Significance
In the fruit fly, ventral appendage (leg) identity is specified by a gene network including Wnt-1/wg, Sp6-9, and Dll, but little is known about the conservation of this network beyond insects. We disrupted Wnt signaling and Sp6-9 in a spider, a member of Chelicerata, the sister group to all remaining arthropods. Our results provide support for the conservation of a leg development gene regulatory network across Arthropoda. Dll has previously been reported to have a role in head segmentation that is restricted to spiders, and we show here that the Sp6-9/Dll cassette has been independently coopted for arachnid head segmentation.
Keywords: Arthropoda, gene regulatory network, Wnt signaling, limb outgrowth, zinc finger
Abstract
The jointed appendages of arthropods have facilitated the spectacular diversity and success of this phylum. Key to the regulation of appendage outgrowth is the Krüppel-like factor (KLF)/specificity protein (Sp) family of zinc finger transcription factors. In the fruit fly, Drosophila melanogaster, the Sp6-9 homolog is activated by Wnt-1/wingless (wg) and establishes ventral appendage (leg) fate. Subsequently, Sp6-9 maintains expression of the axial patterning gene Distal-less (Dll), which promotes limb outgrowth. Intriguingly, in spiders, Dll has been reported to have a derived role as a segmentation gap gene, but the evolutionary origin and regulation of this function are not understood because functional investigations of the appendage-patterning regulatory network are restricted to insects. We tested the evolutionary conservation of the ancestral appendage-patterning network of arthropods with a functional approach in the spider. RNAi-mediated knockdown of the spider Sp6-9 ortholog resulted in diminution or loss of Dll expression and truncation of appendages, as well as loss of the two body segments specified by the early Dll function. In reciprocal experiments, Dll is shown not to be required for Sp6-9 expression. Knockdown of arrow (Wnt-1 coreceptor) disrupted segmentation and appendage development but did not affect the early Sp6-9 expression domain. Ectopic appendages generated in the spider “abdomen” by knockdown of the Hox gene Antennapedia-1 (Antp-1) expressed Sp6-9 comparably to wild-type walking legs. Our results support (i) the evolutionary conservation of an appendage-patterning regulatory network that includes canonical Wnt signaling, Sp6-9, and Dll and (ii) the cooption of the Sp6-9/Dll regulatory cassette in arachnid head segmentation.
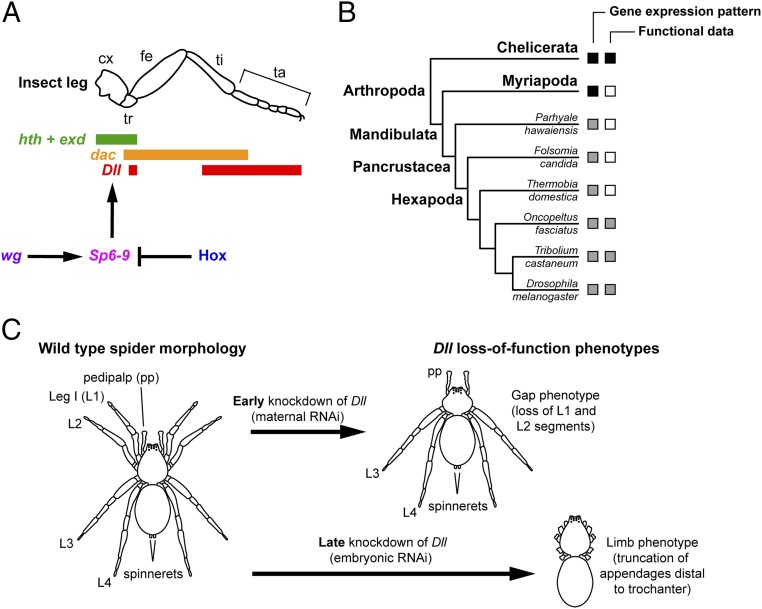
The eponymous jointed leg of Arthropoda has been closely linked to the evolutionary success of this phylum. Nearly every part of the arthropod leg has undergone extensive evolutionary modifications in different lineages, enabling adaptations to various ecological niches and environments (1–3). Arthropod legs are united by the involvement of a conserved suite of four “leg gap genes” to establish the proximodistal (PD) axis (refs. 4–10, reviewed in refs. 11 and 12) (Fig. 1A). The best-studied among them is Distal-less (Dll), the earliest marker of appendage identity, which is required for the development of the distal appendage territory across arthropods (9, 13–18), as well as being associated with body wall outgrowths in other phyla (19, 20).
Fig. 1.
Comparative functional data on arthropod appendage fate specification and PD axis patterning. (A) Regulation of limb gap genes in the walking leg of D. melanogaster. Sp6-9 is required for leg outgrowth and specification of ventral appendage fate. (B) Summary of available gene expression and functional data for Sp6-9 orthologs in Arthropoda (gray squares: previous studies; black squares: this study). (C) Dll has two roles in spider embryogenesis: a head segmentation function and a limb patterning function. Early knockdown via maternal RNAi results in a four-legged phenotype due to the loss of the L1 and L2 segments. Late knockdown via eRNAi results in canonical truncation of appendages distal to the trochanter. cx, coxa; fe, femur; ta, tarsus; ti, tibia; tr, trochanter.
Whereas the regionalization of the PD axis appears to be shared across arthropods, it is not evident whether early specification of leg identity is similarly conserved. In the fruit fly Drosophila melanogaster, two members of the Krüppel-like factor (KLF)/specificity protein (SP) transcription factor gene family were previously implicated in establishing leg fate: buttonhead (btd; orthologous to Sp5) and D-Sp1 (orthologous to Sp6-9; henceforth, “Dmel–Sp6-9”) (21–25). Both are upstream of Dmel-Dll and, in turn, are regulated by the activity of Wnt-1/wingless (wg) and decapentaplegic (dpp) during embryogenesis (26). Subsequently, it was shown that Dmel-btd and Dmel–Sp6-9 have two nonredundant roles in establishing leg fate and promoting leg growth (27); in null mutants of both Dmel-btd and Dmel–Sp6-9, the leg is entirely abolished and wing fate may be induced in ventral tissue. Ectopic expression of both Dmel–Sp6-9 and Dmel-btd can induce wing-to-leg transformations, but Dmel–Sp6-9 has a more important role in D. melanogaster leg fate specification, as Dmel–Sp6-9 can rescue double-null mutants, whereas Dmel-btd cannot. In later development, both Dmel–Sp6-9 and Dmel-btd are required for proper leg growth in larval stages (27, 28).
Beyond D. melanogaster, gene expression surveys of Sp6-9 have demonstrated conservation of expression domains across insects and one crustacean [all members of the same clade, Pancrustacea (29)]; taxonomically broader surveys of wg have shown broadly conserved expression patterns across Arthropoda. By contrast, functional data for this gene network are restricted to insects and one spider (ref. 30 and this study as discussed below). Sp6-9 orthologs have been functionally investigated in two insects, the beetle Tribolium castaneum (31) and the true bug Oncopeltus fasciatus (32) (Fig. 1B). In these two insects, RNAi-mediated knockdown of Sp6-9 orthologs demonstrated that Sp6-9 has a conserved role in appendage growth, but no evidence was obtained for a role of Sp6-9 in specifying leg identity or activating Dll (31, 32). Similarly, functional data for wg are restricted to insects. Consequently, the present understanding of leg-patterning mechanisms reflects an unclear evolutionary scenario regarding the network of interactions between wg, Sp6-9, and Dll, as described in D. melanogaster (Fig. 1 A and B).
A peculiar attribute of Dll in the spider Parasteatoda tepidariorum is that it has two separate functions, the canonical leg-patterning function [phenocopies generated by embryonic RNAi (eRNAi) have distally truncated limbs (16)] and a novel head segmentation function unique to spiders [phenocopies from maternal RNAi lack the body segments corresponding to the first and second walking legs (henceforth, L1 and L2); remaining appendages develop normally (33)] (Fig. 1C). Regulation of this early Dll function is not understood, but a recent work, completed concomitant to the present study, on Sp6-9 in the spider P. tepidariorum has shown that knockdown of Sp6-9 results in the deletion of the L1 and L2 segments, in addition to truncation of the remaining appendages (30). The description of the loss-of-function phenotype of Sp6-9 in the spider is certainly suggestive of a regulatory interaction with Dll, but neither early regulation of Dll by Sp6-9 nor its relationship with Wnt signaling has been tested.
To redress these gaps in the understanding of appendage gene network conservation, we began by assessing incidence and expression of Sp gene family members in Myriapoda and Chelicerata, the two most basally branching groups of arthropods with respect to Pancrustacea (Hexapoda + Crustacea). To test the conservation of the gene network specifying appendage development, and especially how elements of this network relate to the derived role of spider Dll as a gap segmentation gene, we performed RNAi against the wg coreceptor arrow (arr; vertebrate homologs: LRP5 and LRP6), Sp6-9, and Dll in the spider P. tepidariorum. The arr homolog codes for an essential component of the canonical Wnt pathway across Bilateria (34–36) and was specifically selected herein because previous efforts to knock down wg expression directly via RNAi have exhibited variable or limited efficiency in several arthropod species (37, 38), including spiders. By contrast, severe disruption of Wnt signaling by knockdown of arr has been achieved with high penetrance in insects (34, 39, 40).
