Significance
T box riboswitches regulate the expression of amino acid-related genes using a specific uncharged tRNA as their ligand. Previous work demonstrated that the conserved Stem II S-turn motif and Stem IIA/B pseudoknot element of the riboswitch are important for tRNA-dependent regulation; however, their function remained unknown. We identified contacts between these RNA elements and the tRNA ligand, providing evidence that they directly interact with tRNA. This represents an example of tRNA recognition by an S-turn motif or pseudoknot element, contributing to the understanding of alternative roles for tRNA and other RNA–RNA interactions. Furthermore, the novelty of these contacts makes them optimal targets for the ongoing development of antibiotics that target T box riboswitches.
Keywords: T box riboswitch, tRNA, regulation, S-turn, pseudoknot
Abstract
T box riboswitches are RNA regulatory elements widely used by organisms in the phyla Firmicutes and Actinobacteria to regulate expression of amino acid-related genes. Expression of T box family genes is down-regulated by transcription attenuation or inhibition of translation initiation in response to increased charging of the cognate tRNA. Three direct contacts with tRNA have been described; however, one of these contacts is absent in a subclass of T box RNAs and the roles of several structural domains conserved in most T box RNAs are unknown. In this study, structural elements of a Mycobacterium smegmatis ileS T box riboswitch variant with an Ultrashort (US) Stem I were sequentially deleted, which resulted in a progressive decrease in binding affinity for the tRNAIle ligand. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) revealed structural changes in conserved riboswitch domains upon interaction with the tRNA ligand. Cross-linking and mutational analyses identified two interaction sites, one between the S-turn element in Stem II and the T arm of tRNAIle and the other between the Stem IIA/B pseudoknot and the D loop of tRNAIle. These newly identified RNA contacts add information about tRNA recognition by the T box riboswitch and demonstrate a role for the S-turn and pseudoknot elements, which resemble structural elements that are common in many cellular RNAs.
T box riboswitches are cis-acting RNA regulatory elements found in the 5′-untranslated (leader) regions of many essential amino acid-related genes in organisms of the phyla Actinobacteria and Firmicutes (1). Structural rearrangements of the T box riboswitch RNA in response to binding of the cognate uncharged tRNA regulate gene expression at the level of either transcription attenuation (2, 3) or translation initiation (4). High tRNA aminoacylation allows formation of a transcriptional terminator or a sequestrator structure that includes sequences necessary for ribosome binding, resulting in either premature transcription termination or inhibition of translation initiation. When aminoacylation of the cognate tRNA is low, interaction of the uncharged tRNA with the leader RNA stabilizes a competing antiterminator or antisequestrator structure, which allows expression of the downstream gene.
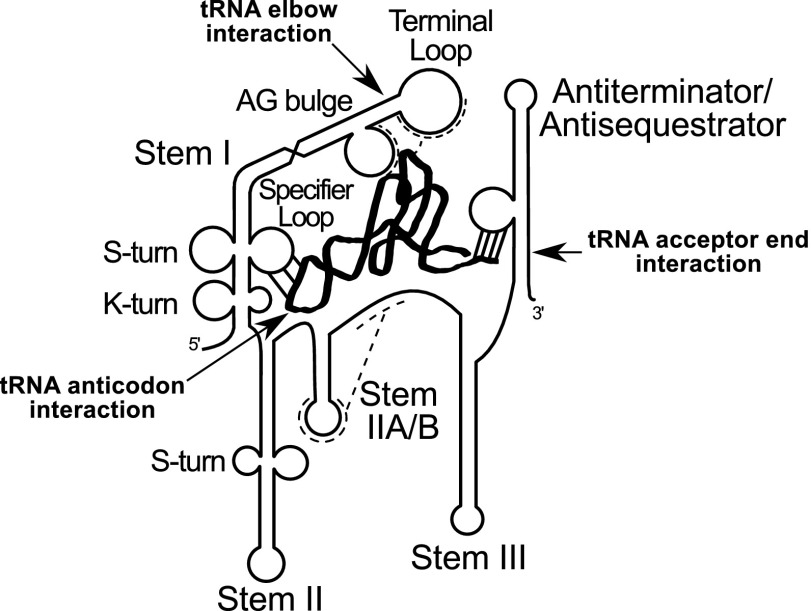
Canonical T box riboswitches contain highly conserved helical domains (Fig. 1) designated Stems I, II, and III, and a pseudoknot element designated Stem IIA/B located between Stem II and Stem III; all of these elements are upstream of the antiterminator or antisequestrator elements (2, 5). Genetic and structural analyses have identified three sites in the T box riboswitch RNA that directly contact the tRNA. The Specifier Sequence, a triplet sequence located in the internal Specifier Loop of Stem I, pairs with the anticodon of either charged or uncharged tRNA and is the major determinant of tRNA-binding specificity (2, 6). The antiterminator or antisequestrator bulge interacts with the acceptor end of uncharged tRNA but not charged tRNA, allowing the riboswitch to monitor the tRNA aminoacylation state (3, 7). A third interaction between the RNA platform at the top of Stem I and the tRNA elbow has been demonstrated only in glyQS genes, which contain a unique sequence arrangement in the Stem I terminal platform (8, 9); this interaction may vary in conformation in other T box RNAs. The glyQS genes lack the Stem II and IIA/B domains, and little information is available about the role of these elements.
Fig. 1.
Generalized model of binding of T box riboswitch RNA to uncharged cognate tRNA (thick black ribbon). Arrows show known tRNA contact points. Major T box riboswitch secondary structural elements are labeled.
A class of T box riboswitches that lack the Stem I terminal platform was recently identified (1, 10). These RNAs, designated Ultrashort (US) Stem I variants, are found in ileS genes, encoding isoleucyl-tRNA synthetases, in many organisms in the phylum Actinobacteria (4). US variants have a truncated Stem I with the Specifier Sequence in the terminal loop and contain all of the other conserved elements found in canonical T box riboswitches, including Stems II and IIA/B. The US RNAs specifically bind cognate tRNAIle in vitro, and binding of uncharged tRNAIle stabilizes an antisequestrator structure and promotes ribosome binding, consistent with regulation at the level of translation initiation (4). Binding of either charged or uncharged tRNAIle stabilizes the initial Stem II and the IIA/B pseudoknot domains. These data provided biochemical evidence for the formation of Stem II and the pseudoknot since most biochemical analyses of T box RNAs utilized glycyl RNAs that lack these elements. Therefore, characterization of US ileS riboswitches provides a unique opportunity to investigate the role of the Stem II and IIA/B domains in vitro.
