Significance
Lysyl oxidases (LOXs) catalyze oxidative deamination of peptidyl lysines on collagen and elastin, generating a highly reactive aldehyde group to initiate intermolecular cross-linking. In humans, this reaction plays fundamental roles in the formation and repair of extracellular matrix fiber networks and the development of connective tissues. Moreover, a relationship between LOX proteins and tissue fibrosis and cancer has been established. Since the identification of prototypic LOX, the atomic structure of LOX protein has remained to be elucidated. Here, we present the high-resolution structure of human lysyl oxidase-like 2 protein. The zinc-bound precursor-state structure provides an important framework for understanding the structure–function relationship of the LOX family and for drug discovery that aims to block LTQ generation of LOX proteins.
Keywords: crystal structure, lysyl oxidase, human LOXL2, precursor state
Abstract
Lysyl oxidases (LOXs), a type of copper- and lysyl tyrosylquinone (LTQ) -dependent amine oxidase, catalyze the oxidative deamination of lysine residues of extracellular matrix (ECM) proteins such as elastins and collagens and generate aldehyde groups. The oxidative deamination of lysine represents the foundational step for the cross-linking of elastin and collagen and thus is crucial for ECM modeling. Despite their physiological significance, the structure of this important family of enzymes remains elusive. Here we report the crystal structure of human lysyl oxidase-like 2 (hLOXL2) at 2.4-Å resolution. Unexpectedly, the copper-binding site of hLOXL2 is occupied by zinc, which blocks LTQ generation and the enzymatic activity of hLOXL2 in our in vitro assay. Biochemical analysis confirms that copper loading robustly activates hLOXL2 and supports LTQ formation. Furthermore, the LTQ precursor residues in the structure are distanced by 16.6 Å, corroborating the notion that the present structure may represent a precursor state and that pronounced conformational rearrangements would be required for protein activation. The structure presented here establishes an important foundation for understanding the structure–function relationship of LOX proteins and will facilitate LOX-targeting drug discovery.
The extracellular matrix (ECM) provides structural and chemical support to resident cells, thereby regulating various cellular functions such as cell adhesion, communication, gene expression, and migration (1–3). Collagen and elastin are the primary structural proteins of the ECM. Starting from intermolecular cross-linking, they gradually assemble to form a functional network compatible for different tissues (4). In humans, cross-linking of ECM proteins is initiated by lysyl oxidase (LOX) (5). The LOX family, which comprises five secreted enzymes, catalyzes the oxidative deamination of the ε-amino group of lysine and hydroxylysine residues on collagen and elastin, generating highly reactive aldehyde that spontaneously condenses with neighboring aldehyde or ε-amino groups to form higher-order cross-linkages (Fig. 1A, Left). Owing to its essential function in ECM formation, deficiency of LOX proteins leads to severe disorders in the development of connective tissue (6, 7). Four additional lysyl oxidase-like proteins (LOXL1–4) have been identified in diverse tissues (8), although their specific functions and substrate preferences in vivo remain to be elucidated.
Fig. 1.
Overall structure of hLOXL2. (A) The reaction and structure of LTQ. (B) Crystal structure of hLOXL2 (residues 318–774, N455Q). The structure is presented in two perpendicular views. The third and fourth SRCR domains are colored cyan and green, respectively; the catalytic domain is colored yellow. The glycosyl groups at Asn644 are shown as black sticks. Zinc and calcium ions are presented as dark blue and green spheres, respectively. All structure figures were prepared using PyMOL (48).
Growing evidence has established the relationship between LOX proteins and various human diseases, such as tissue fibrosis and several types of cancer. Fibrotic diseases are characterized by progressive overproduction of ECM fiber that ultimately leads to organ dysfunction (9). Histopathological studies have indicated the abnormal accumulation of LOX proteins in fibrotic lung and liver, consistent with their role in ECM modeling (10, 11). Inhibition trials targeting LOXL2 have exhibited altered fibrosis progression accompanied with decreased myofibroblasts in animal models (10, 12). Serum LOXL2 level is associated with disease progression of idiopathic pulmonary fibrosis, making it a potential diagnostic biomarker (9, 13). A correlation between LOX proteins and breast tumor progression has also been suggested (14, 15). LOX and LOXL2 are specifically expressed in metastatic breast tumor (16–18) and induce elevated metastasis of noninvasive cancers (17, 19, 20). Inhibition trials targeting LOX and LOXL2 led to reduced tumor migration in cultured cells and animal models (10, 16, 21–23).
LOX proteins and 2,4,5-trihydroxyphenylalanine quinone (TPQ)-dependent copper-containing amine oxidase (CAO) represent two major families of copper- and quinone-containing amine oxidases (8, 24). LOX proteins catalyze the oxidative deamination of peptidyl lysine in addition to small-molecule amines, whereas most CAOs recognize small-molecule amines, and only a few specific CAOs can work with peptidyl lysine (25, 26). Despite their distinct substrate preferences, both families catalyze the deamination of primary amines utilizing a covalently linked quinone cofactor, which derives from posttranslational modification of native residue(s). Quinone generation in both LOX and CAO families, a self-catalyzed process in aerobic conditions, requires a tightly bound copper ion as a cofactor (27–32).