Here, we show that single-copy orthologs of arr and Sp6-9 occur in exemplars of both chelicerates and myriapods. Expression data for representatives of these two subphyla demonstrate that Sp6-9 orthologs are invariably expressed in outgrowing limbs. In strong phenocopies, down-regulation of Ptep–Sp6-9 results in the abrogation of the entire appendage, as well as loss of the L1 and L2 body segments, concomitant to the loss of Ptep-Dll expression. Depletion of Ptep-arr disrupts both body segmentation and appendage growth, in association with depletion of Ptep–Sp6-9 expression in outgrowing legs, suggesting a conserved role for canonical Wnt signaling in segmentation and leg development across arthropods. Critically, depletion of Ptep-arr does not affect the early expression domain of Ptep–Sp6-9 in the presumptive L1 and L2 territory. Our results demonstrate that a conserved gene network patterns appendage development in insects and arachnids, in tandem with the cooption of an Sp6-9/Dll cassette in patterning head segments of arachnids.
Results
Single-Copy Orthologs of arr and Sp6-9 Occur in Myriapods and Arachnids.
The maximum likelihood and Bayesian inference tree topologies recovered the monophyly of the Sp gene family with maximal nodal support (SI Appendix, Fig. S1). The tree topology largely corresponded to previous analyses of the Sp gene family and is consistent with basal divergence of three paralogs in the common ancestor of Metazoa: Sp1-4, Sp5, and Sp6-9. The following differences were recovered in our orthology assignment: The sequence previously identified as Trichoplax adhaerens Sp1-4 was recovered as nested within the KLF13 outgroups, and the putative T. adhaerens Sp5 ortholog was recovered as nested within the Sp1-4 cluster (both placements supported; Dataset S1).
Of the 51 Sp homologs reported herein, a single Sp6-9 ortholog was discovered for the two myriapods (the centipedes Strigamia maritima and Lithobius atkinsoni), the hemimetabolous insect Gryllus bimaculatus, four arachnids (the mite Tetranychus urticae, the harvestman Phalangium opilio, the scorpion Centruroides sculpturatus, and the spider P. tepidariorum), seven pycnogonids (Anoplodactylus insignis, Nymphon molleri, Phoxichilidium tubulariae, Styllopallene sp., Tanystylum orbiculare, Meridinale flava, and Pycnogonum litorale), and the onychophoran Euperipatoides rowelli; two to four Sp6-9 paralogs per species were discovered in the genomes/transcriptomes of three horseshoe crabs. Single-copy Sp1-4 orthologs were discovered for the spider, the scorpion, the harvestman, both centipedes, and all seven pycnogonids; one to three Sp1-4 paralogs were discovered in the horseshoe crabs. Single-copy Sp5 orthologs were discovered for the centipede S. maritima, the horseshoe crab Limulus polyphemus, six pycnogonids, and two onychophorans. No Sp5 orthologs were found in the genomes of the milkweed bug O. fasciatus, the amphipod Parhyale hawaiensis, the mite, the spider, or the scorpion.
Single-copy orthologs of arr were discovered in genomic resources of two myriapods (S. maritima and L. atkinsoni) and one arachnid (P. tepidariorum). Maximal nodal support values were recovered for the placement of the spider sequence within the arr cluster (SI Appendix, Fig. S2).
Expression of Sp6-9 in Exemplars of Chelicerates and Myriapods.
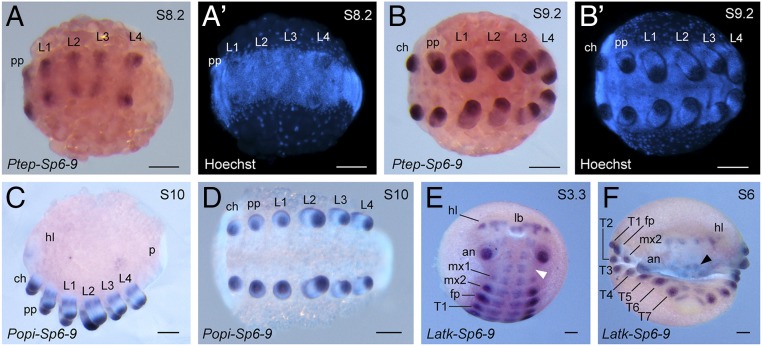
During the formation of limb buds in spider embryogenesis, Ptep–Sp6-9 is detected throughout all limb buds of the prosoma (the anterior tagma, which bears all six limb pairs) by stage 8 (Fig. 2 A and A′). Expression is additionally observed in the head lobes and as faint stripes in the opisthosomal segments (SI Appendix, Fig. S3A). In later stages, expression of Ptep–Sp6-9 becomes heterogeneous in the limb buds of the pedipalps and the walking legs, consisting of a strong distal domain and a weaker, broader proximal domain; expression in the cheliceral limb bud remains homogeneous (Fig. 2 B and B′). Expression is also observed as stripes in the ventral opisthosomal ectoderm of stage 9 embryos (SI Appendix, Fig. S3B). No expression of Ptep–Sp6-9 was detected in the limb bud-derived organs of the opisthosoma (i.e., the primordia of the book lungs, tubular tracheae, and spinnerets; SI Appendix, Fig. S3B). The subdivision of expression domains in the pedipalps and walking legs in elongating appendages reflects comparable dynamics previously reported in a range of insects and a crustacean (29).
Fig. 2.
Expression of Sp6-9 orthologs in Chelicerata and Myriapoda. (A and A′) In the spider, Ptep–Sp6-9 is expressed in all prosomal limb buds. (B and B′) In later stages, Ptep–Sp6-9 is also observed in the ventral neuroectoderm. In all but the cheliceral limb buds, expression is heterogeneous, consisting of a broad proximal ring and a stronger distal expression domain at the terminus of the appendage. (C and D) In limb buds of the harvestman, Popi–Sp6-9 is expressed comparable to the spider. (E and F) In the centipede, Latk–Sp6-9 is strongly expressed in all limb buds, except for the mandibles (white arrowhead). Expression is also visible in the ventral neuroectoderm and the labrum (black arrowhead). A complex expression pattern is observed in the head lobes and developing brain. an, antenna; ch, chelicera; fp, forcipule; hl, head lobe; mx, maxilla; p, posterior terminus; pp, pedipalp; T, trunk leg. (Scale bars: 100 μm.)
Similar expression patterns are observed in the prosoma of the harvestman. Specifically, heterogeneous expression of Popi–Sp6-9 occurs in the limb buds of the pedipalps and walking legs, whereas a single distal domain with tapering proximal expression occurs in the chelicerae (Fig. 2 C and D). In the centipede, Latk–Sp6-9 is expressed in the limb buds of all head and trunk appendages, except for the mandible (Fig. 2E), a pattern congruent with insect and crustacean exemplars (29). Latk–Sp6-9 is additionally detected in the ventral neuroectoderm, the labrum, and as complex domains in the head lobes (Fig. 2 E and F).
In all three species, complementary sense probes were tested as negative controls; no staining was observed in sense controls (SI Appendix, Fig. S4). Taken together, these data demonstrate that expression of the Sp6-9 ortholog is uniformly associated with developing appendages (with the exception of the mandible of insects and myriapods and the spinnerets of spiders) across all surveyed arthropods.
Expression of Ptep-arr.
Throughout spider embryogenesis, Ptep-arr is weakly and ubiquitously expressed in the embryo, comparable to the T. castaneum ortholog of arr (40). Complementary sense probes tested as negative controls showed no staining, suggesting that the expression detected was specific (SI Appendix, Fig. S4).
Ptep–Sp6-9 RNAi Results in Down-Regulation of Ptep-Dll.
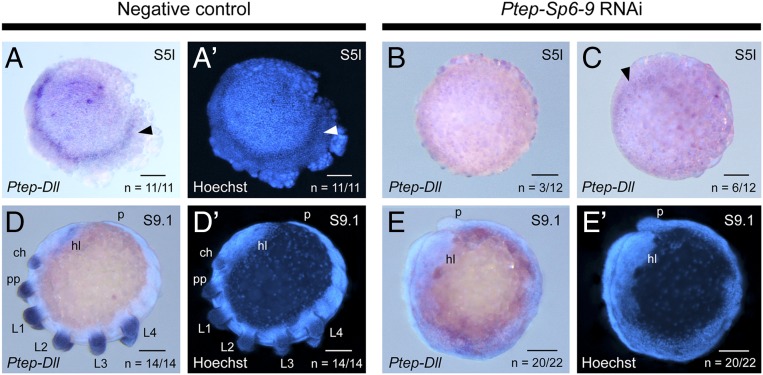
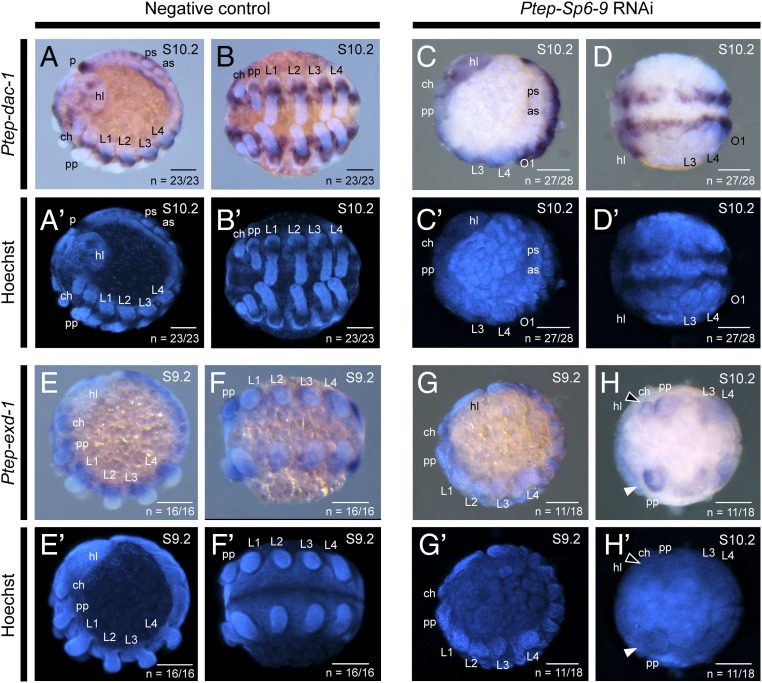
To test whether Ptep–Sp6-9 is required for maintaining Ptep-Dll expression, we studied the function of Ptep–Sp6-9 using maternal RNAi (experimental design and summary are provided in SI Appendix, Fig. S5). In Ptep–Sp6-9 RNAi embryos, 25% (n = 3 of 12) had no detectable Ptep-Dll expression levels using DIG-labeled probes (Fig. 3B) and 50% (n = 6 of 12) of surveyed germ disk-stage embryos had diminished (i.e., barely detectable) Ptep-Dll expression (stage 5l; Fig. 3C). In later stages, 91% of surveyed limb bud-stage embryos (n = 20 of 22) lacked Ptep-Dll expression (stage 9; Fig. 3 D–E′). In embryos exhibiting strong phenocopies (n = 826), the head lobes and tail bud were identifiable, but the labrum and all prosomal appendages were abolished (Fig. 3 E and E′). Expression of Ptep–Sp6-9 was diminished by comparison with negative control embryos in experiments targeting two nonoverlapping fragments of Ptep–Sp6-9 (n = 31 of 31), and similar phenotypic spectra were obtained from knockdown of each fragment (SI Appendix, Figs. S5A and S6), suggesting that the knockdown was specific and on target. In comparison to the classic Dll limb phenotype previously reported in spiders, which results in a deletion of all leg segments distal of the medial dachshund-1 (dac-1) expression domain (16, 33) (Fig. 1C), Ptep–Sp6-9 RNAi truncated limb development more proximally than Dll RNAi, as assessed by expression patterns of the medial PD axis marker dac-1 and the proximal PD axis marker extradenticle-1 (exd-1) (Fig. 4). Severe Ptep–Sp6-9 RNAi phenocopies retained only the Ptep–dac-1 expression associated with neurogenic tissues; the truncated appendages lacked Ptep–dac-1 expression altogether (Fig. 4 C–D′), indicating a deletion more proximal than incurred by Dll RNAi (n = 27 of 28). Consistent with this interpretation, severe Ptep–Sp6-9 RNAi phenocopies expressed the proximal PD axis marker Ptep–exd-1 at the distal termini of truncated appendages, or not at all in a fully abrogated appendage (Fig. 4 G–H′; n = 11 of 18).
