Significance
mRNA treatments represent an exciting approach to cure diseases that cannot be tackled with current therapeutics. However, the delivery of mRNA to target cells remains a challenge, but among the existing alternatives, lipid nanoparticles (LNPs) offer a promising answer to this. Here we determine the structure of LNPs encapsulating mRNA, consisting of a lipid mixture already evaluated in clinical trials. We show that the lipids are not homogeneously distributed across the LNP, and one of the lipids is localized mainly at its surface. The structural information enabled us to design LNPs that successfully modify intracellular protein production in two clinically relevant cell types. Our findings and approach provide a framework for understanding and optimizing vehicles for mRNA delivery.
Keywords: gene therapy, hEPO mRNA, small-angle scattering, adipocytes, hepatocytes
Abstract
The development of safe and efficacious gene vectors has limited greatly the potential for therapeutic treatments based on messenger RNA (mRNA). Lipid nanoparticles (LNPs) formed by an ionizable cationic lipid (here DLin-MC3-DMA), helper lipids (distearoylphosphatidylcholine, DSPC, and cholesterol), and a poly(ethylene glycol) (PEG) lipid have been identified as very promising delivery vectors of short interfering RNA (siRNA) in different clinical phases; however, delivery of high-molecular weight RNA has been proven much more demanding. Herein we elucidate the structure of hEPO modified mRNA-containing LNPs of different sizes and show how structural differences affect transfection of human adipocytes and hepatocytes, two clinically relevant cell types. Employing small-angle scattering, we demonstrate that LNPs have a disordered inverse hexagonal internal structure with a characteristic distance around 6 nm in presence of mRNA, whereas LNPs containing no mRNA do not display this structure. Furthermore, using contrast variation small-angle neutron scattering, we show that one of the lipid components, DSPC, is localized mainly at the surface of mRNA-containing LNPs. By varying LNP size and surface composition we demonstrate that both size and structure have significant influence on intracellular protein production. As an example, in both human adipocytes and hepatocytes, protein expression levels for 130 nm LNPs can differ as much as 50-fold depending on their surface characteristics, likely due to a difference in the ability of LNP fusion with the early endosome membrane. We consider these discoveries to be fundamental and opening up new possibilities for rational design of synthetic nanoscopic vehicles for mRNA delivery.
Recent advances in development of therapeutic modalities based on production of proteins in vivo by administration of mRNA represent a promising approach to treat diseases currently lacking therapeutic alternatives in multiple areas, including among others oncology (1) and cardiology (2). Compared with therapies based on DNA, mRNA has a greater potential for success because it does not need to enter the cellular nucleus, but unfortunately, it is known that mRNA is able to induce an immune response. Work by Kormann et al. (3) has shown that chemically modified mRNA provides tunable protein expression with minimal immune activation. However, the major challenge for these types of treatments is still the delivery of the long, negatively charged nucleic acid into target cells while avoiding degradation (4, 5). Viral vectors have a high transfection efficacy but have the potential of eliciting undesirable immune responses (6). The evaluation of nonviral vectors is particularly topical given the approval by the Recombinant DNA Advisory Committee to allow viral vector-mediated gene therapy to progress into the clinical setting, a decision which has been severely scrutinized (7). Therefore, delivery vehicles based on nonviral cationic species, such as lipids and polymers, have become an attractive alternative because they are able to envelop different types of nucleic acids, deliver them across the cell plasma membrane, and facilitate endosomal escape of the therapeutic molecule (8, 9).
Currently, formulations of siRNA in lipid nanoparticles (LNPs) formed by a cationic ionizable lipid (CIL) (8), cholesterol (Chol), distearoylphosphatidylcholine (DSPC), and a poly(ethylene glycol) (PEG) lipid are showing promising results in different phases of clinical research (10). Development of LNPs based on CILs has been accelerated because such LNPs can be readily produced with high encapsulation efficiency (11) using a microfluidic mixer (12) and show lower toxicity compared with other lipid-based vehicles (10). Recently, Leung et al. (13) showed that mRNA can be efficiently encapsulated in LNP formulations. The effect, however, of the composition of these formulations on transfection efficacy of mRNA remains to be demonstrated.
An important aspect to consider in the design of LNPs is the effect of particle size on activity. Particle size can be controlled by varying PEG lipid content with smaller LNPs being generated at high ratios of PEGylated lipid (11). The relationship between particle size and cellular uptake via endocytosis has been addressed in the past decade for different types of particles through application of both theoretical and experimental approaches (14, 15). Previous studies have shown conflicting results regarding the optimal size of different types of siRNA-containing LNPs (siRNA–LNPs) and their efficiency in reducing protein levels (16–18). These findings indicate that other factors, such as lipid composition and distribution within LNPs, might contribute significantly to the observed effects. From a structural perspective, it has been proposed that the interior of siRNA–LNPs exhibits an internal inverted micellar structure, a so-called L2 phase, based on molecular dynamics simulations (19, 20). The current model also proposes that all of the lipids, except for PEG lipid that sits at the surface, are colocalized across the LNPs (20). These models are, however, not yet supported by empirical experimental data. Knowledge of the structures formed by lipid–nucleic acid complexes becomes essential because it is believed that some structural arrangements more efficiently facilitate endosomal escape by promoting membrane disruption (21). For example, Kim et al. (22) have shown that lipid-based particles with an ordered bicontinuous cubic internal structure have the ability to deliver siRNA with a higher efficiency compared with other lipoplex systems with an internal lamellar arrangement.
In this work, we have investigated the structure, composition, and transfection efficacy of mRNA-containing LNPs (mRNA–LNPs) and demonstrated size-dependent differences with respect to these characteristics. Furthermore, LNP uptake and protein expression were explored for two different cell types, human adipocytes and hepatocytes, which were selected based on their clinical relevance in s.c. and i.v. administration, respectively. The structure of LNPs was determined using a combination of cryogenic transmission electron microscopy (cryo-TEM), dynamic light scattering (DLS), freeze fracture, and small-angle X-ray and neutron scattering (SAXS and SANS, respectively). We determined the lipid distribution within LNPs using isotopic contrast variation in the SANS experiments. The nanoscale structural information obtained was directly applied to master the formulation of LNPs with optimal size and composition, and we successfully demonstrated how this improves the in vitro transfection efficacy in human adipocytes and hepatocytes.
Results
In Vitro Transfection Efficacy of LNPs of Different Size.
LNPs containing human erythropoietin (hEPO) mRNA were formulated with the CIL known as DLin-MC3-DMA. Previously, Jayaraman et al. (23) demonstrated that the most active CILs had pKa values between 6.2 and 6.5, and siRNA–LNPs formulated with DLin-MC3-DMA exhibited the greatest gene-silencing activity among 56 different amino CILs. By applying the protocol of Belliveau et al. (11), the lipid molar compositions of DLin-MC3-DMA:DSPC:Chol:DMPE-PEG2000 was varied in the molar ratio 50:10:40-X:X, with the increase in the PEG lipid content directly compensated by a decrease in the Chol content. Typical diameters of siRNA–LNPs range between 30 and 80 nm when the PEG lipid content is decreased from 5 to 0.5 mol %, respectively (11, 16). In our experiments, the size of mRNA–LNPs was varied between ∼45 and 135 nm (number average diameter, <d>N), by changing the DMPE-PEG2000 content from 3 to 0.25 mol %, respectively. A comparison between the size of empty LNPs and mRNA–LNPs is detailed in Fig. S1. The mRNA–LNPs were significantly larger than empty ones, indicating that mRNA contributes structurally to particle volume and possibly also to the internal structure. Moreover, LNPs formulated with mRNA were found also to be larger than the average values reported for siRNA–LNPs (11, 16). It must be considered that siRNA has generally between 20 and 25 base pairs, whereas hEPO mRNA has 858 nucleotides. Additionally, the preparation protocol has an effect on the structure of the particles. In this case, LNPs were prepared using the fast mixing precipitation protocol, which was done in a microfluidic mixer (12, 13). This process is highly reproducible and likely to be used in scaling-up processes.