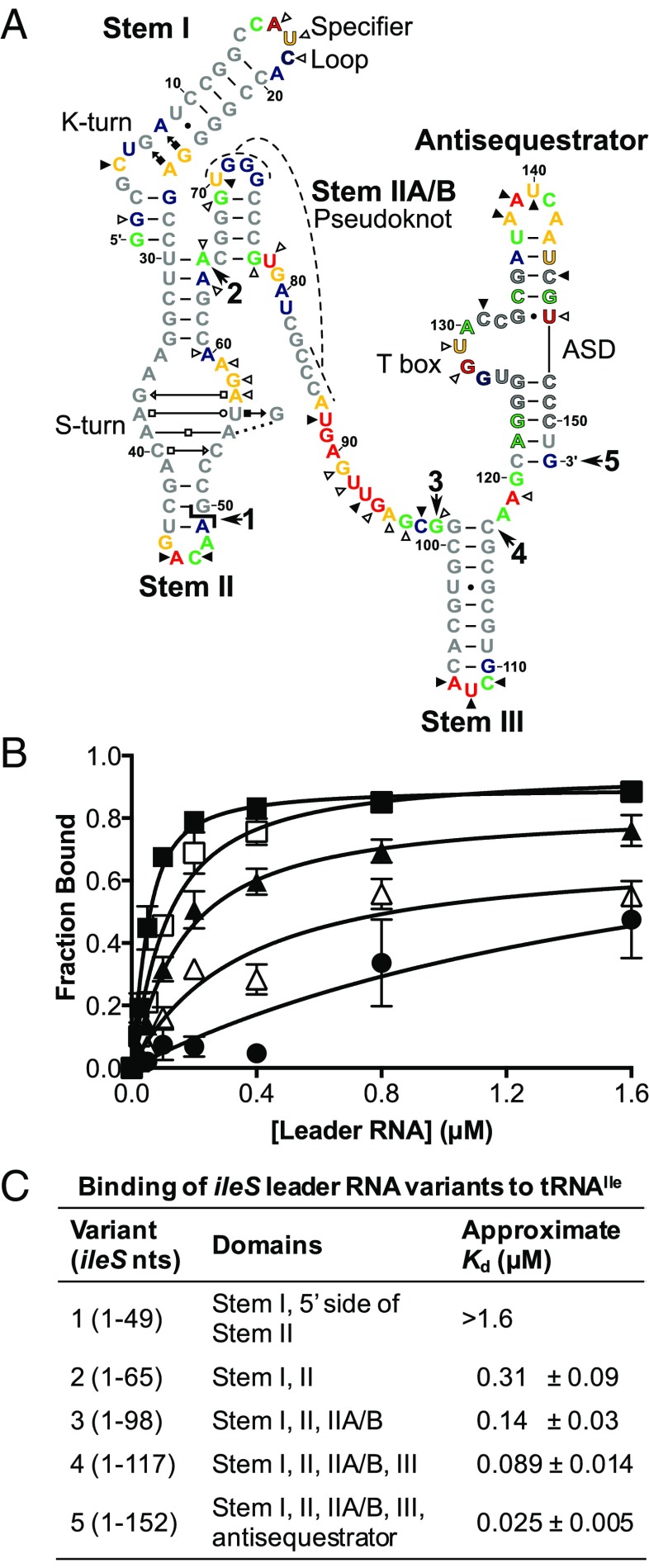
The Stem II domain in most T box riboswitches contains an S-turn motif with the conserved sequences 5′-GAAC-3′ (nucleotides 37–40) on the 5′ side and 5′-AGUA-3′ (nucleotides 53–57) on the 3′ side (Fig. 2A) (1, 5). These sequences are predicted to form a helical structure with consecutive non-Watson–Crick base pairs that introduce an S-shaped bend to the phosphate backbone (11, 12). S-turns are found in ribosomal RNAs, ribozymes, and viral RNAs and often act as a binding site for protein or RNA (13). Mutations in the S-turn motif in the Bacillus subtilis tyrS gene disrupt T box regulation in vivo (5), which suggests that either the primary sequence or the structure formed by the motif is important for interaction with tRNA. The Stem II domain is adjoined by Stem IIA, and nucleotides downstream of Stem IIA base pair with residues in the loop of Stem IIA to form the Stem IIA/B pseudoknot structure, the disruption of which reduces tyrS expression (1, 2, 5).
Fig. 2.
Binding of tRNA to the ileS leader RNA with sequential deletion of conserved structural elements. (A) M. smegmatis ileS RNA predicted secondary structure. Major conserved structural elements are labeled. Predicted non-Watson–Crick interactions are indicated by Leontis–Westhof symbols (40). Outlined nucleotides denote conserved primary sequences: Specifier Sequence (A16–C18), T box sequence (A122–G135), ASD (U144–C150). The  symbol marks the position of the split in the bipartite RNA. Bold numbers with arrows indicate the 3′ nucleotide in the variants (1: nucleotides 1–49; 2: nucleotides 1–65; 3: nucleotides 1–98; 4: nucleotides 1–117; 5: nucleotides 1–152). Colors indicate SHAPE reactivity in the absence of tRNAIle: red, highly reactive; gold, reactive; green, moderately reactive; blue, minimally reactive; gray, unreactive. Triangles indicate changes in reactivity upon interaction with cognate tRNAIle: open, less reactive (relative reactivity ≤ −0.1); solid, more reactive (relative reactivity ≥0.1). ASD, anti-Shine-Dalgarno. (B) Binding of tRNAIle to ileS RNA variants. Radioactively labeled tRNA was incubated with increasing concentrations of ileS RNA and passed through a size-exclusion filter, followed by quantification of the retained tRNA. Variant 1, filled circles (R2 = 0.61); variant 2, open triangles (R2 = 0.90); variant 3, filled triangles (R2 = 0.92); variant 4, open squares (R2 = 0.94); variant 5, filled squares (R2 = 0.93). Error bars denote SEM, n ≥ 3. (C) Approximate Kd values for binding of tRNAIle to ileS RNA variants.
symbol marks the position of the split in the bipartite RNA. Bold numbers with arrows indicate the 3′ nucleotide in the variants (1: nucleotides 1–49; 2: nucleotides 1–65; 3: nucleotides 1–98; 4: nucleotides 1–117; 5: nucleotides 1–152). Colors indicate SHAPE reactivity in the absence of tRNAIle: red, highly reactive; gold, reactive; green, moderately reactive; blue, minimally reactive; gray, unreactive. Triangles indicate changes in reactivity upon interaction with cognate tRNAIle: open, less reactive (relative reactivity ≤ −0.1); solid, more reactive (relative reactivity ≥0.1). ASD, anti-Shine-Dalgarno. (B) Binding of tRNAIle to ileS RNA variants. Radioactively labeled tRNA was incubated with increasing concentrations of ileS RNA and passed through a size-exclusion filter, followed by quantification of the retained tRNA. Variant 1, filled circles (R2 = 0.61); variant 2, open triangles (R2 = 0.90); variant 3, filled triangles (R2 = 0.92); variant 4, open squares (R2 = 0.94); variant 5, filled squares (R2 = 0.93). Error bars denote SEM, n ≥ 3. (C) Approximate Kd values for binding of tRNAIle to ileS RNA variants.