Fig. 3.
Ptep–Sp6-9 is required for activation of Ptep-Dll expression. (A and A′) Expression of Ptep-Dll in early stages of wild-type spiders occurs as a ring corresponding to segments L1 and L2 (arrowhead). Expression of Ptep-Dll is lost (B) or diminished (C; arrowhead) in Ptep–Sp6-9 RNAi embryos. (D and D′) Expression of Ptep-Dll in limb bud stages of wild-type spiders occurs in all prosomal appendages and in the head lobes. (E and E′) In strong phenocopies from Ptep–Sp6-9 RNAi, all appendages are abolished and no Ptep-Dll expression is detected. ch, chelicera; hl, head lobe; pp, pedipalp. (Scale bars: 100 μm.)
Fig. 4.
Ptep–Sp6-9 RNAi causes severe truncation of appendages. (A–D′) Expression of dac-1, a medial limb territory marker. As inferred from expression of Ptep–dac-1 in wild-type embryos (A–B′), strong Ptep–Sp6-9 RNAi phenocopies undergo truncations more proximally than the trochanter, resulting in the loss of the entire Ptep–dac-1 domain in all appendages (C–D′); only the expression in the ventral ectoderm is seen in strong phenocopies (D and D′). (E–H′) Expression of exd-1, a proximal limb territory marker. (E–F′) Wild-type expression of Ptep–exd-1 spans the proximal-most segments and the body wall. (G and G′) Intermediate phenocopies exhibit Ptep–exd-1 in the termini of truncated appendages. (H and H′) Severe Ptep–Sp6-9 RNAi phenocopies either retain Ptep–exd-1 only at the distal remnant of truncated appendages (black arrowhead) or lose this territory entirely (white arrowhead). as, anterior spinneret; ch, chelicera; hl, head lobe; p, posterior terminus; pp, pedipalp; ps, posterior spinneret; O1, first opisthosomal segment. (Scale bars: 100 μm.)
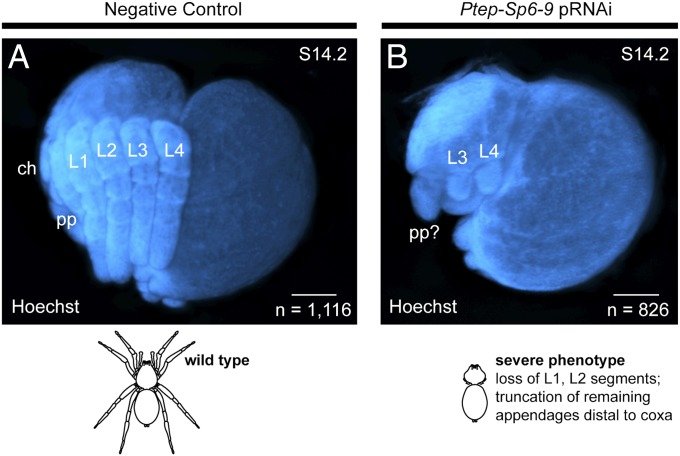
The head segmentation function of Ptep-Dll specifically affects the L1 and L2 segments (33). To test whether the segmentation phenotype incurred by Ptep–Sp6-9 is distinguishable from that of Ptep-Dll, we examined the expression of the segment polarity gene Ptep–engrailed-1 (en-1) in intermediate phenocopies (n = 256), where some truncated appendages were identifiable but a segment gap defect was still evident. In these embryos, we observed that Ptep–en-1 expression was lost in the two segments corresponding to the L1 and L2 segments (SI Appendix, Fig. S7), which is consistent with loss of Ptep-Dll expression in those segments during segmentation of the germband (Fig. 3 B and C). Moreover, the morphology of severe Ptep–Sp6-9 RNAi phenocopies in older stages shows a truncation of the prosoma incurred by the loss of two body segments, as well as truncation of all remaining appendages up to the proximal-most leg segment, the coxa (n = 826; Fig. 5). These phenotypes overlap the union of the two known Dll loss-of-function phenotypes in spiders (Fig. 1B), in addition to the more proximal truncation of the PD axis.
Fig. 5.
Severe Ptep–Sp6-9 RNAi embryos recapitulate the fusion of early and late Dll loss-of-function phenotypes. (A) In a wild-type embryo at stage 14, all six pairs of appendages are visible in the lateral view. (B) In a severe Ptep–Sp6-9 RNAi phenotype, the embryo exhibits both a head segmentation phenotype as well as truncation of all appendages distal to the coxa. While these embryos never survive to hatching, interpretations of the phenotype are shown as line drawings of hypothetical adult counterparts to convey phenotypic effects. ch, chelicera; pp, pedipalp. (Scale bars: 100 μm.)
In the weakest subset of phenocopies, we observed shorter appendages in comparison to negative control embryos, but without any segmentation defects or appendage truncation (n = 497; SI Appendix, Fig. S8). The morphology of embryos in this phenotypic spectrum is comparable to Sp6-9 RNAi phenocopies in T. castaneum and O. fasciatus, wherein appendage segments were reduced in length and/or fused but entire appendages were not abolished (31, 32).
Ptep-Dll Is Not Required for the Proximal Ptep–Sp6-9 Expression Domain in Appendages.
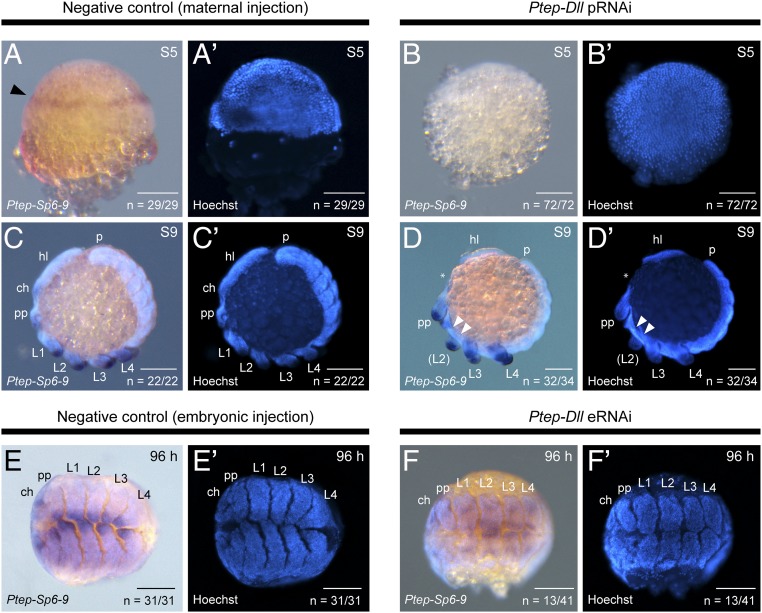
In the fruit fly D. melanogaster, Sp6-9 has been shown to be upstream of Dll. We performed both maternal and eRNAi against Ptep-Dll and assayed phenocopies for Ptep–Sp6-9 expression. Maternal knockdown recapitulated the early function of Ptep-Dll as a head segmentation gene (loss of L1 and L2 segments), with concomitant loss of the ring of Ptep–Sp6-9 expression in the germ disk, in comparison to wild-type embryos (Fig. 6 A–B′; n = 72 of 72) The transience of this knockdown results in wild-type appendages on the remaining prosomal segments. Accordingly, Ptep-Dll parental RNAi (pRNAi) phenocopies showed typical expression of Ptep–Sp6-9 in the remaining appendages (chelicerae, pedipalps, and L3 and L4 legs), as well as elsewhere in the embryo (Fig. 6 C–D′; n = 32 of 34). As the L1 and L2 territory is deleted upon knockdown of either Ptep-Dll or Ptep–Sp6-9, we are presently unable to infer the regulatory relationship between the gene pair in early embryogenesis.
