Significance
In part due to concerns over greenhouse gas levels, interest in enzymes that fix carbon dioxide is considerable. The 2-oxoacid:ferredoxin oxidoreductases are essential enzymes in three of six of the known pathways of biological CO2 fixation. Only one member of this enzyme superfamily has been extensively characterized by spectroscopy, kinetic analyses, and computation, and that is the pyruvate:ferredoxin oxidoreductase (PFOR) from the model acetogen Moorella thermoactica. Remarkably, electron transfer between cofactors in PFOR is gated by coenzyme A (CoA) such that CoA binding accelerates electron transfer from 4 min to 1 ms. Here we present the structure of this well-characterized PFOR and reveal the binding mode of CoA with the accompanying domain movements and conformational rearrangements.
Keywords: pyruvate:ferredoxin oxidoreductase, thiamine pyrophosphate, coenzyme A, gated electron transfer, carbon dioxide fixation
Abstract
Pyruvate:ferredoxin oxidoreductase (PFOR) is a microbial enzyme that uses thiamine pyrophosphate (TPP), three [4Fe-4S] clusters, and coenzyme A (CoA) in the reversible oxidation of pyruvate to generate acetyl-CoA and carbon dioxide. The two electrons that are generated as a result of pyruvate decarboxylation are used in the reduction of low potential ferredoxins, which provide reducing equivalents for central metabolism, including the Wood–Ljungdahl pathway. PFOR is a member of the 2-oxoacid:ferredoxin oxidoreductase (OFOR) superfamily, which plays major roles in both microbial redox reactions and carbon dioxide fixation. Here, we present a set of crystallographic snapshots of the best-studied member of this superfamily, the PFOR from Moorella thermoacetica (MtPFOR). These snapshots include the native structure, those of lactyl-TPP and acetyl-TPP reaction intermediates, and the first of an OFOR with CoA bound. These structural data reveal the binding site of CoA as domain III, the function of which in OFORs was previously unknown, and establish sequence motifs for CoA binding in the OFOR superfamily. MtPFOR structures further show that domain III undergoes a conformational change upon CoA binding that seals off the active site and positions the thiolate of CoA directly adjacent to the TPP cofactor. These structural findings provide a molecular basis for the experimental observation that CoA binding accelerates catalysis by 105-fold.
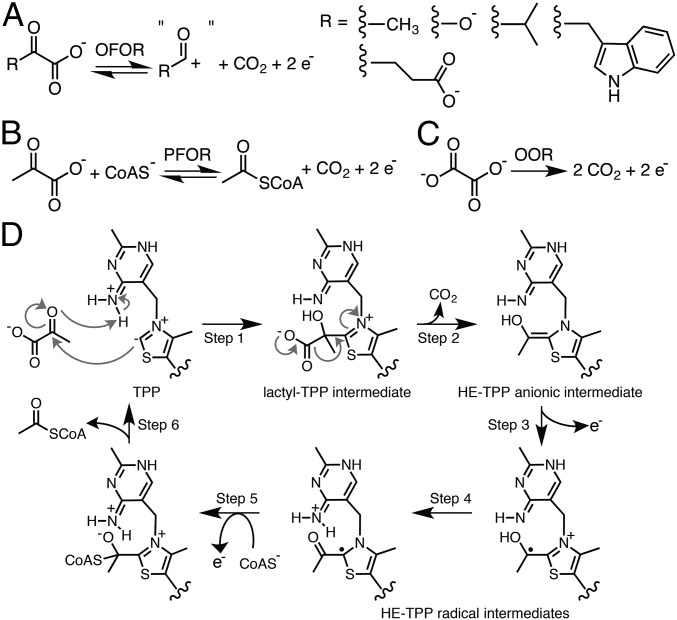
The 2-oxoacid:ferredoxin oxidoreductases (OFORs, EC 1.2.7.11) play key roles in both microbial redox chemistry and carbon dioxide fixation (1). Found in three of the six autotrophic pathways for CO2 fixation, members of this ancient enzyme family are ubiquitous in archaea and common in bacteria (1–4). OFOR reactions are typically reversible. In one direction, these enzymes reduce electron-transport proteins called ferredoxins using electrons derived from the oxidative decarboxylation of 2-oxoacids (Fig. 1A) (3, 4). These electrons are low potential (∼ −500 mV) and thus can be used to drive some of the more challenging of reactions performed by microbes, such as the reduction of CO2 and N2 (5, 6). In the reverse direction, OFORs use low-potential electrons to fix CO2, thereby synthesizing a variety of 2-oxoacids. This enzymatic chemistry requires multiple cofactors: thiamine pyrophosphate (TPP) acts as a potent nucleophile, forming adducts with the 2-oxoacid substrates, and enzyme-bound [4Fe-4S] clusters deliver electrons into or out of the active site. In all but one case (7), coenzyme A (CoA) is also required. Classified by the substrate specificities, OFORs include pyruvate:ferredoxin oxidoreductase (PFOR, Fig. 1B) (3), 2-oxoglutarate:ferredoxin oxidoreductase (OGOR) (8), 2-ketoisovalerate:ferredoxin oxidoreductase (VOR) (2, 9), indopyruvate:ferredoxin oxidoreductase (IOR) (10, 11), and oxalate oxidoreductase (OOR, Fig. 1C) (7, 12–14).
