Significance
In recent years, adeno-associated virus (AAV) has attracted considerable attention as a result of its success as a gene therapy vector. However, several aspects of its biology remain elusive. Given that AAV vectors mimic the latent phase of the viral life cycle, defining the mechanisms involved in the regulation of AAV latency is of particular importance. Our studies demonstrate that epigenetic processes are involved in the regulation of viral latency and reveal virus–host interactions and helper functions that are aimed at counteracting the epigenetic repression of the viral genome during the lytic phase of the viral life cycle. These observations will inform the design of future AAV vector technologies.
Keywords: adeno-associated virus, latency, KAP1, Rep, PP1-NIPP1
Abstract
Adeno-associated virus (AAV) is a small human Dependovirus whose low immunogenicity and capacity for long-term persistence have led to its widespread use as vector for gene therapy. Despite great recent successes in AAV-based gene therapy, further improvements in vector technology may be hindered by an inadequate understanding of various aspects of basic AAV biology. AAV is unique in that its replication is largely dependent on a helper virus and cellular factors. In the absence of helper virus coinfection, wild-type AAV establishes latency through mechanisms that are not yet fully understood. Challenging the currently held model for AAV latency, we show here that the corepressor Krüppel-associated box domain-associated protein 1 (KAP1) binds the latent AAV2 genome at the rep ORF, leading to trimethylation of AAV2-associated histone 3 lysine 9 and that the inactivation of KAP1 repression is necessary for AAV2 reactivation and replication. We identify a viral mechanism for the counteraction of KAP1 in which interference with the KAP1 phosphatase protein phosphatase 1 (PP1) by the AAV2 Rep proteins mediates enhanced phosphorylation of KAP1-S824 and thus relief from KAP1 repression. Furthermore, we show that this phenomenon involves recruitment of the NIPP1 (nuclear inhibitor of PP1)–PP1α holoenzyme to KAP1 in a manner dependent upon the NIPP1 FHA domain, identifying NIPP1 as an interaction partner for KAP1 and shedding light on the mechanism through which PP1 regulates cellular KAP1 activity.
Adeno-associated virus serotype 2 (AAV2) is a small ssDNA parvovirus that has evolved a unique biphasic life cycle in which replication is dependent on both cellular host factors and coinfection by a helper virus such as adenovirus (Ad5) or herpesvirus (HSV-1) (1). Unable to replicate autonomously, infection by AAV2 alone leads to the establishment of latency either through long-term episomal persistence (2) or through preferential integration of the viral genome into specific sites in the human genome (3–5). The current model for AAV2 latency states that the viral genome is silenced through simultaneous binding of the viral p5 promoter by the AAV2 master regulator Rep and the cellular transcription factors YY1 and MLTF and that the induction of AAV2 gene expression upon helper virus coinfection occurs through interactions between these factors and the helper factor Ad5 E1A or HSV-1 ICP0 (6–8). Despite the widely accepted view that latent AAV2 is silenced exclusively through the binding of Rep and YY1 to p5, evidence that the viral genome assumes a chromatinized configuration shortly after infection (9, 10) suggests a role for epigenetic modification in the establishment of latency and/or transcriptional regulation. The epigenetic landscape of AAV remains unknown, however.
Recent years have seen a rapidly expanding interest in the AAV field as a result of its widespread use as a vector for gene therapy. Despite great recent successes in AAV-based gene therapy (11–16), further improvements may be hindered by an inadequate understanding of various aspects of basic AAV biology. Given that AAV vectors likely mimic the latent phase of the viral life cycle, defining the mechanisms involved in the regulation of AAV latency is of particular importance for the future design and safety of improved vectors. In this study, we sought to gain insight into the regulation of AAV latency by using a screening approach known as “BioID” (17) to identify interaction partners for the AAV2 replication (Rep) proteins. BioID exploits the fusion of the promiscuous biotin ligase BirA* to a bait protein to trigger proximity-dependent biotinylation of neighboring proteins, thus allowing the identification of a much broader scope of protein associations than achievable with conventional affinity purification.
Screens were performed using each of the four related Rep isoforms—Rep78, Rep68, Rep52, and Rep40—which together orchestrate every aspect of the viral life cycle. The large Rep proteins (Rep78/68) consist of an origin-binding domain (OBD) containing site-specific DNA binding and endonuclease activity (18) and an ATPase domain with helicase activity (19) and are necessary for viral replication, integration, and transcriptional regulation (20–22). The small Rep proteins (Rep52/40) share only the ATPase domain and are involved mostly in viral packaging and transcriptional regulation (23, 24). In addition, Rep52 and Rep78 share a C-terminal zinc finger (ZNF) domain implicated in several protein interactions (25, 26). Our BioID screen identified the transcriptional corepressor Krüppel-associated box domain-associated protein 1 (KAP1/TRIM28/TIF1-β) as an interaction partner of the Rep proteins and led to the discovery that Rep78 and Rep52 interact with a protein complex containing KAP1, protein phosphatase 1 (PP1), and nuclear inhibitor of PP1 (NIPP1) to counteract KAP1-mediated repression of the latent viral genome.
Results
The Latent AAV2 Genome Is Repressed Through KAP1 Recruitment to the rep ORF and Subsequent Histone Methylation.
For the BioID screen, BirA* was fused to the N terminus of each of the four Rep isoforms, and the BirA*–Rep fusion proteins were expressed in 293T cells in the presence of free biotin. Biotinylated proteins were affinity purified 48 h after transfection and analyzed by LC-MS/MS. Identified in lysates from each of the four screens was the corepressor KAP1, which acts to form transcriptionally repressive heterochromatin through the recruitment of chromatin-modifying proteins such as the histone methyltransferase SETDB1 and the NuRD histone deacetylase complex containing CHD3 (27, 28). Peptides identified for KAP1 are shown in Table S1. Several known interaction partners of the Rep proteins were also identified by BioID (Table S1), lending support to the quality and coherence of our results. The well-documented function of KAP1 as a mediator of heterochromatin (29, 30) combined with a growing body of evidence demonstrating a role for KAP1 in the regulation of viral elements (31–34) led us to focus our efforts on this candidate. With the exception of Rep68, the physical interaction between each of the Rep proteins and KAP1 was confirmed using biotinylation and immunoprecipitation assays (Fig. S1). Given that Rep40 represents the only shared domain between the Rep proteins, these results suggest that the central Rep ATPase domain is sufficient to mediate a Rep–KAP1 interaction but that the C-terminal ZNF domain of Rep78 is necessary to stabilize this interaction in the presence of an N-terminal OBD.