Fig. 6.
Effects of Ptep-Dll knockdown on Ptep–Sp6-9 expression. (A and A′) Negative control embryo at stage 5 in a lateral view showing wild-type expression of Ptep–Sp6-9 in the presumptive L1 and L2 territory. (B and B′) Ptep-Dll pRNAi phenocopy in a dorsal view showing absence of Ptep-Sp6-9 expression; the germ disk has been dissected away from the yolk. (C and C′) Negative control embryo at stage 9 showing wild-type expression of Ptep–Sp6-9. (D and D′) Ptep-Dll pRNAi phenocopies lack the L1 and/or L2 segments (left side of the five-legged mosaic embryo shown here; the right side retains three pairs of legs), but the remaining appendages exhibit wild-type expression of Ptep–Sp6-9. The asterisk indicates damage incurred to chelicera during specimen mounting. (E–F′) Embryos recovered from eRNAi experiments retain the full complement of legs 96 h after injection but undergo the canonical distal truncation phenotype (compare F and F′ with C and C′ and E and E′), resulting in the deletion of the distal-most expression territory of Ptep–Sp6-9. (F and F′) Ptep–Sp6-9 is detected in the proximal region of the appendages. ch, chelicera; hl, head lobe; p, posterior terminus; pp, pedipalp. (Scale bars: 100 μm.)
Embryonic knockdown recapitulated the canonical function of Dll as a PD axis gene; a proportion of surviving Ptep-Dll eRNAi phenocopies exhibited truncated appendages. In these phenocopies, Ptep–Sp6-9 expression is still detected in the proximal territory of the appendages, but the strongest expression domain at the distal terminus is greatly reduced upon truncation (Fig. 6 F and F′; n = 13 of 41). As with the L1 and L2 region in early embryogenesis, we cannot establish the regulatory relationship between Ptep-Dll and Ptep–Sp6-9 in the deleted region of the appendage axis.
Ptep-arr RNAi Disrupts Anteroposterior Segmentation and Appendage Development.
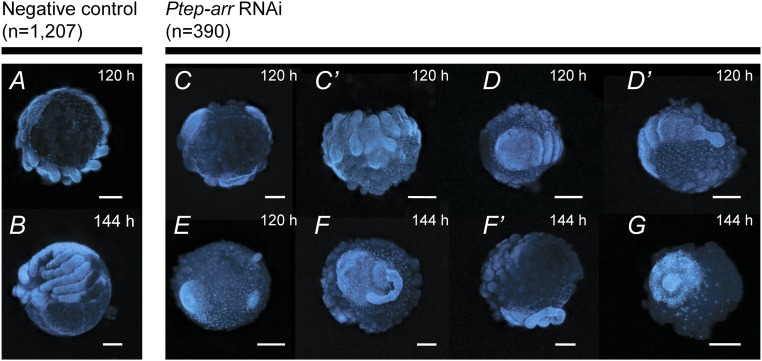
To test whether canonical Wnt signaling has a conserved role in leg development, we studied the function of the Wnt-1 coreceptor Ptep-arr using maternal RNAi (the experimental design and a summary are provided in SI Appendix, Fig. S9). Loss-of-function phenocopies from Ptep-arr RNAi underwent disruption of segmentation and appendage development (n = 390), as inferred from morphology (Fig. 7) and expression assays for the segment polarity genes wg (n = 8 of 12) and en-1 (Fig. 8 A–D′; n = 7 of 7). These phenocopies were smaller than wild-type counterparts, consisting only of small, rudimentary heads in severe cases, and always lacked appendages [comparable to severe arr RNAi phenocopies of T. castaneum (39)]. As landmarks for the anteroposterior (AP) axis, we assayed the expression of the anterior head marker orthodenticle-1 (otd-1) and the pedipalpal segment marker Hox gene labial-1 (lab-1). Strong phenocopies showed regionalization of the AP axis (expression stripes of otd-1 and lab-1 in the anterior germband), despite loss of segmentation (Fig. 8 E–H′).
Fig. 7.
Phenotypic spectrum of Ptep-arr RNAi phenocopies, visualized with Hoechst staining. (A and B) Negative control embryos at 120 h and 144 h (reference time points for this experiment). (C) Mild phenocopy in a lateral view, showing complete development of the prosoma and a partial opisthosoma. (C′) Ventral view of the embryo shown in C, showing defects of segmentation and limb development. A moderate phenocopy without limbs shows underdeveloped head lobes (D) and truncated opisthosoma (D′). (E) Strong phenocopy exhibiting failure of germ disk condensation. (F) Severe phenocopy exhibiting lack of prosomal segmentation, lack of appendages, and defects in AP axis orientation. (F′) Lateral view of embryo shown in F, showing posterior delamination of the germband from the yolk. (G) Severe phenotype; we interpret the entire embryo to constitute the germ disk remnant. (Scale bars: 100 μm.)
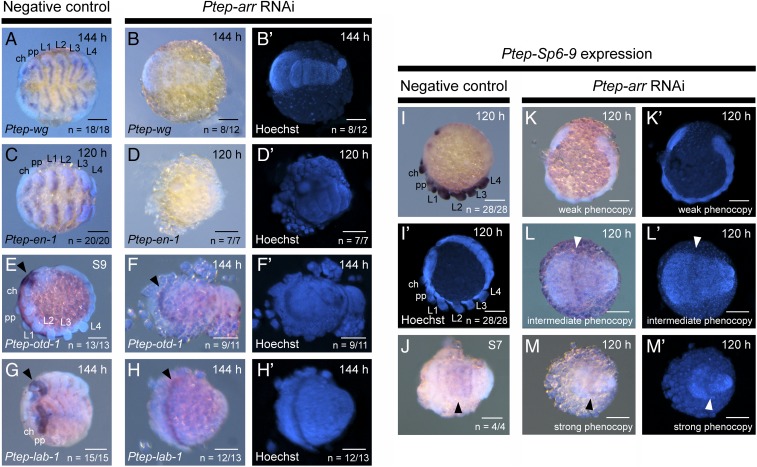
Fig. 8.
Depletion of Ptep-arr reveals conserved aspects of Wnt signaling in insects and spiders. (A) Wild-type expression of Ptep-wg in negative control embryo at 144 h. (B and B′) In Ptep-arr RNAi embryos, segmentation defects are observed throughout the germband, no appendages are formed, and Ptep-wg expression is not detected. (C) Wild-type expression of Ptep–en-1 in negative control embryos at 120 h. (D and D′) In Ptep-arr RNAi embryos, Ptep–en-1 expression is not detected. (E) Wild-type expression of Ptep–otd-1 shows a broad expression domain in the precheliceral territory (arrowhead), as well as in the ventral midline. (F and F′) Ptep-arr RNAi embryos demonstrate regionalization of the anterior germband, showing a typical stripe of anterior Ptep–otd-1 expression (arrowhead). No expression is observed in the ventral midline at 144 h, suggesting either developmental delay or neurogenic defects, or both. (G) In wild-type embryos, Ptep–lab-1 is most strongly expressed in the pedipalpal segment (arrowhead). (H and H′) Ptep-arr RNAi phenocopies retain a stripe of Ptep–lab-1 expression in the anterior territory (arrowhead), suggesting regionalization of the anterior germband. Expression of Ptep–Sp6-9 in negative control (I , I′, and J) and Ptep-arr RNAi embryos (K–M′) is illustrated. Weak phenocopies from Ptep-arr RNAi bear incorrectly oriented germbands and diminished Ptep–Sp6-9 expression (K and K′), whereas strong phenocopies retain only a medial stripe of Ptep–Sp6-9 (L–M′), which presumably corresponds to the segmental expression of Ptep–Sp6-9 in earlier stages of wild-type embryos (J). ch, chelicera; pp, pedipalp. (Scale bars: 100 μm.)
To examine the regulatory relationship between Wnt signaling and Sp6-9, we assayed Ptep–Sp6-9 expression in a range of Ptep-arr RNAi phenocopies (Fig. 8 I–M′). Weak Ptep-arr RNAi phenocopies exhibited irregularly distributed and truncated limb buds showing greatly diminished Ptep–Sp6-9 expression, in addition to posterior segmentation defects (Fig. 8 K and K′). In severe Ptep-arr RNAi phenocopies lacking segments and legs, Ptep–Sp6-9 expression was never detected in ventral tissues corresponding to appendage primordia (n = 11 of 11). Intriguingly, we did detect expression of Ptep–Sp6-9 in the presumptive L1 and L2 territory that is comparable to the broad expression band in wild-type embryos at germband stages (stage 7) in a subset of phenotypes (Fig. 8 L–M′; n = 7 of 11). We interpret this result to mean that Ptep-arr (and, by extension, Wnt activity) is required for activation of Ptep–Sp6-9 in the appendage primordia, but not necessarily head regionalization.
Ectopic Spider Legs Induced by Hox RNAi Express Ptep–Sp6-9.