Fig. 1.
OFOR reactions and PFOR reaction scheme. (A) General scheme of OFOR reactions. (B) Reaction catalyzed by PFOR. (C) Reaction catalyzed by OOR. (D) Simplified mechanism of pyruvate oxidation by PFOR. See text for discussion.
Structural data are limited for this superfamily. The only available structures are of the PFOR from Desulfovibrio africanus (DaPFOR) (15–17), the OOR from Moorella thermoacetica (MtOOR) (12, 13), and two OFORs from Sulfolobus tokodaii (StOFOR), the physiological substrate of which has not been established (18). Very few family members have been extensively characterized. The most well-studied OFOR is the one described here, the PFOR from the model acetogen M. thermoactica (MtPFOR) (19–23). PFORs oxidize pyruvate to generate acetyl-CoA and carbon dioxide or, alternatively, catalyze the synthesis of pyruvate from acetyl-CoA and carbon dioxide (Fig. 1B). The oxidation of pyruvate by PFOR is required to connect glycolysis and the Wood–Ljungdahl pathway of reductive acetogenesis (24, 25) (SI Appendix, Fig. S1A), whereas the pyruvate synthesis reaction is an integral step in the reductive TCA cycle (26). Although newly discovered, MtOOR has quickly become one of the better-characterized family members (Fig. 1C). OORs oxidize oxalate to form two molecules of CO2, providing both low-potential electrons and CO2 for acetate synthesis by the Wood–Ljungdahl pathway (SI Appendix, Fig. S1B). The unusual feature of OOR is that it is the only known OFOR that does not require CoA.
Our current mechanistic understanding of enzymes in the OFOR superfamily comes from the biochemical, spectroscopic, and computational characterization of MtPFOR (19–23). Briefly, pyruvate oxidation begins with the deprotonation of the C2 of TPP by N4′ of TPP to form a carbanion (27), followed by a nucleophilic attack of the resulting C2 ylide on the 2-oxo carbon of pyruvate and protonation of the oxyanion by N4′ of TPP (27), forming a lactyl-TPP covalent intermediate (17, 28) (Fig. 1D, step 1). The TPP adduct then undergoes decarboxylation (Fig. 1D, step 2) and one-electron transfer (Fig. 1D, step 3) to one of the three enzyme-bound [4Fe-4S] clusters (23), forming a hydroxyethylidene-TPP (HE-TPP) radical intermediate (19, 29, 30).
The HE-TPP radical intermediate, which has been extensively studied by electron paramagnetic resonance (EPR) experiments in various PFORs (22, 29–31), is quite stable in the absence of CoA (half-life is ∼4 min) (21). CoA binding accelerates the rate of electron transfer from the HE-TPP radical intermediate into the [4Fe-4S] cluster electron transport chain by an impressive 105-fold (Fig. 1D, step 5), the molecular basis of which has been the topic of considerable interest and study (14, 21, 22, 31). Electron transfer from this TPP-bound intermediate through the internal electron transfer chain of [4Fe-4S] clusters to a ferredoxin, bound to the surface of PFOR, results in the formation of an acetyl-TPP intermediate. The reaction cycle is completed following acetylation of CoA and product release (Fig. 1D, step 6). Although all OFORs, except for OOR, require CoA for activity, no structural data on CoA binding have been available until now. Here we present the structure of an OFOR with CoA bound, the structure of the well-characterized PFOR from M. thermoacetica. We further consider the mechanistic implications of these structural data for both CoA-dependent OFORs and CoA-independent OORs.