The working mechanism of CAOs has been largely elucidated by systematic crystallographic and biochemical studies (24). Despite its fundamental role in ECM modeling and multiple disease relationships, the structure of the LOX family remains elusive. Sequence analysis shows that LOX and CAO families share no obvious homology. The LOX family members have diverse N-terminal domains followed by a conserved catalytic domain. LOX and LOXL1 contain an N-terminal propeptide, which is removed through proteolytic cleavage in the ECM during protein maturation (5). LOXL2–4 each consists of four scavenger receptor cysteine-rich (SRCR) domains, which have been suggested to mediate the interactions of cell-surface and ECM proteins (33).
The catalytic domain of LOX proteins harbors the copper-binding motif and the functional quinone group, which has been identified as lysyl tyrosylquinone (LTQ) generated through posttranslational cross-linkage between specific lysine and tyrosine residues (Fig. 1A, Right) (34). A working mechanism similar to that of the CAO family has been proposed for LOXs (5, 8, 35). Nevertheless, the distant homology between their catalytic domains suggests potentially different structural organization in LOX proteins, which may confer a distinct mechanism for LTQ formation and substrate recognition.
Here we present the crystal structure of human LOXL2 in a zinc-bound precursor state at 2.4-Å resolution, which establishes the framework for mechanistic investigation of the structural–functional relationship of LOX proteins.
Results
Overall Structure of Human LOXL2.
Details of the protein preparation and crystallization can be found in Materials and Methods. Briefly, human LOXL2 (hLOXL2) residues 26–774, comprising the four N-terminal SRCR domains followed by the catalytic domain, were transiently expressed in mammalian HEK293F cells and secreted into medium. A single-point mutation (N455Q) was introduced to eliminate glycosylation on the fourth SRCR. hLOXL2-N455Q secrets well in the HEK293F system despite a previous report on the importance of glycosylation at Asn455 for hLOXL2 secretion in the Drosophila S2 expression system (36). Limited proteolysis of the purified protein yielded a fragment (residues 318–774) consisting of the third and fourth SRCR domains and the intact catalytic domain. To ensure protein homogeneity and to simplify protein purification, we engineered a specific cutting site for the caspase DrICE right before residue 318. The cleavage-resulted segment (residues 318–774, N455Q), after further purification, gave rise to well-diffracting crystals. The structure was solved through a combination of molecular replacement and osmium-based single-wavelength anomalous dispersion (Os-SAD). The atomic model comprising residues 322–762 was built and refined to 2.4-Å resolution.
In each asymmetric unit (ASU), two hLOXL2 molecules are observed, which are nearly superimposable with an rmsd of 0.80 Å. Intermolecular association is mediated mainly through the SRCR4 domains and, to a minor extent, through SRCR4 and the opposing SRCR3 (Fig. S1). Sparse intermolecular hydrogen bonds (H-bonds) have been observed between SRCR3 and SRCR4. Despite the observation of two molecules in a single ASU, hLOXL2 is revealed as being in a monomeric state in solution.
The overall structure of hLOXL2 exhibits a triangular shape, with SRCR3, SRCR4, and the catalytic domain each occupying a corner of the triangle (Fig. 1B). The N-linked glycan, which is conserved among LOXL2–4, is located on the catalytic domain peripheral to the triangular plane. All 22 cysteine residues are engaged in disulfide bonds that are distributed throughout the structure and stabilize local conformations.
The group A SRCR domain is characterized by three disulfide bonds in a conserved pattern (33). Structural alignment of the third and fourth SRCR domains of hLOXL2 with that of the human Mac-2–binding protein (M2BP-SRCR) show evident similarities (Fig. S2A). All display an identical disulfide bond pattern and similar folding, featuring a single α-helix surrounded by β-strands, confirming the prediction that the SRCR domains in hLOXL2 belong to group A.
In the resolved structure, the SRCR4 domain shows no association with the catalytic domain and merely contacts SRCR3 with the H-bonds between Tyr333 on SRCR3 and Gly481 and Glu545 on SRCR4. In contrast, nine H-bonds mediate extensive interactions between SRCR3 and the catalytic domain (Fig. S2B). Gln383 and Gly331 on SRCR3 and Asn606 and Gln608 on the catalytic domain form three H-bonds, while another six are formed between Gly345/Arg327/Trp347 on SRCR3 and Ser753/Gly750/Ser751/Phe752 on the C terminus of the catalytic domain, involving both main chains and side chains (Fig. S2B).
Architecture of the Catalytic Domain.