The Stem III domain is present in most T box riboswitches but is not conserved in size or sequence and is not sensitive to mutation (1, 14). An RNA polymerase (RNAP) pause site was identified in Stem III in the B. subtilis glyQS gene, but its functional role is unclear (14). It was recently observed that Stem III stacks on the antiterminator in the glyQS riboswitch, stabilizing the tRNA-bound state to facilitate readthrough (15, 16).
In this study, the role of the conserved elements of the Mycobacterium smegmatis ileS T box riboswitch in tRNAIle binding was analyzed by mutational, cross-linking, and selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) studies. These studies revealed two interactions between the ileS RNA and tRNAIle. The Stem II S-turn element interacts with the tRNAIle T arm and the Stem IIA/B pseudoknot interacts with the tRNAIle D loop. This study presents biochemical evidence for these contacts and identifies an aspect of the molecular basis for tRNA recognition by the riboswitch that is likely to be conserved in the majority of T box RNAs. We also provide a biological role in tRNA recognition for the S-turn and pseudoknot RNA elements, which are found in many cellular RNAs.
Results
Sequential Deletion of Conserved Structural Elements in ileS Leader RNA Causes Decreased tRNAIle Binding.
M. smegmatis ileS leader RNA variants with sequential deletions of the antisequestrator, Stem III, Stem IIA/B, and the 3′ side of Stem II (variants 1–4 in Fig. 2A) were generated to assess their contribution to tRNAIle binding. Sequential deletion progressively lowered tRNA binding compared with the complete construct (variant 5; Fig. 2B) and increased the approximate Kd (Fig. 2C). Binding to the noncognate B. subtilis tRNAGly was below the limit of detection for all five leader RNA constructs (Fig. S1), indicating that the observed interaction with tRNAIle is specific. These results show that the Stem II, IIA/B, and III domains contribute to tRNAIle-binding affinity.
Stem II and IIA/B Undergo Structural Changes upon Interaction with tRNAIle.
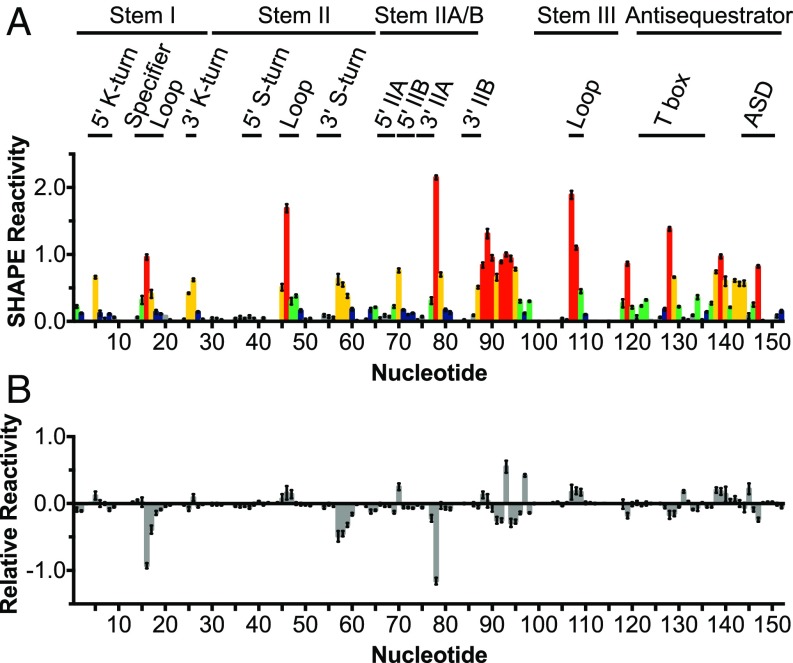
tRNA-dependent structural changes in ileS leader RNA were assessed using SHAPE (17). Local nucleotide flexibility was determined by the addition of N-methylisotoic anhydride, which forms 2′-adducts with unconstrained or flexible nucleotides more readily than with those constrained by base-pairing or other structural interactions. The location of the adducts was determined by primer extension inhibition. In the absence of tRNA, reactive nucleotides of Stem I were confined to C5, G25, and A26 (K-turn motif) and the Specifier Loop (Figs. 2A and 3A). In Stem II, reactive nucleotides were found in the 3′ side of the S-turn motif (A57–A59) and in the terminal loop. In Stem IIA/B, reactive nucleotides were found in Stem IIB (U70 and A87) and in the 3′ of Stem IIA (G77–G79, turn between pseudoknot helices). Nucleotides U88–G98 in the linker region between Stem IIA/B and Stem III were highly reactive, while in Stem III, the only reactive nucleotides were found in the terminal loop. In the antisequestrator, nucleotides in the bulge (part of the T box sequence) and terminal loop were highly reactive. These results are consistent with the predicted secondary structure (Fig. 2A) and indicate that the main structural elements form in the absence of the tRNA ligand.
Fig. 3.
tRNAIle-induced changes in the S-turn and pseudoknot elements of the M. smegmatis ileS leader RNA. (A) ileS RNA was incubated with NMIA or DMSO, followed by an RT inhibition reaction. SHAPE reactivity is the difference between the frequency of RT stops at each nucleotide in NMIA vs. DMSO samples. Colors indicate SHAPE reactivity as in Fig. 2A. Error bars denote SEM, n = 9. The ileS RNA nucleotidesare numbered below the x axis and nucleotides that participate in structural motifs are denoted. ASD, anti-Shine-Dalgarno; NMIA, N-methylisotoic anhydride. (B) Relative reactivity of M. smegmatis ileS leader RNA in the presence of tRNAIle. The ileS RNA was cofolded with 10-fold excess cognate tRNAIle before SHAPE experiments carried out as described in A. Relative reactivity is the difference in SHAPE reactivity in the presence of tRNAIle vs. the absence of tRNA. Error bars denote SEM, n = 9.