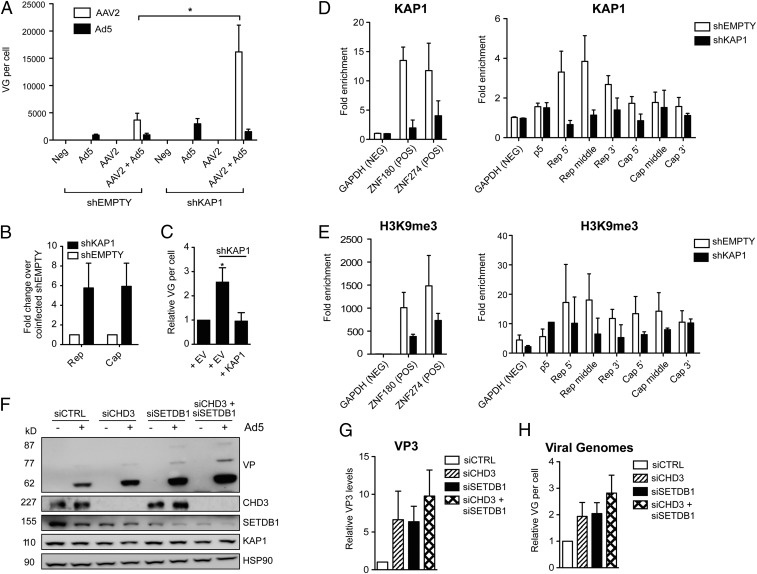
To explore the possible significance of the Rep–KAP1 interaction in the AAV life cycle, we next performed viral replication experiments in cells depleted for KAP1. 293T cells were transduced with lentiviral vectors expressing either an shRNA targeting the 3′ UTR of KAP1 (shKAP1) or the corresponding empty vector (shEMPTY). Forty-eight hours later, cells were infected with Ad5 alone or AAV2 alone or were coinfected with Ad5 and AAV2 to initiate productive replication. Viral replication and transcription were analyzed ∼42 h after infection or when cells displayed optimal cytopathic effect. AAV2 replication was undetectable in either KAP1-depleted or control cells in the absence of Ad5; this was expected, as AAV2 is known to be dependent on several helper factors to initiate replication. In the context of coinfection, however, a five- to sixfold enhancement in AAV2 genome replication, transcription, and protein expression was observed in KAP1-depleted cells compared with controls (Fig. 1 A and B and Fig. S2), Importantly, complementation of KAP1-depleted cells with exogenous KAP1 restored baseline levels of AAV2 replication and protein expression (Fig. 1C and Fig. S2).
Fig. 1.
The latent AAV2 genome is repressed through KAP1 recruitment to the rep ORF and subsequent histone methylation. (A–C) AAV2 replication in control (shEMPTY) or KAP1-depleted (shKAP1) 293T cells. (A) Viral genome (VG) replication. (B) Rep and cap transcripts. Transcript levels represent fold changes over control cells infected with AAV2 + Ad5. Data are reported as mean ± SEM, n = 5. (C) AAV2 replication in KAP1-depleted cells complemented with exogenous KAP1 or an empty vector (EV) control. Data are reported as mean ± SEM, n = 4. (D) ChIP-qPCR performed on control or KAP1-depleted 293T cells infected with AAV2 (100 IU per cell) using anti-KAP1 antibody or IgG. (Left) GAPDH was used as a negative control, and the ZNF genes ZNF180 and ZNF274 were used as positive controls. NEG, negative; POS, positive. (Right) Purified chromatin was analyzed by qPCR using primers for the viral p5 promoter or various regions of the rep and cap ORFs. (E) ChIP-qPCR was performed as described above using anti-H3K9me3 antibody. Values represent fold enrichment over IgG and are reported as mean ± SEM for three independent experiments. (F–H) AAV2 replication and protein expression in 293T cells depleted for CHD3 (siCHD3) and/or SETDB1 (siSETDB1). (F) AAV2 capsid (VP) protein expression and depletion of CHD3 and SETDB1 analyzed by Western blotting. (G) Quantification of VP3 levels using ImageJ software. (H) Viral genome replication analyzed by real-time qPCR. Values are reported as mean ± SEM, n = 4. Statistical significance was determined by unpaired t test, *P < 0.05.
We next asked if KAP1 could be repressing AAV2 through its recruitment to the viral genome and subsequent formation of heterochromatin. We performed KAP1-specific ChIP on 293T cells 2 d after infection with AAV2 alone and analyzed the purified chromatin by qPCR using primers specific for various regions of the AAV2 genome. GAPDH was used as a negative control, and two ZNF genes, ZNF180 and ZNF274, were used as positive controls (35). KAP1 binding was detected across rep, particularly at the 5′ and middle regions (Fig. 1D and Fig. S3) corresponding with the genomic location of the AAV2 p19 promoter. Binding was accordingly lost in KAP1-depleted cells, confirming that the observed signal was KAP1 dependent. To determine the functional significance of this binding, we also performed ChIP-qPCR for trimethylated histone 3 lysine 9 (H3K9me3), a known marker for KAP1-mediated repression. H3K9me3 was enriched across the AAV2 genome (Fig. 1E and Fig. S3), spreading downstream from KAP1-binding sites in rep. Furthermore, this enrichment appeared to be KAP1 dependent, as H3K9 trimethylation was lost in KAP1-depleted cells at a ratio similar to that observed for controls ZNF180 and ZNF274. Importantly, depletion of CHD3 and SETDB1, two members of the KAP1 repressive complex (27, 28), independently and cooperatively led to an enhancement in AAV2 replication and protein expression (Fig. 1 F–H). Taken together, these data strongly suggest that KAP1 represses AAV2 through the binding of AAV2 rep and the subsequent recruitment of histone- and chromatin-modifying proteins, which then act together to methylate AAV2-associated H3K9.
Inactivation of KAP1 Repression Through Phosphorylation of Serine 824 Is Necessary to Support AAV2 Transcriptional Activation and Lytic Replication.
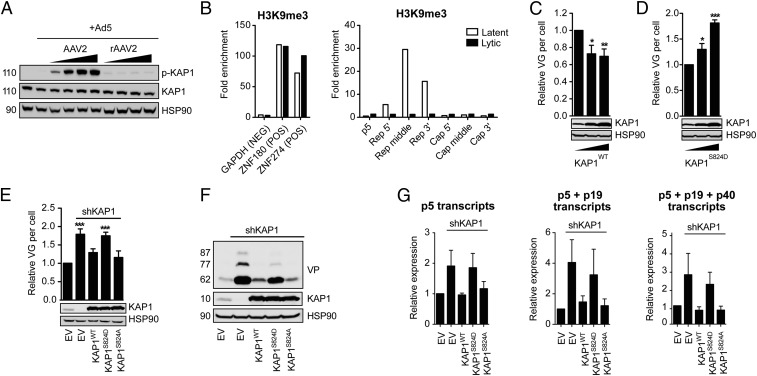
Upon DNA damage, ataxia-telangiectasia mutated (ATM)-dependent phosphorylation of KAP1 at serine 824 (p-KAP1-S824) results in the release of the repressive complex, relaxation of heterochromatin, and relief of transcriptional repression (36, 37). We questioned whether AAV2 replication was associated with KAP1-S824 phosphorylation, which would suggest a requirement for the inactivation of KAP1 corepressor activity. We first explored this possibility by monitoring levels of phosphorylated KAP1-S824 in cells infected with increasing concentrations of either AAV2 or recombinant AAV2 (rAAV2) in the presence of Ad5. rAAV2 is comprised of only the viral inverted terminal repeats (ITRs) flanking a GFP transgene cassette and as such is replication defective. However, the ITRs are known to recruit components of the Mre11/Rad50/NBS1 (MRN) complex, the principal mediator of ATM activation, and could potentially trigger KAP1-S824 phosphorylation (38). We therefore used rAAV2 to control for the input of these structures.
A clear dose-dependent increase in KAP1-S824 phosphorylation was observed in 293T cells coinfected with Ad5 and increasing concentrations of AAV2 (Fig. 2A). This was not observed in the presence of rAAV2, which is replication defective and therefore additionally requires Rep and Cap in trans for replication (Fig. 2A), suggesting that active AAV2 replication, and not simply the viral structure represented by rAAV2, is necessary to trigger KAP1-S824 phosphorylation. In addition, H3K9me3-specific ChIP performed in cells coinfected with AAV2 and Ad5 revealed a complete loss of H3K9 trimethylation on lytic AAV2 genomes, indicating that release of the KAP1 repressive complex is necessary to support lytic replication (Fig. 2B). The observation that AAV2 replication levels are reduced in cells overexpressing wild-type KAP1 (KAP1WT) (Fig. 2C) and are enhanced with overexpression of the dominant negative phospho-mimetic KAP1-S824D mutant (KAP1S824D) (Fig. 2D) further supports a functional role for KAP1-S824 phosphorylation in AAV2 replication. Furthermore, KAP1-depleted cells complemented with KAP1S824D supported enhanced levels of AAV2 replication and protein expression, which were comparable to those observed in KAP1-depleted cells, whereas complementation with the phosphoablatant mutant KAP1S824A restored replication and protein expression to baseline levels (Fig. 2 E and F).
