Significance
Bacterially produced small molecules are indispensable leads in the development of antibiotics, anticancer therapeutics, or immunomodulators. To unveil novel aspects in the biosynthetic potential of bacteria, a consideration of the ecological context in which the adapted producers thrive is extremely insightful. Here, we describe two natural products produced by Pseudomonas sp. QS1027, a bacterium that resides in the vicinity of the bacterial predator Dictyostelium discoideum. The two metabolites are jessenipeptin, a nonribosomal cyclic lipopeptide, and mupirocin, a known polyketide antibiotic. Both compounds are quorum-sensing regulated and display potent synergistic inhibitory activity against clinically relevant methicillin-resistant Staphylococcus aureus (MRSA).
Keywords: nonribosomal peptides, Pseudomonas, structure elucidation, social amoebae, synergy
Abstract
Investigating microbial interactions from an ecological perspective is a particularly fruitful approach to unveil both new chemistry and bioactivity. Microbial predator–prey interactions in particular rely on natural products as signal or defense molecules. In this context, we identified a grazing-resistant Pseudomonas strain, isolated from the bacterivorous amoeba Dictyostelium discoideum. Genome analysis of this bacterium revealed the presence of two biosynthetic gene clusters that were found adjacent to each other on a contiguous stretch of the bacterial genome. Although one cluster codes for the polyketide synthase producing the known antibiotic mupirocin, the other cluster encodes a nonribosomal peptide synthetase leading to the unreported cyclic lipopeptide jessenipeptin. We describe its complete structure elucidation, as well as its synergistic activity against methicillin-resistant Staphylococcus aureus, when in combination with mupirocin. Both biosynthetic gene clusters are regulated by quorum-sensing systems, with 3-oxo-decanoyl homoserine lactone (3-oxo-C10-AHL) and hexanoyl homoserine lactone (C6-AHL) being the respective signal molecules. This study highlights the regulation, richness, and complex interplay of bacterial natural products that emerge in the context of microbial competition.
The endeavor to expand our access to nature’s chemical diversity is a matter of utmost urgency as most anticancer agents or antibiotics are somehow derived from natural products (1, 2). The advent of multidrug resistance—both in infectious diseases and cancer—calls for new potent bioactive natural products. In particular, the use of combinations of the latter has received much attention due to beneficial synergistic effects, as well as reduced resistance rates (3).
By virtue of their exquisite biosynthetic machineries, microorganisms are exceptionally prolific producers of bioactive and therapeutically useful natural products (1, 2, 4). Traditional screening approaches, in which bacteria are isolated and cultured under a few different conditions, have led to the identification of a large number of bioactive natural products. Although this approach was fruitful during the golden age of antibiotics (1950s and 60s), it now suffers from a major drawback: the prohibitively large rediscovery rate of already known compounds (3, 5). If microorganisms are considered within their ecological context, however, the identification of novel microbial natural products can be greatly enhanced. A detailed understanding of certain ecological niches can thus lead to new chemistry, when brute force screening approaches fail to unveil the true metabolic potential of selected bacteria. In particular, microorganisms that are found in association with other organisms have led to the discovery of unforeseen structural diversity and interesting bioactivities. Examples are numerous (reviewed in refs. 6–8) and to name but a few they include: wasp–bacteria (9), ant–bacteria (10), beetle–bacteria (6, 11), bee–bacteria (12), sponge–bacteria (13), and bacteria–algae interactions (14, 15).
In the last few years, research has shown that amoebae–bacteria interactions are particularly rich sources of natural products (16–18). Associations between amoebae and bacteria range from highly antagonistic, when amoebae feed on bacteria or bacteria kill amoebae (17, 18), to highly mutualistic, in the case of primitive farming (16, 19). However, bacteria are often loosely associated with amoebae and little is known about this nominally commensal relationship. Amoebae are highly bacterivorous organisms that can devour large amounts of bacteria, with ingestion rates up to 300 h−1 for the model organism Dictyostelium discoideum (20, 21). It is not surprising that amoebae exert a tremendous evolutionary selection pressure on bacteria that share the same habitat. Thus, it is expected that amoebae-associated bacteria may have adapted to surviving in the vicinity of the highly bacterivorous predator. Various mechanisms for bacterial survival have been described: the formation of strong biofilms can hinder predation, enhanced swarming motility may allow for evasion, and some bacteria are even able to survive within these predators (22). Additionally, the production of secondary metabolites provides a very efficient means for bacteria to combat amoebae, as well as competing bacteria, and thus survive and even thrive in hostile environments.
Herein, we provide a detailed account on one such example where a bacterium isolated from the close vicinity of the amoebal predator D. discoideum, led to the identification of an antimicrobially active cyclic lipopeptide (CLP). Bioinformatics and molecular biological analyses of the biosynthetic gene cluster allowed inference about its regulation and hinted at the cooperative biological activity with coproduced mupirocin.