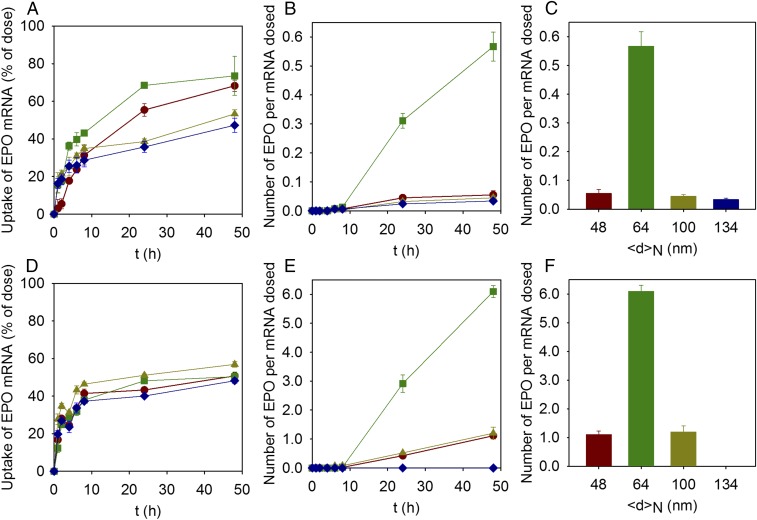
Fig. 1 shows LNP-mediated uptake of hEPO encoding mRNA in human adipocytes (Fig. 1 A–C) and induced pluripotent stem cell (iPSC)-derived hepatocytes (Fig. 1 D–F), as a function of LNP size and time postdosing. The uptake of LNPs was tracked using 3H-labeled DSPC as a marker. Although there was no significant size-dependent difference in LNP uptake in hepatocytes, adipocytes exhibited slight variations in uptake based on particle size. In adipocytes, LNPs with a number average diameter of ∼65 nm were taken up more readily compared with smaller and larger average-sized LNPs (Fig. 1 A and D). Furthermore, we found that LNP uptake was initiated almost instantaneously (within the first hour) following dosing with the uptake kinetics displaying a plateau after 24 h in adipocytes, whereas in hepatocytes the plateau was reached already after 8 h (Fig. 1 A and D). We also observed that although not all LNPs were taken up, about 50% were located intracellularly 48 h postdosing, measured by the disappearance of the 3H-DSPC radioactivity from the extracellular medium, irrespective of particle size or cell type (Fig. 1 A and D).
Fig. 1.
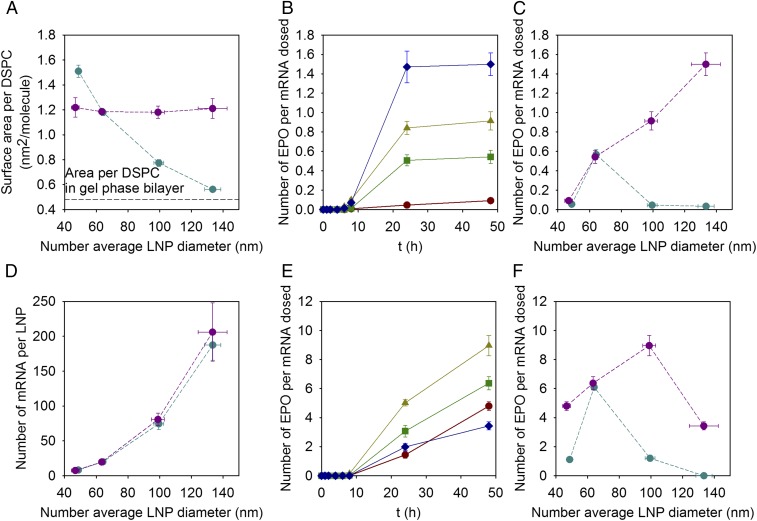
Cellular uptake and cellular protein production for LNPs of different sizes prepared with the standard variation of size approach. (A and D) Uptake of LNPs expressed as the percent of hEPO mRNA dosed as a function of time in (A) adipocytes and (D) hepatocytes for LNPs with lipid molar compositions of DLin-MC3-DMA:DSPC:Chol:DMPE-PEG2000 in the ratio 50:10:40-X:X for X: 3% (<d>N = 48 nm; red circles), 1.5% (<d>N = 64 nm; green squares), 0.5% (<d>N = 100 nm; yellow triangles), and 0.25% (<d>N = 134 nm; blue diamonds). Lines are to guide the eye. (B and E) Number of hEPO produced per mRNA dosed as a function of time in (B) adipocytes and (E) hepatocytes for LNPs with the same lipid composition as A and D. Lines are to guide the eye. (C and F) Number of hEPO produced per mRNA dosed after 48 h of dosing (C) adipocytes and (F) hepatocytes for LNPs with the same lipid composition as A and D. The experiments were done in the presence of 1% human serum. Values are means ± SEM (n = 3).
The kinetic data show that no hEPO was detected before 8 h in any of the cell types, independently of LNP size (Fig. 1 B and E); however, LNPs with a number average diameter of ∼65 nm (1.5 mol % of DMPE-PEG2000) showed significant hEPO production (Fig. 1 B, C, E, and F) after 20 h. Notably, the protein expression was more than 10-fold higher in hepatocytes, ∼6 hEPO protein molecules per dosed mRNA, than in adipocytes, ∼0.6 hEPO protein molecules per dosed mRNA. Because there are negligible size-dependent differences in LNP uptake, our data thus indicate that the limiting step for protein production is release of the mRNA from endosomes (endosomal escape) and that this process per se is critically dependent on LNP particle size and cell type.
LNP Structure and Composition.
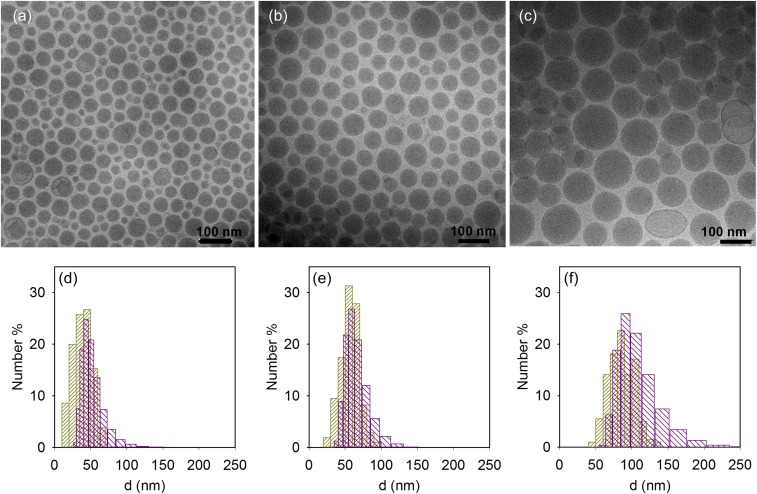
Fig. 2 shows electron micrographs of hEPO mRNA–LNPs, which were formulated as previously described for the in vitro studies by varying the content of PEG lipid and Chol. We found that mRNA–LNPs are spherical and have an electron-dense core as reported previously by Leung et al. (19). Some vesicles were also found in the sample with largest LNPs, whereas some of the smallest LNPs have slight defects or “blebs” as described by Leung et al. (13). The size distribution was also determined from the cryo-TEM images and correlated well with the average size and distribution as determined using DLS. It is important to note that although low polydispersity has been reported (11), there was still a slight distribution in size and that the level of distribution was, in itself, dependent on the average size of the particle population. In fact, the overlap of the size distributions for the different preparations (Fig. 2 D–F) suggests that the dramatic effects observed in hEPO protein production (Fig. 1) might be due either to structural differences between the different preparations or that only LNPs with a size very close to 65 nm lead to efficient endosomal escape. This finding motivated an investigation into the internal structure of mRNA–LNPs to understand the remarkable difference in protein expression as a function of the size of the LNPs.
Fig. 2.
Cryo-TEM characterization of the LNPs. (A–C) Cryo-TEM images and (D–F) size number distribution of LNPs with lipid molar compositions of DLin-MC3-DMA:DSPC:Chol:DMPE-PEG2000 in the ratio 50:10:40-X:X for X: (A and D) 3, (B and E) 1.5, and (C and F) 0.5. The size distribution corresponds to the DLS measurements (purple) and cryo-TEM image analysis (yellow).