Results
Structure of MtPFOR Provides a View of a Canonical OFOR.
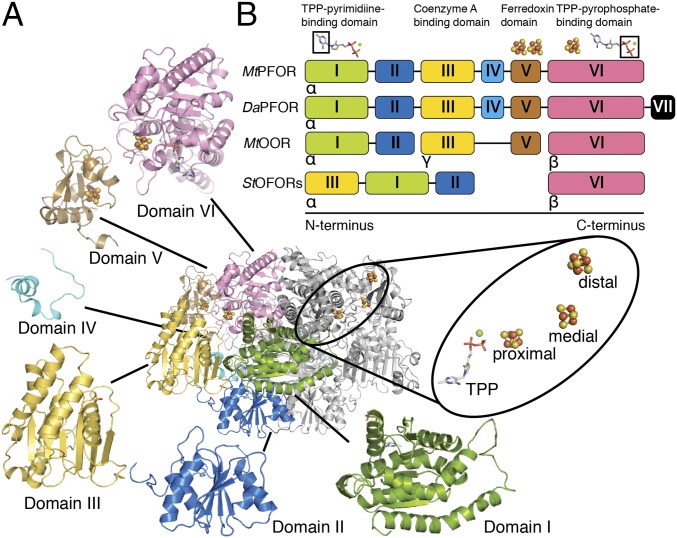
MtPFOR was crystallized anaerobically and its structure was determined to 2.60-Å resolution (SI Appendix, Tables S1 and S2). Three MtPFOR homodimers are found in the asymmetric unit (ASU). Each MtPFOR monomer is composed of six domains (I–VI, Fig. 2A), three [4Fe-4S] clusters, and one TPP molecule (Fig. 2A, Inset). Domains I and VI are found in all TPP-dependent enzymes; domain II is also found in various TPP-dependent enzyme families, whereas domain III is present only in OFORs (32). The overall fold (Fig. 2A) and relative positions of cofactors (Fig. 2A, Inset) in MtPFOR are conserved among the structurally characterized OFORs (SI Appendix, Figs. S2 and S3), despite their differences in domain arrangements and oligomer states (Fig. 2B).
Fig. 2.
The overall structure of MtPFOR and domain arrangements of structurally characterized OFORs. (A) Ribbon figure of the MtPFOR α2 homodimer. The monomeric subunit on the Left is colored by domain (see B for color); the monomeric subunit on the Right is colored gray. Each monomeric subunit is composed of six domains. Domain I (green, residues 2–256) binds the pyrimidine moiety of TPP and forms part of the dimer interface. Domain II (blue, residues 257–414), which is known as a transketolase C-terminal (TKC) domain, also forms part of dimer interface. Domain III (yellow, residues 415–630) is a CoA-binding domain, the function of which has been unclear until this work. Domain IV (cyan, residues 631–665) is a structured linker that connects domain III and domain V. Domain V (brown, residues 666–782) adopts a ferredoxin fold that accommodates the [4Fe-4S] clusters that are distal and medial to the TPP molecule. Domain VI (pink, residues 783–1171) binds the pyrophosphate moiety of TPP and the proximal [4Fe-4S] cluster. The redox active cofactors are shown in the oval Inset. (B) The domain arrangements of structurally characterized OFORs. MtPFOR and DaPFOR are α2 homodimers with six and seven domains, respectively. MtOOR is a (αγβ)2 dimer of heterotrimers in which each trimer forms a catalytic unit composed of five domains. Both structurally characterized StOFORs are (αβ)2 dimers of heterodimers in which each dimer forms a catalytic unit composed of four domains. Domains I–III of StOFORs are arranged as domain III–I–II from N terminus to C terminus, which is different from the other three cases in which domains are arranged in numerical order. The functions of the domains are indicated.