The addition of cognate tRNAIle resulted in numerous changes in reactivity (Figs. 2A and 3B); nucleotides that experienced a change in reactivity of ≥0.1 are highlighted in Fig. 2A. Reactivity was reduced in the Specifier Loop (nucleotides A16–C18), which is consistent with base-pairing of the Specifier Sequence with the tRNAIle anticodon. Additionally, the reactivity of C5 increased, which may indicate tRNA-induced changes to the kink-turn structure. Reduced reactivity was also observed in the junction between Stems I and II (nucleotides G2, A64, and A65), suggesting that the structural integrity of this region increases upon interaction with tRNAIle. In Stem II, reduced reactivity was observed on the 3′ side of the S-turn motif (nucleotides A57–A60), which suggests either protection of this region by direct interaction with nucleotides in tRNAIle or tRNA-induced structural changes. The reduced reactivity of the nucleotides 3′ of Stem IIA (G77 and U78), together with the increased reactivity of U70 and U88 and decreased reactivity of G69, suggest that tRNA binding stabilizes the pseudoknot, which is consistent with the previous observation that tRNAIle binding stabilizes the element (4). Most of the nucleotides in the linker region (U88–G98) became less reactive, suggesting that this region is more constrained upon tRNA binding or protected by interactions with the tRNA. Two nucleotides (U93 and C97) in the linker became more reactive in the presence of tRNA; these nucleotides may be constrained such that the 2′-hydroxyl is more accessible. The decreased reactivity of A119 suggests that the base of the antisequestrator is more constrained, consistent with the observation of stacking between Stem III and the antisequestrator in the tRNA-bound state (15, 16). In the antisequestrator bulge, the reactivity of nucleotides G128 and U129 decreased, consistent with direct base-pairing with the acceptor end of uncharged tRNA, and the reactivity of C131 increased, consistent with the previous observation that the flexibility of these nucleotides changed upon tRNA binding (6). Additionally, the reactivity of C145 increased and the reactivity of U147 decreased, suggesting that tRNAIle binding changes the structural integrity of this helix. The increased reactivity of loop nucleotides in Stem II (A46, C47), Stem III (A107, U108, C109), and the antisequestrator (A138, A139, and U140) is likely due to reduced reactivity of other positions. The addition of noncognate B. subtilis tRNAGly did not change the reactivity profile of the ileS RNA (Fig. S2), indicating that the observed changes are specific to tRNAIle.
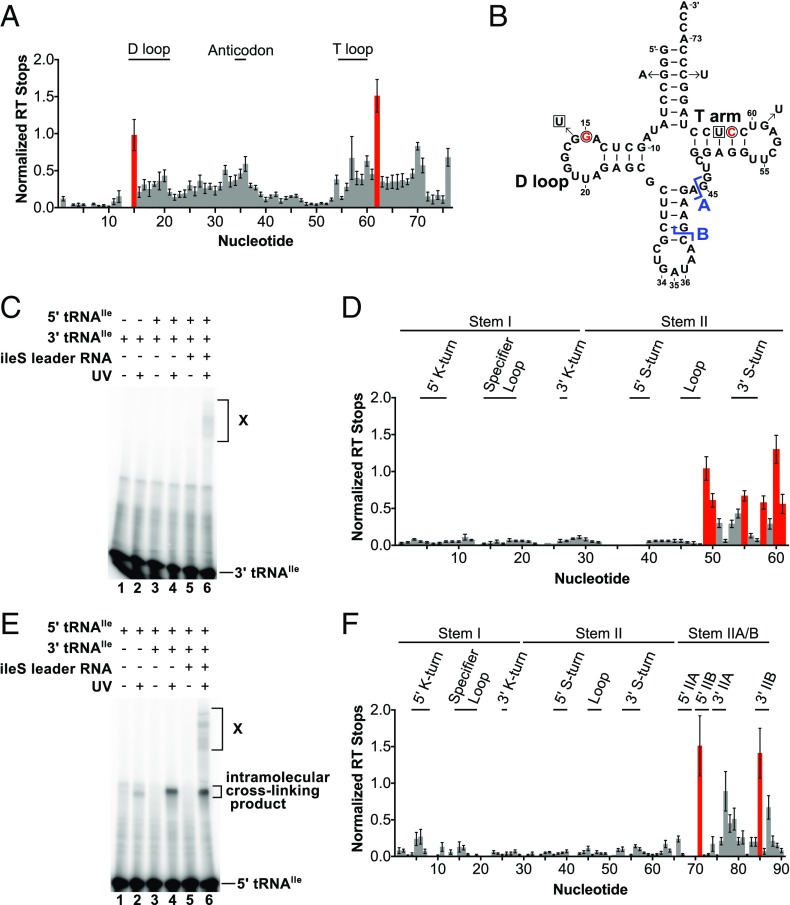
ileS Leader RNA Cross-Links to tRNAIle.
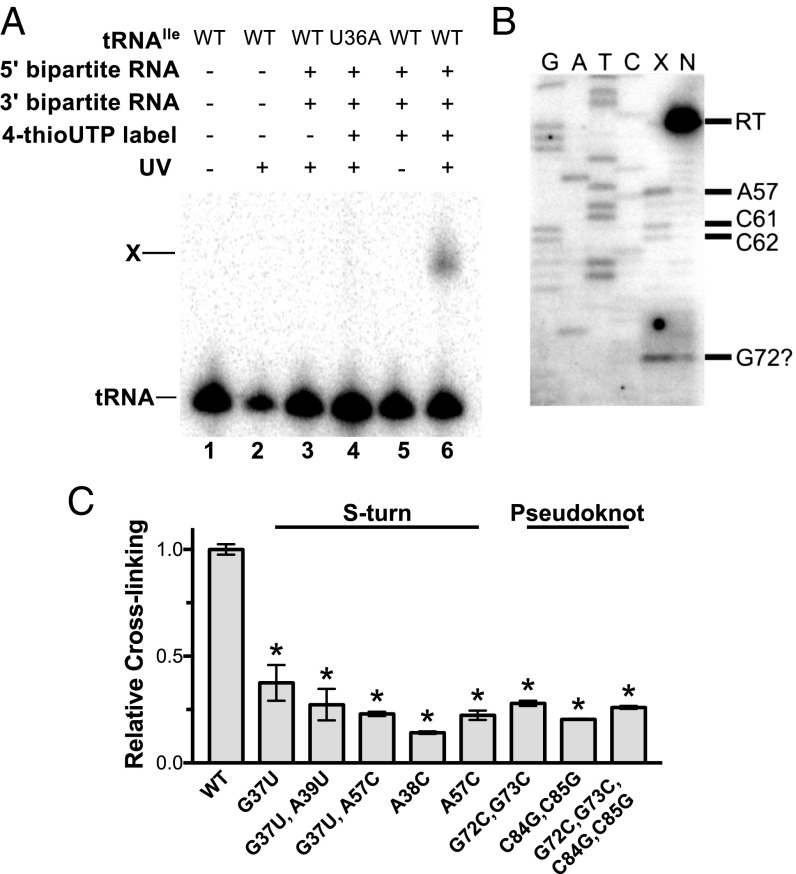
Possible contacts between the Stem II or Stem IIA/B domains and tRNA were investigated by cross-linking experiments using 4-thio-UTP, a UTP analog that generally does not disrupt RNA structure (18). The sulfur in 4-thio-U forms covalent bonds with functional groups within a 3-Å radius upon UV exposure (18). A bipartite leader RNA was generated to allow incorporation of the 4-thio-UTP specifically into the 3′ side of Stem II and Stem IIA/B (nucleotides 50–98), resulting in the substitution of 4-thio-UTP at nucleotides 56, 70, 78, 81, 88, 92, and 93. The bipartite RNA exhibited a small decrease in tRNAIle binding (Kd = 0.35 µM vs. 0.14 µM for the bipartite vs. unimolecular RNAs, respectively).