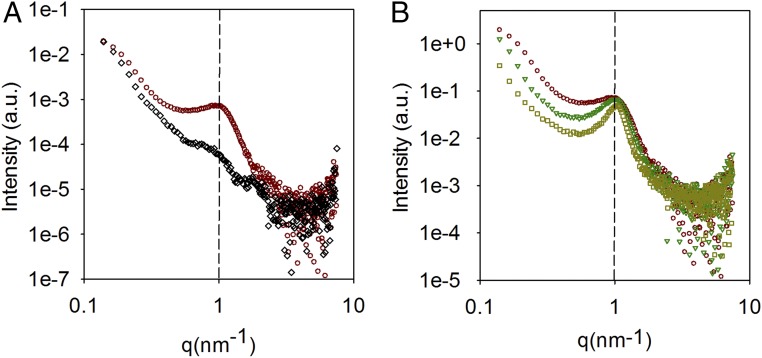
Fig. 3 show SAXS data for LNPs and provides clear evidence for an internal structure of mRNA–LNPs. Notably, the structure is particularly pronounced in the presence of mRNA; that is, the structural peak is almost not present in the LNP sample without mRNA. Furthermore, from Fig. 3B, the structure in mRNA–LNPs seems to be size invariant, manifested by the fact that all of the curves show a common peak at q ∼ 1 nm−1, which corresponds to a correlation distance of ∼6 nm. The peak becomes more pronounced as the PEG lipid content decreases; that is, the size of the LNP increases, but the position along the q axis is essentially the same. Because the presence of only one peak does not allow a conclusion regarding the type of internal arrangement, it is not obvious if this internal structure is crucial for the functional response observed previously as a function of the size of the mRNA–LNPs. Thus, further analysis of the lipid distribution on the LNPs was performed using the isotropic contrast variation method in SANS.
Fig. 3.
SAXS characterization of LNPs. (A) Small-angle X-ray scattering data for LNPs without mRNA with lipid molar compositions of DLin-MC3-DMA:DSPC:Chol:DMPE-PEG2000 in the ratio 50:10:40-X:X for X: 1.5% (<d>N = 42 nm; black diamonds) and mRNA containing LNPs for X: 3% (<d>N = 48 nm; red circles). (B) Small-angle X-ray scattering data for mRNA containing LNPs for X: 3% (<d>N = 48 nm; red circles), 1.5% (<d>N = 64 nm; green triangles) and 0.5% (<d>N = 100 nm; yellow squares). The dashed black vertical line corresponds to the position of the structural peak.
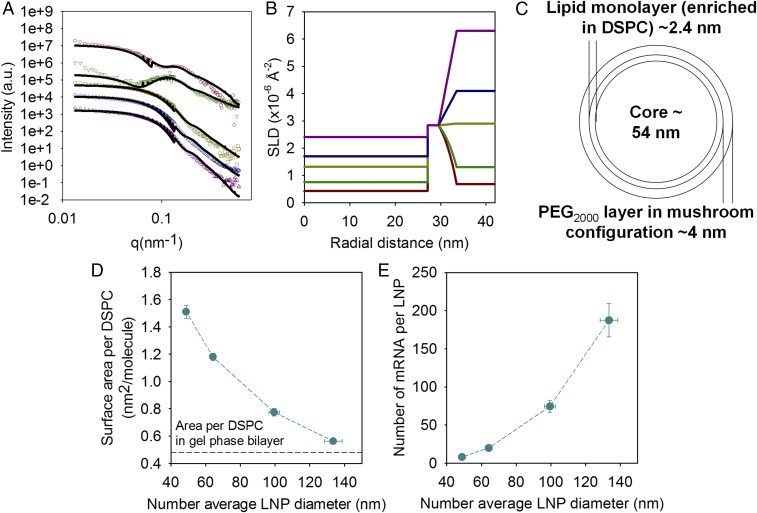
Fig. 4A shows SANS data of LNPs containing 1.5 mol % of PEG lipid and deuterated DSPC and Chol in solvents with different D2O/H2O ratios. The scattering only from samples with mRNA in buffers with the lowest D2O content (18% and 27%, respectively), where the scattering length density (SLD) of DSPC has the largest difference with the SLD of the solvent compared with the other lipids, shows a distinct bump in the low-q region, whereas the data for the highest D2O content do not. This type of scattering curve is similar to those reported for SANS measurements using isotopic contrast variation of core–shell colloidal systems that are sterically stabilized by PEG grafts (24). Simultaneous fitting of the data in Fig. 4A using a core and two shells model agrees best with the experimental results with a minimum number of fitted parameters. Fig. 4B shows the SLD profiles as a function of the distance to the center of the LNP, and Fig. 4C gives a schematic representation of the model used in the scattering analysis. In this model, the LNP is formed by a core containing DLin-MC3-DMA, Chol, water, and mRNA. The core is surrounded by a layer of ∼2.4 nm in thickness, which corresponds well to a monolayer containing the DSPC and a fraction of the CIL, Chol, and the DMPE portion of the PEG lipid. The second layer is ∼4 nm in thickness, which is commensurate with a PEG lipid layer in a mushroom configuration (25). This concurs well with the hypothesis proposed by Belliveau et al. (11) that PEG lipids are mainly located on the surface of LNPs and that a larger relative proportion of PEG lipids allows higher surface-to-volume ratios and consequently smaller particle sizes.
Fig. 4.
Characterization of the lipid distribution within the LNPs, their surface composition, and number of mRNA per LNPs. (A) SANS data (symbols) for mRNA containing LNPs with a lipid molar compositions of DLin-MC3-DMA:DSPC:Chol:DMPE-PEG2000 in the ratio 50:10:38.5:1.5 with deuterated DSPC and Chol in 18% (red circles), 27% (green inverted triangles), 50% (yellow squares), 68% (blue diamonds), and 100% (purple triangles) D2O buffer. The solid lines correspond to the best fit using the multishell model with exponential decay. Only the data at low q are plotted, and the intensity of the samples in the upper curves has been offset for clarity. (B) Scattering length density (SLD) profiles as a function of the distance to the center of the LNPs corresponding to the fits of the data in A. (C) Schematic representation of the lipid distribution in the LNP according to the SLD radial profile. (D) Area per DSPC molecule and (E) number of mRNA per LNP as a function of the LNP size for LNPs with lipid molar compositions of DLin-MC3-DMA:DSPC:Chol:DMPE-PEG2000 in the ratio 50:10:40-X:X. The horizontal line in D corresponds to the reported value of the area per DSPC in the gel phase (26). Lines are to guide the eye. Values are means ± SEM (n = 3).
The fact that the same core and two-shell model captures the scattering under several contrast conditions suggests that the SLD profiles in Fig. 4B are quite reasonable. Based on these SLD profiles, the composition of the system can be elucidated. The SLD of the solvent is given by the H2O/D2O ratio and the SLD of the PEG layer varies as it extends into the solvent. The SLD of the core is constant inside the LNP, but it changes as a function of the D2O content. The SLD of mRNA also increases as the volume fraction of D2O increases because it has labile hydrogens that can be exchanged for deuterium (see values of SLD in Table S1). Using the fitted values for the SLDs of the core as a function of the D2O content and the known SLD values of the LNP components (Table S1) with the established lipid:mRNA mole ratio, it is possible to calculate the water content in the LNP, according to
where v is the volume fraction of the various components. The estimated water volume fraction was 24 (±2)%. The SLD of the lipid monolayer remains constant independently of solvent, which confirms that no solvent is present in this layer as expected. This layer contains all of the DSPC but also DLin-MC3-DMA, Chol, and the DMPE region of the PEG lipid. Provided that all DSPC is located in the monolayer that surrounds the LNP core, it is possible to calculate the surface area per DSPC for this lipid composition, which is on average 1.2 nm2 per molecule. It has been reported that DSPC in the gel phase has an area per molecule of 0.48 nm2 (26), which is smaller than the estimated area per DSPC for all of the LNP sizes (Fig. 4D). The calculated surface area per DSPC as a function of the PEG lipid content in the LNP (Fig. 4D), assuming all of the DSPC is at the surface and a volume fraction of water of 24%, shows that the area per DSPC decreases as the particle size increases. That means that as the particle size increases, the DSPC content at the surface increases, whereas the one corresponding to the other lipid components decreases. This observation is consistent with the observed formation of pure DSPC vesicles in Fig. 2C for the larger LNPs. Furthermore, taking into account the data regarding water content and lipid distribution in the LNPs, it is possible to estimate the number of mRNA molecules per LNP as a function of their size (Fig. 4E).