The catalytic domain of hLOXL2 contains 14 β-strands and three α-helices. DALI search (37) of the catalytic domain structure against the Protein Data Bank (PDB) resulted in a list of proteins with distinct functions, with the highest-scored structure being a domain of β-galactosidase (residues 181–305) from Bifidobacterium bifidum (PDB ID code: 5dmy) (Fig. S3A). Based on structural alignment, the superimposable and nonoverlapping regions are defined as the core and the extra segments (ES1–3), respectively (Fig. 2A).
Fig. 2.
Structure of the catalytic domain of hLOXL2. (A) Overall structure of the hLOXL2 catalytic domain. The β-sandwich structure that can be superimposed with that of β-galactosidase is defined as the structure core and is colored yellow; the nonoverlapping regions, colored orange, are defined as extra segments (ES1–3). Disulfide bonds are shown as sticks. Bound zinc and calcium are displayed as spheres. (B) Topology of the catalytic domain. The structure core and ES regions are color-coded as in A. The LTQ precursor and the His residues involved in copper binding are indicated.
The core exhibits a sandwich structure constituted by three β-sheets, with the largest one consisting of β8, 7, 12, 13, and 14 (Fig. 2B). A disulfide bond between Cys732 and Cys746 helps stabilize one edge of this β-sheet. Two smaller β-sheets formed by β2, 3, and 11 and by β1, 4, 10, and 9, respectively, are intervened by α1 and flexible loops. A disulfide bond between Cys663 and Cys685 fixes the end of β9 with β10, which hosts Tyr689, one of the crucial LTQ precursor residues. Two flexible loops reside between β8 and β9 and between β9 and β10, which are linked by an additional disulfide bridge between Cys673 and Cys657 (Fig. 2A). A sphere-shaped density is enclosed by β1 and the loop between β12 and β13, coordinated by oxygen from six highly conserved residues in LOX family: Asn727, Asn728, Asp724, Glu722, Asp549, and Leu550 (Fig. S3B). This octahedral coordination pattern is consistent with a bound calcium ion, which might play a role in local structure stabilization.
Among the three ES segments that surround the core of the catalytic domain (Fig. 2 A and B), ES1 (residues 568–580) contains a bent α-helix (α2), which links to the core through a disulfide bridge between Cys579 and Cys695. The major structure element of ES2 (residues 614–634) is a β-hairpin (β5 and β6), which accommodates the highly conserved copper-binding motif (Fig. 2B and Fig. S3C). ES2 is disulfide-bonded to ES1 through Cys625 and Cys573, which may stabilize this crucial β-hairpin. ES3 (residues 745–774) is at the C terminus of the catalytic domain and is responsible for the interaction with the SRCR3 domain (Fig. S2B).
Human LOX family members possess a conserved catalytic domain (5). Sequence alignment reveals a high conservation in ES1, ES2, and most regions of the structure core, implying a similar architecture among LOX proteins, which is exhibited in the ConSurf figure (Fig. S4 A and B). Notably, the putative active site of hLOXL2 involving LTQ precursors and the copper-binding motif is extraordinarily conserved in the LOX family, as is consistent with their crucial role in functionality (Fig. S4 A and C).
The Copper-Binding Site of hLOXL2 Is Occupied by Zinc.
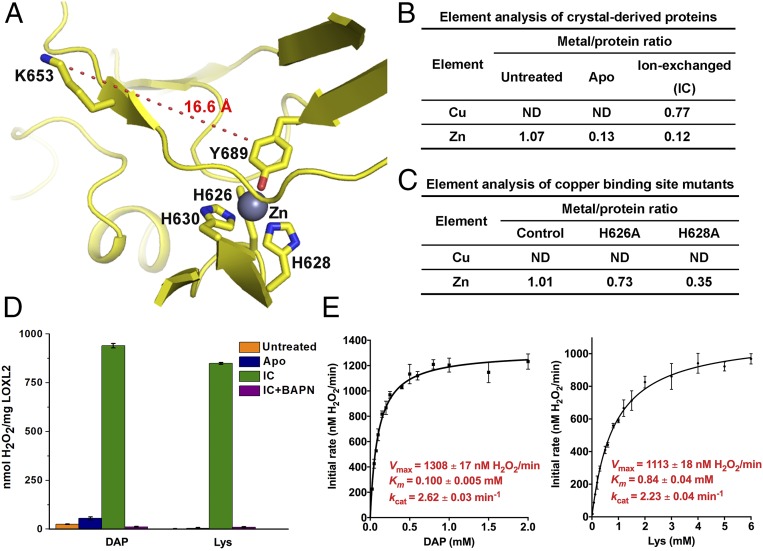
An electron density was unambiguously observed in the predicted copper-binding site, indicating the presence of a metal ion, although copper was excluded throughout protein expression, purification, and crystallization (Fig. 3A and Fig. S3C). To identify the bound metal, hundreds of hLOXL2 crystals were collected, carefully washed, dissolved, and subjected to element analysis. The crystal-derived hLOXL2 is indeed copper free but contains a high abundance of zinc (Fig. 3B, untreated sample) that may come from the mammalian cell medium. Consistent results were obtained from fluorescent scans of hLOXL2 crystals at both the zinc and copper edges (Fig. S5). To further corroborate the structural assignment of a zinc ion to this site, the binding-site residues were mutated, and the resulting mutants were subjected to element analysis. Decreased zinc content was detected (Fig. 3C), confirming that the copper-binding site is occupied by zinc in the structure.
