.neuron.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zakeri B, et al. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci USA. 2012;109:E690–E697. doi: 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah NH, Muir TW. Inteins: Nature’s gift to protein chemists. Chem Sci (Camb) 2014;5:446–461. doi: 10.1039/C3SC52951G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kane PM, et al. Protein splicing converts the yeast TFP1 gene product to the 69-kD subunit of the vacuolar H(+)-adenosine triphosphatase. Science. 1990;250:651–657. doi: 10.1126/science.2146742. [DOI] [PubMed] [Google Scholar]
- 11.Hirata R, et al. Molecular structure of a gene, VMA1, encoding the catalytic subunit of H(+)-translocating adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J Biol Chem. 1990;265:6726–6733. [PubMed] [Google Scholar]
- 12.Wu H, Hu Z, Liu XQ. Protein trans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803. Proc Natl Acad Sci USA. 1998;95:9226–9231. doi: 10.1073/pnas.95.16.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwai H, Züger S, Jin J, Tam P-H. Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. FEBS Lett. 2006;580:1853–1858. doi: 10.1016/j.febslet.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 14.Carvajal-Vallejos P, Pallissé R, Mootz HD, Schmidt SR. Unprecedented rates and efficiencies revealed for new natural split inteins from metagenomic sources. J Biol Chem. 2012;287:28686–28696. doi: 10.1074/jbc.M112.372680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zettler J, Schütz V, Mootz HD. The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Lett. 2009;583:909–914. doi: 10.1016/j.febslet.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Zhang S, Ma C, Chalfie M. Combinatorial marking of cells and organelles with reconstituted fluorescent proteins. Cell. 2004;119:137–144. doi: 10.1016/j.cell.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 17.Bedbrook CN, et al. Genetically encoded spy peptide fusion system to detect plasma membrane-localized proteins in vivo. Chem Biol. 2015;22:1108–1121. doi: 10.1016/j.chembiol.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong SSC, Kotera I, Mills E, Suzuki H, Truong K. Split-intein mediated re-assembly of genetically encoded Ca(2+) indicators. Cell Calcium. 2012;51:57–64. doi: 10.1016/j.ceca.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Dong Y, Gou Y, Li Y, Liu Y, Bai J. Synaptojanin cooperates in vivo with endophilin through an unexpected mechanism. eLife. 2015;4:1–50. doi: 10.7554/eLife.05660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei X, Potter CJ, Luo L, Shen K. Controlling gene expression with the Q repressible binary expression system in Caenorhabditis elegans. Nat Methods. 2012;9:391–395. doi: 10.1038/nmeth.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 22.Lints R, Hall DH. 2009. Male neuronal support cells, overview. WormAtlas, ed Herndon LA, 10.3908/wormatlas.2.9.
- 23.Avery L, Horvitz HR. Pharyngeal pumping continues after laser killing of the pharyngeal nervous system of C. elegans. Neuron. 1989;3:473–485. doi: 10.1016/0896-6273(89)90206-7. [DOI] [PubMed] [Google Scholar]
- 24.Raizen DM, Lee RY, Avery L. Interacting genes required for pharyngeal excitation by motor neuron MC in Caenorhabditis elegans. Genetics. 1995;141:1365–1382. doi: 10.1093/genetics/141.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pokala N, Liu Q, Gordus A, Bargmann CI. Inducible and titratable silencing of Caenorhabditis elegans neurons in vivo with histamine-gated chloride channels. Proc Natl Acad Sci USA. 2014;111:2770–2775. doi: 10.1073/pnas.1400615111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng H, Hope IA. The Caenorhabditis elegans homeobox gene ceh-19 is required for MC motorneuron function. Genesis. 2013;51:163–178. doi: 10.1002/dvg.22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pereira L, et al. A cellular and regulatory map of the cholinergic nervous system of C. elegans. eLife. 2015;4:1–46. doi: 10.7554/eLife.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sassone-Corsi P. The cyclic AMP pathway. Cold Spring Harb Perspect Biol. 2012;4:a011148. doi: 10.1101/cshperspect.a011148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim S, Govindan JA, Tu ZJ, Greenstein D. SACY-1 DEAD-Box helicase links the somatic control of oocyte meiotic maturation to the sperm-to-oocyte switch and gamete maintenance in Caenorhabditis elegans. Genetics. 2012;192:905–928. doi: 10.1534/genetics.112.143271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song BM, Avery L. Serotonin activates overall feeding by activating two separate neural pathways in Caenorhabditis elegans. J Neurosci. 2012;32:1920–1931. doi: 10.1523/JNEUROSCI.2064-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trojanowski NF, Nelson MD, Flavell SW, Fang-Yen C, Raizen DM. Distinct mechanisms underlie quiescence during two Caenorhabditis elegans sleep-like states. J Neurosci. 2015;35:14571–14584. doi: 10.1523/JNEUROSCI.1369-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Sieburth D. PKA controls calcium influx into motor neurons during a rhythmic behavior. PLoS Genet. 2013;9:e1003831. doi: 10.1371/journal.pgen.1003831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Correll LA, Woodford TA, Corbin JD, Mellon PL, McKnight GS. Functional characterization of cAMP-binding mutations in type I protein kinase. J Biol Chem. 1989;264:16672–16678. [PubMed] [Google Scholar]
- 34.Bacaj T, Shaham S. Temporal control of cell-specific transgene expression in Caenorhabditis elegans. Genetics. 2007;176:2651–2655. doi: 10.1534/genetics.107.074369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Churgin MA, He L, Murray JI, Fang-Yen C. Efficient single-cell transgene induction in Caenorhabditis elegans using a pulsed infrared laser. G3 (Bethesda) 2013;3:1827–1832. doi: 10.1534/g3.113.007682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singhal A, Shaham S. Infrared laser-induced gene expression for tracking development and function of single C. elegans embryonic neurons. Nat Commun. 2017;8:14100. doi: 10.1038/ncomms14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis MW, Morton JJ, Carroll D, Jorgensen EM. Gene activation using FLP recombinase in C. elegans. PLoS Genet. 2008;4:e1000028. doi: 10.1371/journal.pgen.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voutev R, Hubbard EJA. A “FLP-Out” system for controlled gene expression in Caenorhabditis elegans. Genetics. 2008;180:103–119. doi: 10.1534/genetics.108.090274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kage-Nakadai E, et al. A conditional knockout toolkit for Caenorhabditis elegans based on the Cre/loxP recombination. PLoS One. 2014;9:e114680. doi: 10.1371/journal.pone.0114680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoier EF, Mohler WA, Kim SK, Hajnal A. The Caenorhabditis elegans APC-related gene apr-1 is required for epithelial cell migration and Hox gene expression. Genes Dev. 2000;14:874–886. [PMC free article] [PubMed] [Google Scholar]
- 41.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boulin T, Etchberger JF, Hobert O. Reporter gene fusions. WormBook. 2006:1–23. doi: 10.1895/wormbook.1.106.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perez F, Granger BE. IPython: A system for interactive scientific computing. Comput Sci Eng. 2007;9:21–29. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.