Fig. 2.
Phosphorylation of KAP1-S824 is necessary for AAV2 transcription and replication. (A) p-KAP1-S824 in 293T cells infected with 10–10,000 genome-containing particles (gcp) per cell of either AAV2 or rAAV2 in the presence of Ad5. (B) ChIP-qPCR performed as described in Fig. 1 D and E in 293T cells infected with AAV2 (100 IU per cell) with or without Ad5; n = 1. (C and D) AAV2 genome replication in 293T cells overexpressing KAP1WT (C) or KAP1S824D (D). Values are reported as mean ± SEM, n = 4. (E and F) AAV2 replication in control cells and KAP1-depleted cells reconstituted with KAP1WT, KAP1S824D, or KAP1S824A or treated with an empty vector (EV) control. (E) Viral genome replication. Values are reported as mean ± SEM, n = 4. (F) AAV2 capsid (VP) and KAP1 protein levels. (G) AAV2 transcription in control cells and KAP1-depleted cells complemented with KAP1WT, KAP1S824D, or KAP1S824A or treated with an empty vector (EV) control 16 h after infection with AAV2 (1 IU per cell) alone. Values are reported as mean ± SEM, n = 4. Statistical significance was determined by unpaired t test, *P < 0.05, **P < 0.01, ***P < 0.001.
To determine whether KAP1-S824 phosphorylation constitutes a viral reactivation switch, we looked at the effect of KAP1-S824 phosphorylation on basal expression levels from the three AAV promoters in the absence of helper virus coinfection. AAV transcription during latency is virtually undetectable, and furthermore it has been well established that a helper factor such as Ad5 E1A is essential for activation of the p5 promoter (6). To observe measurable effects on basal AAV2 transcription, these experiments were therefore necessarily performed in 293T cells, which are transformed with AdV E1/E2 and thus can support extremely low levels of AAV transcription without helper virus coinfection. Cells depleted for KAP1 and reconstituted with KAP1WT, KAP1S824D, or KAP1S824A were infected with AAV2 alone and harvested 14 h postinfection for transcriptional analysis. Both KAP1-depleted cells and cells reconstituted with KAP1S824D supported two- to sixfold greater levels of basal transcription from the three AAV2 promoters, while complementation with KAP1WT and KAP1S824A restored transcription to negligible levels (Fig. 2G). Taken together, these data suggest that relief from KAP1 repression through phosphorylation is necessary to allow for the E1A-mediated reactivation of latent AAV2.
Rep52 and Rep78 Mediate Phosphorylation of KAP1-S824 Through Interactions with PP1.
We next performed a time course of lytic replication to determine if phosphorylation of KAP1-S824 could be related to the onset of Rep expression. Substantially elevated levels of p-KAP1-S824 were apparent 18 h after infection with Ad5 and AAV2, correlating well with the onset of Rep expression and leading us to ask if the Rep proteins might be directly modulating KAP1 activity (Fig. 3A). To address this, cells expressing various mutants of the Rep proteins were monitored for p-KAP1-S824 levels. The large Rep proteins possess endonuclease activity shown to trigger DNA damage (25, 39), and all four Rep proteins share a helicase domain with the potential to disrupt DNA as well. To minimize the possibility of DNA damage response (DDR)-dependent induction of p-KAP1-S824 via ATM, endonuclease mutants (Y156F) of Rep68 and Rep78 and catalytic ATPase mutants (K340H) of all four Rep proteins were used (40, 41). Substantially elevated levels of p-KAP1-S824 were apparent in the presence of both Rep52 and Rep78, independently of either endonuclease or ATPase activity (Fig. 3B). Basal levels of p-KAP1-S824 were also visible with Rep40 and Rep68 but were three- to sixfold lower than for Rep52 and Rep78. These data suggest that the Rep proteins, and in particular Rep52 and Rep78, actively mediate phosphorylation of KAP1-S824 via an unknown pathway, independently of their ability to cause DNA damage. This idea was further supported by transfection and infection experiments performed in the presence of an ATM inhibitor (ATMi), which demonstrated that ATM activation is not necessary for the observed Rep-mediated phosphorylation of KAP1-S824 (Fig. S4). Given that Rep52 and Rep78 share a C-terminal ZNF domain not present in Rep40 or Rep68, we suspected this region might also be important for the phosphorylation of KAP1-S824. Expression of a series of C-terminal truncation mutants in which the Rep52/Rep78 ZNF domain was progressively removed (Fig. S5) completely abrogated phosphorylation of KAP1-S824 while having no effect on the Rep–KAP1 interaction (Fig. S5), suggesting that the Rep proteins act through an intermediary protein(s).
Fig. 3.
Rep52 and Rep78 mediate phosphorylation of KAP1-S824 through interactions with PP1. (A) p-KAP1-S824 in 293T cells infected with Ad5 alone or coinfected with Ad5 and either AAV2 or rAAV2 [1,000 genome-containing particles (gcp) per cell] monitored at 4, 18, 24, and 42 h post infection. (B) p-KAP1-S824 in 293T cells expressing the indicated Rep proteins. Values are reported as mean ± SEM, n = 3. (C) GFP-trap experiment performed in 293T cells expressing Rep52 and GFP-tagged PP1α, PP1β, PP1γ, or a GFP control. (D–F) AAV2 replication and transcription at different time points in 293T cells depleted for PP1α (siPP1α), PP1β (siPP1β), or both (siPP1α/β). (D) PP1 depletion analyzed by Western blotting. (E) AAV2 genome replication. Values are reported as mean ± SEM, n = 7. (F) Transcription from the three AAV2 promoters. Values are reported as mean ± SEM, n = 4. (G) Coimmunoprecipitation of FLAG-tagged proteins from 293T cells expressing FLAG-PP1α or a FLAG-GFP control and the indicated Rep proteins. (H) Immunoblot of p-KAP1-S824 in 293T cells transfected with either Rep52 or Rep52K372A. Protein levels were quantified using ImageJ software. Values are reported as mean ± SEM, n = 3. Statistical significance was determined by unpaired t test, *P < 0.05, **P < 0.01, ***P < 0.001. EV, empty vector; IP, immunoprecipitation.
Potential cellular factors that could be interacting with Rep to control the phosphorylation state of KAP1-S824 include PP1 and its specific regulators. Upon completion of DNA repair, basal levels of p-KAP1-S824 are restored through the combined activities of PP1α/β and protein phosphatase 4 (42, 43). Several regulatory subunits of PP1 were identified as interaction partners for Rep alongside KAP1 in our original BioID screen, leading us to hypothesize that the Rep proteins could be interfering with this pathway and antagonizing PP1 activity. Coimmunoprecipitation experiments in cells expressing Rep52 and GFP-tagged PP1-α, -β, or -γ indicated a physical interaction between Rep52, PP1α, and PP1γ (Fig. 3C). Furthermore, AAV2 lytic replication and transcription were enhanced with depletion of PP1α but not PP1β (Fig. 3 D–F), supporting observations from the coimmunoprecipitation as well as a role for PP1 antagonism in the AAV life cycle. Interestingly, this effect was most pronounced at early time points indicating that PP1 might act predominantly during early infection events, such as transcriptional activation.