Fig. 3.
The zinc-bound hLOXL2 structure may represent a precursor state. (A) Close-up view of the putative catalytic site of hLOXL2. The LTQ precursors and copper-binding site residues are displayed as sticks; the zinc ion is shown as a sphere. Distances between residues are shown by a dashed line. (B) Element analysis of the crystal-derived hLOXL2. Aliquots of hLOXL2 protein dissolved from crystals were treated with the indicated conditions and subjected to element analysis to determine metal content. Apo, hLOXL2 treated with 1,10-phenanthroline to remove zinc; IC, apo hLOXL2 treated with copper for ion exchange; untreated, the dissolved hLOXL2 proteins without metal chelator treatment. Protein concentrations were calculated based on the sulfur element quantified in the same experiments. (C) Element analysis of the copper-binding site–mutated hLOXL2 variants reveals decreased zinc content. The boundaries of the tested variants are identical to those for crystallization. Control, residues 318–774 with N455Q. (D) Functional assessment of the hLOXL2 samples purified from crystals. The crystal-derived hLOXL2 samples in B were subjected to enzymatic assay. IC+BAPN, 50 μM BAPN was added to the assay to inhibit enzymatic activity. (E) Michaelis–Menten curve and kinetic parameters of ion-exchanged hLOXL2. The initial rates of oxidative deamination were determined using ion-exchanged hLOXL2 (residues 318–774 with N455Q) and were plotted versus a range of DAP (Left) or Lys (Right) concentrations. The Michaelis–Menten curve was fitted, and Km and kcat were calculated. All error bars represent SDs from three independent experiments. Please refer to Materials and Methods for details of the enzymatic assays.
In the hLOXL2 structure, zinc is coordinated by the side chains of His626, His628, His630, and Tyr689 (Fig. 3A and Fig. S3C), exhibiting a tetrahedral geometry. Among these, Tyr689 is one of the LTQ precursors. In the apo structure of CAOs bound with zinc or copper, the metal ion was bound in similar geometry by three His residues and the precursor tyrosine of TPQ (31, 32), indicating conserved metal coordination among precursor LOXs and apo CAOs.
The hLOXL2 Structure Represents a Zinc-Bound Precursor State.
Previous studies have established the essential role of copper in LTQ generation (28). As such, occupation of the copper-binding site by zinc may inhibit quinone generation and block hLOXL2 in a precursor state, as experimentally shown in CAO proteins (29, 30). To verify this hypothesis, we performed ion exchange in the zinc-bound hLOXL2 and investigated both enzymatic activity and LTQ generation of the copper-loaded protein. As the glycosylation mutation N455Q appears to have no effect on the enzymatic activity of hLOXL2, all the protein variants described hereafter contain this single-point mutation unless otherwise indicated (Fig. S6A).
As expected, the enzymatic assay with zinc-bound hLOXL2 resulted in undetectable activity (Fig. 3 B and D, untreated sample), suggesting a potential precursor state in the current structure. Removal of zinc had no obvious effect on hLOXL2 activity. Intriguingly, ion exchange or copper loading robustly stimulated hLOXL2 activity [Fig. 3 B and D, apo and ion exchange (IC) samples], which was undoubtedly suppressed by β-aminopropionitrile (BAPN), a known inhibitor of the LOX family. Determination of the Michaelis constant (Km) and kcat further corroborates the substrate affinity and catalytic turnover rate of copper-activated hLOXL2 (Fig. 3E). Moreover, ion exchange failed to enhance the activity of copper-binding site mutants (Fig. S6B). Collectively, the enzymatic assays reinforce the role of copper in activating hLOXL2 and functionally support that the current structure is in a zinc-bound precursor state.
An unexpected feature of the putative active site of hLOXL2 is the 16.6-Å distance between Lys653 and Tyr689 (Fig. 3A), the LTQ precursors of hLOXL2 (36). The high-quality density map ensures the accuracy of local model building for Lys653 and Tyr689. In addition, neither residue is involved in the packing interface, ruling out possible disturbance of crystal packing. The structural observation suggests either that the recombinant protein used for crystallization is incapable of LTQ formation or that a pronounced conformational change from the current structure is required for protein maturation.