Results
Isolation of a D. discoideum–Associated Bacterium and Genome Analysis.
We purified bacteria from fruiting bodies of D. discoideum (Fig. 1) isolated from soil and deer dung collected at Mountain Lake Biological Station in Virginia. We focused on bacteria, which were transiently associated with amoebae, and tested them for grazing resistance using a D. discoideum edibility assay. Specifically, a plaque assay was used in which amoebae are added to a lawn of bacteria on solid growth medium (SM/5) (23). Edible bacteria were identified by the appearance of characteristic amoebal grazing plaques (Fig. 1). Only bacteria that proved to be inedible were further tested by adding various amounts of comestible Klebsiella pneumoniae. We identified one strain (QS1027), which prevented amoebal grazing, even in the presence of an excess of K. pneumoniae—an indication for the production of amoebicidal compounds.
Fig. 1.
Bacterial strain Pseudomonas sp. QS1027 was isolated from Dictyostelium discoideum fruiting bodies. QS1027 is not a food source to the amoeba from which it was isolated.
To infer about the metabolic potential of this strongly antagonistic bacterial strain, we sequenced its genomic DNA using Next Generation Illumina sequencing. A draft genome was assembled into 82 contigs with a total size of circa 7.48 Mbp and a GC content of 61%. Preliminary sequence analysis based on three genes (16S rRNA, gyrB, rpoD) showed similarity to Pseudomonas jessenii. Subsequently, the genome of Pseudomonas sp. QS1027 (hereafter referred to as QS1027) was mined for putative biosynthetic gene clusters (BGCs) using antiSMASH (24). The latter predicted one polyketide synthase (pks) gene cluster and several nonribosomal peptide synthetase (nrps) gene clusters.
We found a stretch of about 150 kbp that harbored two BGCs, the pks and a truncated nrps cluster, intersected by a short stretch of regulatory genes (Fig. 2). The pks BGC was predicted to code for the mupirocin biosynthetic machinery. The antibiotic mupirocin, also known as pseudomonic acid, is produced by a type I PKS along with additional tailoring enzymes encoded on a 75-kbp stretch (25, 26).
Fig. 2.
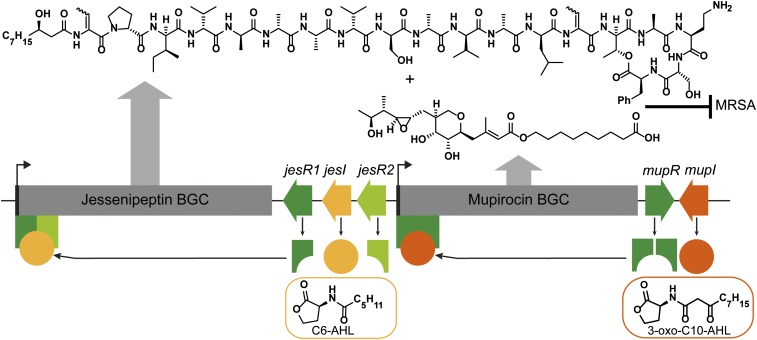
(A) Gene architecture of the jessenipeptin and mupirocin BGCs, which are found adjacent to each other on the genome of QS1027. (B) Domain organization of the jessenipeptin BGC based on the antiSMASH prediction. The predicted A-domain substrates as well as the C-domain specificities are shown. CS, starter C domain; C/E, dual condensation–epimerization domain: LCL, C domains that condense two l-amino acids.
The nrps BGC adjacent to the mupirocin cluster appeared to span over the contig boundaries. Gap closing between nrps clusters that ended/started at the contig boundary via PCR and Sanger sequencing of the amplicons allowed us to obtain the complete nrps BGC. In total, 19 modules were present, each containing a condensation (C), adenylation (A), and thiolation (T) domain. Additionally, the presence of a starter C domain indicated the presence of an N-terminal fatty acid moiety, suggesting a lipopeptide consisting of 19 amino acids may be produced by this NRPS.
Identification, Isolation, and Structure Elucidation of Mupirocin and Jessenipeptin.
To identify the natural products associated with the two adjacent BGCs, we combined analytical chemistry methods with molecular biology and in silico approaches. Since one BGC was predicted to code for mupirocin biosynthetic genes (27), we used commercially available mupirocin to identify whether QS1027 produces this polyketide, when cultured under different conditions. We identified a peak in the HPLC profile of crude QS1027 extract with both the same retention time and the same MS profile as bona fide mupirocin. Generation of a gene disruption mutant (Δmup) by deletion of a gene fragment coding for a keto synthase domain in the presumed mupirocin BGC led to the disappearance of the mupirocin peak in the HPLC profile of Δmup culture extract (SI Appendix, Figs. S19 and S22). Thus, we unambiguously linked the mupirocin BGC to the production of mupirocin in QS1027 (Fig. 2).