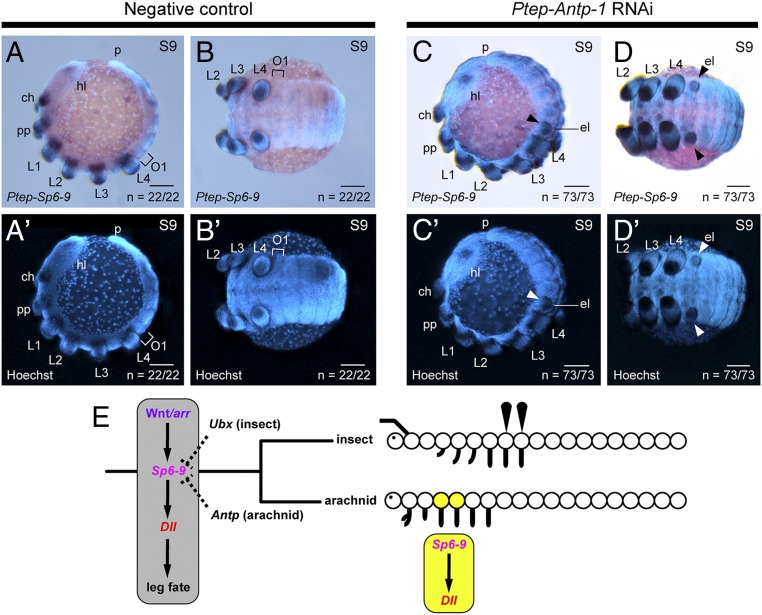
In the fruit fly, the regulation of Sp homologs and Dll in D. melanogaster is modulated by the trunk Hox gene Ultrabithorax (Ubx), with Ubx loss-of-function mutants expressing Sp genes in the ectopic appendage on the first abdominal segment (26). Notably, the identity of the anterior-most Hox gene that represses leg identity is not the same in insects (Ubx) and arachnids [Antennapedia (Antp) (41)]. To test whether convergence in Hox gene function (repression of leg development) is correlated with convergent integration of posterior Hox genes in the appendage-patterning GRN, we replicated the knockdown of Ptep–Antp-1 to generate 10-legged spider embryos (41) and assayed them for expression of Ptep–Sp6-9. The small ectopic appendages of the first opisthosomal segment in Ptep–Antp-1 RNAi embryos expressed Ptep–Sp6-9 comparable to wild-type prosomal limb buds during embryogenesis (Fig. 9; n = 73 of 73). These data are consistent with the prediction of convergent assembly of the insect and arachnid GRNs, wherein Sp homologs mediate the regulation of Dll by Hox signaling (26).
Fig. 9.
Ectopic legs of 10-legged spiders express Ptep–Sp6-9. Expression of Ptep–Sp6-9 in a wild-type embryo in lateral (A and A′) and ventral (B and B′) views is illustrated. Note the absence of limb buds and Ptep–Sp6-9 expression in the first opisthosomal segment (O1; bracket). (C–D′) Ptep–Antp-1 RNAi embryos have a fifth pair of legs (arrowheads) on the O1 segment, which expresses Ptep–Sp6-9 throughout the limb axis. (E) Parsimonious inference of a conserved gene subnetwork that regulates appendage development in the common ancestor of insects and arachnids (in gray), with independent integration of different Hox inputs in descendant lineages (dashed lines). Cooption of the Sp6-9/Dll cassette into head segmentation of arachnids is shown in yellow. ch, chelicera; el, ectopic leg; hl, head lobe; p, posterior terminus; pp, pedipalp. (Scale bars: 100 μm.)
Discussion
A Conserved Appendage-Patterning Gene Network in Insects and Arachnids.
Various workers have examined the patterning of the limb PD axis across Arthropoda via a combination of gene expression surveys (e.g., refs. 4, 8, 11, 18, and 42–45) and functional studies (e.g., refs. 6, 7, 9, 15, 16, 18, and 46–48). By comparison with the regionalization of the PD axis, evolution of the GRN underlying specification of arthropod leg fate is poorly understood from a functional standpoint (Fig. 1 A and B). As examples, dpp and wg both play a role in establishing positional information along the PD axis, and dpp also patterns dorsal fate in ventral appendages of D. melanogaster. However, dpp expression in other surveyed arthropods is not comparable to the patterns described in D. melanogaster; thus, dpp may not serve the same role in appendage development across Arthropoda (11, 49, 50). In spiders, functional interrogation via maternal RNAi has demonstrated a role for Ptep-dpp in AP axial patterning during early embryogenesis, but its role in appendage patterning has not been explored, likely due to the severity of Ptep-dpp RNAi defects (51). Similarly, no comparative functional data exist for wg orthologs outside of winged insects, and these vary widely in penetrance and phenotypic spectrum (37, 38, 49, 52). As an example, in the cricket Gryllus bimaculatus, Gbim-wg eRNAi resulted in only transient and early diminution of Gbim-wg expression, followed by wild-type expression by onset of limb bud stages, and corresponding wild-type morphology of hatchlings from all injected embryos (38).
To test for evolutionary conservation of leg-patterning mechanisms across arthropods, we identified the regulatory subnetwork formed by Wnt signaling, Sp homologs, and Dll as a key target for functional comparison, and focused on arachnids for reasons of phylogenetic significance and limited representation of functional data. Our results constitute an instance of systemic disruption of Wnt-1/Wg signaling in a noninsect arthropod (Fig. 8 B and B′), which we achieved by targeting the downstream coreceptor arr. In contrast to previous efforts to knock down wg, RNAi against arr results in a highly comparable phenotypic spectrum in insects and arachnids, wherein both segmentation and appendage development are disrupted and leg-patterning genes are not activated in the developing appendages. These results support a common requirement for Wnt activity for leg patterning in the common ancestor of insects and arachnids.
A discrepancy of function of Sp6-9 has previously been observed across the three available insect data points (Fig. 1B). In two cases (the true bug O. fasciatus and the beetle T. castaneum), Sp6-9 orthologs were linked to allometric growth by RNAi datasets, but not specification of leg fate (as in double–loss-of-function mutants of btd and Sp6-9 in D. melanogaster). RNAi data must be interpreted with caution, as it can be difficult to demonstrate the lack of a gene’s function without assessing knockdown efficiency. Apropos, it was noted in the O. fasciatus study that incomplete penetrance could not be ruled out as an alternative explanation, as Ofas–Sp6-9 RNAi embryos retained Ofas–Sp6-9 expression, but those data were not shown (32). In both the T. castaneum and recent P. tepidariorum studies, verification of Sp6-9 knockdown was not assessed at all (30, 31). Incidentally, we attempted Gbim–Sp6-9 maternal RNAi in this study [following the protocols of Takagi et al. (53)] but obtained results similar to a previous experiment on Gbim-wg, with 100% of late-stage embryos bearing normal appendages (38).
Our results demonstrate that Ptep–Sp6-9 is required for appendage development in spiders, as inferred from the truncation of all prosomal appendages in strong Ptep–Sp6-9 RNAi phenocopies, with concomitant loss of Ptep–Sp6-9 expression. The reciprocal experiments could be taken to mean that Ptep-Dll (both early and late functions) may not be required for Ptep–Sp6-9 expression in developing appendages; this interpretation would be consistent with evolutionary conservation of the network established for D. melanogaster leg development, in which Sp6-9 is upstream of Dll. Furthermore, the invariable association of Sp6-9 ortholog expression with developing appendages in previously undersampled parts of arthropod phylogeny (Myriapoda and Chelicerata) supports our inference of conserved Sp6-9 dynamics in the arthropod common ancestor (Fig. 10).
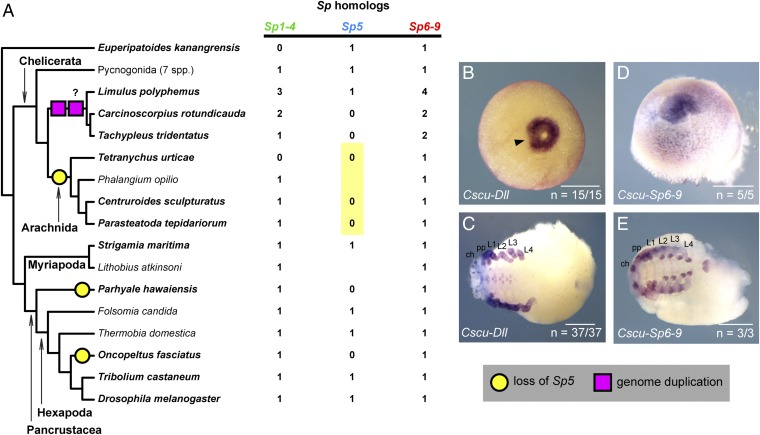
Fig. 10.
Evolutionary dynamics of the Sp gene family across Panarthropoda. (A) Incidence of Sp homologs across surveyed arthropod model systems reveals absence of Sp5 orthologs in genomes of Chelicerata, as well as some pancrustacean species. Boldfaced terminals indicate taxa with genomes. Yellow circles denote inferred losses of Sp5 (i.e., absence from genomes; absences from transcriptomes are not scored as losses). Pink squares denote the single or twofold whole-genome duplication inferred previously in horseshoe crabs (65). (B) Wild-type expression of Cscu-Dll in an early-stage embryo of the scorpion. In the earliest stages where a blastopore is detectable, Cscu-Dll is expressed as a ring surrounding the blastopore. The arrowhead marks a discontinuity in the ring of expression. (C) In later stages, the same riboprobe reveals the expected expression pattern of Dll orthologs in the distal territories of all appendages, in the ventral ectoderm, and in the telson. (D) Wild-type expression of Cscu–Sp6-9 in early stages is comparable to Cscu-Dll expression in equivalent stages. (E) In later stages, expression of Cscu–Sp6-9 in the limbs, ventral ectoderm, and telson is comparable to that of other arachnids at equivalent stages. ch, chelicera; pp, pedipalp. (Scale bars: B and C, 1,000 μm.)