Using the conserved PP1 consensus binding sequence [KR][X0–1][VI]{P}[FW] (44) as our guideline, we identified one putative noncanonical binding site in the Rep ATPase domain (372KMVIW376), partially overlapping with the Walker B motif (Fig. S6). Coimmunoprecipitation experiments using FLAG-tagged PP1α confirmed a physical interaction between Rep52 and PP1α (Fig. 3G). However, mutation of the first lysine in the putative binding site (K372A) did not affect this interaction (Fig. 3G), indicating this region may not represent a true PP1-binding site. Alternatively, it is possible that this region acts together with the Rep ZNF domain to bind PP1. Interestingly however, the K372A mutation completely abrogated Rep-mediated phosphorylation of KAP1-S824 (Fig. 3H) without affecting the ability of Rep to regulate the AAV2 p5 promoter (45) or to interact with KAP1 (Fig. S6). There is precedence indicating that a functional interaction between PP1 and its regulatory subunits is based on multiple points of interaction, only one of which necessarily consists of the conserved binding site. Furthermore, it has been demonstrated that mutation of only one interaction site may be sufficient to abolish the regulation of PP1 while being insufficient to abolish the physical association between PP1 and its regulatory component (46, 47). Therefore it is possible that the mutation of additional, as yet unidentified, interaction sites in Rep would be necessary to disrupt Rep–PP1 binding, while mutation of only the PP1-binding site is sufficient to abolish regulation of PP1 by Rep.
The Rep Proteins Interfere with the Formation of a KAP1–NIPP1–PP1 Complex to Inhibit KAP1-S824 Dephosphorylation by PP1.
The free PP1 catalytic subunit has exceptionally broad substrate specificity. PP1 activity is therefore regulated through its interactions with numerous PP1-interacting proteins (PIPs), which can inhibit PP1 or act as substrate specifiers (48). To further define a potential mechanism for the above observations, we next looked at the nuclear inhibitor of PP1 (NIPP1/PPP1R8), a major regulator of PP1 that was identified alongside KAP1 in our BioID screen. NIPP1 contains three functional domains: (i) an N-terminal forkhead-associated (FHA) protein interaction domain, (ii) a central PP1-binding domain containing the consensus PP1-binding motif, and (iii) a multifunctional C-terminal domain that binds RNA, has endoribonuclease activity, and inhibits PP1 activity via an unknown mechanism (46, 49, 50). Cross-linked GFP-trap experiments in cells expressing Rep52 with various NIPP1 mutants (Fig. 4A) fused to eGFP revealed that KAP1 and Rep are recruited to NIPP1 via the N-terminal FHA domain, forming a complex with the NIPP1–PP1α holoenzyme (Fig. 4B). PP1α has previously been shown to form a constitutive unit with KAP1 at certain promoters (42). Our results, however, suggest that in this instance the interaction between KAP1 and PP1 is mediated through NIPP1. Recruitment was dependent upon the NIPP1 FHA domain, which binds hyperphosphorylated proteins through a dipeptide motif consisting of phospho-threonines followed by a proline. Although no function has yet been attributed to it, one such motif exists at threonine 541 of KAP1, suggesting that it may be directly recruited to NIPP1 via the FHA domain. Levels of p-KAP1-S824 were elevated in cells overexpressing NIPP1-WT compared with cells expressing the PP1-binding mutant NIPP1-RATA, suggesting that the inhibition of PP1 by NIPP1 has a role in KAP1 phosphorylation (Fig. 4 C and D). Surprisingly, p-KAP1-S824 levels were also elevated in the presence of NIPP1-FHAm (FHA binding mutant). It is possible that the residual binding observed between KAP1 and NIPP1-FHAm (Fig. 4B) is sufficient to support the regulation of phosphorylation. Alternatively, overexpression of NIPP1-FHAm may act to constitutively inhibit the nuclear pool of PP1. In addition, NIPP1-WT and NIPP1-FHAm, but not NIPP1-RATA, enhanced Rep-mediated phosphorylation of KAP1-S824 (Fig. 4 E and F), supporting the idea that this pathway is exploited by Rep to maintain enhanced levels of p-KAP1-S824 during infection.
Fig. 4.
Rep proteins interfere with the NIPP1–PP1 complex to antagonize KAP1-S824 dephosphorylation. (A) Schematic representation of NIPP1 mutants. The N-terminal FHA domain is shown in blue. FHAm denotes the FHA-binding mutant, which is unable to recruit hyperphosphorylated proteins. The central PP1-binding domain containing the consensus PP1-binding site RVTF is shown in yellow. RATA denotes the PP1-binding mutant. The C-terminal PP1 interaction and inhibitory domain are shown in green, and the RNA-binding region is shown in lime. (B) Cross-linked GFP-trap experiment performed in cells expressing Rep52 and each of the GFP-tagged NIPP1 constructs or a GFP control. (C) Immunoblot of p-KAP1-S824 in cells expressing NIPP1-WT, -RATA, -FHAm, or the empty vector (EV) control. (D) Quantification of p-KAP1-S824 levels in C. Values are reported as mean ± SEM, n = 4. (E) Immunoblot of p-KAP1-S824 in cells expressing Rep52 and NIPP1-WT, -RATA, -FHAm, or an empty vector (EV) control. (F) Quantification of p-KAP1-S824 levels in E. Values are reported as mean ± SEM, n = 5. (G) Cross-linked GFP trap performed in cells expressing NIPP1-WT-GFP and T7-tagged Rep52, Rep52K372A, or a Renilla control. Statistical significance was determined by unpaired t test, *P < 0.05.
To further explore the role of Rep in this pathway, we performed cross-linked GFP-trap experiments in cells expressing NIPP1-WT-eGFP with either wild-type Rep52 or the phosphorylation-deficient Rep52-K372A (Fig. 4G). Interestingly, recruitment of KAP1 to NIPP1 appeared to be entirely independent of Rep52, while the abundance of PP1α in the complex was significantly decreased in the presence of Rep52 but not Rep52-K372A. The loss of PP1α was associated with increased p-KAP1-S824 and a concomitant loss of SETDB1, supporting the hypothesis that Rep52 mediates phosphorylation of KAP1-S824 through PP1 interference to counteract KAP1 repression. Furthermore, as loss of PP1α from the complex was observed with Rep52 but not Rep52-K372A, these results suggest that the K372A mutation may in fact interfere with the Rep-PP1 interaction in the context of endogenous PP1, even though we did not observe an effect with overexpressed FLAG-PP1α (Fig. 3G).
Targeting of KAP1 as a Helper Function for AAV2 Replication.
Given that basal levels of Rep expression during latency are not sufficient to counteract KAP1 and that depletion of KAP1 alone is not sufficient to trigger AAV2 transcription and replication, we hypothesized that AAV2 helper viruses might act as a biological switch necessary to allow the up-regulation of rep expression before the onset of KAP1 phosphorylation. KAP1 protein levels were significantly depleted in 293T and HeLa cells infected with increasing concentrations of Ad5 and were restored in the presence of 5 μM of the proteasome inhibitor MG132 (Fig. S7). Interestingly, Ad5 E1B55K has been shown to interfere with KAP1 SUMOylation during early infection (51), presenting the possibility that hypoSUMOylated KAP1 may be targeted by the cell for proteosomal degradation. Similar results were also observed for HSV-1, another AAV helper virus (Fig. S7), suggesting that KAP1 targeting may represent an unknown helper function for AAV2 replication necessary to release the viral genome from its latent state.
Discussion
Significant breakthroughs in AAV vector design have previously been derived from an enhanced understanding of basic AAV biology. However, many aspects of the AAV life cycle still remain elusive. Of particular relevance to AAV vectors, whose biology may mimic that of latent viral genomes, the nature and contribution of epigenetic marks to the genome organization and temporal gene regulation of wild-type AAV are not yet known. In the present study, we employed a BioID strategy to screen for interaction partners of the AAV2 Rep proteins in an effort to elucidate the mechanisms involved in the establishment of and release from AAV2 latency. This approach led to the discovery that the transcriptional corepressor KAP1 interacts with three of the four Rep proteins and is recruited to the latent AAV2 genome, where it mediates transcriptional repression through the formation of heterochromatin.