To identify the product of the putative NRPS, we proceeded to inactivate the respective BGC by generating in-frame deletion of the gene fragment coding for the A domain of the first module (Δjes). Comparison of HPLC profiles of culture extracts of QS1027 and the deletion mutant led to the identification of a peak absent in the deletion mutant (SI Appendix, Fig. S22) that was present in the wild type. To isolate the corresponding natural product, culture conditions were optimized to increase the yield of the presumed lipopeptide. The producing strain was fermented on a 30-L scale in HL/5 medium. Extraction of the conditioned medium with ethyl acetate and chromatographic purification of the crude extract led to the isolation of 150-mg homogeneous material. High-resolution mass spectrometry (HR-MS) measurements revealed a pseudomolecular ion peak with m/z 1906.1073 [M + H]+, which is consistent with a molecular formula of C91H148N20O24.
By comparing databases, including SciFinder, Reaxys, and Norine (28), we found no previously reported molecule with this formula; thus we termed this compound jessenipeptin. A bioinformatics prediction of the A-domain specificities (presuming that the colinearity rule is obeyed) provided a first assessment of the primary sequence of jessenipeptin and indicated the presence of an N-terminal fatty acid moiety (24, 29). An A-domain analysis, however, did not allow us to distinguish between threonine or 2,3-dehydro-2-aminobutyric acid (Dhb) as threonine can be dehydrated to yield Dhb (30). Predicting the presence of functional epimerases also failed, hence the absolute configuration of the amino acids of jessenipeptin could not be predicted. The same problem was previously described for the structure prediction of syringopeptin, a different but related CLP (31).
A combination of MS/MS studies and chemical degradation followed by derivatizations allowed us to determine the amino acid sequence of jessenipeptin (SI Appendix, Figs. S4–S8), as well as the nature of the N-terminal fatty acid. The latter was identified as (3R)-hydroxy decanoic acid by total hydrolysis of jessenipeptin followed by conversion of the fatty acid in its methyl and Mosher ester. Comparison of the resulting ester with corresponding synthetic standards using GC-MS allowed for an unambiguous characterization (Fig. 3 and SI Appendix, Fig. S17) (32). In addition, derivatization of the hydrolyzed amino acids with Marfey’s reagent (33) confirmed the presence of the following amino acids: 2× l-Ala, 4× d-Ala, 1× l-Dab (2,4-Diaminobutyric acid), 1× d-Ile, 1× d-Leu, 1× l-Phe, 1× d-Pro, 2× d-Ser, 1× d-allo-Thr, and 3× d-Val. A domain analysis, however, predicted the presence of three Thr residues. This hinted at the fact that two Thr were further modified, e.g., to yield Dhb.
Fig. 3.
(A) Our approach for the structure elucidation of jessenipeptin. (B) GC-MS traces of derivatized fatty acid moiety (top trace) and traces of the corresponding synthetic (R)- and (S)-configured 3-hydroxy decanoic acid derivatives (bottom trace). The fatty acid derivative obtained from jessenipeptin coelutes with the respective (R)-configured synthetic counterpart. (C) Scandium triflate-mediated peptide cleavage yields fragments F1 and F2. A single l-alanine is present in F1. (D) Stable-isotope labeling results in shifts of one mass unit for fragments containing l-alanine in MS/MS experiments. The MS spectra of unlabeled jessenipeptin are depicted in green; the corresponding spectra of the labeled peptide are depicted in red. The mass shifts observed in two indicative fragments (m/z 776 and 507) are shown.
Although MS/MS data allowed the placement of most of the residues in the correct order, assigning their absolute configuration proved difficult. In particular, the positions of the four d-alanine and two l-alanine residues in jessenipeptin could not be resolved. We addressed this problem using a combination of partial hydrolysis and stable isotope labeling experiments.
A recent publication by Ni et al. (34) describes a scandium(III) triflate-mediated peptide hydrolysis method, which displays selectivity for cleavage of amide bonds adjacent to serine or threonine residues. Hence, we were able to obtain fragment F1, which was purified by HPLC. To our surprise, we were able to obtain another fragment, F2, in which cleavage occurred between the two alanine residues at position six and seven (Fig. 3). Total hydrolysis of fragment F1 and subsequent derivatization using Marfey’s method clearly indicated the presence of l- and d-alanine in a 1:2 ratio. In fragment F2, however, we could only identify the presence of d-alanine. This allowed us to assign the configuration of alanine at position seven as l (Fig. 3 and SI Appendix, Fig. S16).