To examine LTQ formation, redox-cycling staining (38) was employed. Strong staining emerges in the protein of the ion-exchanged sample (Fig. S7, Upper Left) but not in the untreated sample or in LTQ precursor mutants (Fig. S7, Upper Right), indicating the emergence of a covalently linked redox cofactor associated with Lys653 and Tyr689. In addition, staining is undetectable in copper-binding site mutants, supporting the crucial role of copper in the induction of this cofactor. Taken together, the redox-cycling staining results are consistent with the formation of LTQ in copper-activated hLOXL2.
A previous study of endogenous LOX or the LTQ model compound revealed absorption of ultraviolet-visible spectroscopy (UV-Vis) with the λmax around 504 nm, which shifted to 454 nm when LTQ was labeled by phenylhydrazine (34). After ion exchange, activated hLOXL2 exhibited an obvious pink color, suggesting the emergence of a chromophore. To evaluate its spectrometry behavior, UV-Vis scans were performed with hLOXL2 variants. Copper-activated hLOXL2 exhibited the characteristic absorption of LTQ around 510 nm, which was eliminated by Lys653 or Tyr689 substitution (Fig. 4A, Left), suggesting the formation of LTQ between these residues. Consistently, parallel scans with phenylhydrazine-labeled hLOXL2 exhibit characteristic absorption around 455 nm (Fig. 4A, Right), supporting the notion that LTQ is the chromophore formed during hLOXL2 activation.
Fig. 4.
Validation of quinone formation in activated hLOXL2. (A) The UV-Vis scan revealed the characteristic absorption of LTQ in activated hLOXL2 but not in the indicated mutants. hLOXL2 proteins with (Right) or without (Left) phenylhydrazine labeling were subjected to the UV-Vis scan. All samples shown in this figure are based on the construct of residues 318–774 (N455Q) unless otherwise indicated. IC, the indicated variants were treated for ion exchange as in Fig. 3B before analysis. Please refer to Materials and Methods for assay details. (B) Mass spectrometric analysis of activated hLOXL2 identified the LTQ-containing peptide. Untreated and ion-exchanged hLOXL2 were labeled with phenylhydrazine and subjected to MS analysis after trypsin digestion. The phenylhydrazine-labeled LTQ-containing peptide was identified only in ion-exchanged hLOXL2 and not in the untreated protein. Cysteine residues modified by carbamidomethyl group are marked with a black star. (Inset) The azo tautomer of phenylhydrazine-labeled LTQ.
Finally, mass spectrum analysis identified the peptide cross-linked through Lys653 and Tyr689 exclusively in activated hLOXL2 (Fig. 4B). The calculated mass of the cross-linker strictly matches phenylhydrazine-labeled LTQ.
Overall, our biochemical studies support the generation of LTQ accompanying copper activation. Therefore, the present structure represents a zinc-bound precursor state, and pronounced conformational changes would be required for LTQ formation.
Discussion
Copper- and quinone-dependent amine oxidases are classified into two nonhomologous families: CAO and LOX (8, 24). CAOs are typically homodimeric enzymes. Each monomer comprises N-terminal α/β domains followed by a catalytic domain, which folds into a large antiparallel β-sandwich structure. Two β-hairpin arms extended from the catalytic domain, hug its opposing partner, and help stabilize the homodimeric architecture (Fig. S8A). TPQ and the copper-binding site reside on opposite β-sheets of the sandwich and are buried in a substrate channel (Fig. S8B) (39, 40).
Compared with CAOs, the hLOXL2 structure reveals a monomeric state. In addition, the current hLOXL2 structure shares no overall similarity with that of CAOs, as indicated by structural alignments. The catalytic domain of LOXL2 folds into a small β-sandwich core, which is surrounded by three extra segments and is stabilized by extensive disulfide bridges. In the present hLOXL2 structure, the copper-binding site is occupied by zinc, which blocks protein activation. Zinc is coordinated by Tyr689 and His residues from a successive β-hairpin peripheral to structure core. Contrarily, one of the histidines in the copper-binding motif of CAOs is recruited from a distant position in the primary sequence. Nevertheless, the tetrahedral geometry of metal coordination at an active site is similar among precursor LOXL2 and apo CAOs (Figs. S3C and S8C). Notably, in the structure of holoCAO with an “off-copper” TPQ conformation, TPQ no longer acts as a copper ligand, leaving its C5 atom accessible for substrate attack (Fig. S8B). In this conformation, copper is coordinated by the conserved histidines and two water molecules (axial and equatorial water), exhibiting a square pyramidal geometry (24, 39). Metal coordination as well as the conformation of LTQ in the activated LOXL2 requires further investigation.