To gain a better understanding of the structural origin of the peak in the SAXS data in Fig. 3, we focused on the components that according to the SANS data primarily contribute to the core of the LNP, i.e., DLin-MC3-DMA, Chol, and mRNA. An experiment was designed to replicate the core phase of the LNPs in which these components were dialyzed against a buffer pH 3:ethanol mixture in a 3:1 volume ratio, followed by dialysis against PBS buffer (pH 7.4). The solvents were chosen to emulate the formation of LNPs using the microfluidic mixer and the subsequent dialysis. We also selected polyadenylic acid (polyA), an adenosine monophosphate homopolymer, as a model for mRNA based on simplification of structure and consistent biophysical properties. Fig. 5 A and B show the scattering data of the bulk phase in the presence and absence of polyA for both buffer conditions.
Fig. 5.
Core structure of the LNPs and cholesterol solubility. (A and B) SAXS data for DLin-MC3-DMA:Chol mixtures in 50:38.5 mol ratio in the presence of polyA (red curve) and absence (black curve), which have been dialyzed against (A) citrate buffer:ethanol 3:1 volume mixture and (B) PBS buffer. The dashed black vertical lines correspond to the position of the structural peaks, and the pink continuous lines correspond to cholesterol monohydrate crystals. For all of the SAXS data, the intensity of the samples in the upper curves has been offset for clarity. (C) Measured light scattering intensity as a function of the mole percentage of cholesterol for the addition of cholesterol nanocrystals to a DLin-MC3-DMA emulsion in citrate buffer:ethanol 3:1 volume mixture. The solid line is a fit to the data with high cholesterol content. The solubility is estimated from extrapolating the line to zero intensity. (D and E) TEM images for DLin-MC3-DMA:Chol mixtures in 50:38.5 mol ratio in the presence of polyA, which have been dialyzed against (D) citrate buffer:ethanol 3:1 volume mixture and (E) PBS buffer. Insets correspond to a higher magnification of the image. (F) Schematic representation of the proposed disordered inverse hexagonal phase inside the mRNA–LNPs core.
The SAXS data of all of the bulk samples showed two characteristic peaks at q values of ∼1.86 and 3.71 nm−1 that correspond to Bragg spacings of 3.38 and 1.69 nm, respectively. These peaks have been identified previously as Chol monohydrate crystals, present in Chol/lipid mixtures by Huang et al. (27). To understand the origin of these peaks, we used light scattering to estimate the solubility of Chol nanosuspensions in DLin-MC3-DMA emulsions in the buffer pH 3:ethanol mixture (Fig. 5C). The measured solubility of Chol corresponded to a DLin-MC3-DMA:Chol molar ratio of 76:24, which is below the commonly employed ratio for the preparation of LNPs of ∼56:44. Because the highest percentage of the Chol nanocrystals were of the anyhdrated form, the solubility of the monohydrate form could be calculated by extrapolating from the work of Jandacek et al. (28), who report the solubility of anhydrate and monohydrate Chol in triolein at 21 °C and 37 °C. Based on their data, the solubility of monohydrate Chol corresponds to a DLin-MC3-DMA:Chol molar ratio of 82:18. We found that decreasing the amount of Chol in the samples resulted in a decrease in the intensity of the Chol monohydrate peaks (Fig. S2). SAXS and SANS data of LNPs did not indicate the presence of Chol monohydrate crystals. Two-dimensional Chol crystal domains can, however, form in lipid bilayer membranes (29), and the presence of Chol crystals at the LNP surface cannot therefore be definitively ruled out.
Regarding the structure of the bulk phase of the LNPs, the presence of the three peaks in a q-position ratio of 1:√3:√4 is an indication of the formation of an inverted hexagonal phase, a so-called H2 phase. This shows that the LNP structure at low pH is formed by water and/or water–mRNA rigid cylinders packed in a hexagonal arrangement. The first peak appears at q = 1.17 nm−1 and corresponds to a cylinder center-to-center distance of 6.2 nm. In fact, the micrographs of LNPs in this solution of pH 3 with 25% ethanol (Fig. S3) show that the particles are not spherical but more irregular, which is consistent with this type of arrangement. The scattering data from the bulk samples in PBS are strikingly similar to the empty and mRNA–LNPs of comparable size and solvent conditions (Figs. 3A and 5B) manifesting the effect mRNA has on the structure. The sample containing polyA in PBS shows one sharp structural peak at the same position as the first peak of the sample at low pH suggesting a disordered inverse hexagonal phase type structure also at neutral pH.
Fig. 5 D and E show electron micrographs from the bulk phases with polyA at low and neutral pH. The images were taken upon freeze fracture preparation of the samples. Fig. 5D clearly exhibits channels that correspond to the hexagonal phase rods. These rods have a separation distance of ∼6 nm, which corresponds well to the distance calculated from SAXS. Fig. 5E shows a more disordered phase that upon closer look appear to be distorted channels with a similar distance as observed at low pH. The combination of the SAXS data and the TEM images of the LNPs’ bulk phase at neutral pH are further evidence that the LNP cores are not structured in a multilamellar or inversed micellar phase conformation, as it has been proposed previously for siRNA–LNPs (19, 20). A schematic representation of the proposed internal structure is shown in Fig. 5F. The sample without polyA, however, has a significantly different structure manifested as a broad bump.
Optimizing the Design of LNPs.
The structural analysis revealed that the internal structure of the mRNA–LNPs corresponds to a disordered inversed hexagonal phase, irrespectively of their size. However, this does not explain why LNPs with a number average size of ∼65 nm and surface area per DSPC of 1.2 nm2 gave the maximum protein expression in both adipocytes and hepatocytes. The structural information therefore suggests that surface composition of LNPs plays a major role in transfection efficacy. By varying the lipid composition but maintaining the DLin-MC3-DMA:Chol molar ratio, we formulated LNPs that had different sizes but a constant surface area per DSPC molecule at 1.2 nm2 (Fig. 6A). These LNPs were applied in in vitro cellular experiments, comparing them to LNPs with variable surface area per DSPC molecule.
Fig. 6.
The surface composition and size of LNPs regulates cellular production. (A) Area per DSPC molecule and (D) number of mRNA per LNP as a function of the LNP size for LNPs with variable (blue) and constant (purple) surface composition. The horizontal line corresponds to the reported value of the area per DSPC in the gel phase (26). (B and E) Number of hEPO produced per mRNA dosed as a function of time in (B) adipocytes and (E) hepatocytes for LNPs with constant surface composition and different size: <d>N = 47 nm (red circles), <d>N = 64 nm (green squares), <d>N = 99 nm (yellow triangles), and <d>N = 133 nm (blue diamonds). (C and F) Number of hEPO expressed per mRNA dosed after 48 h of dosing (C) adipocytes and (F) hepatocytes for LNPs with variable (blue) and constant (purple) surface composition. Lines are to guide the eye. The experiments were done in the presence of 1% human serum. Values are means ± SEM (n = 3).
The uptake of the LNPs was, in general terms, consistent with previous results with respect to demonstration that uptake per se is not dependent on particle size albeit being slightly lower than in previous experiments (Fig. S4). In addition, despite the slightly lower uptake, the number of hEPO protein molecules generated per mRNA molecule did not change for LNPs with an average size of ∼65 nm, and in fact, it showed an excellent reproducibility considering that the experiments were carried out with new cells and new LNPs and a few months apart. Furthermore, this indicates that the limiting step in transfection is not the particle uptake but instead the release of mRNA from the endosomal compartment. Notably, it was observed that the ability of particles with a constant surface area per DSPC to facilitate hEPO protein generation differed from the previously investigated LNPs (Fig. 6 B, C, E, and F). When the surface area per DSPC was kept at 1.2 nm2, the LNP transfection efficiency increased markedly with increasing particle size, in both human adipocytes and hepatocytes. In the case of human adipocytes, protein expression was almost three times higher for the largest LNPs with a constant surface area per DSPC, compared with the best performing LNPs (∼65 nm) with a variable surface area. In hepatocytes, protein generation also increased 1.4 times with increased particle size, ∼100 nm vs. ∼65 nm, although a decrease in expression was observed for the largest particles. It should, however, be pointed out that the protein expression for the largest LNPs (<d>N = 130 nm) in both adipocytes and hepatocytes was increased significantly, about 50-fold in the adipocytes, using 1.2 nm2 per DSPC. Moreover, from previous experiments in hepatocytes the protein expression was below the limit of detection, so the improvement was significant also in hepatocytes. The calculated number of hEPO mRNAs per LNP as a function of size does not change with the surface area per DSPC (Fig. 6D). It is therefore clear through these data that both surface composition and size of LNPs determine transfection efficacy in vitro in both cell types.