To locate the remaining l-alanine, we made use of stable isotope labeling. Feeding deuterium labeled alanine (l-alanine-2-d1) allows one to determine whether epimerization of l-Ala to d-Ala has occurred (35). This method relies on a characteristic feature of bacterial nonribosomal peptides, whereby d-amino acids are typically installed by first incorporating the respective l-amino acid, followed by an epimerization step. A deuterium at the α-position of the amino acid will be exchanged for a proton, if epimerization takes place. The mass difference between a proton and a deuterium can easily be monitored by MS/MS measurements. By cultivating QS1027 in minimal medium, supplemented with l-alanine-2-d1, we were able to observe virtually quantitative incorporation of two deuterium labels into jessenipeptin. MS/MS measurements of the respective labeled jessenipeptin showed the two l-alanine residues to be located at positions 7 and 16 (Fig. 3 and SI Appendix, Figs. S13–S15), which is in accordance with the results from Marfey’s analysis of fragment F1 and F2.
Since the theoretical mass for a linear peptide, based on amino acids and the fatty acid found by MS/MS experiments, differed by 18 mass units from the observed mass, the presence of a lactone ring was assumed. Treatment of jessenipeptin with aqueous sodium hydroxide yielded a compound with a mass matching the calculated mass for the linear peptide. To determine the size and position of the lactone ring, we masked the free hydroxyl groups in jessenipeptin as silyl ethers, using tert-butyldimethylsilyl triflate (TBS-OTf). The derivatized peptide was analyzed by MS/MS, revealing that either threonine-15 or serine-18 lacks TBS-groups, suggesting lactone formation between one of these residues and the C terminus. The 1H,1H-correlation spectroscopy experiments showed a strong downfield shift for the β-proton of Thr-15, which provides evidence for the lactone ring closure as shown in Fig. 3 (SI Appendix, Fig. S9) (36). Thus, jessenipeptin falls into the class of cyclic lipodepsipeptides, most of which bear intriguing bioactivities (37). Cyclic lipopeptides produced by Pseudomonas species are typically classified into six groups (38). Jessenipeptin can be placed in the tolaasin group. Members of this group are CLPs with 19–25 amino acids, and they contain lactone rings consisting of five amino acids. Furthermore, they often bear unusual amino acids such as Dab and Dhb. All these features are met by jessenipeptin.
Biological Activity and Synergy of Mupirocin and Jessenipeptin.
Since QS1027 was isolated from the fruiting bodies of D. discoideum, we evaluated the amoebicidal activities of jessenipeptin and mupirocin. Growth inhibition or killing of D. discoideum was determined in a liquid culture assay. Although mupirocin was inactive against D. discoideum, jessenipeptin exhibited an IC50 (D. discoideum) of 4 μg/mL (2 μM). We then tested whether D. discoideum could graze on Δmup, Δjes, or ΔmupΔjes. None of these strains were edible to D. discoideum. It is conceivable that other secondary metabolites of QS1027 may constitute additional virulence factors. Interestingly, however, a broad antimicrobial screen revealed jessenipeptin to be highly active against MRSA as well as other pathogenic Gram-positive bacteria (SI Appendix, Table S1). As both mupirocin and jessenipeptin appear to have a similar range of activity and are produced by the same organism, we tested for synergistic antimicrobial effects against Enterococcus faecalis, Bacillus subtilis, MRSA, and D. discoideum. Indeed, we could show strong synergy between mupirocin and jessenipeptin against B. subtilis and most importantly against MRSA. We determined the fractional inhibitory concentrations of a variety of mupirocin/jessenipeptin mixtures. Fig. 4 displays the isobolograms that show synergistic activity for mupirocin and jessenipeptin.
Fig. 4.
Mupirocin and jessenipeptin act synergistically against MRSA (A) and B. subtilis (B). Two compounds are considered to act synergistically, if the sum of their fractional inhibitory concentrations (FIC) is below 0.5 (light blue area). Strong synergy is present if the sum of the FICs is below 0.25 (dark blue area). Compound ratios are shown in brackets (wt/wt, jessenipeptin:mupirocin).
Regulation of Mupirocin and Jessenipeptin Production.
Since luxI/luxR-type regulatory genes intersect both the mupirocin and jessenipeptin BGC, we investigated their role in the production of mupirocin and jessenipeptin. These luxR/luxI-type genes typically code for an acyl homoserine lactone (AHL)-based quorum-sensing (QS) system, with the luxI gene coding for an autoinducer synthase and the luxR coding for the cognate AHL-binding response regulator (39–41). Furthermore, we could also identify a set of luxR- and luxI-type genes within the mupirocin BGC. Hence, we investigated whether these luxI/luxR genes code for QS systems. In particular, we addressed the question of the nature of the respective AHLs and whether the systems display any cross talk.