We note that the phenotypic spectrum observed in limbs of Ptep–Sp6-9 RNAi embryos spans the loss-of-function phenotypes in D. melanogaster, as well as the range of outgrowth phenotypes reported in D. melanogaster, T. castaneum, and O. fasciatus knockdown experiments. Therefore, a possible explanation for the discrepancy in Sp6-9 knockdown experiments is that allometric growth phenotypes observed in T. castaneum and O. fasciatus reflect incomplete penetrance of Sp6-9 RNAi and, in turn, incomplete knockdown of Dll expression in these developing appendages. This hypothesis could be tested in the future via CRISPR-Cas9–mediated mutagenesis in T. castaneum and O. fasciatus.
Cooption of the Sp6-9/Dll Module in Head Segmentation of Arachnids.
Among metazoans, Dll and Sp transcription factors play critical roles in the development of several tissues. Previous phylogenetic inferences have supported the presence of at least three Sp gene family members in the common ancestor of Metazoa (29, 54). Subsequent divergences of the nine paralogs present in vertebrates (Sp1 to Sp9) are likely attributable to twofold whole-genome duplication in the vertebrate common ancestor (55). Within the Sp6-9 orthogroup, at least two vertebrate paralogs, Sp8 and Sp9, regulate Fgf8 expression and outgrowth of the apical ectodermal ridge in the mouse, chick, and zebrafish (56, 57). Hypomorphic mutants of Sp8 lose expression of various appendage markers, including Dlx [vertebrate Dll ortholog (55)]. While fewer comparative data are available among spiralians, in the planarian Schmidtea mediterranea, RNAi against Smed-Dlx or Smed–Sp6-9, followed by excision of the head, resulted in the inability to regenerate eyes as well as other tissues (58). Recently, it was shown that regenerating appendages in the annelid Platynereis dumerilii express Pdum–Sp6-9, Pdum-Dll, and orthologs of other limb-patterning genes (59). In some cases, the spatial relationships of these annelid genes are comparable to those in developing arthropod appendages, but their functions remain unknown. Thus, a Sp6-9/Dll regulatory cassette has been reported in various roles and lineages across Bilateria (27, 60).
Given the recently described novel role of Ptep-Dll as a gap segmentation gene in spiders, our experiments with Ptep–Sp6-9 are poised to address whether this phenomenon constitutes a new case of cooption of an Sp/Dll cassette. In our Ptep–Sp6-9 RNAi experiments, the union of both the Ptep-Dll maternal RNAi (head segmentation) phenotype and the eRNAi (distal limb truncation) phenotype in the strong Ptep–Sp6-9 RNAi phenocopies (Figs. 1C and 5) was associated with the diminution or complete loss of Ptep-Dll expression in the relevant stages of embryogenesis (Fig. 3). These data support a model of activation and maintenance of Ptep-Dll by Ptep–Sp6-9 for both the head segmentation and leg-patterning functions of Dll. Furthermore, we observed that the head segmentation domain of Ptep–Sp6-9 was not lost in severe Ptep-arr RNAi phenocopies, which bear part of a regionalized AP axis but never develop segments or appendages in later developmental stages. This result suggests the independence of the Ptep–Sp6-9/Dll gap gene function from Wnt signaling, and is hypothesized herein to be due to cooption of an ancient Sp/Dll gene cassette.
Loss of Sp5 is Characteristic of Arachnids.
Why would the Sp6-9/Dll cassette be recruited for this function in an arachnid? One possibility may be that Sp6-9 fulfills the role of the Sp5/btd ortholog of insects. In D. melanogaster, btd is one of the classically known gap segmentation genes, and expression surveys of btd orthologs support evolutionary conservation of this head segmentation role, at least in the common ancestor of Mandibulata (29, 61). The outstanding question then is whether Sp5 could also retain this function in Chelicerata.
By comparison with Sp6-9, Sp5 lacks the same breadth of functional data points, obviating clear polarization of gene function on a tree topology even within Pancrustacea. As an alternative approach to inferring evolution of Sp5 function, we mapped the evolutionary losses of Sp5 across Panarthropoda (with the assumption that loss of critical Sp5 functions was rescued through their cooption by other genes). Our survey of recently sequenced genomes and developmental transcriptomes of various Panarthropoda pinpoints the evolutionary loss of Sp5 in the common ancestor of the four arachnids we surveyed (spider, scorpion, mite, and harvestman). The presence of an Sp5 ortholog in Onychophora, one centipede, one horseshoe crab, and six sea spider species supports the inference that Sp5 was present in the common ancestor of panarthropods and also of chelicerates, and that a shared loss of Sp5 likely occurred in the common ancestor of arachnids. The absence of Sp5 in the genomes of O. fasciatus and P. hawaiensis is interpreted to constitute independent loss events (Fig. 10).
If we interpret the shared absence of arachnid Sp5 to mean that an Sp6-9/Dll cassette could have replaced the role of Sp5 in the common ancestor of arachnids, then we should expect to find evidence for gap gene-like expression for Sp6-9 and Dll in other arachnids as well. To test this prediction, we surveyed expression of Sp6-9 and Dll in early embryogenesis of the scorpion C. sculpturatus, following our previous approach to the study of this species (62) (harvestman embryos proved too fragile to examine at equivalent stages). Consistent with our prediction, we discovered that before the germband stage, Cscu–Sp6-9 and Cscu-Dll are expressed as a ring around the blastopore, which subsequently splits at one end, precisely as in spiders (Fig. 10 B and D).
While functional tools do not exist for scorpions, this datum accords with the interpretation that the cooption of the Sp6-9/Dll cassette into head segmentation occurred before the divergence of spiders from other arachnids. Future tests of this evolutionary scenario should emphasize expression surveys of Sp5 orthologs in Xiphosura and Pycnogonida to establish gap gene-like expression patterns in tandem with knockdown experiments of Sp6-9 in mites and harvestmen.
Methods
Bioinformatics and Phylogenetic Analysis.
Orthologs of Sp gene family members were identified in genomes of S. maritima (63), C. sculpturatus (64), P. tepidariorum (64), L. polyphemus (65), and E. rowelli (66) and from transcriptomes of P. opilio (67), L. atkinsoni (this study), Carcinoscorpius rotundicauda (65), Tachypleus tridentatus (65), Peripatopsis capensis (67), and seven pycnogonids. In addition, the genomes of O. fasciatus (68) and P. hawaiensis (69) were examined for the incidence of an Sp5 ortholog, which had not been found previously (29, 30, 32). For all searches, D. melanogaster D-Sp1 [National Center for Biotechnology Information (NCBI) accession no. ABW09374.2], P. hawaiensis Sp6-9 (NCBI accession no. CBH30981.1), and D. melanogaster arr (NCBI accession no. NP524737.2) were initially used as peptide sequence queries in BLAST searches, and hits with an e-value <10−5 were retained. All putative orthologs were verified using reciprocal BLAST searches.
Sequences previously compiled by Schaeper et al. (29) were downloaded from the GenBank database, and new putative orthologs were added for alignment. Multiple sequence alignment was conducted de novo with MUSCLE v.3.8.31 (70). Outgroup sequences used to root the tree consisted of KLF-9/13 orthologs of Nematostella vectensis (XP_001624390.1), T. castaneum (EEZ98378.1), Danio rerio (NP_001070240.1), and Mus musculus (NP_067341.2). For the Sp tree, we inferred phylogenies both with and without the KLF-9/13 outgroups and both with and without masking of ambiguously aligned sites using GBlocks v.091b (71) with parameters as specified in our previous work (64); all alignments are provided as Datasets S1–S3. For the LRP gene tree, the alignment was constructed anew, using LRP4 orthologs and a megalin sequence as outgroups. Due to the paucity of ambiguously aligned sites, the LRP alignment was not treated with GBlocks (Dataset S4).
Phylogenetic reconstruction of amino acid alignments consisted of maximum likelihood analysis with RAxML v.8.0 (72) under the LG + Γ model, with 250 independent starts and 250 bootstrap resampling replicates, and with Bayesian inference analysis with MrBayes v.3.2 under a mixed + Γ model (73). Four runs, each with four chains and a default distribution of chain temperatures, were run for 5 × 106 generations, with sampling every 5,000th iteration. Command files for phylogenetic analyses are provided as Datasets S5 and S6. Tree files are provided as Datasets S7–S10. Convergence was independently assessed using average split frequency and with Tracer v. 1.6. As a conservative treatment, 106 generations (20%) were discarded as burn-in.
Cloning of Orthologs and Probe Synthesis.
Fragments of Ptep–Sp6-9 were amplified using standard PCR protocols and cloned using a TOPO TA Cloning Kit using One Shot Top10 chemically competent Escherichia coli (Invitrogen) following the manufacturer’s protocol, and their PCR product identities were verified via sequencing with M13 universal primers. All gene-specific primers sequences are provided in SI Appendix, Table S1.
Embryo Collection, Fixation, and in Situ Hybridization.
Animals were maintained, and embryos fixed and assayed for gene expression, following established or minimally modified protocols, as detailed previously (62, 74). PCRs for generating riboprobe templates, synthesis of digoxin-labeled probes, and preservation of embryos all followed our recently detailed procedures (74). Probes were used at a concentration of 30–50 ng/μL. Sense probes were always developed for the same duration as complementary antisense probes. Completion of staining lasted 0.5–6 h at room temperature. Images were taken using a Nikon SMZ25 fluorescence stereomicroscope mounted with a DS-Fi2 digital color camera driven by Nikon Elements software.
Double-Stranded RNA Synthesis and RNAi in P. tepidariorum.
Double-stranded RNA (dsRNA) was synthesized following the manufacturer’s protocol using a MEGAscript T7 kit (Ambion/Life Technologies) from amplified PCR product. The quality of dsRNA was checked, and the concentration was adjusted as described previously (74).