Discussion
LNPs have proven to have potential as delivery vehicles for mRNA in therapeutic treatments. Recently, Pardi et al. (30) reported delivery of LNPs containing luciferase mRNA in vitro (human embryonic kidney and dendritic cells) and in vivo through different administration routes (e.g., i.v. and s.c.). Patel et al. (31) found that modulation of the mTOR pathway could help to enhance or inhibit the LNP-mediated mRNA delivery. Our work focused on revealing the structure and composition of mRNA–LNPs, and based on the data, we propose and apply methods for rational design of LNPs to optimize their performance.
The most striking observation in this work is that the phospholipid DSPC is mainly located at the surface of mRNA–LNPs. The determinant for DSPC localization is still unclear, but the formation of an inverse hexagonal phase between oppositely charged DLin-MC3-DMA and mRNA could be attributed to the segregation of this phospholipid because the curvature of this phase is not favorable for DSPC. The effect of surface area per DSPC on protein expression is also remarkable, considering that this lipid is zwitterionic and forms a lamellar phase on its own. Thus, DSPC should not contribute toward endosomal escape. Other components at the LNP surface must, therefore, favor this mechanism, especially DLin-MC3-DMA and Chol because it is presumed that the PEG lipid will be shed before cellular uptake. It follows that the DLin-MC3-DMA, which is practically uncharged at physiological pH, can become ionized once endocytosed and entrained in the early endosome compartment. Because these early endosomes are expected to contain negatively charged phosphatidylserine lipids, one may speculate that the LNP cargo might escape by LNPs adhering to and fusing with the early endosomal membrane through electrostatic means. Similar mechanism has been discussed previously by Safinya and coworkers (32). The concomitant decrease of DLin-MC3-DMA at the surface for the variable surface composition LNPs would be expected as the amount of DSPC increases in this layer, indicated by the small surface area per DSPC as the LNP size increases, approaching almost a pure DSPC monolayer. With larger LNPs containing more mRNA, this scenario would explain the dramatic shift in hEPO production between LNPs of variable and constant surface composition in Fig. 6. Additionally, we found that the solubility of Chol in the bulk phase was low, and this might suggest an enrichment of Chol and potentially the formation of Chol crystalline domains at the LNP surface. We are currently investigating the presence of these crystals on LNPs because they may contribute to increased endosomal escape. Additionally, the low lipid packing found at the surface of the LNP could support the idea of Chol crystals.
The combined X-ray and neutron scattering data and the electron micrographs obtained from the bulk phase and the disperse phase indicate that the internal structure of mRNA–LNPs under neutral pH conditions is a disordered inverse hexagonal phase, independent of LNP particle size. From the data in Fig. 5 A and D, the DLin-MC3-DMA/Chol system, in the absence of RNA, clearly forms a regular inverse hexagonal H2 phase at the LNP preparation conditions, pH 3 with 25% ethanol. At this condition, DLin-MC3-DMA is expected to be fully ionized (apparent pKa 6.44) (23). The addition of RNA to the LNPs, in pH 3 and 25% ethanol, decreases the order because the second-order peaks become less pronounced. This can be understood as follows: RNA (with its counterions), located inside the aqueous cylinders, screens the charges of the DLin-MC3-DMA, thereby making cylinders more flexible, i.e., decreasing their persistence length. At neutral conditions (PBS, pH 7.4, data in Fig. 5 B and E), DLin-MC3-DMA will be less ionized, and the nonionic fraction (expected to be significant at this pH) will to some extent act as a solvent, leading to a further decrease of the order in the system. In fact, this phase, when observed by polarized microscopy, is isotropic, which is also consistent with the almost perfect spherical shape of the LNPs (Fig. 2).
Considering the observed water volume fraction of water (24%) and the calculated volume fraction of RNA (3%), this system is rather concentrated with respect to aqueous mRNA containing cylinders. Currently, based on the lack of validated scattering theories we are not able to discriminate between a disordered inverse hexagonal phase and a concentrated wormlike inverse micellar system (with significant excluded volume interactions). We conclude that in the event of the fusion of a LNP with an oppositely charged membrane, e.g., in the endosomal compartment, the mRNA in either case will be able to diffuse (by reptation) out from the LNPs, thereby being released. Likewise, Koltover et al. (33) previously showed that cationic liposome–DNA complexes that form inverse hexagonal phases have a higher transfection efficacy compared with complexes with a lamellar structure. This may explain the potential superior transfection efficiency of LNPs in contrast to other lipid-based delivery vectors. It does not, however, explain the effect of size on the transfection efficacy.
Regarding the in vitro cell experiments, an important observation based on data reported in this work is that the difference in cellular uptake of LNPs of varying size is rather modest. Gao et al. (14) and Zhang et al. (15) have proposed theoretical models to explain size-dependent endocytosis of nanoparticles. Both studies concluded that the optimal particle size for uptake was ∼50–60 nm. It was suggested that smaller particles were taken up less efficiently due to the energy penalty of the membrane bending, whereas uptake of larger particles is limited by the rate of receptor diffusion within the plane of the plasma membrane to the site of nanoparticle uptake (14, 15). These models are in agreement with the results relating to uptake of mRNA–LNPs with variable surface composition in adipocytes, but they fail to explain the results specifically in hepatocytes. It is not clear if the results in hepatocytes could be attributed to wider optimal size distribution for uptake or other alternative uptake mechanisms. Moreover, it is important to emphasize that although these nanoparticles have low polydispersity, there is an evident size distribution within each size category of LNPs. This is something that is common for all formulations as shown in Fig. 2.
There are contradictory results in the literature regarding the optimal particle size of LNPs for RNA delivery. In previous work, Akinc et al. (18) prepared lipidoid–siRNA formulations with constant lipid composition but differing in particle size by extruding particles through membranes with different pore sizes. They found that in vivo efficacy of their lipidoid–siRNA formulation increased as the particle size decreased, and they argued that smaller particles access hepatocytes more effectively. In contrast, Bao et al. (17) showed that the gene silencing activity of siRNA–LNPs was inversely correlated with the amount of PEG lipid. Thus, the largest LNPs with 0.5 mol % of PEG-DMA perform the best, and this was attributed to a better endosome disrupting ability. Recently, Chen et al. (34) investigated hepatic gene silencing potency of siRNA–LNPs in vivo following i.v. administration. In their work, LNPs with a number-average diameter of 78 nm resulted in the highest gene silencing, compared with smaller and larger LNPs. The authors also showed that the potency of LNPs is related to the content of cationic lipid. In contrast, we report data that demonstrate that particle size alone is not a determinant for the effects of the transfected nucleic acid. Instead, we demonstrate that there is a critical dependence on both particle size and particle surface composition with the latter being behind most of the effect in particle sizes greater than ∼65 nm in size.
Our results indicate that the limiting step in transfection efficacy is the release of mRNA from the (early) endosomal compartment because the uptake of LNPs is not correlated with their optimal size/composition for protein expression. Gilleron et al. (35) estimated that only 1–2% of siRNAs delivered by LNPs escape from endosomes into the cytosol. They proposed that LNPs concentrate in an endosomal compartment that shares characteristics of early and late endosomes. Wittrup et al. (36) have shown that the release of siRNA formulated in lipoplexes and LNPs from the maturing endosomes occurs within 5–15 min of endocytosis. In their work, furthermore, gene knockdown also occurred a few hours after release, whereas in our work we do not detect protein production until 8 h after the administration of LNPs. Therefore, efforts must be focused on designing LNPs that can escape effectively from the early endosome compartment.
In summary, we showed that mRNA–LNPs are able to transfect human adipocytes and hepatocytes in vitro. The data generated regarding the location of the different lipids across the LNP allowed for the design and formulation of a delivery vehicle with improved performance. The increase in protein production is critical because it allows for administration of lower doses, which in turn could contribute to minimize of undesirable toxic side effects from the LNPs (10). In addition, the structural information reported in this work is of importance for future efforts to develop safer delivery vehicles by designing nanoparticles with specific internal structures and surface compositions, resulting in improved bioperfomance of mRNA–LNPs.