To identify the nature of the QS signals as well as their role in the regulation of mupirocin and jessenipeptin production, we generated in-frame deletion mutants of these five regulatory genes, as well as combinations thereof in QS1027. Deletions of any of the three regulatory genes luxR1, luxR2, and luxI led to a complete suppression of jessenipeptin production, yet mupirocin production was left unaltered. Thus, we termed these regulatory genes jesI, jesR1, and jesR2. JesR2 has a predicted N-terminal autoinducer binding domain as well as a C-terminal DNA-binding helix-turn-helix (HTH) motif, whereas JesR1 only bears the C-terminal DNA-binding domain. The fact that both JesRs are required for the production of jessenipeptin hints at the formation of a possible heterodimeric JesR1/JesR2 complex, which upon binding of the cognate AHL activates the jessenipeptin BGC. Although the formation of LuxR homodimers is documented (42, 43), heterodimer formation is typically associated with inactivation rather than activation. Episomal complementation of ΔjesR1 and ΔjesR2 with the respective genes led to a restoration of jessenipeptin production. Jessenipeptin production in the ΔjesI mutant could also be restored by supplementation of the growth medium with C6-AHL (a variety of AHLs were tested, and the response to C6-AHL was the largest, followed by that to C8-AHL; SI Appendix, Fig. S23). This QS signal is also identifiable in the WT supernatant by liquid chromatography (LC)-MS analysis and was found to be absent in the ΔjesI mutant (SI Appendix, Fig. S21).
Analogously, we identified the QS signal associated with the luxI/luxR-type genes in the mupirocin BGC (from here on forward referred to as mupI/mupR). Both the ΔmupR and the ΔmupI mutant did not produce any mupirocin and jessenipeptin production remained unaltered. Episomal complementation of ΔmupR with the respective gene restored mupirocin production (SI Appendix, Fig. S28). Supplementation of the growth medium of ΔmupI mutant with 3-oxo-C10 AHL led to the production of mupirocin (SI Appendix, Fig. S26), and this signal molecule was also identified in the supernatant of WT, but was absent in ΔmupI mutant strain (SI Appendix, Fig. S20). These data are in accordance with previous reports that mupirocin is under QS control by 3-oxo-C10 AHL (44, 45).
These results indicate the existence of two independent QS systems, which regulate the production of jessenipeptin and mupirocin, respectively (Fig. 5). To investigate whether the production of either of the two secondary metabolites affects production of the other metabolite, we compared the production of mupirocin in the Δjes mutant and vice versa. However, no change was observed in the production of either mupirocin or jessenipeptin (SI Appendix, Fig. S22). We can conclude that production of either secondary metabolite does not affect production of the other one.
Fig. 5.
Two independent QS systems regulate the production of jessenipeptin and mupirocin, which synergistically inhibit the growth of MRSA.
Discussion
The coevolution of bacteria and bacterivorous amoebae in a confined space imposes strong selection pressures upon both organisms. Hence, some bacteria have evolved efficient defense strategies to evade amoebal grazing (17, 18, 22). In particular, the production of amoebicidal secondary metabolites by the bacteria can ensure the survival of the bacteria in the vicinity of its predator. Moreover, it is advantageous for bacteria to live in the vicinity of a bacterivorous organism due to the reduction in bacterial competitors for the grazing-resistant bacteria. The latter gain greater access to resources that need not be shared between other bacterial species. Bacteria isolated from the habitat of amoebae are believed to display chemical diversity that may be larger than in the absence of bacterivores. Hence, we specifically interrogated the biosynthetic capacities of amoeba-associated bacteria. We were particularly interested in bacteria that did not display a close mutualistic relationship with amoebae [such as farmed bacteria (19)] but rather those transiently associated with amoebae. We isolated bacteria from the fruiting bodies of D. discoideum, which clearly indicated that a direct amoebae–bacteria contact had occurred and that these bacteria were able to evade amoeabal predation. A particular strain was further analyzed due to its strongly amoebicidal phenotype. Genome sequencing and subsequent bioinformatics analysis revealed the strain to belong to the genus Pseudomonas. A large number of BGCs could be identified in the genome of the Gram-negative γ-proteobacterium, and the unusual presence of two BGCs intersected by a few regulatory genes caught our attention. We identified one BGC cluster to code for the biosynthetic machinery of the antibiotic mupirocin. Interestingly, not many strains that produce mupirocin have been described to date (46). The other BGC coded for a nonribosomal peptide synthetase, which was found to produce the previously unreported cyclic lipopeptide jessenipeptin. The structure of the latter was fully elucidated by a combination of chemical, analytical, and bioinformatics methods. The large CLP consists of 19 amino acids including d- and l-amino acids as well as nonproteinogenic amino acids and a (3R)-hydroxy decanoic acid residue. The absolute configuration of most amino acids in jessenipeptin could be determined via Marfey’s method (33). The presence of multiple d- and l-alanines, however, rendered its structure elucidation difficult. A recent study showed that scandium(III) triflate promotes selective cleavage of a peptide adjacent to a serine or threonine residue by means of an N,O-acyl rearrangement (34). This was the key to fragment jessenipeptin, which tremendously facilitated subsequent structure elucidation. Eventually, feeding experiments with α-deutero l-alanine (35) combined with MS/MS analyses enabled us to determine the position of all d- and l-alanine residues within jessenipeptin. Hence, we fully elucidated the structure of jessenipeptin. Since QS1027 was isolated from the habitat of social amoebae, we tested jessenipeptin and mupirocin with respect to amoebicidal activity against the predator D. discoideum. Although mupirocin was inactive against D. discoideum, jessenipeptin exhibited a strong amoebicidal activity (IC50 = 4 μg/mL). Importantly, however, both mupirocin and jessenipeptin were highly active against MRSA with minimal inhibitory concentration values of 0.2 and 3.12 μg/mL, respectively.