Incubation of radioactively labeled tRNAIle with the 4-thio-U bipartite leader RNA followed by UV irradiation resulted in the appearance of an RNA band with reduced gel migration (Fig. 4A, lane 6), dependent on the presence of the ileS RNA and 4-thio-UTP incorporation into the 3′ leader RNA fragment. Introduction of a tRNA anticodon U36A mutation known to disrupt the Specifier Sequence–anticodon interaction (4) resulted in loss of the slowly migrating RNA band (Fig. 4A, lane 4), indicating that its formation is dependent on specific recognition of the tRNA anticodon. The slowly migrating RNA was therefore considered a leader RNA–tRNA cross-linked product.
Fig. 4.
Bipartite ileS RNA cross-links to tRNAIle via the Stem II S-turn motif and the pseudoknot element. (A) Cross-linking of tRNAIle to bipartite ileS RNA. The 4-thio-U–substituted 3′ RNA fragment was cofolded with the 5′ RNA fragment in the presence of radioactively labeled tRNAIle. . RNAs were exposed to UV light as indicated and separated on a denaturing gel. tRNA, non–cross-linked tRNA; U36A, anticodon mutant; X, cross-linked tRNA. (B) Primer extension inhibition mapping of the cross-link position. A 32P-labeled oligonucleotide complementary to 3′-terminal sequences (nucleotides 83–98) of the ileS RNA was used in reactions with the cross-linked product (lane X) and non–cross-linked leader RNA (lane N). (C) Cross-linking efficiency of mutant leader RNAs relative to the WT RNA. Error bars denote SEM, n ≥ 3. *P< 0.0001 between WTand mutant RNAs as detected by two-sided Dunnett’s test.
The ileS Leader RNA Cross-Links to tRNAIle via the S-Turn Motif in Stem II and the Stem IIA/B Pseudoknot Element.
The observation that formation of the ileS leader RNA–tRNAIle cross-linked product was dependent on 4-thio-UTP incorporation into the 3′ ileS fragment indicates that at least one uridine in that region participates in cross-linking. Primer extension inhibition reactions were carried out to identify the uridine(s) that participate in the tRNA cross-linking based on reverse transcriptase (RT) stops (at the nucleotide 3′ of the cross-link position) only in cross-linked RNA. An RT stop was observed at nucleotide A57, which corresponds to a cross-link at nucleotide U56, in the Stem II S-turn (Fig. 2A), which was the only uridine observed to inhibit primer extension (Fig. 4B, lane X). Primer extension inhibition products at C61 and C62 correspond to RNA cross-linking at nucleotides A60 and C61, respectively (Fig. 4B, lane X), and are likely to represent intramolecular cross-links. An additional product near G72 is more abundant in the cross-linked vs. non–cross-linked samples, but the identity of this primer extension inhibition product is unclear due to sequence compression caused by sequential guanine nucleotides in the RNA (G71–G73). This product may be a result of RNA cross-linking at nucleotide U70 in the pseudoknot element, which exhibited increased SHAPE reactivity upon the addition of tRNAIle (Fig. 2A).
Mutational analysis of the S-turn motif was used to investigate its possible role in the interaction with tRNAIle. An A38C mutation, predicted to disrupt the conserved base triple (A38–G55–U56; Fig. 2A), resulted in the largest decrease in cross-linking (Fig. 4C). A38 is distant from the proposed cross-linking site (U56) in the primary sequence but is predicted to be in close proximity in the S-turn structure. Other mutations in the 5′ (G37U and G37U, A38U) and 3′ (A57C) sides of the motif also reduced formation of the cross-linked product, supporting the model that the tRNA interaction is dependent on the S-turn element.
G37 is predicted to interact with A57 in a non-Watson–Crick base pair (Fig. 2A) that is isosteric to a U–C pairing (19). The combination of the G37U and A57C substitutions therefore was predicted to increase cross-linking in comparison with the single G37U and A57C mutant RNAs. However, no increase in cross-linking was observed (Fig. 4C), which indicates either failure to form the S-turn structure in the isosteric mutant or a requirement for a G·A base pair for the tRNA interaction.
Mutational analysis also was used to determine if cross-linking to tRNAIle requires the Stem IIA/B pseudoknot element. Mutations in the 5′ (G72C, G73C) or 3′ (C84G, C85G) sides of Stem IIB resulted in decreased cross-linking (Fig. 4C), indicating that Stem IIB is important for the tRNA interaction. Complementary mutations (G72C, G73C, C84G, C85G) predicted to restore base-pairing did not increase cross-linking (Fig. 4C). These data indicate failure to form the pseudoknot in the complementary mutant, the formation of an alternative structure, or the importance of primary sequence of Stem IIB for the contact with tRNAIle. Previous analysis of the B. subtilis tyrS T box riboswitch indicated that conserved primary sequence in the pseudoknot element plays an essential role for riboswitch function (5).
D Loop and T Arm of tRNAIle Cross-Link to ileS Leader RNA.
Primer extension inhibition reactions followed by capillary electrophoresis were used to identify cross-linked positions in tRNAIle (Fig. 5A). G3A and C70U complementary mutations were introduced into the acceptor stem of tRNAIle to facilitate progress of RT through a stretch of G-C base pairs found in this region (Fig. 5B). This modification did not alter the binding affinity or cross-linking efficiency (Fig. S3). UV-dependent primer extension inhibition products corresponding to G15 (in the D loop) and C62 (in the T arm) were observed (Fig. 5 A and B). A significant level of background was also observed, which is likely to be due to nonspecific cross-linking with the U nucleotides in the linker region. This result indicates that the D loop and T arm of tRNAIle directly contact ileS RNA.
Fig. 5.