A surprise from the putative active site of hLOXL2 structure is the 16.6-Å distance between the side chains of Lys653 and Tyr689, precursors of LTQ. Nevertheless, a UV-Vis scan and mass spectrum analysis consistently supported the emergence of LTQ in activated hLOXL2, suggesting that conformational changes would be required during LTQ generation. The approaching of Lys653 to Tyr689 is likely to be involved in the conformational change. Interspace surrounded by ES1, ES2, and β8 might support the approaching of Lys653 to Tyr689. Consistent with this speculation, the local sequence surrounding β8 is highly conserved among the LOX family (Fig. S4 B and C). In contrast, Tyr689 resides on one of the β-sheets of the structure core and is involved in copper coordination. Limited structural flexibility and dependence on copper might restrict the movement of Tyr689 during LTQ generation. Currently, the precise mechanism of the conformational changes is inconclusive. The elucidation of the active-state structure of LOXs is crucial for understanding these processes.
In most CAO structures, the copper-binding motif and TPQ are buried in a substrate cavity adaptive to small-molecule amines (Fig. S8B) (39, 40). The size and shape of these cavities are believed to be significant for substrate accessibility and preference. Unusually, Pichia pastoris lysyl oxidase (PPLO) and Aspergillus nidulans amine oxidase (ANAO) possess a relatively broad substrate funnel compatible with peptidyl lysine (25, 26). LOX proteins are characterized by the deamination of the lysine side chain on protein substrates. This functional role requires an accessible active site for substrate attachment. In the hLOXL2 structure, the side chain of Tyr689 lies on the protein surface. Located on ES2 peripheral to the structure core, the copper-binding motif is also solvent exposed. These structural features appear adaptive to the access of oxygen and peptidyl lysine during LTQ modification and deamination.
In summary, the hLOXL2 structure presented here enables a preliminary understanding of the structural–functional relationship of LOX proteins. Importantly, this precursor-state structure might provide information for the development of drugs that aim to inhibit LTQ generation and LOXs maturation. However, an in-depth understanding of LTQ generation as well as substrate recognition is restricted by the precursor state of the current structure. To better understand the working mechanism of LOXs, further structural studies should focus on capturing these proteins in a more functional state.
Materials and Methods
Materials used in this study and detailed protocols are described in SI Materials and Methods.
Construct Design and Protein Preparation.
To express the N-tagged hLOXL2, a six-His tag was engineered between the signal peptide and hLOXL2 cDNA (GenBank accession number: DQ895929.2) encoding residues 26–774. The N455Q mutation was introduced to eliminate the glycosylation on the fourth SRCR domain. To obtain the protein segment of residues 318–774 (N455Q), a DEVD coding sequence was introduced right before Ala318 of hLOXL2 (N455Q) to form the DrICE protease-cutting site. All further mutations were introduced based on this construct.
hLOXL2 constructs were transiently expressed in mammalian HEK293F cells and purified by a Ni- nitrilotriacetic acid (Ni-NTA) affinity column. The protein segment of residues 318–774 (N455Q) was released through on-column DrICE digestion and purified by Source Q and mono Q anion-exchange chromatography (GE Healthcare).
Crystallization, Data Collection, and Structure Determination.
Crystals of hLOXL2 (residues 318–774 with N455Q) were grown at 18 °C using the hanging-drop vapor-diffusion method. The buffer yielding the best crystals contained 28% PEG2000, 100 mM Li2SO4, and 100 mM Hepes (pH 7.6). Native datasets were collected at the Shanghai Synchrotron Radiation Facility (SSRF) BL17U beamline and were processed with HKL2000 (41). SAD datasets were collected at the Spring-8 BL41XU beamline and were processed with XDS (42). Phase was solved by a combination of molecular replacement taking the SRCR domain homolog structure (PDB ID code: 1by2) as a search model and Os-SAD. Heavy atom positions were determined with SHELXD (43), and the initial phase was generated with PHASER (44). The model was built in Coot (45) and refined with PHENIX (46).
Enzymatic Assay.
The activity of hLOXL2 was measured by detecting the H2O2 released during deamination using the HRP-coupled Amplex Red assay (47). Briefly, the reaction was started by adding hLOXL2 to 120 μL of reaction mixture containing 50 mM sodium borate (pH 8.0), 15 mM substrate, 5 μM Amplex Red, and 0.2 U/mL HRP. Fluorescence was recorded at 37 °C over time with an excitation wavelength of 525 nm and an emission wavelength of 598 nm and was transformed to H2O2 concentration using a standard curve. All activity measurements were carried out in triplicate, and SDs were calculated. Details are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Protein Chemistry Facility at the Center of Biomedical Analysis of Tsinghua University for mass spectrum analysis. This work was supported by Ministry of Science and Technology Grant 2014ZX09507003006 (to Y.S.) and National Natural Science Foundation of China Grants 31130002 and 31321062 (to Y.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank (PDB), www.wwpdb.org (PDB ID code: 5ZE3).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720859115/-/DCSupplemental.