With both the jessenipeptin and the mupirocin BGC being on a contiguous stretch on the genome we wondered if their biosynthetic products acted synergistically. Indeed, mupirocin and jessenipeptin displayed strong synergistic effects against MRSA. Mupirocin is an isoleucyl-tRNA synthetase inhibitor (47), and many CLP interact with the cell membrane to form pores (48). It is possible that in this case synergy is caused by jessenipeptin increasing the cytosolic concentration of mupirocin.
Both BGCs were only intersected by a few regulatory genes of the luxI/R family. This led to the question of whether both BGCs were coregulated. Generation of in-frame gene deletions of either of the two jesR or the jesI gene led to the suppression of jessenipeptin production, yet mupirocin was still produced. LuxI/luxR systems typically code for QS systems. In this case C6-AHL was identified as the QS signal associated with jessenipeptin production. The BGC of mupirocin also revealed luxI/R genes. Deletion of either of these genes led to the suppression of mupirocin production, while jessenipeptin production was unaltered. We identified 3-oxo-C10-AHL as the QS signal. This was also reported as the QS signal in the only well-characterized mupirocin producer (26). Thus, two independent QS systems are involved in the production of mupirocin and jessenipeptin. Interestingly, in P. aeruginosa two similar QS systems with C4-AHL and 3-oxo-C12 form a hierarchical QS system, which is not the case for the jessenipeptin–mupirocin QS system.
Conclusion
In summary, our study highlights the structural and functional diversity of natural products that can be obtained from microbial associations. Genome mining and detailed structural analysis led to the identification and structure determination of the nonribosomal peptide jessenipeptin, whose BGC is adjacent to that of mupirocin. Both secondary metabolites are QS regulated and display synergistic activity against MRSA. As the nature of amoebae–bacteria interactions is multifaceted, we can expect this diversity to be reflected in an equally rich bacterial secondary metabolome, which enables bacteria to adapt to coexisting with amoebae.
Materials and Methods
We propagated Pseudomonas sp. QS1027 in Luria Bertani (LB) liquid or on solid (supplemented with 1.5%, wt/wt, agar) medium (Carl Roth) or SM/5 medium (Formedium) (both solid and liquid) at 28 or 22 °C depending on the assay.
Supplementary Material
Acknowledgments
We thank A. Perner and H. Heinecke for MS and NMR measurements. We also thank C. Weigel for performing antimicrobial assays and F. Kloss for useful discussions. We are grateful for financial support from the Leibniz Association. This work was supported by Deutsche Forschungsgemeinschaft Grants STA1431/2-1 and SFB1127. An Aventis Foundation Ph.D. fellowship (to M.K.) is acknowledged. M.G.-A. acknowledges financial support from the ERC for a Marie Skłodowska-Curie Individual Fellowship (IF-EF) Project reference 700036.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the DDBJ/ENA/GenBank database (accession no. PHSU00000000, the version described in this paper is version PHSU01000000).
This article contains supporting information online at Www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721790115/-/DCSupplemental.