Our findings bear interesting parallels with observations made for the regulation of both KSHV and CMV latency by KAP1. Recruitment of KAP1 to the CMV genome leads to H3K9me3 deposition across various lytic genes, while latency-associated genes remain free from repressive marks (32). Similarly, latency-associated nuclear antigen (LANA)-mediated recruitment of KAP1 to the KSHV genome is essential for the shutdown of lytic gene expression during the early stages of infection (52). Here, we observed KAP1-dependent deposition of H3K9me3 across the latent AAV2 genome, spreading downstream from KAP1 recruitment sites near the p19 promoter in rep in a manner consistent with the mechanism of long-range heterochromatin spreading shown to establish KAP1-mediated repression of KRAB–ZFP clusters (53). Interestingly, no enrichment for KAP1 or H3K9me3 was detected at the p5 promoter, a region whose transactivating activity is necessary for initiation from all three viral promoters. It will be interesting to determine whether this region is protected from repressive marks to ensure rapid and dynamic regulation of p5 upon reactivation.
In agreement with a role for KAP1 as a repressor of latent AAV2, both KAP1 depletion and expression of a repression-deficient phosphomimetic KAP1-S824D mutant resulted in enhanced lytic replication, transcription, and protein expression in cells coinfected with AAV2 and Ad5. This effect was not observed in cells infected with AAV2 alone, however, reflecting the dependency of AAV2 on various helper factors to initiate replication and suggesting that KAP1 repression provides a second layer of regulation, the antagonism of which is necessary but not sufficient for reactivation. Similar observations were made for both KSHV and CMV. KSHV replication was enhanced, but not triggered, by KAP1 depletion both in the context of induced KRta expression and hypoxia-induced KSHV reactivation (31, 54). Similarly, TNFα-mediated NF-κB induction was necessary for the full reactivation of CMV upon KAP1 depletion (32). It is well established that reactivation of AAV2 is dependent upon interactions between helper factors such as Ad5 E1A and cellular factors YY1 and MLTF bound to the p5 promoter region. However, our observation that levels of basal AAV2 transcription in latently infected 293T cells were enhanced in cells either depleted for KAP1 or reconstituted with KAP1-S824D strongly indicates that KAP1 binding additionally serves to silence the viral genome through histone methylation, even while the p5 promoter may be minimally activated by low levels of E1A. We therefore propose that AAV2 reactivation is dependent upon the removal of repressive H3K9me3 from the viral genome to render it transcriptionally competent and to allow for E1A-mediated activation of p5 and thus transactivation of all three viral promoters.
It is also interesting to note that KSHV, CMV, and AAV2, episomal viruses with the ability to modulate KAP1 activity, are clearly capable of replication without the need for KAP1 depletion, suggesting the intriguing possibility that these viruses have domesticated KAP1 repression to their advantage. It is possible, for example, that heterochromatinization allows latent episomes to better evade immune recognition or may prevent deleterious recombination events and/or genome degradation. Upon reactivation, KSHV directly mediates phosphorylation of KAP1 via its viral kinase vPk (31), and, although which CMV protein is responsible has not yet been established, phosphorylation of KAP1-S824 was observed only in cells that were also positive for CMV IE antigens (32). Here we show that AAV2 Rep52 and Rep78 mediate the inactivation of KAP1 repression by inhibiting its dephosphorylation by the phosphatase PP1α. Rep expression reduced the abundance of PP1α from a complex comprised of KAP1, SETDB1, PP1α, and NIPP1, and this was dependent upon the presence of an intact putative PP1-binding site in Rep, suggesting that Rep52 achieves enhanced phosphorylation of KAP1-S824 by sequestering PP1 from the complex.
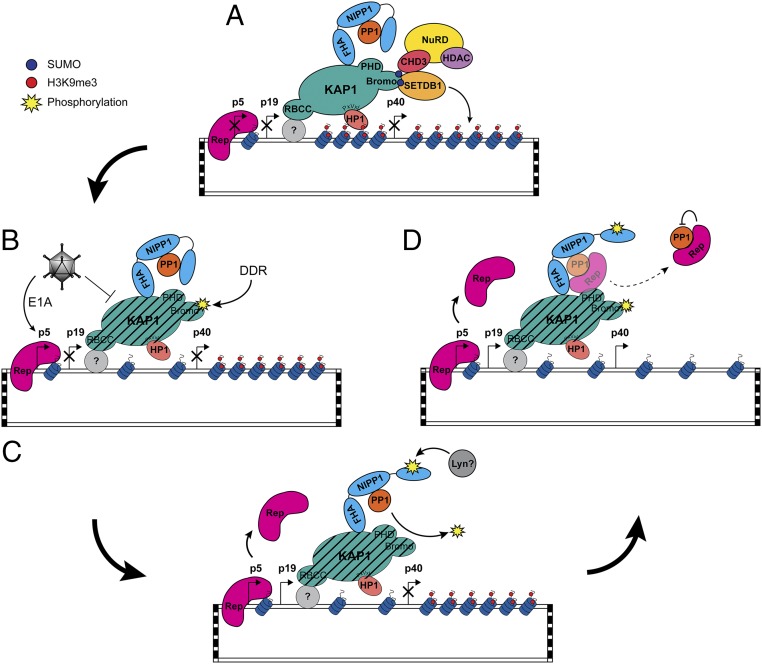
Although we have shown that Rep can mediate the phosphorylation of KAP1 independently from ATM activation, this does not exclude a role for the activation of effectors of the DDR in the context of a productive infection. In fact, the nature of the mechanism outlined above presupposes an initial trigger for KAP1 phosphorylation, and both helper virus and productive AAV2 infections are known to trigger robust activation of DDR proteins (55, 56). Taken together, these data suggest a two-part mechanism in which helper factors or the DDR upon viral infection trigger the initial phosphorylation of KAP1, a signal then potentiated through Rep-mediated antagonism of PP1 during lytic replication. We envision that the inactive NIPP1–PP1 holoenzyme forms a constitutive regulatory unit with KAP1, effectively serving to tether inactive PP1 to KAP1 for rapid regulation (Fig. S8). NIPP1 does not systematically inhibit PP1 activity, however. Rather, inhibition is dependent upon interactions between PP1 bound to the central domain of NIPP1 and the inhibitory C-terminal domain of NIPP1, which additionally has RNA binding and endoribonuclease activity (46, 47). This interaction can be interrupted through the simultaneous phosphorylation of tyrosine 335 in the C-terminal domain and binding of RNA, effectively activating the PP1–NIPP1 holoenzyme (46). Upon initiation of a DDR, when KAP1 is rapidly phosphorylated, NIPP1 inhibition of PP1 may be inactivated through RNA-binding and phosphorylation of the C-terminal region of NIPP1 by the DDR-responsive tyrosine kinase Lyn (46), allowing PP1 to rapidly restore basal levels of p-KAP1-S824. The role of the Rep proteins might then be to sequester PP1 or to compete for binding to sustain high levels of p-KAP1-S824 and support lytic infection (Fig. 5 and Fig. S8).
Fig. 5.