D loop and T arm of the tRNAIle cross-link to the ileS leader RNA. (A) Primer extension inhibition to map cross-link positions in the tRNAIle. The bipartite ileS RNA with 4-thio-U substituted for U in the 3′ fragment was cofolded with G3A, C70U tRNAIle. RNAs were exposed to UV light, and primer extension was performed using a fluorescently labeled oligonucleotide complementary to the 3′ end of the tRNA construct. Normalized RT stops are the difference between the frequency of RT stops at each nt in the cross-linked vs. non–cross-linked samples. Red bars indicate UV-dependent primer extension inhibition products. Error bars denote SEM, n = 8. (B) Predicted secondary structure of tRNAIle. The nucleotides that cross-link to the leader RNA are red and circled. The blue  symbol indicates the positions of the splits in the bipartite tRNAs (A: 5′ 1–44, 3′ 45–76; B: 5′ 1–39, 3′ 40–76); the sites of 4-thio-U incorporation are boxed, and arrows indicate the modifications made to the tRNA sequence. (C) Cross-linking of the tRNAIle T arm to the leader RNA. The 3′ fragment of bipartite tRNA A with 4-thio-U at position 63 was radioactively labeled and cofolded with the 5′ fragment of tRNA A in the presence and absence of ileS RNA, exposed to UV light, and separated on a denaturing gel. X, intermolecular cross-linked RNA. (D) Primer extension inhibition analysis of the cross-linked RNA from C was performed as in A. (E) Cross-linking of the tRNAIle D arm to leader RNA. The 5′ fragment of bipartite tRNA B with 4-thio-U at position 16 was radioactively labeled and cofolded with the G59U 3′ fragment of tRNA B and cross-linking analysis was carried out as in C. (F) Primer extension inhibition analysis was carried out as described in A.
symbol indicates the positions of the splits in the bipartite tRNAs (A: 5′ 1–44, 3′ 45–76; B: 5′ 1–39, 3′ 40–76); the sites of 4-thio-U incorporation are boxed, and arrows indicate the modifications made to the tRNA sequence. (C) Cross-linking of the tRNAIle T arm to the leader RNA. The 3′ fragment of bipartite tRNA A with 4-thio-U at position 63 was radioactively labeled and cofolded with the 5′ fragment of tRNA A in the presence and absence of ileS RNA, exposed to UV light, and separated on a denaturing gel. X, intermolecular cross-linked RNA. (D) Primer extension inhibition analysis of the cross-linked RNA from C was performed as in A. (E) Cross-linking of the tRNAIle D arm to leader RNA. The 5′ fragment of bipartite tRNA B with 4-thio-U at position 16 was radioactively labeled and cofolded with the G59U 3′ fragment of tRNA B and cross-linking analysis was carried out as in C. (F) Primer extension inhibition analysis was carried out as described in A.
To determine where the tRNA T arm and D loop contact the ileS RNA, 4-thio-UTP was specifically incorporated at either U63 or G16 in separate bipartite tRNA molecules (Fig. 5B). The observed affinities of the bipartite tRNAs for the leader RNA were comparable to the intact tRNA (bipartite vs. unimolecular RNAs for tRNA A, Kd = 0.43 µM vs. 0.14 µM; and for tRNA B, Kd = 0.19 µM vs. 0.089 µM). To maintain proper folding of the tRNA with 4-thio-UTP at nt 16, a complementary G59U mutation was introduced due to covariation of nucleotide 16 with nucleotide 59 in the phylum Actinobacteria (20) and formation of hydrogen bonds with nucleotide 59 in the variable pocket of other tRNAs (21, 22).
Formation of the cross-linked complex was visualized by radioactively labeling the 4-thio-UTP–containing tRNA fragment. In the presence of the leader RNA, an intermolecular cross-linked product (indicated by a higher molecular weight band, labeled “X” in Fig. 5 C and E, lane 6) was detected after exposure to UV light. An additional UV-dependent lower molecular weight product was observed when the tRNA included 4-thio-UTP at nt 16 (Fig. 5E, lanes 2, 4, and 6). This product was independent of the leader RNA (Fig. 5E, lanes 2 and 4), suggesting that it represents an intramolecular cross-linking product.
The cross-linked product was used in primer extension inhibition reactions. Placement of 4-thio-UTP at nucleotide 63 (T arm) yielded products corresponding mainly to nucleotides on the 3′ side of Stem II (A49, G50, G55, G58, A60, and C61; Fig. 5D). G55 participates in the S-turn base triple and G58, A60, and C61 are located just 3′ of the S-turn motif. The primer extension inhibition products at A49 and G50 are likely to represent intramolecular cross-links. With 4-thio-UTP in the D loop (nucleotide 16), products were observed at G71 and C85 (Fig. 5F), which are in Stem IIB of the pseudoknot element. These nucleotides are predicted to be in close proximity when the pseudoknot is formed (Fig. 2A). These results demonstrate a direct contact between the T arm of tRNAIle and the Stem II S-turn motif as well as between the D loop of tRNAIle and the IIA/B pseudoknot of the ileS T box RNA.
Discussion
Previous biochemical and structural studies of T box RNA–tRNA interactions have focused primarily on glyQS genes, which lack Stem II and the IIA/B pseudoknot. We recently demonstrated specific tRNAIle binding to ileS US T box RNAs from Actinobacteria, which contain the elements missing in glyQS T box RNAs, and observed tRNAIle-dependent stabilization of Stems II and IIA/B (4). Deletion of the antisequestrator element from an ileS T box RNA was predicted to decrease tRNA binding because the antisequestrator directly binds the acceptor end of tRNA (4). Reduced tRNA binding after deletion of Stem III is consistent with the proposed role of Stem III in stabilization of the tRNA-binding pocket in glyQS RNAs (14). Deletion of Stem IIA/B and the 3′ side of Stem II also decreased tRNA binding, which is consistent with results that indicate the importance of these elements for B. subtilis tyrS gene expression in vivo (5). Using SHAPE, we demonstrated that the addition of tRNAIle alters the structure of the Stem II S-turn element and the Stem IIA/B pseudoknot. These elements were further investigated for their potential participation in a direct interaction with tRNA.
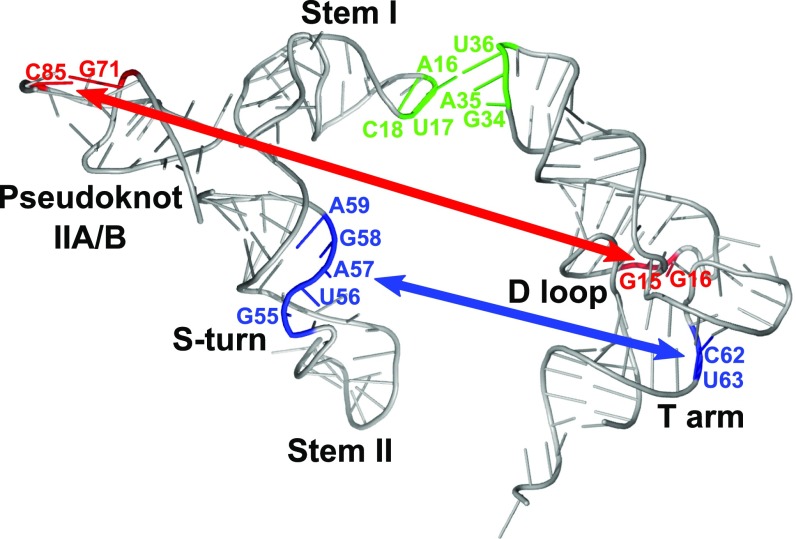
Cross-linking experiments identified two direct contacts with tRNA, one between the S-turn motif and the T arm of tRNAIle and the other between the Stem IIA/B pseudoknot and the D loop of tRNAIle (Fig. 6). High conservation of both structural elements (with the exception of glycyl genes; ref. 1) and their sensitivity to mutation (5) suggest that these newly discovered tRNA contacts may occur in other T box riboswitches that contain Stem II and Stem IIA/B, although this remains to be demonstrated.