Maternal RNAi was performed in virgin spider females (sisters from the same clutch) with injections every other day along the lateral surface of the opisthosoma, for a total of four injections. The dsRNA was injected at a concentration of 2.5 μg/μL, and 5 μg of dsRNA was delivered at each injection (total of 20 μg). Females were fed the first day after the final injection and mated within 24 h of the first (Ptep-arr RNAi) or final (Ptep–Sp6-9 RNAi) injection. Each set of pRNAi experiments was accompanied by a set of negative controls, which were injected with an identical volume of 1× Tribolium injection buffer. To rule out off-target effects, dsRNA was synthesized for injection as two nonoverlapping Ptep–Sp6-9 fragments of similar size (727 bp and 816 bp), with each injected into five females. Phenotypes were scored by severity, as described above; raw counts are reported in SI Appendix, Tables S2 and S3. Development was followed until stage 14, and embryos were periodically fixed and scored. Efficiency of knockdown was verified using in situ hybridization. An identical procedure was used to perform maternal RNAi against Ptep–Antp-1, Ptep-Dll, and Ptep-arr (SI Appendix, Fig. S9).
eRNAi against Ptep-Dll followed the original report of this procedure (33), with an 819-bp fragment. The first clutch of a newly mated female was obtained and divided into four sets of 100. One-quarter of the embryos were injected under halocarbon-700 oil with 1× Tribolium injection buffer, and the remaining 300 embryos were injected with Ptep-Dll-dsRNA; both solutions were mixed with a 1:20 dilution of rhodamine dextran for visualization. Embryos were reared for 4 d, and a subset of surviving embryos was assayed for Ptep–Sp6-9 expression.
Supplementary Material
Acknowledgments
Comments from the editors and two anonymous reviewers greatly refined the ideas and experiments presented. Sea spider egg clutches were collected by Georg Brenneis for sequencing of developmental transcriptomes. Carlos Santibañez López assisted with scorpion gene expression assays. Access to the onychophoran draft genome was kindly provided by Georg Mayer and Stephen Richards. Edits from Jesús A. Ballesteros, Guilherme Gainett, Gonzalo Giribet, Carlos E. Santibañez López, and Andrew Z. Ontano were incorporated into the manuscript. This material is based on work supported by the National Science Foundation under Grant IOS-1552610.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. N.H.P. is a guest editor invited by the Editorial Board.
Data deposition: All genomic resources have been deposited in Genbank (accession nos. MG857586–MG857620).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720193115/-/DCSupplemental.
References
- 1.Cisne JL. Evolution of the world fauna of aquatic free-living arthropods. Evolution. 1974;28:337–366. doi: 10.1111/j.1558-5646.1974.tb00757.x. [DOI] [PubMed] [Google Scholar]
- 2.Tiegs OW, Manton SM. The evolution of the Arthropoda. Biol Rev Camb Philos Soc. 1958;33:255–333. [Google Scholar]
- 3.Waloszek D, Chen J, Maas A, Wang X. Early Cambrian arthropods—New insights into arthropod head and structural evolution. Arthropod Struct Dev. 2005;34:189–205. [Google Scholar]
- 4.Panganiban G, Nagy L, Carroll SB. The role of the Distal-less gene in the development and evolution of insect limbs. Curr Biol. 1994;4:671–675. doi: 10.1016/s0960-9822(00)00151-2. [DOI] [PubMed] [Google Scholar]
- 5.Panganiban G, Sebring A, Nagy L, Carroll S. The development of crustacean limbs and the evolution of arthropods. Science. 1995;270:1363–1366. doi: 10.1126/science.270.5240.1363. [DOI] [PubMed] [Google Scholar]
- 6.Dong PD, Chu J, Panganiban G. Proximodistal domain specification and interactions in developing Drosophila appendages. Development. 2001;128:2365–2372. doi: 10.1242/dev.128.12.2365. [DOI] [PubMed] [Google Scholar]
- 7.Dong PDS, Dicks JS, Panganiban G. Distal-less and homothorax regulate multiple targets to pattern the Drosophila antenna. Development. 2002;129:1967–1974. doi: 10.1242/dev.129.8.1967. [DOI] [PubMed] [Google Scholar]
- 8.Prpic N-M, Tautz D. The expression of the proximodistal axis patterning genes Distal-less and dachshund in the appendages of Glomeris marginata (Myriapoda: Diplopoda) suggests a special role of these genes in patterning the head appendages. Dev Biol. 2003;260:97–112. doi: 10.1016/s0012-1606(03)00217-3. [DOI] [PubMed] [Google Scholar]
- 9.Angelini DR, Kaufman TC. Functional analyses in the hemipteran Oncopeltus fasciatus reveal conserved and derived aspects of appendage patterning in insects. Dev Biol. 2004;271:306–321. doi: 10.1016/j.ydbio.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Sharma PP, Schwager EE, Extavour CG, Giribet G. Evolution of the chelicera: A dachshund domain is retained in the deutocerebral appendage of Opiliones (Arthropoda, Chelicerata) Evol Dev. 2012;14:522–533. doi: 10.1111/ede.12005. [DOI] [PubMed] [Google Scholar]
- 11.Angelini DR, Kaufman TC. Insect appendages and comparative ontogenetics. Dev Biol. 2005;286:57–77. doi: 10.1016/j.ydbio.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 12.Estella C, Voutev R, Mann RS. A dynamic network of morphogens and transcription factors patterns the fly leg. Curr Top Dev Biol. 2012;98:173–198. doi: 10.1016/B978-0-12-386499-4.00007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen SM, Brönner G, Küttner F, Jürgens G, Jäckle H. Distal-less encodes a homoeodomain protein required for limb development in Drosophila. Nature. 1989;338:432–434. doi: 10.1038/338432a0. [DOI] [PubMed] [Google Scholar]
- 14.Cohen SM. Specification of limb development in the Drosophila embryo by positional cues from segmentation genes. Nature. 1990;343:173–177. doi: 10.1038/343173a0. [DOI] [PubMed] [Google Scholar]
- 15.Prpic N-M, Wigand B, Damen WG, Klingler M. Expression of dachshund in wild-type and Distal-less mutant Tribolium corroborates serial homologies in insect appendages. Dev Genes Evol. 2001;211:467–477. doi: 10.1007/s004270100178. [DOI] [PubMed] [Google Scholar]
- 16.Schoppmeier M, Damen WG. Double-stranded RNA interference in the spider Cupiennius salei: The role of Distal-less is evolutionarily conserved in arthropod appendage formation. Dev Genes Evol. 2001;211:76–82. doi: 10.1007/s004270000121. [DOI] [PubMed] [Google Scholar]
- 17.Angelini DR, Smith FW, Jockusch EL. Extent with modification: Leg patterning in the beetle Tribolium castaneum and the evolution of serial homologs. G3. 2012;2:235–248. doi: 10.1534/g3.111.001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma PP, Schwager EE, Giribet G, Jockusch EL, Extavour CG. Distal-less and dachshund pattern both plesiomorphic and apomorphic structures in chelicerates: RNA interference in the harvestman Phalangium opilio (Opiliones) Evol Dev. 2013;15:228–242. doi: 10.1111/ede.12029. [DOI] [PubMed] [Google Scholar]
- 19.Panganiban G, et al. The origin and evolution of animal appendages. Proc Natl Acad Sci USA. 1997;94:5162–5166. doi: 10.1073/pnas.94.10.5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janssen R, Eriksson BJ, Budd GE, Akam M, Prpic N-M. Gene expression patterns in an onychophoran reveal that regionalization predates limb segmentation in pan-arthropods. Evol Dev. 2010;12:363–372. doi: 10.1111/j.1525-142X.2010.00423.x. [DOI] [PubMed] [Google Scholar]
- 21.Wimmer EA, Jäckle H, Pfeifle C, Cohen SM. A Drosophila homologue of human Sp1 is a head-specific segmentation gene. Nature. 1993;366:690–694. doi: 10.1038/366690a0. [DOI] [PubMed] [Google Scholar]
- 22.Wimmer EA, Simpson-Brose M, Cohen SM, Desplan C, Jäckle H. Trans- and cis-acting requirements for blastodermal expression of the head gap gene buttonhead. Mech Dev. 1995;53:235–245. doi: 10.1016/0925-4773(95)00439-8. [DOI] [PubMed] [Google Scholar]
- 23.Wimmer EA, Frommer G, Purnell BA, Jäckle H. buttonhead and D-Sp1: A novel Drosophila gene pair. Mech Dev. 1996;59:53–62. doi: 10.1016/0925-4773(96)00575-8. [DOI] [PubMed] [Google Scholar]
- 24.Wimmer EA, Cohen SM, Jäckle H, Desplan C. buttonhead does not contribute to a combinatorial code proposed for Drosophila head development. Development. 1997;124:1509–1517. doi: 10.1242/dev.124.8.1509. [DOI] [PubMed] [Google Scholar]
- 25.Schöck F, Purnell BA, Wimmer EA, Jäckle H. Common and diverged functions of the Drosophila gene pair D-Sp1 and buttonhead. Mech Dev. 1999;89:125–132. doi: 10.1016/s0925-4773(99)00215-4. [DOI] [PubMed] [Google Scholar]
- 26.Estella C, Rieckhof G, Calleja M, Morata G. The role of buttonhead and Sp1 in the development of the ventral imaginal discs of Drosophila. Development. 2003;130:5929–5941. doi: 10.1242/dev.00832. [DOI] [PubMed] [Google Scholar]
- 27.Estella C, Mann RS. Non-redundant selector and growth-promoting functions of two sister genes, buttonhead and Sp1, in Drosophila leg development. PLoS Genet. 2010;6:e1001001. doi: 10.1371/journal.pgen.1001001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Córdoba S, Requena D, Jory A, Saiz A, Estella C. The evolutionarily conserved transcription factor Sp1 controls appendage growth through Notch signaling. Development. 2016;143:3623–3631. doi: 10.1242/dev.138735. [DOI] [PubMed] [Google Scholar]