Model for the release of AAV2 from KAP1-mediated latency. (A) Incoming AAV2 genomes undergo second-strand synthesis, concatamerization, and chromatinization upon nuclear entry. KAP1 is recruited to the rep ORF via an unknown binding partner where it forms a scaffold for the recruitment of SETDB1 and CHD3, leading to the methylation of AAV2-associated histones. NIPP1 is recruited to KAP1 via the FHA domain, where it serves to tether inactive PP1 to KAP1. (B and C) Upon coinfection, KAP1 repression is partially lifted through several potential mechanisms: helper-mediated degradation of KAP1 as we have observed, interference with KAP1 SUMOylation as observed by others (51), and/or phosphorylation of KAP1-S824 triggered by initial helper- or AAV-mediated stress response (B), allowing up-regulation of rep by Ad5 E1A (C). Unknown cellular factors/RNA binding inactivate NIPP1, allowing PP1 to restore baseline levels of p-KAP1-S824. (D) Sequestration of PP1 by Rep sustains enhanced levels of phosphorylated KAP1-S824 to support lytic replication.
This work demonstrates not only an example of PP1 targeting by a parvovirus but also an example of PP1 targeting for the purpose of regulating KAP1 activity. It will be interesting to determine whether this mechanism might extend to other members of the parvovirus family or if known viral targets of KAP1, such as KSHV and CMV, might also manipulate this pathway to achieve relief from KAP1 repression. PP1 has previously been shown to form a constitutive unit with KAP1 at the p21 promoter, where it is thought to set a basal transcription rate as well as to rapidly restore KAP1 corepressor function after ATM activation by regulating KAP1-S824 phosphorylation (42). There is no evidence for free cellular pools of PP1, however. Rather, PP1 constitutes the catalytic unit of a large array of multisubunit holoenzymes, which regulate and target PP1 activity to prevent uncontrolled protein dephosphorylation and cell death (48). It has yet to be determined what PIP is responsible for regulating PP1 activity toward KAP1. Our finding that KAP1, PP1, and NIPP1 exist as a complex sheds light on this question and suggests that PP1 may be targeted to KAP1 via the NIPP1 FHA domain and then is dynamically regulated by NIPP1 to maintain homeostatic levels of phosphorylated KAP1-S824.
This work further presents evidence that AAV2 latency is regulated in part through the epigenetic modification of its genome, challenging the long-standing model for AAV latency whereby the viral genome is silenced exclusively through binding of the p5 promoter by the cellular factors YY1 and MLTF and the Rep proteins (6, 57). The findings presented here have additional relevance for gene therapy, as our data highlight the possibility that current production helper plasmids may lack helper genes that may be critical for navigating host responses. Understanding the epigenetic control of AAV may also shed light on the intriguing and unexplained resistance of AAV gene-therapy vectors to host shut-off and will undoubtedly contribute to understanding the consequences of integrating wild-type and recombinant viruses. In addition, the recent controversial discovery of AAV2 sequences in human liver tumors (58) has caused some to call into question the safety of rAAV vectors and has highlighted the need to further explore AAV2 regulation of latency. These findings may thus provide key insights into the impact and contribution of AAV2 latency on the development of human diseases.
Methods
Cell Lines and Viruses.
293T HEK cells and HeLa human cervical epithelial cells were obtained from the American Tissue Culture Collection (ATCC). Cells were cultured in DMEM (Invitrogen) supplemented with 10% FBS (Invitrogen) plus 1% penicillin/streptomycin (Sigma) and were tested for mycoplasma once per month.
AAV2 and human Ad5 were produced and purified as previously described (59).
BioID Screening.
Ten 10-cm dishes of 293T cells per BirA*-Rep construct were transfected using 6 μg DNA and 50 μL polyethylenimine (PEI) (Polysciences Inc.) in 500-μL serum-free (SF) medium per dish. Six hours posttransfection, the medium was replaced with fresh DMEM + 10% FBS, and D-Biotin (Life Technologies) was added to a final concentration of 100 μM. Forty-eight hours posttransfection, cells were harvested for LC-MS/MS analysis as previously described (17). Mass spectrometry was performed by the KCL Proteomics Facility at Denmark Hill.
Immunoprecipitation.
293T cells were transfected in a six-well format with 200 ng of Rep-expressing constructs and 250 ng of FLAG-PP1α or FAG-GFP using 8 μL PEI in 80 μL of SF medium. Forty-eight hours after transfection, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer [50 mM Tris (pH 8), 150 mM NaCl, 2 mM EDTA (pH 8), 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1× protease inhibitors], and lysates were incubated with 2 μg anti-FLAG (F7425; Sigma) for 1.5 h on a rotator (PTR-60; Grant-bio) at 4 °C. Forty microliters for of protein G agarose beads (Pierce) were added and incubated a further 3 h. Beads were washed four times in RIPA buffer, and proteins were eluted from beads by boiling at 95 °C for 10 min in 60 μL of 2× Laemmli buffer.
Cross-Linking Immunoprecipitation.
293T cells were transfected in a six-well format with 200 ng of Rep-expressing constructs and 750 ng of FLAG-GFP, FLAG-KAP1, or GFP-NIPP1 using 8 μL PEI in 80 μL of SF medium. Forty-eight hours after transfection cells were fixed in 350 μL 0.05% formaldehyde for 10 min at 37 °C and were then quenched in 350 μL 0.125 M glycine (pH 7) for 5 min at room temperature before being lysed in 500 μL of cross-linking immunoprecipitation buffer [150 mM NaCl, 10 mM Hepes (pH 7), 6 mM MgCl2, 2 mM DTT, 10% glycerol, 1× protease inhibitors, 200 μM sodium orthovanadate] on ice for 10 min. Lysates were subjected to three 10-s cycles of sonication (Branson Sonifier 250), output ∼2, and clarified at 1,000 × g for 10 min at 4 °C. Forty microliters of protein G agarose beads (Pierce) per sample were incubated with 2 μg FLAG antibody (F7425; Sigma) for 1.5 h on a rotator at 4 °C before being added to the cell lysates and incubated a further 3–4 h. Beads were harvested and washed four times in RIPA buffer, and cross-links were reversed in 25 μL of reverse cross-link buffer (10 mM EDTA, 5 mM DTT, 1% SDS) at 65 °C for 45 min. Proteins were eluted from beads by adding 3 μL of 2× Laemmli SDS buffer and heating at 95 °C for 10 min. For GFP-trap experiments, 293T cells were transfected with 800 ng of Rep- and/or NIPP1-expressing constructs. Twenty-four hours after transfection, cells were harvested as described above, and cell lysates were incubated with 30 μL NHS-activated Sepharose (GE Healthcare) covalently linked to GFP nanobodies. Beads were harvested as described above.
Infections.
For KAP1 depletion, 293T cells were transduced with a lentiviral vector expressing either a hairpin targeting the 3′ UTR of KAP1 or the corresponding empty vector 48 h before infection with AAV2/Ad5. Cells were infected at ∼80% confluency with 10 IU of AAV2 per cell unless stated otherwise in approximately two-fifths of the normal well volume. Two hours after AAV2 infection, Ad5 was added at a multiplicity of infection (MOI) of 2 pfu per cell, and medium was replaced with fresh DMEM + 10% FBS 1 h after Ad5 infection. Cells were harvested for qPCR, RT-qPCR, or Western blot ∼42 h after infection or when they displayed optimal cytopathic effect (CPE). Optimal CPE is defined by cells that display a rounded and enlarged phenotype as opposed to the normal star-shaped morphology of HEK293T cells and are beginning to detach but still appear bright and healthy. Cells that had completely lifted by the time of harvest were deemed too advanced in the infection cycle and were excluded from analysis.
ChIP-qPCR.