Materials and Methods
Materials.
The ionizable cationic lipid O-(Z,Z,Z,Z-heptatriaconta-6,9,26,29-tetraem-19-yl)-4-(N,N-dimethylamino)butanoate (DLin-MC3-DMA) was synthesized at AstraZeneca. The 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC) was obtained from Avanti Polar Lipids, 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethyleneglycol)-2000] (DMPE-PEG2000) was obtained from NOF Corporation, and Cholesterol (Chol) was obtained from Sigma–Aldrich. Deuterated DSPC (d83) and Chol (d7) were obtained from Avanti Polar Lipids. The 3H-labeled DSPC was synthesized at AstraZeneca. Erythropoietin (EPO) mRNA (858 nucleotides) ARCA capped modified with 5-methylcytidine and pseudouridine was purchased from TriLink Biotechnologies. Polyadenylic acid (PolyA) was purchased from Sigma–Aldrich and had a molecular weight range between 600 and 4,000 bases according to analysis by agarose gel electrophoresis in formamide performed by the manufacturer. Hydrogeneous PBS (1 mM KH2PO4, 155 mM NaCl, and 3 mM Na2HPO4 0.7H2O, pH 7.4) was obtained from Life Technologies, whereas deuterated PBS was prepared with D2O from Sigma–Aldrich. Citrate buffer was purchased from Teknova, and HyClone RNase free water was obtained from GE Healthcare Cell Culture.
LNP Preparation and Characterization.
LNPs were prepared using the microfluidic setup described in detail elsewhere (12). Briefly, stocks of lipids were dissolved in ethanol and mixed in the appropriate molar ratios to obtain a lipid concentration of 12.5 mM (1.85 mg/mL). In the experiments that required radiolabeled DSPC, 1.8 nmol radiolabeled 3H-DSPC was added per formulation, which corresponds to a radioactivity of 5 MBq per formulation. mRNA was diluted in RNase free 50 mM citrate buffer pH 3.0 to obtain a mRNA:lipid weight ratio of 10:1 (CIL:nucleotide 3:1 molar ratio). Empty LNPs were also prepared using 50 mM citrate buffer as the aqueous phase. The aqueous and ethanol solutions were mixed in a 3:1 volume ratio using a microfluidic apparatus NanoAssemblr, from Precision NanoSystems Inc., at a mixing rate of 12 mL/min. LNPs were dialyzed overnight against 500× sample volume using Slide-A-Lyzer G2 dialysis cassettes from Thermo Scientific with a molecular weight cutoff of 10 K. For SAXS, SANS, and cryo-TEM measurements, the particles were concentrated after dialysis to ∼90 mg/mL of lipid using Amicon ultracentrifugation filters.
The size of LNPs was determined by DLS measurements using a Zetasizer Nano ZS from Malvern Instruments Ltd. The number- and volume-weighted particle size distributions were calculated using a particle refractive index of 1.45. The particle concentration (number of LNPs per volume), surface area, and mRNA molecules per LNPs were computed using known molecular volumes (Table S1) and the particle composition, including the water content. The encapsulation and concentration of mRNA were determined using the RiboGreen assay. The encapsulation in all of the samples was typically 90–99%. All of the samples were prepared within a week of use to ensure the chemical stability of the components and no history-dependent in vitro efficacy.
Preparation of Bulk Phases of LNPs.
DLin-MC3-DMA and Chol were dissolved in ethanol and mixed in the appropriate molar ratio to a lipid concentration of 50 mM (23 mg/mL). A total of 1.385 mL of the lipid mixtures were placed in Slide-A-Lyzer G2 dialysis cassettes from Thermo Scientific with a molecular weight cutoff of 10 K with polyA (0.415 mL of 16.4 mg/mL polyA in water, 3:1 CIL:nucleotide molar ratio) to a total final volume of 1.8 mL. The samples were mixed inside the dialysis cassette and placed rapidly in the dialysis buffer. The final concentrations of the lipid mixture and polyA were 21.1 and 2.9 mg/mL, respectively. The samples were dialyzed initially for 2 d against 900 mL (×500 volume of the sample) 50 mM citrate buffer pH 3:ethanol 3:1 by volume mixture, followed by dialysis for 2 d against 900 mL of PBS buffer. The supernatant was removed from the cassettes, and the solid dispersions were extracted and characterized using SAXS.
Patient Consent.
Samples of adipose tissues were collected from patients undergoing elective surgery at Sahlgrenska University Hospital in Gothenburg, Sweden. All study subjects received written and oral information before giving written informed consent for the use of the tissue. The studies were approved by The Regional Ethical Review Board in Gothenburg, Sweden.
Isolation and Culture of Human Adipose-Derived Stem Cells.
Human s.c. white adipose tissue was obtained from healthy women undergoing elective fat removal. Human adipose-derived stem cells (hASCs) were isolated from the stromal vascular fraction as described in Bartesaghi et al. (37) and cultured in a growth medium DMEM/Ham’s F-12 with 10% FBS, 10 mM Hepes, 33 µM biotin, 17 µM pantothenate, 1 nM Fibroblast growth factor (bFGF), all from Sigma–Aldrich, 50 U/mL penicillin and 50 µg/mL streptomycin at 37 °C, 5% CO2 in air with 80% humidity. For adipocyte differentiation, 90% confluent cells were treated with DMEM/F12 with 3% FCS (PAA; Gold) supplemented with 1 µM dexamethasone (Sigma), 500 µM 3-isobutyl-1-methyxanthine (IBMX; Sigma), and 100 nM insulin. To promote white adipogenesis, 1 µM pioglitazone (Pio) was included in the differentiation medium. Media was changed every other day during proliferation and differentiation, until fully differentiated (day 14). hASCs were tested for and free of mycoplasma.
Culture of iPSC-Derived Hepatocytes.
iPSC-derived hepatocytes (iCell Hepatocytes; Cellular Dynamics International) were cultured in line with the manufacturers protocols. Human hepatocytes were validated by hepatic morphology, hepatic protein staining, and hepatic Cytochrome P450 activity (for details, see https://cellulardynamics.com/). Briefly, cells were thawed rapidly and transferred to prewarmed (37 °C) Kryothaw media (KryoThaw-I-250B; SciKon). Viable cells were isolated by centrifugation at 110 × g for 10 min. Pelleted cells were resuspended in room temperature RPMI medium (Life Technologies) containing 1:50 dilution of B27 (Life Technologies), 20 ng/mL Oncostatin M (R&D Systems), 1 µM dexamethasone (Fisher Scientific), and 25 µg/mL Gentamicin (Life Technologies). Cells were plated at 300,000 cells/cm2 in 96-well Biocoat plates (Becton Dickinson) in the above described media at room temperature with the media further supplemented with 250 µg/mL Matrigel (Becton Dickinson) and transferred to an incubator (37 °C, 5% CO2). Following a 4-h incubation, the medium was substituted to RPMI media containing 1:50 dilution of B27, 20 ng/mL Oncostatin M, 0.1 µM dexamethasone, and 25 µg/mL Gentamicin (Life Technologies) and also containing 250 µg/mL Matrigel as before. Twenty-four hours later, media was substituted to Williams E medium (Life Technologies) containing 1:25 dilution of Hepatocyte maintenance mixture B (Life Technologies) and 0.1 µM Dexamethasone. Media was exchanged every 24 h using the latter described media over the next 4 d to allow maturation before subsequent experimental procedures. The iPSC derived hepatocytes have been extensively characterized based on morphology and protein markers definitive for hepatocytes (including, e.g., albumin and cytochrome P450 enzyme) as well as functionality representative of hepatic biology. iPSC were tested for and free of mycoplasma.
LNP Uptake and hEPO Quantification.
Fully differentiated primary white adipocytes and iPSC-derived hepatocytes were treated with 125 ng of radiolabeled mRNA–LNPs in the presence of 1% human serum. LNP uptakes were quantified using a microbeta luminescence counter (PerkinElmer) as function of time. Uptake of LNPs is expressed as the percent of hEPO mRNA dosed as a function of time. Human Erythropoietin Quantikine IVD ELISA Kit (R&D System) was used to quantify the hEPO levels in culture media collected at several time points post-NP transfection. Data are reported as number of hEPO expressed per mRNA dosed as a function of time or after 48 h of initial dosing. The data correspond to triplicates, which is a standard choice when it comes to biological replicates in in vitro cell biology experiments. The experiments also showed high reproducibility, which enables SD calculations.