References
- 1.Ringer P, Colo G, Fässler R, Grashoff C. Sensing the mechano-chemical properties of the extracellular matrix. Matrix Biol. 2017;64:6–16. doi: 10.1016/j.matbio.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Wang JH-C, Thampatty BP, Lin J-S, Im H-J. Mechanoregulation of gene expression in fibroblasts. Gene. 2007;391:1–15. doi: 10.1016/j.gene.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sedlmeier G, Sleeman JP. Extracellular regulation of BMP signaling: Welcome to the matrix. Biochem Soc Trans. 2017;45:173–181. doi: 10.1042/BST20160263. [DOI] [PubMed] [Google Scholar]
- 4.Handorf AM, Zhou Y, Halanski MA, Li WJ. Tissue stiffness dictates development, homeostasis, and disease progression. Organogenesis. 2015;11:1–15. doi: 10.1080/15476278.2015.1019687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moon H-J, Finney J, Ronnebaum T, Mure M. Human lysyl oxidase-like 2. Bioorg Chem. 2014;57:231–241. doi: 10.1016/j.bioorg.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mäki JM, et al. Inactivation of the lysyl oxidase gene Lox leads to aortic aneurysms, cardiovascular dysfunction, and perinatal death in mice. Circulation. 2002;106:2503–2509. doi: 10.1161/01.cir.0000038109.84500.1e. [DOI] [PubMed] [Google Scholar]
- 7.Mäki JM, et al. Lysyl oxidase is essential for normal development and function of the respiratory system and for the integrity of elastic and collagen fibers in various tissues. Am J Pathol. 2005;167:927–936. doi: 10.1016/S0002-9440(10)61183-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finney J, Moon H-J, Ronnebaum T, Lantz M, Mure M. Human copper-dependent amine oxidases. Arch Biochem Biophys. 2014;546:19–32. doi: 10.1016/j.abb.2013.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenbloom J, Ren S, Macarak E. New frontiers in fibrotic disease therapies: The focus of the Joan and Joel Rosenbloom Center for Fibrotic Diseases at Thomas Jefferson University. Matrix Biol. 2016;51:14–25. doi: 10.1016/j.matbio.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 10.Barry-Hamilton V, et al. Allosteric inhibition of lysyl oxidase-like-2 impedes the development of a pathologic microenvironment. Nat Med. 2010;16:1009–1017. doi: 10.1038/nm.2208. [DOI] [PubMed] [Google Scholar]
- 11.Aumiller V, et al. Comparative analysis of lysyl oxidase (like) family members in pulmonary fibrosis. Sci Rep. 2017;7:149. doi: 10.1038/s41598-017-00270-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikenaga N, et al. Selective targeting of lysyl oxidase-like 2 (LOXL2) suppresses hepatic fibrosis progression and accelerates its reversal. Gut. 2017;66:1697–1708. doi: 10.1136/gutjnl-2016-312473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chien JW, et al. Serum lysyl oxidase-like 2 levels and idiopathic pulmonary fibrosis disease progression. Eur Respir J. 2014;43:1430–1438. doi: 10.1183/09031936.00141013. [DOI] [PubMed] [Google Scholar]
- 14.Cano A, Santamaría PG, Moreno-Bueno G. LOXL2 in epithelial cell plasticity and tumor progression. Future Oncol. 2012;8:1095–1108. doi: 10.2217/fon.12.105. [DOI] [PubMed] [Google Scholar]
- 15.Wang T-H, Hsia S-M, Shieh T-M. Lysyl oxidase and the tumor microenvironment. Int J Mol Sci. 2016;18:E62. doi: 10.3390/ijms18010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moreno-Bueno G, et al. Lysyl oxidase-like 2 (LOXL2), a new regulator of cell polarity required for metastatic dissemination of basal-like breast carcinomas. EMBO Mol Med. 2011;3:528–544. doi: 10.1002/emmm.201100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirschmann DA, et al. A molecular role for lysyl oxidase in breast cancer invasion. Cancer Res. 2002;62:4478–4483. [PubMed] [Google Scholar]
- 18.Peinado H, et al. A molecular role for lysyl oxidase-like 2 enzyme in snail regulation and tumor progression. EMBO J. 2005;24:3446–3458. doi: 10.1038/sj.emboj.7600781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollosi P, Yakushiji JK, Fong KSK, Csiszar K, Fong SFT. Lysyl oxidase-like 2 promotes migration in noninvasive breast cancer cells but not in normal breast epithelial cells. Int J Cancer. 2009;125:318–327. doi: 10.1002/ijc.24308. [DOI] [PubMed] [Google Scholar]
- 20.Akiri G, et al. Lysyl oxidase-related protein-1 promotes tumor fibrosis and tumor progression in vivo. Cancer Res. 2003;63:1657–1666. [PubMed] [Google Scholar]