- 29.Schaeper ND, Prpic N-M, Wimmer EA. A clustered set of three Sp-family genes is ancestral in the Metazoa: Evidence from sequence analysis, protein domain structure, developmental expression patterns and chromosomal location. BMC Evol Biol. 2010;10:88. doi: 10.1186/1471-2148-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Königsmann T, Turetzek N, Pechmann M, Prpic N-M. Expression and function of the zinc finger transcription factor Sp6-9 in the spider Parasteatoda tepidariorum. Dev Genes Evol. 2017;227:389–400. doi: 10.1007/s00427-017-0595-2. [DOI] [PubMed] [Google Scholar]
- 31.Beermann A, Aranda M, Schröder R. The Sp8 zinc-finger transcription factor is involved in allometric growth of the limbs in the beetle Tribolium castaneum. Development. 2004;131:733–742. doi: 10.1242/dev.00974. [DOI] [PubMed] [Google Scholar]
- 32.Schaeper ND, Prpic N-M, Wimmer EA. A conserved function of the zinc finger transcription factor Sp8/9 in allometric appendage growth in the milkweed bug Oncopeltus fasciatus. Dev Genes Evol. 2009;219:427–435. doi: 10.1007/s00427-009-0301-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pechmann M, et al. Novel function of Distal-less as a gap gene during spider segmentation. PLoS Genet. 2011;7:e1002342. doi: 10.1371/journal.pgen.1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wehrli M, et al. arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature. 2000;407:527–530. doi: 10.1038/35035110. [DOI] [PubMed] [Google Scholar]
- 35.Tamai K, et al. LDL-receptor-related proteins in Wnt signal transduction. Nature. 2000;407:530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- 36.He X, Semenov M, Tamai K, Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: Arrows point the way. Development. 2004;131:1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- 37.Angelini DR, Kaufman TC. Functional analyses in the milkweed bug Oncopeltus fasciatus (Hemiptera) support a role for Wnt signaling in body segmentation but not appendage development. Dev Biol. 2005;283:409–423. doi: 10.1016/j.ydbio.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 38.Miyawaki K, et al. Involvement of Wingless/Armadillo signaling in the posterior sequential segmentation in the cricket, Gryllus bimaculatus (Orthoptera), as revealed by RNAi analysis. Mech Dev. 2004;121:119–130. doi: 10.1016/j.mod.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 39.Bolognesi R, Fischer TD, Brown SJ. Loss of Tc-arrow and canonical Wnt signaling alters posterior morphology and pair-rule gene expression in the short-germ insect, Tribolium castaneum. Dev Genes Evol. 2009;219:369–375. doi: 10.1007/s00427-009-0299-3. [DOI] [PubMed] [Google Scholar]
- 40.Beermann A, Prühs R, Lutz R, Schröder R. A context-dependent combination of Wnt receptors controls axis elongation and leg development in a short germ insect. Development. 2011;138:2793–2805. doi: 10.1242/dev.063644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khadjeh S, et al. Divergent role of the Hox gene Antennapedia in spiders is responsible for the convergent evolution of abdominal limb repression. Proc Natl Acad Sci USA. 2012;109:4921–4926. doi: 10.1073/pnas.1116421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams TA. Distalless expression in crustaceans and the patterning of branched limbs. Dev Genes Evol. 1998;207:427–434. doi: 10.1007/s004270050133. [DOI] [PubMed] [Google Scholar]
- 43.Jockusch EL, Williams TA, Nagy LM. The evolution of patterning of serially homologous appendages in insects. Dev Genes Evol. 2004;214:324–338. doi: 10.1007/s00427-004-0412-6. [DOI] [PubMed] [Google Scholar]
- 44.Pechmann M, Prpic N-M. Appendage patterning in the South American bird spider Acanthoscurria geniculata (Araneae: Mygalomorphae) Dev Genes Evol. 2009;219:189–198. doi: 10.1007/s00427-009-0279-7. [DOI] [PubMed] [Google Scholar]
- 45.Barnett AA, Thomas RH. The expression of limb gap genes in the mite Archegozetes longisetosus reveals differential patterning mechanisms in chelicerates. Evol Dev. 2013;15:280–292. doi: 10.1111/ede.12038. [DOI] [PubMed] [Google Scholar]
- 46.Ronco M, et al. Antenna and all gnathal appendages are similarly transformed by homothorax knock-down in the cricket Gryllus bimaculatus. Dev Biol. 2008;313:80–92. doi: 10.1016/j.ydbio.2007.09.059. [DOI] [PubMed] [Google Scholar]
- 47.Mito T, et al. Divergent and conserved roles of extradenticle in body segmentation and appendage formation, respectively, in the cricket Gryllus bimaculatus. Dev Biol. 2008;313:67–79. doi: 10.1016/j.ydbio.2007.09.060. [DOI] [PubMed] [Google Scholar]
- 48.Sharma PP, et al. A conserved genetic mechanism specifies deutocerebral appendage identity in insects and arachnids. Proc Biol Sci. 2015;282:20150698. doi: 10.1098/rspb.2015.0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ober KA, Jockusch EL. The roles of wingless and decapentaplegic in axis and appendage development in the red flour beetle, Tribolium castaneum. Dev Biol. 2006;294:391–405. doi: 10.1016/j.ydbio.2006.02.053. [DOI] [PubMed] [Google Scholar]
- 50.Pechmann M, Khadjeh S, Sprenger F, Prpic N-M. Patterning mechanisms and morphological diversity of spider appendages and their importance for spider evolution. Arthropod Struct Dev. 2010;39:453–467. doi: 10.1016/j.asd.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 51.Akiyama-Oda Y, Oda H. Axis specification in the spider embryo: dpp is required for radial-to-axial symmetry transformation and sog for ventral patterning. Development. 2006;133:2347–2357. doi: 10.1242/dev.02400. [DOI] [PubMed] [Google Scholar]
- 52.Refki PN, Khila A. Key patterning genes contribute to leg elongation in water striders. Evodevo. 2015;6:14. doi: 10.1186/s13227-015-0015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takagi A, et al. Functional analysis of the role of eyes absent and sine oculis in the developing eye of the cricket Gryllus bimaculatus. Dev Growth Differ. 2012;54:227–240. doi: 10.1111/j.1440-169X.2011.01325.x. [DOI] [PubMed] [Google Scholar]
- 54.Presnell JS, Schnitzler CE, Browne WE. KLF/SP transcription factor family evolution: Expansion, diversification, and innovation in eukaryotes. Genome Biol Evol. 2015;7:2289–2309. doi: 10.1093/gbe/evv141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bell SM, et al. Sp8 is crucial for limb outgrowth and neuropore closure. Proc Natl Acad Sci USA. 2003;100:12195–12200. doi: 10.1073/pnas.2134310100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawakami Y, et al. Sp8 and Sp9, two closely related buttonhead-like transcription factors, regulate Fgf8 expression and limb outgrowth in vertebrate embryos. Development. 2004;131:4763–4774. doi: 10.1242/dev.01331. [DOI] [PubMed] [Google Scholar]
- 58.Lapan SW, Reddien PW. dlx and sp6-9 control optic cup regeneration in a prototypic eye. PLoS Genet. 2011;7:e1002226. doi: 10.1371/journal.pgen.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grimmel J, Dorresteijn AWC, Fröbius AC. Formation of body appendages during caudal regeneration in Platynereis dumerilii: Adaptation of conserved molecular toolsets. Evodevo. 2016;7:10. doi: 10.1186/s13227-016-0046-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 61.Janssen R, Budd GE, Damen WGM. Gene expression suggests conserved mechanisms patterning the heads of insects and myriapods. Dev Biol. 2011;357:64–72. doi: 10.1016/j.ydbio.2011.05.670. [DOI] [PubMed] [Google Scholar]
- 62.Sharma PP, Schwager EE, Extavour CG, Wheeler WC. Hox gene duplications correlate with posterior heteronomy in scorpions. Proc Biol Sci. 2014;281:20140661. doi: 10.1098/rspb.2014.0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chipman AD, et al. The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLoS Biol. 2014;12:e1002005. doi: 10.1371/journal.pbio.1002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwager EE, et al. The house spider genome reveals an ancient whole-genome duplication during arachnid evolution. BMC Biol. 2017;15:62. doi: 10.1186/s12915-017-0399-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kenny NJ, et al. Ancestral whole-genome duplication in the marine chelicerate horseshoe crabs. Heredity (Edinb) 2016;116:190–199. doi: 10.1038/hdy.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mayer G, et al. 2017 Velvet Worm Genome Project, Baylor College of Medicine. Available at https://www.hgsc.bcm.edu/arthropods/velvet-worm-genome-project. Accessed January 25, 2018.
- 67.Sharma PP, et al. Phylogenomic interrogation of arachnida reveals systemic conflicts in phylogenetic signal. Mol Biol Evol. 2014;31:2963–2984. doi: 10.1093/molbev/msu235. [DOI] [PubMed] [Google Scholar]
- 68.Vargas Jentzsch IM, et al. 2015 Oncopeltus fasciatus Official Gene Set OGS_v1.1 for genome assembly Oncopeltus fasciatus v1.0. Available at https://data.nal.usda.gov/dataset/oncopeltus-fasciatus-official-gene-set-v11_128. Accessed March 16, 2018.
- 69.Kao D, et al. The genome of the crustaceanParhyale hawaiensis, a model for animal development, regeneration, immunity and lignocellulose digestion. eLife. 2016;5:e20062. doi: 10.7554/eLife.20062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 72.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Setton EVW, et al. Expression and function of spineless orthologs correlate with distal deutocerebral appendage morphology across Arthropoda. Dev Biol. 2017;430:224–236. doi: 10.1016/j.ydbio.2017.07.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.