Small-Angle X-Ray Scattering.
SAXS experiments were performed using a Ganesha 300XL instrument from SAXSLAB Aps. This instrument is equipped with a Genix 3D microfocus source generating X-rays of a wavelength λ = 0.154 nm. The method measures the scattered intensity on a 2D 300 K Pilatus detector as a function of the momentum transfer, q. The setup employed for the measurements was a two-pinhole system with a beam stop size of 2 mm and a sample-to-detector distance of 350 mm, which leads to a q range of 0.14 < q (nm−1) < 7.53. The measurements of LNPs were carried out with reusable quartz capillaries of 1.5 mm diameter from Hilgenberg GmbH. The concentration of the samples for these measurements was 10–12 mg/mL of lipid. Each sample was measured for 2 h. The data presented are background subtracted, where the background corresponds to the buffer measured in the same capillary. The samples corresponding to the bulk phases were measured for 30 min in a thermostated sandwich holder with disposable mica windows. All of the samples were placed in a thermostated block connected to a circulating water bath to maintain the temperature at 25 °C.
Small-Angle Neutron Scattering.
SANS experiments were performed using the KWS-2 instrument operated by Jülich Centre for Neutron Science (JCNS) at Forschungs-Neutronenquelle Heinz Maier-Leibnitz (38). These measurements are based on the interactions of neutrons with matter, which are scattered by the atomic nuclei. The contrast of a specific material is known as the scattering length density (SLD). Table S1 shows the SLD for neutrons and X-rays of the different lipids and RNA.
Three different instrument setups were employed: 1.2- and 7.8-m sample-to-detector distance with a neutron wavelength λ = 0.5 nm and 20-m sample-to-detector distance with λ = 1 nm. These configurations cover the q range of 0.0133 < q (nm−1) < 4.11. The measurements were done in Hellma quartz disk-shaped (“banjo”) cuvettes of 1- and 2-mm path length. LNPs were diluted in the appropriate solvent ratio of D2O/H2O to a final concentration of 3 mg/mL. The samples were placed in a thermostated block to maintain the temperature at 25 °C. The measurement time was adjusted between 0.5 and 3 h depending on the recorded scattering intensity. The data were background subtracted (solvent) and corrected for empty cell and transmission.
Small-Angle Scattering Data Evaluation.
The analyses of the small-angle scattering (SAS) data were performed using the software SasView. The lipid composition across the LNP was determined using the sample with the lipid molar composition DLin-MC3-DMA:DSPC:Chol:DMPE-PEG2000 of 50:10:38.5:1.5, using deuterated DSPC and Chol. The SANS data from the isotopic contrast variation experiments were fitted using the OnionExpShellModel that describes the form factor of a core–multishell sphere where the SLD of the shell can have an exponential decay. The parameters to fit were the thickness of the lipid monolayer and PEG layer, the radius of the core, the SLD of the core and the lipid monolayer, and the exponential decay factor for the SLD of the PEG layer. These parameters were optimized using the simultaneous fitting option with the complex ParkMC fit engine.
Cryogenic Transmission Electron Microscopy of LNPs.
Cryo-TEM experiments were performed as described by Bello et al. (39). A small drop of a concentrated LNP formulation (lipid concentration of 10–20 mg/mL) was placed on a polymer-coated carbon-reinforced copper grid. The grid was bloated with a filter paper to remove the excess of liquid to form a thin film. The samples were vitrified rapidly in liquid ethane at −180 °C to prevent crystallization of water. The preparation was done immediately before examination, and they were kept below −165 °C during the experiment. Cryo-TEM measurements were performed using a Zeiss Libra 120 instrument from Carl Zeiss Nano Technology System operated at 80 kV. The images were recorded with a CCD camera from TRS GmbH and processed with iTEM software from Olympus Soft Imaging Solutions GmbH. The size distributions were calculated from three high-quality images for each LNP sample (>100 particles for each LNP formulation).
Freeze Fracture of Bulk Phases.
Small amounts of the bulk phase samples were placed on small gold cups and rapidly frozen in liquid propane. Three samples were prepared at each freeze fracture preparation, and five freeze fracture preparations were performed, i.e., 15 areas to image. The rapidly frozen bulk phase samples were then transferred into a precooled (−170 °C) vacuum chamber of a Balzer BAF 400 freeze etching system and put under vacuum immediately. The samples were tempered to −105 °C before the freeze fracture, and then they were etched to enhance the structure at the newly fractured surface through sublimation of water by placing a cold trap above the sample. Subsequently, they were rotary shadowed by first platina at an angle of 45° followed by carbon at an angle of 80°. The platina and carbon were evaporated on the fractured surface under vacuum at −105 °C. The thicknesses of the deposited metals were controlled with a quartz crystal monitor, which gave a thickness of the evaporated platina layer of 2 nm. The samples were cleaned from the metal replicas using solvents and acid. The clean replicas were then placed on 400 mesh copper grids before analysis in a transmission electron microscope LEO 906 E at an accelerating voltage of 80 kV.
Solubility of Chol Nanocrystals in DLin-MC3-DMA Emulsion.
A DLin-MC3-DMA emulsion was prepared with a concentration of 11.45 mg/mL in a 50-mM citrate buffer pH 3:ethanol mixture 3:1 by volume with 1.9 mg/mL of DMPE-PEG2000 as stabilizer. The sample was placed on an ultrasonic bath (Elma Transsonic Bath T460/H) for ∼30 min followed by sonication in a focused ultrasonic bath (Covaris S2) at high intensity for 20 min at 18 °C. The emulsion had an average size of 150 ± 10 nm as measured by DLS (zetasizer Nano ZS; Malvern Instruments Ltd). Chol nanocrystals were manufactured by wet milling. A suspension of 9.2 wt % Chol with 2 wt % of DMPE-PEG2000 in milli-Q water was sonicated in an ultrasonic bath for 30 min, stirred over night at room temperature, and sonicated again before milling. Wet milling of 550 μL of sample was performed using a Fritsch Planetary Micromill P7 equipped with 1.2 mL milling bowls and 0.6–0.8 mm milling beads of zirconium oxide, at 44 × g, for 4 × 30 min, with 15 min pauses in between. The sample was extracted from the milling vessel, and the beats were rinsed four times with 550-μL aliquots of milli-Q water. The final concentration of Chol and DMPE-PEG2000 was determined by HPLC. Chol nanosuspension had an average size of 160 nm as measured using a Mastersizer 2000 (Malvern Instruments Ltd.).
The solubility of Chol at 25 °C in the DLin-MC3-DMA emulsion in 50 mM citrate buffer, pH 3:ethanol 3:1 by volume mixture was determined by recording the intensity of the light scattered with a luminescence spectrometer (Perkin–Elmer LS 55) as described by Lindfors et al. (40). The solubility was determined by a extrapolating a linear fit to the high-concentration data to the line of zero intensity. All of the samples were prepared with a fixed concentration of DLin-MC3-DMA (1 mM) and varying concentration of Chol (between 0.05 and 1 mM), and they were left stirring overnight. The mixtures were further diluted by a factor of 100 before the measurements. The emission and excitation wavelengths were set to 700 nm, and the scattered intensity was recorded at an angle of 90°.
Supplementary Material
Acknowledgments
We are grateful to Joachim Rädler (Ludwig Maximilians Universität München) for fruitful discussion regarding the SAXS data and Fredrik Höök (Chalmers University of Technology) for his valuable comments on the manuscript. We acknowledge Marc Obiols-Rabasa (Lund University) for his help with the SAXS measurements, Gunnel Karlsson (Lund University) and Jonny Eriksson (Uppsala University) for their help with the cryo-TEM experiments, and Annika Altskär (Research Institutes of Sweden) for her help with the freeze fracture measurements. We also acknowledge Johan Broddefalk (AstraZeneca) for the synthesis of the cationic ionizable lipid, Cecilia Ericsson (AstraZeneca) for the synthesis of the radiolabeled phospholipid, Eva K. Carlsson (AstraZeneca) for her contribution in developing the in vitro assays, and Anke Marx (AstraZeneca) for performing X-ray powder diffraction measurements. This work is based upon experiments performed at the KWS-2 instrument operated by JCNS at Heinz Maier-Leibnitz Zentrum, Garching, Germany. The SAS data analysis benefitted from SasView software, originally developed by the Data Analysis for Neutron Scattering Experiments project under NSF Award DMR-0520547.