Cells were cross-linked in their medium in 1% formaldehyde (10 min at room temperature) and quenched with 0.125 M glycine (5 min at room temperature) before being lysed (1 × 108 cells/mL) in 50 mM Tris⋅HCl (pH 8), 10 mM EDTA, 1% SDS, 1× protease inhibitors for 10 min on ice. Lysates were sonicated to obtain 200- to 500-bp fragments (15 30-s cycles with 90-s intervals, output ∼2). Ten milliliters of lysates were used to assess sonication efficiency by reverse cross-linking for 15 min at 95 °C and then incubation with RNase A for 30 min at 37°. DNA was extracted and visualized on a 1.5% agarose gel. The remaining lysates were clarified at 16,000 × g for 10 min at 4°. The equivalent of 2 × 106 cells was diluted 25-fold in RIPA buffer [50 mM Tris (pH 8), 150 mM NaCl, 2 mM EDTA (pH 8), 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1× protease inhibitors] and precleared with 80 mL protein G agarose beads (preblocked in 0.1 mg/mL BSA for 30 min) for 2 h on a rotator at 4 °C. For the immunoprecipitation, antibodies were added to lysates and incubated with antibody for 1 h on a rotator at 4 °C [5 mg IgG (ab37415; Abcam), 4 mg H3K9me3 (ab8893; Abcam), 1 mg KAP1 (ab10483; Abcam)] before the addition of 80 mL of preblocked beads and overnight incubation as above. Beads were harvested and washed four times in RIPA buffer, four times in high-salt wash [20 mM Tris⋅HCl (pH 8), 1 mM EDTA, 500 mM NaCl, 0.5% Nonidet P-40, 1× protease inhibitors], and four times in TE buffer [10 mM Tris⋅HCl (pH 8), 1 mM EDTA] and were eluted in 160 mL of elution buffer (100 mM HaHCO3, 1% SDS) for 15 min at 30 °C. Cross-links were reversed by adding NaCl to a final concentration of 0.2 M and overnight incubation at 67 °C. Eluates were then incubated with 2 mL RNase A (10 mg/mL) and 2 mL proteinase K (20 mg/mL) at 45° for 1 h. DNA was extracted using a PCR purification kit (Qiagen) and analyzed by qPCR using primers specific for GAPDH, ZNF180, ZNF274, or various regions of the AAV2 genome. Purified chromatin was diluted 10-fold and quantified by real-time PCR using the SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich) for qPCR using an ABI PRISM system (Applied Biosystems). Primer sequences are listed in Table S2. Cycle threshold (CT) values for 10% input were adjusted by subtracting 3.322 cycles to correct for the 10-fold dilution factor (https://www.thermofisher.com/uk/en/home/life-science/epigenetics-noncoding-rna-research/chromatin-remodeling/chromatin-immunoprecipitation-chip/chip-analysis.html). Percent input was then calculated as 100 × 2−(CT of adjusted 10% input − CT of ChIP-ed DNA). The percent input for each antibody was then normalized to values for IgG to calculate final fold enrichment.
Supplementary Material
Acknowledgments
We thank M. Bardelli, G. Berger, R. Galão, T. Foster, and S. Pickering for technical assistance and C. Swanson for insightful comments and support. This work was supported by UK Medical Research Council Grants 1001764 (to R.M.L.) and MR/N022890/1 (to E.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721883115/-/DCSupplemental.
References
- 1.Henckaerts E, Linden RM. Adeno-associated virus: A key to the human genome? Future Virol. 2010;5:555–574. doi: 10.2217/fvl.10.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schnepp BC, Jensen RL, Chen CL, Johnson PR, Clark KR. Characterization of adeno-associated virus genomes isolated from human tissues. J Virol. 2005;79:14793–14803. doi: 10.1128/JVI.79.23.14793-14803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petri K, et al. Presence of a trs-like motif promotes Rep-mediated wild-type adeno-associated virus type 2 integration. J Virol. 2015;89:7428–7432. doi: 10.1128/JVI.00426-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janovitz T, et al. High-throughput sequencing reveals principles of adeno-associated virus serotype 2 integration. J Virol. 2013;87:8559–8568. doi: 10.1128/JVI.01135-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hüser D, Gogol-Döring A, Chen W, Heilbronn R. Adeno-associated virus type 2 wild-type and vector-mediated genomic integration profiles of human diploid fibroblasts analyzed by third-generation PacBio DNA sequencing. J Virol. 2014;88:11253–11263. doi: 10.1128/JVI.01356-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang LS, Shi Y, Shenk T. Adeno-associated virus P5 promoter contains an adenovirus E1A-inducible element and a binding site for the major late transcription factor. J Virol. 1989;63:3479–3488. doi: 10.1128/jvi.63.8.3479-3488.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geoffroy MC, Epstein AL, Toublanc E, Moullier P, Salvetti A. Herpes simplex virus type 1 ICP0 protein mediates activation of adeno-associated virus type 2 rep gene expression from a latent integrated form. J Virol. 2004;78:10977–10986. doi: 10.1128/JVI.78.20.10977-10986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi Y, Seto E, Chang LS, Shenk T. Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 9.Marcus-Sekura CJ, Carter BJ. Chromatin-like structure of adeno-associated virus DNA in infected cells. J Virol. 1983;48:79–87. doi: 10.1128/jvi.48.1.79-87.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Penaud-Budloo M, et al. Adeno-associated virus vector genomes persist as episomal chromatin in primate muscle. J Virol. 2008;82:7875–7885. doi: 10.1128/JVI.00649-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nathwani AC, et al. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N Engl J Med. 2014;371:1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bainbridge JWB, et al. Long-term effect of gene therapy on Leber’s congenital amaurosis. N Engl J Med. 2015;372:1887–1897. doi: 10.1056/NEJMoa1414221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett J, et al. Safety and durability of effect of contralateral-eye administration of AAV2 gene therapy in patients with childhood-onset blindness caused by RPE65 mutations: A follow-on phase 1 trial. Lancet. 2016;388:661–672. doi: 10.1016/S0140-6736(16)30371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendell JR, et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N Engl J Med. 2017;377:1713–1722. doi: 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- 15.George LA, et al. Hemophilia B gene therapy with a high-specific-activity factor IX variant. N Engl J Med. 2017;377:2215–2227. doi: 10.1056/NEJMoa1708538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rangarajan S, et al. AAV5–factor VIII gene transfer in severe hemophilia A. N Engl J Med. 2017;377:2519–2530. doi: 10.1056/NEJMoa1708483. [DOI] [PubMed] [Google Scholar]
- 17.Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Im DS, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 19.Im DS, Muzyczka N. Partial purification of adeno-associated virus Rep78, Rep52, and Rep40 and their biochemical characterization. J Virol. 1992;66:1119–1128. doi: 10.1128/jvi.66.2.1119-1128.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chejanovsky N, Carter BJ. Mutagenesis of an AUG codon in the adeno-associated virus rep gene: Effects on viral DNA replication. Virology. 1989;173:120–128. doi: 10.1016/0042-6822(89)90227-4. [DOI] [PubMed] [Google Scholar]