Fig. 6.
Structural model of the ileS leader RNA–tRNA interactions. 3D models of M. smegmatis ileS Stem I+II RNA and Stem IIA/B RNA were generated using RNAComposer (41, 42) and aligned to depict a model of the leader RNA. Nucleotides involved in leader RNA–tRNA interactions are highlighted on the model and the tRNA crystal structure (Protein Data Bank: 1EHZ); the previously identified Specifier Sequence–anticodon interaction is shown in green, the D loop–pseudoknot interaction is shown in red, and the T arm–Stem II S-turn interaction is shown in blue.
S-turn motifs in other RNAs often participate in binding of proteins and RNA (12, 23–25) and are important for ribosome assembly, translation, and viral replication and packaging (12, 19, 26–28). S-turn formation depends not only on base–base interactions but also on base–phosphate backbone interactions (29). The G37U and A57C single mutations reduced cross-linking, as predicted, but the G37U, A57C double substitution, which was predicted to introduce an isosteric pair at the 37–57 position, did not restore cross-linking efficiency (Fig. 4C); the G37U, A57C double mutant is not expected to maintain the base–phosphate backbone interaction in the structure, which may be responsible for failure to restore cross-linking.
Pseudoknot elements are essential for a range of molecular processes (e.g., translation, ribosomal frameshifting, and viral replication) (30–33), and many other riboswitches, including the fluoride, S-box, and purine riboswitches (34–36), use pseudoknot structures in their ligand-binding pocket (37). Pseudoknots contain two intercalating helices and are typically stabilized by stacking and non-Watson–Crick base-pairing interactions (37, 38). The mutations that disrupt Stem IIB (G72C, G73C or C84G, C85G) reduced cross-linking as expected; however, the complementary mutations (G72C, G73C, C84G, C85G), predicted to restore base-pairing, did not restore the cross-linking efficiency (Fig. 4C). These changes may destabilize the pseudoknot element by changing the non–base-pairing interactions, which would explain the observed decrease in the cross-linking efficiency.
In addition to U56 (S-turn) and G71 (pseudoknot), primer extension inhibition products corresponding to positions A60 and C61 of the leader RNA were observed in the cross-linked complex (Fig. 4A). This may be the result of intramolecular cross-linking to U56, as A60 and C61 are likely to be located in close proximity to U56 when the RNA is properly folded. Since both A60 and C61 cross-linked to the tRNA with 4-thio-U at position 63 (Fig. 5D) and A60 exhibited decreased SHAPE reactivity in the presence of tRNAIle (Figs. 2A and 3B), these nucleotides may be important for interaction with the tRNA T arm; however, the exact nature of this interaction remains unknown.
In the absence of a structure that shows the entire tRNA binding pocket formed by a full-length T box RNA including both Stem II and IIA/B, biochemical studies are essential for the determination of the molecular details of T box riboswitch gene regulation. Although the importance of the Stem II and IIA/B elements of the riboswitch has been demonstrated by genetic analyses (5), their roles in regulation and possible interactions with tRNA were unknown. The current study adds features to molecular recognition of tRNA by the T box riboswitch and provides biochemical characterization of the Stem II and IIA/B elements. This study also presents an example of interactions between an S-turn motif and the T arm of tRNA and between a pseudoknot element and the tRNA D loop. Further investigation of these interactions will benefit the understanding of T box riboswitch gene regulation and may provide information applicable to other RNA systems that use S-turn or pseudoknot elements. These results also add to the understanding of regulation of essential genes found in many clinically important species, including Mycobacterium tuberculosis, which contains an ileS leader sequence that closely resembles that of M. smegmatis.
Materials and Methods
RNAs were transcribed and purified using standard protocols. The tRNA binding assays were performed as in ref. 4 using 10K filters. The SHAPE protocol was adapted from refs. 17 and 39, and the cross-linking protocol was modified from ref. 18. For details, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Plant-Microbe Genomics Facility at The Ohio State University for their assistance with capillary electrophoresis, Chris Woltjen for technical assistance, and members of T.M.H.’s laboratory for helpful discussions. This work was supported by NIH Institute of General Medical Sciences Grant R01 GM047823 (to T.M.H.) and Training Grant T32 GM086252 (to J.K.F.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721254115/-/DCSupplemental.
References
- 1.Gutiérrez-Preciado A, Henkin TM, Grundy FJ, Yanofsky C, Merino E. Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol Mol Biol Rev. 2009;73:36–61. doi: 10.1128/MMBR.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993;74:475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- 3.Grundy FJ, Rollins SM, Henkin TM. Interaction between the acceptor end of tRNA and the T box stimulates antitermination in the Bacillus subtilis tyrS gene: A new role for the discriminator base. J Bacteriol. 1994;176:4518–4526. doi: 10.1128/jb.176.15.4518-4526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherwood AV, Grundy FJ, Henkin TM. T box riboswitches in Actinobacteria: Translational regulation via novel tRNA interactions. Proc Natl Acad Sci USA. 2015;112:1113–1118. doi: 10.1073/pnas.1424175112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rollins SM, Grundy FJ, Henkin TM. Analysis of cis-acting sequence and structural elements required for antitermination of the Bacillus subtilis tyrS gene. Mol Microbiol. 1997;25:411–421. doi: 10.1046/j.1365-2958.1997.4851839.x. [DOI] [PubMed] [Google Scholar]
- 6.Yousef MR, Grundy FJ, Henkin TM. Structural transitions induced by the interaction between tRNAGly and the Bacillus subtilis glyQS T box leader RNA. J Mol Biol. 2005;349:273–287. doi: 10.1016/j.jmb.2005.03.061. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, Ferré-D’Amaré AR. Direct evaluation of tRNA aminoacylation status by the T-box riboswitch using tRNA-mRNA stacking and steric readout. Mol Cell. 2014;55:148–155. doi: 10.1016/j.molcel.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Ferré-D’Amaré AR. Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature. 2013;500:363–366. doi: 10.1038/nature12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grigg JC, et al. T box RNA decodes both the information content and geometry of tRNA to affect gene expression. Proc Natl Acad Sci USA. 2013;110:7240–7245. doi: 10.1073/pnas.1222214110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vitreschak AG, Mironov AA, Lyubetsky VA, Gelfand MS. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA. 2008;14:717–735. doi: 10.1261/rna.819308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leontis NB, Westhof E. A common motif organizes the structure of multi-helix loops in 16S and 23S ribosomal RNAs. J Mol Biol. 1998;283:571–583. doi: 10.1006/jmbi.1998.2106. [DOI] [PubMed] [Google Scholar]