References
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clardy J, Fischbach MA, Walsh CT. New antibiotics from bacterial natural products. Nat Biotechnol. 2006;24:1541–1550. doi: 10.1038/nbt1266. [DOI] [PubMed] [Google Scholar]
- 3.Brown ED, Wright GD. Antibacterial drug discovery in the resistance era. Nature. 2016;529:336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- 4.Clardy J, Walsh C. Lessons from natural molecules. Nature. 2004;432:829–837. doi: 10.1038/nature03194. [DOI] [PubMed] [Google Scholar]
- 5.Lewis K. Platforms for antibiotic discovery. Nat Rev Drug Discov. 2013;12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 6.Scott JJ, et al. Bacterial protection of beetle-fungus mutualism. Science. 2008;322:63. doi: 10.1126/science.1160423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beemelmanns C, Guo H, Rischer M, Poulsen M. Natural products from microbes associated with insects. Beilstein J Org Chem. 2016;12:314–327. doi: 10.3762/bjoc.12.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piel J. Metabolites from symbiotic bacteria. Nat Prod Rep. 2009;26:338–362. doi: 10.1039/b703499g. [DOI] [PubMed] [Google Scholar]
- 9.Kroiss J, et al. Symbiotic Streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat Chem Biol. 2010;6:261–263. doi: 10.1038/nchembio.331. [DOI] [PubMed] [Google Scholar]
- 10.Oh D-C, Poulsen M, Currie CR, Clardy J. Dentigerumycin: a bacterial mediator of an ant-fungus symbiosis. Nat Chem Biol. 2009;5:391–393. doi: 10.1038/nchembio.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piel J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc Natl Acad Sci USA. 2002;99:14002–14007. doi: 10.1073/pnas.222481399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Müller S, et al. Paenilamicin: structure and biosynthesis of a hybrid nonribosomal peptide/polyketide antibiotic from the bee pathogen Paenibacillus larvae. Angew Chem Int Ed Engl. 2014;53:10821–10825. doi: 10.1002/anie.201404572. [DOI] [PubMed] [Google Scholar]
- 13.Wilson MC, et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature. 2014;506:58–62. doi: 10.1038/nature12959. [DOI] [PubMed] [Google Scholar]
- 14.Seyedsayamdost MR, Wang R, Kolter R, Clardy J. Hybrid biosynthesis of roseobacticides from algal and bacterial precursor molecules. J Am Chem Soc. 2014;136:15150–15153. doi: 10.1021/ja508782y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seyedsayamdost MR, Case RJ, Kolter R, Clardy J. The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat Chem. 2011;3:331–335. doi: 10.1038/nchem.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stallforth P, et al. A bacterial symbiont is converted from an inedible producer of beneficial molecules into food by a single mutation in the gacA gene. Proc Natl Acad Sci USA. 2013;110:14528–14533. doi: 10.1073/pnas.1308199110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klapper M, Götze S, Barnett R, Willing K, Stallforth P. Bacterial alkaloids prevent amoebal predation. Angew Chem Int Ed Engl. 2016;55:8944–8947. doi: 10.1002/anie.201603312. [DOI] [PubMed] [Google Scholar]
- 18.Götze S, et al. Structure, biosynthesis, and biological activity of the cyclic lipopeptide anikasin. ACS Chem Biol. 2017;12:2498–2502. doi: 10.1021/acschembio.7b00589. [DOI] [PubMed] [Google Scholar]
- 19.Brock DA, Douglas TE, Queller DC, Strassmann JE. Primitive agriculture in a social amoeba. Nature. 2011;469:393–396. doi: 10.1038/nature09668. [DOI] [PubMed] [Google Scholar]
- 20.Steinert M, Heuner K. Dictyostelium as host model for pathogenesis. Cell Microbiol. 2005;7:307–314. doi: 10.1111/j.1462-5822.2005.00493.x. [DOI] [PubMed] [Google Scholar]
- 21.Chisholm RL, Firtel RA. Insights into morphogenesis from a simple developmental system. Nat Rev Mol Cell Biol. 2004;5:531–541. doi: 10.1038/nrm1427. [DOI] [PubMed] [Google Scholar]
- 22.Matz C, Kjelleberg S. Off the hook–how bacteria survive protozoan grazing. Trends Microbiol. 2005;13:302–307. doi: 10.1016/j.tim.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Froquet R, Lelong E, Marchetti A, Cosson P. Dictyostelium discoideum: a model host to measure bacterial virulence. Nat Protoc. 2009;4:25–30. doi: 10.1038/nprot.2008.212. [DOI] [PubMed] [Google Scholar]
- 24.Weber T, et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuller AT, et al. Pseudomonic acid: an antibiotic produced by Pseudomonas fluorescens. Nature. 1971;234:416–417. doi: 10.1038/234416a0. [DOI] [PubMed] [Google Scholar]
- 26.Thomas CM, Hothersall J, Willis CL, Simpson TJ. Resistance to and synthesis of the antibiotic mupirocin. Nat Rev Microbiol. 2010;8:281–289. doi: 10.1038/nrmicro2278. [DOI] [PubMed] [Google Scholar]