- 21.Chang J, et al. Pre-clinical evaluation of small molecule LOXL2 inhibitors in breast cancer. Oncotarget. 2017;8:26066–26078. doi: 10.18632/oncotarget.15257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erler JT, et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 23.Cox TR, et al. The hypoxic cancer secretome induces pre-metastatic bone lesions through lysyl oxidase. Nature. 2015;522:106–110. doi: 10.1038/nature14492. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Klema VJ, Wilmot CM. The role of protein crystallography in defining the mechanisms of biogenesis and catalysis in copper amine oxidase. Int J Mol Sci. 2012;13:5375–5405. doi: 10.3390/ijms13055375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duff AP, et al. The crystal structure of Pichia pastoris lysyl oxidase. Biochemistry. 2003;42:15148–15157. doi: 10.1021/bi035338v. [DOI] [PubMed] [Google Scholar]
- 26.McGrath AP, et al. Structure and activity of Aspergillus nidulans copper amine oxidase. Biochemistry. 2011;50:5718–5730. doi: 10.1021/bi200555c. [DOI] [PubMed] [Google Scholar]
- 27.Klinman JP, Bonnot F. Intrigues and intricacies of the biosynthetic pathways for the enzymatic quinocofactors: PQQ, TTQ, CTQ, TPQ, and LTQ. Chem Rev. 2014;114:4343–4365. doi: 10.1021/cr400475g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bollinger JA, Brown DE, Dooley DM. The Formation of lysine tyrosylquinone (LTQ) is a self-processing reaction. Expression and characterization of a Drosophila lysyl oxidase. Biochemistry. 2005;44:11708–11714. doi: 10.1021/bi0504310. [DOI] [PubMed] [Google Scholar]
- 29.Matsuzaki R, Fukui T, Sato H, Ozaki Y, Tanizawa K. Generation of the topa quinone cofactor in bacterial monoamine oxidase by cupric ion-dependent autooxidation of a specific tyrosyl residue. FEBS Lett. 1994;351:360–364. doi: 10.1016/0014-5793(94)00884-1. [DOI] [PubMed] [Google Scholar]
- 30.Cai D, Williams NK, Klinman JP. Effect of metal on 2,4,5-trihydroxyphenylalanine (topa) quinone biogenesis in the Hansenula polymorpha copper amine oxidase. J Biol Chem. 1997;272:19277–19281. doi: 10.1074/jbc.272.31.19277. [DOI] [PubMed] [Google Scholar]
- 31.Kim M, et al. X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase. Nat Struct Biol. 2002;9:591–596. doi: 10.1038/nsb824. [DOI] [PubMed] [Google Scholar]
- 32.Chen Z, et al. Crystal structure at 2.5 A resolution of zinc-substituted copper amine oxidase of Hansenula polymorpha expressed in Escherichia coli. Biochemistry. 2000;39:9709–9717. doi: 10.1021/bi000639f. [DOI] [PubMed] [Google Scholar]
- 33.Hohenester E, Sasaki T, Timpl R. Crystal structure of a scavenger receptor cysteine-rich domain sheds light on an ancient superfamily. Nat Struct Biol. 1999;6:228–232. doi: 10.1038/6669. [DOI] [PubMed] [Google Scholar]
- 34.Wang SX, et al. A crosslinked cofactor in lysyl oxidase: Redox function for amino acid side chains. Science. 1996;273:1078–1084. doi: 10.1126/science.273.5278.1078. [DOI] [PubMed] [Google Scholar]
- 35.Lucero HA, Kagan HM. Lysyl oxidase: An oxidative enzyme and effector of cell function. Cell Mol Life Sci. 2006;63:2304–2316. doi: 10.1007/s00018-006-6149-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu L, et al. Post-translational modifications of recombinant human lysyl oxidase-like 2 (rhLOXL2) secreted from Drosophila S2 cells. J Biol Chem. 2013;288:5357–5363. doi: 10.1074/jbc.C112.421768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 38.Paz MA, Flückiger R, Boak A, Kagan HM, Gallop PM. Specific detection of quinoproteins by redox-cycling staining. J Biol Chem. 1991;266:689–692. [PubMed] [Google Scholar]
- 39.Li R, Klinman JP, Mathews FS. Copper amine oxidase from Hansenula polymorpha: The crystal structure determined at 2.4 A resolution reveals the active conformation. Structure. 1998;6:293–307. doi: 10.1016/s0969-2126(98)00033-1. [DOI] [PubMed] [Google Scholar]
- 40.Parsons MR, et al. Crystal structure of a quinoenzyme: Copper amine oxidase of Escherichia coli at 2 A resolution. Structure. 1995;3:1171–1184. doi: 10.1016/s0969-2126(01)00253-2. [DOI] [PubMed] [Google Scholar]
- 41.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 42.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 44.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 47.Palamakumbura AH, Trackman PC. A fluorometric assay for detection of lysyl oxidase enzyme activity in biological samples. Anal Biochem. 2002;300:245–251. doi: 10.1006/abio.2001.5464. [DOI] [PubMed] [Google Scholar]
- 48.Schrödinger, LLC 2010. The PyMOL Molecular Graphics System (Schrödinger, San Diego), Version 1.5.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.