- 12.Leontis NB, Stombaugh J, Westhof E. Motif prediction in ribosomal RNAs lessons and prospects for automated motif prediction in homologous RNA molecules. Biochimie. 2002;84:961–973. doi: 10.1016/s0300-9084(02)01463-3. [DOI] [PubMed] [Google Scholar]
- 13.Owens RA, Baumstark T. Structural differences within the loop E motif imply alternative mechanisms of viroid processing. RNA. 2007;13:824–834. doi: 10.1261/rna.452307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grundy FJ, Henkin TM. Kinetic analysis of tRNA-directed transcription antitermination of the Bacillus subtilis glyQS gene in vitro. J Bacteriol. 2004;186:5392–5399. doi: 10.1128/JB.186.16.5392-5399.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chetnani B, Mondragón A. Molecular envelope and atomic model of an anti-terminated glyQS T-box regulator in complex with tRNAGly. Nucleic Acids Res. 2017;45:8079–8090. doi: 10.1093/nar/gkx451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang X, Michnicka M, Zhang Y, Wang Y-X, Nikonowicz EP. Capture and release of tRNA by the T-loop receptor in the function of the T-box riboswitch. Biochemistry. 2017;56:3549–3558. doi: 10.1021/acs.biochem.7b00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): Quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 18.Harris ME, Christian EL. RNA crosslinking methods. Methods Enzymol. 2009;468:127–146. doi: 10.1016/S0076-6879(09)68007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong X, et al. Tertiary structural and functional analyses of a viroid RNA motif by isostericity matrix and mutagenesis reveal its essential role in replication. J Virol. 2006;80:8566–8581. doi: 10.1128/JVI.00837-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan PP, Lowe TM. GtRNAdb: A database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ladner JE, et al. Structure of yeast phenylalanine transfer RNA at 2.5 Å resolution. Proc Natl Acad Sci USA. 1975;72:4414–4418. doi: 10.1073/pnas.72.11.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi H, Moore PB. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: A classic structure revisited. RNA. 2000;6:1091–1105. doi: 10.1017/s1355838200000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lancaster L, Lambert NJ, Maklan EJ, Horan LH, Noller HF. The sarcin-ricin loop of 23S rRNA is essential for assembly of the functional core of the 50S ribosomal subunit. RNA. 2008;14:1999–2012. doi: 10.1261/rna.1202108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hausner T-P, Atmadja J, Nierhaus KH. Evidence that the G2661 region of 23S rRNA is located at the ribosomal binding sites of both elongation factors. Biochimie. 1987;69:911–923. doi: 10.1016/0300-9084(87)90225-2. [DOI] [PubMed] [Google Scholar]
- 25.Moazed D, Robertson JM, Noller HF. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature. 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- 26.Correll CC, et al. Crystal structure of the ribosomal RNA domain essential for binding elongation factors. Proc Natl Acad Sci USA. 1998;95:13436–13441. doi: 10.1073/pnas.95.23.13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clever JL, Parslow TG. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto H, et al. Structure of the mammalian 80S initiation complex with initiation factor 5B on HCV-IRES RNA. Nat Struct Mol Biol. 2014;21:721–727. doi: 10.1038/nsmb.2859. [DOI] [PubMed] [Google Scholar]
- 29.Havrila M, Réblová K, Zirbel CL, Leontis NB, Šponer J. Isosteric and nonisosteric base pairs in RNA motifs: Molecular dynamics and bioinformatics study of the sarcin-ricin internal loop. J Phys Chem B. 2013;117:14302–14319. doi: 10.1021/jp408530w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giedroc DP, Cornish PV. Frameshifting RNA pseudoknots: Structure and mechanism. Virus Res. 2009;139:193–208. doi: 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deiman BA, Koenen AK, Verlaan PW, Pleij CW. Minimal template requirements for initiation of minus-strand synthesis in vitro by the RNA-dependent RNA polymerase of turnip yellow mosaic virus. J Virol. 1998;72:3965–3972. doi: 10.1128/jvi.72.5.3965-3972.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jopling CL, Spriggs KA, Mitchell SA, Stoneley M, Willis AE. L-Myc protein synthesis is initiated by internal ribosome entry. RNA. 2004;10:287–298. doi: 10.1261/rna.5138804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfingsten JS, Costantino DA, Kieft JS. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006;314:1450–1454. doi: 10.1126/science.1133281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serganov A, et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 36.Ren A, Rajashankar KR, Patel DJ. Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature. 2012;486:85–89. doi: 10.1038/nature11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peselis A, Serganov A. Structure and function of pseudoknots involved in gene expression control. Wiley Interdiscip Rev RNA. 2014;5:803–822. doi: 10.1002/wrna.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pleij CW, Rietveld K, Bosch L. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 1985;13:1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mortimer SA, Weeks KM. Time-resolved RNA SHAPE chemistry: Quantitative RNA structure analysis in one-second snapshots and at single-nucleotide resolution. Nat Protoc. 2009;4:1413–1421. doi: 10.1038/nprot.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Popenda M, et al. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012;40:e112. doi: 10.1093/nar/gks339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purzycka KJ, et al. Automated 3D RNA structure prediction using the RNAComposer method for riboswitches. Methods Enzymol. 2015;553:3–34. doi: 10.1016/bs.mie.2014.10.050. [DOI] [PubMed] [Google Scholar]
- 43.McDaniel BA, Grundy FJ, Henkin TM. A tertiary structural element in S box leader RNAs is required for S-adenosylmethionine-directed transcription termination. Mol Microbiol. 2005;57:1008–1021. doi: 10.1111/j.1365-2958.2005.04740.x. [DOI] [PubMed] [Google Scholar]
- 44.Fuchs RT, Grundy FJ, Henkin TM. S-adenosylmethionine directly inhibits binding of 30S ribosomal subunits to the SMK box translational riboswitch RNA. Proc Natl Acad Sci USA. 2007;104:4876–4880. doi: 10.1073/pnas.0609956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu C, et al. Crystal structures of the SAM-III/SMK riboswitch reveal the SAM-dependent translation inhibition mechanism. Nat Struct Mol Biol. 2008;15:1076–1083. doi: 10.1038/nsmb.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karabiber F, McGinnis JL, Favorov OV, Weeks KM. QuShape: Rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA. 2013;19:63–73. doi: 10.1261/rna.036327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGinnis JL, Duncan CDS, Weeks KM. High-throughput SHAPE and hydroxyl radical analysis of RNA structure and ribonucleoprotein assembly. Methods Enzymol. 2009;468:67–89. doi: 10.1016/S0076-6879(09)68004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.