- 21.Surosky RT, et al. Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome. J Virol. 1997;71:7951–7959. doi: 10.1128/jvi.71.10.7951-7959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pereira DJ, Muzyczka N. The cellular transcription factor SP1 and an unknown cellular protein are required to mediate Rep protein activation of the adeno-associated virus p19 promoter. J Virol. 1997;71:1747–1756. doi: 10.1128/jvi.71.3.1747-1756.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King JA, Dubielzig R, Grimm D, Kleinschmidt JA. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 2001;20:3282–3291. doi: 10.1093/emboj/20.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kyöstiö SR, et al. Analysis of adeno-associated virus (AAV) wild-type and mutant Rep proteins for their abilities to negatively regulate AAV p5 and p19 mRNA levels. J Virol. 1994;68:2947–2957. doi: 10.1128/jvi.68.5.2947-2957.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berthet C, Raj K, Saudan P, Beard P. How adeno-associated virus Rep78 protein arrests cells completely in S phase. Proc Natl Acad Sci USA. 2005;102:13634–13639. doi: 10.1073/pnas.0504583102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Pasquale G, Stacey SN. Adeno-associated virus Rep78 protein interacts with protein kinase A and its homolog PRKX and inhibits CREB-dependent transcriptional activation. J Virol. 1998;72:7916–7925. doi: 10.1128/jvi.72.10.7916-7925.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schultz DC, Friedman JR, Rauscher FJ., 3rd Targeting histone deacetylase complexes via KRAB-zinc finger proteins: The PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001;15:428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivanov AV, et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell. 2007;28:823–837. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sripathy SP, Stevens J, Schultz DC. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol Cell Biol. 2006;26:8623–8638. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang PC, et al. Kruppel-associated box domain-associated protein-1 as a latency regulator for Kaposi’s sarcoma-associated herpesvirus and its modulation by the viral protein kinase. Cancer Res. 2009;69:5681–5689. doi: 10.1158/0008-5472.CAN-08-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rauwel B, et al. Release of human cytomegalovirus from latency by a KAP1/TRIM28 phosphorylation switch. eLife. 2015;4 doi: 10.7554/eLife.06068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rowe HM, et al. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res. 2013;23:452–461. doi: 10.1101/gr.147678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 35.Frietze S, O’Geen H, Blahnik KR, Jin VX, Farnham PJ. ZNF274 recruits the histone methyltransferase SETDB1 to the 3′ ends of ZNF genes. PLoS One. 2010;5:e15082. doi: 10.1371/journal.pone.0015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodarzi AA, Kurka T, Jeggo PA. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol. 2011;18:831–839. doi: 10.1038/nsmb.2077. [DOI] [PubMed] [Google Scholar]
- 37.Li X, et al. Role for KAP1 serine 824 phosphorylation and sumoylation/desumoylation switch in regulating KAP1-mediated transcriptional repression. J Biol Chem. 2007;282:36177–36189. doi: 10.1074/jbc.M706912200. [DOI] [PubMed] [Google Scholar]
- 38.Schwartz RA, et al. The Mre11/Rad50/Nbs1 complex limits adeno-associated virus transduction and replication. J Virol. 2007;81:12936–12945. doi: 10.1128/JVI.01523-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt M, Afione S, Kotin RM. Adeno-associated virus type 2 Rep78 induces apoptosis through caspase activation independently of p53. J Virol. 2000;74:9441–9450. doi: 10.1128/jvi.74.20.9441-9450.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis MD, Wu J, Owens RA. Mutational analysis of adeno-associated virus type 2 Rep68 protein endonuclease activity on partially single-stranded substrates. J Virol. 2000;74:2936–2942. doi: 10.1128/jvi.74.6.2936-2942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chejanovsky N, Carter BJ. Mutation of a consensus purine nucleotide binding site in the adeno-associated virus rep gene generates a dominant negative phenotype for DNA replication. J Virol. 1990;64:1764–1770. doi: 10.1128/jvi.64.4.1764-1770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, et al. SUMOylation of the transcriptional co-repressor KAP1 is regulated by the serine and threonine phosphatase PP1. Sci Signal. 2010;3:ra32. doi: 10.1126/scisignal.2000781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfeifer GP. Protein phosphatase PP4: Role in dephosphorylation of KAP1 and DNA strand break repair. Cell Cycle. 2012;11:2590–2591. doi: 10.4161/cc.21234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meiselbach H, Sticht H, Enz R. Structural analysis of the protein phosphatase 1 docking motif: Molecular description of binding specificities identifies interacting proteins. Chem Biol. 2006;13:49–59. doi: 10.1016/j.chembiol.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 45.Dutheil N, et al. Adeno-associated virus Rep represses the human integration site promoter by two pathways that are similar to those required for the regulation of the viral p5 promoter. J Virol. 2014;88:8227–8241. doi: 10.1128/JVI.00412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beullens M, et al. The C-terminus of NIPP1 (nuclear inhibitor of protein phosphatase-1) contains a novel binding site for protein phosphatase-1 that is controlled by tyrosine phosphorylation and RNA binding. Biochem J. 2000;352:651–658. [PMC free article] [PubMed] [Google Scholar]
- 47.O’Connell N, et al. The molecular basis for substrate specificity of the nuclear NIPP1:PP1 holoenzyme. Structure. 2012;20:1746–1756. doi: 10.1016/j.str.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verbinnen I, Ferreira M, Bollen M. Biogenesis and activity regulation of protein phosphatase 1. Biochem Soc Trans. 2017;45:89–99. doi: 10.1042/BST20160154. [DOI] [PubMed] [Google Scholar]
- 49.Jagiello I, Beullens M, Stalmans W, Bollen M. Subunit structure and regulation of protein phosphatase-1 in rat liver nuclei. J Biol Chem. 1995;270:17257–17263. doi: 10.1074/jbc.270.29.17257. [DOI] [PubMed] [Google Scholar]
- 50.Jagiello I, et al. NIPP-1, a nuclear inhibitory subunit of protein phosphatase-1, has RNA-binding properties. J Biol Chem. 1997;272:22067–22071. doi: 10.1074/jbc.272.35.22067. [DOI] [PubMed] [Google Scholar]
- 51.Bürck C, et al. KAP1 is a host restriction factor that promotes HAdV E1B-55K SUMO modification. J Virol. 2015;90:930–946. doi: 10.1128/JVI.01836-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun R, Liang D, Gao Y, Lan K. Kaposi’s sarcoma-associated herpesvirus-encoded LANA interacts with host KAP1 to facilitate establishment of viral latency. J Virol. 2014;88:7331–7344. doi: 10.1128/JVI.00596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Groner AC, et al. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet. 2010;6:e1000869. doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang L, et al. Inhibition of KAP1 enhances hypoxia-induced Kaposi’s sarcoma-associated herpesvirus reactivation through RBP-Jκ. J Virol. 2014;88:6873–6884. doi: 10.1128/JVI.00283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwartz RA, Carson CT, Schuberth C, Weitzman MD. Adeno-associated virus replication induces a DNA damage response coordinated by DNA-dependent protein kinase. J Virol. 2009;83:6269–6278. doi: 10.1128/JVI.00318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Collaco RF, Bevington JM, Bhrigu V, Kalman-Maltese V, Trempe JP. Adeno-associated virus and adenovirus coinfection induces a cellular DNA damage and repair response via redundant phosphatidylinositol 3-like kinase pathways. Virology. 2009;392:24–33. doi: 10.1016/j.virol.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seto E, Shi Y, Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature. 1991;354:241–245. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- 58.Nault J-C, et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat Genet. 2015;47:1187–1193. doi: 10.1038/ng.3389. [DOI] [PubMed] [Google Scholar]
- 59.Zeltner N, Kohlbrenner E, Clément N, Weber T, Linden RM. Near-perfect infectivity of wild-type AAV as benchmark for infectivity of recombinant AAV vectors. Gene Ther. 2010;17:872–879. doi: 10.1038/gt.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morita E, Arii J, Christensen D, Votteler J, Sundquist WI. Attenuated protein expression vectors for use in siRNA rescue experiments. Biotechniques. 2012;0:1–5. doi: 10.2144/000113909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grimm D, Kern A, Rittner K, Kleinschmidt JA. Novel tools for production and purification of recombinant adenoassociated virus vectors. Hum Gene Ther. 1998;9:2745–2760. doi: 10.1089/hum.1998.9.18-2745. [DOI] [PubMed] [Google Scholar]
- 62.Hörer M, et al. Mutational analysis of adeno-associated virus Rep protein-mediated inhibition of heterologous and homologous promoters. J Virol. 1995;69:5485–5496. doi: 10.1128/jvi.69.9.5485-5496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.