- 27.Shanks RMQ, Caiazza NC, Hinsa SM, Toutain CM, O’Toole GA. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol. 2006;72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pupin M, et al. Norine: A powerful resource for novel nonribosomal peptide discovery. Synth Syst Biotechnol. 2016;1:89–94. doi: 10.1016/j.synbio.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ziemert N, et al. The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS One. 2012;7:e34064. doi: 10.1371/journal.pone.0034064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grgurina I, Mariotti F. Biosynthetic origin of syringomycin and syringopeptin 22, toxic secondary metabolites of the phytopathogenic bacterium Pseudomonas syringae pv. syringae. FEBS Lett. 1999;462:151–154. doi: 10.1016/s0014-5793(99)01528-8. [DOI] [PubMed] [Google Scholar]
- 31.Scholz-Schroeder BK, Soule JD, Gross DC. The sypA, sypS, and sypC synthetase genes encode twenty-two modules involved in the nonribosomal peptide synthesis of syringopeptin by Pseudomonas syringae pv. syringae B301D. Mol Plant Microbe Interact. 2003;16:271–280. doi: 10.1094/MPMI.2003.16.4.271. [DOI] [PubMed] [Google Scholar]
- 32.Jenske R, Vetter W. Highly selective and sensitive gas chromatography-electron-capture negative-ion mass spectrometry method for the indirect enantioselective identification of 2- and 3-hydroxy fatty acids in food and biological samples. J Chromatogr A. 2007;1146:225–231. doi: 10.1016/j.chroma.2007.01.102. [DOI] [PubMed] [Google Scholar]
- 33.Marfey P. Determination of D-amino acids. II. Use of a bifunctional reagent, 1,5-difluoro-2,4-dinitrobenzene. Carslberg Res Commun. 1984;49:591–596. [Google Scholar]
- 34.Ni J, Sohma Y, Kanai M. Scandium(iii) triflate-promoted serine/threonine-selective peptide bond cleavage. Chem Commun (Camb) 2017;53:3311–3314. doi: 10.1039/c6cc10300f. [DOI] [PubMed] [Google Scholar]
- 35.Bode HB, et al. Determination of the absolute configuration of peptide natural products by using stable isotope labeling and mass spectrometry. Chemistry. 2012;18:2342–2348. doi: 10.1002/chem.201103479. [DOI] [PubMed] [Google Scholar]
- 36.Nutkins JC, et al. Structure determination of tolaasin, an extracellular lipodepsipeptide produced by the mushroom pathogen Pseudomonas tolaasii paine. J Am Chem Soc. 1991;113:2621–2627. [Google Scholar]
- 37.Süssmuth RD, Mainz A. Nonribosomal peptide synthesis-principles and prospects. Angew Chem Int Ed Engl. 2017;56:3770–3821. doi: 10.1002/anie.201609079. [DOI] [PubMed] [Google Scholar]
- 38.Gross H, Loper JE. Genomics of secondary metabolite production by Pseudomonas spp. Nat Prod Rep. 2009;26:1408–1446. doi: 10.1039/b817075b. [DOI] [PubMed] [Google Scholar]
- 39.Keller L, Surette MG. Communication in bacteria: an ecological and evolutionary perspective. Nat Rev Microbiol. 2006;4:249–258. doi: 10.1038/nrmicro1383. [DOI] [PubMed] [Google Scholar]
- 40.Lazdunski AM, Ventre I, Sturgis JN. Regulatory circuits and communication in Gram-negative bacteria. Nat Rev Microbiol. 2004;2:581–592. doi: 10.1038/nrmicro924. [DOI] [PubMed] [Google Scholar]
- 41.Venturi V. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol Rev. 2006;30:274–291. doi: 10.1111/j.1574-6976.2005.00012.x. [DOI] [PubMed] [Google Scholar]
- 42.Fuqua C, Greenberg EP. Listening in on bacteria: acyl-homoserine lactone signalling. Nat Rev Mol Cell Biol. 2002;3:685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 43.Zhu J, Winans SC. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc Natl Acad Sci USA. 2001;98:1507–1512. doi: 10.1073/pnas.98.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.El-Sayed AK, Hothersall J, Thomas CM. Quorum-sensing-dependent regulation of biosynthesis of the polyketide antibiotic mupirocin in Pseudomonas fluorescens NCIMB 10586. Microbiology. 2001;147:2127–2139. doi: 10.1099/00221287-147-8-2127. [DOI] [PubMed] [Google Scholar]
- 45.Hothersall J, et al. Manipulation of quorum sensing regulation in Pseudomonas fluorescens NCIMB 10586 to increase mupirocin production. Appl Microbiol Biotechnol. 2011;90:1017–1026. doi: 10.1007/s00253-011-3145-2. [DOI] [PubMed] [Google Scholar]
- 46.Matthijs S, et al. Antimicrobial properties of Pseudomonas strains producing the antibiotic mupirocin. Res Microbiol. 2014;165:695–704. doi: 10.1016/j.resmic.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 47.Hughes J, Mellows G. Inhibition of isoleucyl-transfer ribonucleic acid synthetase in Escherichia coli by pseudomonic acid. Biochem J. 1978;176:305–318. doi: 10.1042/bj1760305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider T, Müller A, Miess H, Gross H. Cyclic lipopeptides as antibacterial agents - potent antibiotic activity mediated by intriguing mode of actions. Int J Med Microbiol. 2014;304:37–43. doi: 10.1016/j.ijmm.2013.08.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.