Footnotes
Conflict of interest statement: M.Y.A., T.K., S.B., S.W., X.W., A.J.K, A.D., and L.L. are employed by AstraZeneca R&D Gothenburg.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720542115/-/DCSupplemental.
References
- 1.Wang Y, et al. Systemic delivery of modified mRNA encoding herpes simplex virus 1 thymidine kinase for targeted cancer gene therapy. Mol Ther. 2013;21:358–367. doi: 10.1038/mt.2012.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zangi L, et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat Biotechnol. 2013;31:898–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kormann MSD, et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 4.Yin H, et al. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 5.Dowdy SF. Overcoming cellular barriers for RNA therapeutics. Nat Biotechnol. 2017;35:222–229. doi: 10.1038/nbt.3802. [DOI] [PubMed] [Google Scholar]
- 6.Nayerossadat N, Maedeh T, Ali PA. Viral and nonviral delivery systems for gene delivery. Adv Biomed Res. 2012;1:27. doi: 10.4103/2277-9175.98152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anonymous Gene-therapy trials must proceed with caution. Nature. 2016;534:590. doi: 10.1038/534590a. [DOI] [PubMed] [Google Scholar]
- 8.Semple SC, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 9.Junquera E, Aicart E. Recent progress in gene therapy to deliver nucleic acids with multivalent cationic vectors. Adv Colloid Interface Sci. 2016;233:161–175. doi: 10.1016/j.cis.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Barros SA, Gollob JA. Safety profile of RNAi nanomedicines. Adv Drug Deliv Rev. 2012;64:1730–1737. doi: 10.1016/j.addr.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Belliveau NM, et al. Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Mol Ther Nucleic Acids. 2012;1:e37. doi: 10.1038/mtna.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhigaltsev IV, et al. Bottom-up design and synthesis of limit size lipid nanoparticle systems with aqueous and triglyceride cores using millisecond microfluidic mixing. Langmuir. 2012;28:3633–3640. doi: 10.1021/la204833h. [DOI] [PubMed] [Google Scholar]
- 13.Leung AKK, Tam YYC, Chen S, Hafez IM, Cullis PR. Microfluidic mixing: A general method for encapsulating macromolecules in lipid nanoparticle systems. J Phys Chem B. 2015;119:8698–8706. doi: 10.1021/acs.jpcb.5b02891. [DOI] [PubMed] [Google Scholar]
- 14.Gao H, Shi W, Freund LB. Mechanics of receptor-mediated endocytosis. Proc Natl Acad Sci USA. 2005;102:9469–9474. doi: 10.1073/pnas.0503879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang S, Li J, Lykotrafitis G, Bao G, Suresh S. Size-dependent endocytosis of nanoparticles. Adv Mater. 2009;21:419–424. doi: 10.1002/adma.200801393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S, et al. Development of lipid nanoparticle formulations of siRNA for hepatocyte gene silencing following subcutaneous administration. J Control Release. 2014;196:106–112. doi: 10.1016/j.jconrel.2014.09.025. [DOI] [PubMed] [Google Scholar]
- 17.Bao Y, et al. Effect of PEGylation on biodistribution and gene silencing of siRNA/lipid nanoparticle complexes. Pharm Res. 2013;30:342–351. doi: 10.1007/s11095-012-0874-6. [DOI] [PubMed] [Google Scholar]
- 18.Akinc A, et al. Development of lipidoid-siRNA formulations for systemic delivery to the liver. Mol Ther. 2009;17:872–879. doi: 10.1038/mt.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leung AKK, et al. Lipid nanoparticles containing siRNA synthesized by microfluidic mixing exhibit an electron-dense nanostructured core. J Phys Chem C Nanomater Interfaces. 2012;116:18440–18450. doi: 10.1021/jp303267y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rozmanov D, Baoukina S, Tieleman DP. Density based visualization for molecular simulation. Faraday Discuss. 2014;169:225–243. doi: 10.1039/c3fd00124e. [DOI] [PubMed] [Google Scholar]
- 21.Dan N, Danino D. Structure and kinetics of lipid-nucleic acid complexes. Adv Colloid Interface Sci. 2014;205:230–239. doi: 10.1016/j.cis.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 22.Kim H, Leal C. Cuboplexes: Topologically active siRNA delivery. ACS Nano. 2015;9:10214–10226. doi: 10.1021/acsnano.5b03902. [DOI] [PubMed] [Google Scholar]
- 23.Jayaraman M, et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew Chem Int Ed Engl. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zackrisson M, Stradner A, Schurtenberger P, Bergenholtz J. Small-angle neutron scattering on a core-shell colloidal system: A contrast-variation study. Langmuir. 2005;21:10835–10845. doi: 10.1021/la051664v. [DOI] [PubMed] [Google Scholar]
- 25.Marsh D, Bartucci R, Sportelli L. Lipid membranes with grafted polymers: Physicochemical aspects. Biochim Biophys Acta. 2003;1615:33–59. doi: 10.1016/s0005-2736(03)00197-4. [DOI] [PubMed] [Google Scholar]
- 26.Kenworthy AK, Hristova K, Needham D, McIntosh TJ. Range and magnitude of the steric pressure between bilayers containing phospholipids with covalently attached poly(ethylene glycol) Biophys J. 1995;68:1921–1936. doi: 10.1016/S0006-3495(95)80369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J, Buboltz JT, Feigenson GW. Maximum solubility of cholesterol in phosphatidylcholine and phosphatidylethanolamine bilayers. Biochim Biophys Acta. 1999;1417:89–100. doi: 10.1016/s0005-2736(98)00260-0. [DOI] [PubMed] [Google Scholar]
- 28.Jandacek RJ, Webb MR, Mattson FH. Effect of an aqueous phase on the solubility of cholesterol in an oil phase. J Lipid Res. 1977;18:203–210. [PubMed] [Google Scholar]
- 29.Varsano N, Fargion I, Wolf SG, Leiserowitz L, Addadi L. Formation of 3D cholesterol crystals from 2D nucleation sites in lipid bilayer membranes: Implications for atherosclerosis. J Am Chem Soc. 2015;137:1601–1607. doi: 10.1021/ja511642t. [DOI] [PubMed] [Google Scholar]
- 30.Pardi N, et al. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J Control Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel S, et al. Boosting intracellular delivery of lipid nanoparticle-encapsulated mRNA. Nano Lett. 2017;17:5711–5718. doi: 10.1021/acs.nanolett.7b02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan C-L, et al. Endosomal escape and transfection efficiency of PEGylated cationic liposome-DNA complexes prepared with an acid-labile PEG-lipid. Biomaterials. 2012;33:4928–4935. doi: 10.1016/j.biomaterials.2012.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koltover I, Salditt T, Rädler JO, Safinya CR. An inverted hexagonal phase of cationic liposome-DNA complexes related to DNA release and delivery. Science. 1998;281:78–81. doi: 10.1126/science.281.5373.78. [DOI] [PubMed] [Google Scholar]
- 34.Chen S, et al. Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J Control Release. 2016;235:236–244. doi: 10.1016/j.jconrel.2016.05.059. [DOI] [PubMed] [Google Scholar]
- 35.Gilleron J, et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 36.Wittrup A, et al. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat Biotechnol. 2015;33:870–876. doi: 10.1038/nbt.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartesaghi S, et al. Thermogenic activity of UCP1 in human white fat-derived beige adipocytes. Mol Endocrinol. 2015;29:130–139. doi: 10.1210/me.2014-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radulescu A, Pipich V, Frielinghaus H, Appavou MS. KWS-2, the high intensity/wide Q -range small-angle neutron diffractometer for soft-matter and biology at FRM II. J Phys Conf Ser. 2012;351:012026. [Google Scholar]
- 39.Bello G, et al. Characterization of the aggregates formed by various bacterial lipopolysaccharides in solution and upon interaction with antimicrobial peptides. Langmuir. 2015;31:741–751. doi: 10.1021/la503267k. [DOI] [PubMed] [Google Scholar]
- 40.Lindfors L, et al. Amorphous drug nanosuspensions. 2. Experimental determination of bulk monomer concentrations. Langmuir. 2006;22:911–916. doi: 10.1021/la052367t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.