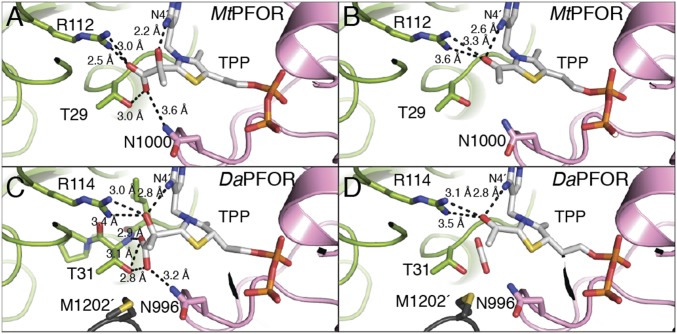
The active site of MtPFOR shares high similarity with that of DaPFOR (overall sequence identity is 64%). Three pyruvate-binding residues discovered in DaPFOR—Thr31, Arg114, and Asn996 (Fig. 3C and SI Appendix, Fig. S4B)—are also present in MtPFOR (Fig. 3A and SI Appendix, Fig. S4A). The active sites of PFORs, MtOOR, and StOFORs are also similar but are tailored for different substrates (SI Appendix, Fig. S4). Structural data show that MtOOR uses two positively charged residues, Arg31α and Arg109α (SI Appendix, Fig. S4C), to bind oxalate, a dicarboxylic acid, whereas DaPFOR uses Thr31 and Arg114 to bind pyruvate, a monocarboxylic acid. The physiological substrate(s) for StOFORs are not known, but the enzymes are active on 2-oxoglutarate, a dicarboxylic acid like oxalate. Although no substrate-bound structure is available, it is interesting to note that Thr29/31 in PFOR (Thr257α in StOFOR2) and Arg112/114 in PFOR (Arg344α in StOFOR2) are conserved, but like MtOOR, there is an additional positively charged residue in StOFOR’s active site (Lys49β) (SI Appendix, Fig. S4D). Thus, the active site of StOFOR appears to be designed to bind a dicarboxylic acid but one of a different size than oxalate.
Fig. 3.
Active-site comparisons for intermediates in MtPFOR and DaPFOR. (A) Lactyl-TPP intermediate in MtPFOR. The thiazolium ring is planar, and the carbon–carbon bond between C2 of TPP and the lactyl group is 1.6 Å as expected for a carbon–carbon bond. (B) Acetyl-TPP adduct in MtPFOR. The thiazolium ring is planar, and the carbon–carbon bond between C2 of TPP and the acetyl group is 1.5 Å as expected for a carbon–carbon bond. (C) Lactyl-TPP intermediate captured in the structure of DaPFOR [PDB ID code 2C3P (17)] has a nonplanar aromatic thiazolium ring. The carbon–carbon bond between C2 of TPP and the lactyl group is 1.9 Å. (D) Putative HE-TPP intermediate in the structure of DaPFOR [PDB ID code 2C3Y (17)] has a nonplanar aromatic thiazolium ring. The carbon–carbon bond between C2 of TPP and the acetyl group is long for a carbon–carbon bond at 1.8 Å.
OFORs display differences in active-site solvent accessibility (SI Appendix, Fig. S5). MtPFOR and StOFORs have open active sites, whereas those of DaPFOR and MtOOR are sequestered from solvent, at least in some of the states that have been observed crystallographically. In the case of DaPFOR, a domain that is unique to OFORs from the Desulfovibrio genus (domain VII) blocks solvent access to the neighboring subunit’s active site (15). For MtOOR, a domain III loop called the “plug loop,” which is unique to OORs, seals the active site, and Glu154γ from this loop hydrogen-bonds to active-site residue Arg31α (12, 13).
The Structures of MtPFOR with Lactyl-TPP Intermediate and Acetyl-TPP Adduct.
To obtain structures of MtPFOR with substrates and/or intermediates bound, we carried out crystal-soaking experiments. In particular, crystals were soaked in pyruvate for 15 min and 12 h, and structures were determined to 3.0-Å and 3.2-Å resolution, respectively (SI Appendix, Tables S1 and S2). These soaking experiments did not change the overall structure of MtPFOR, with 902 Cα of one monomer displaying rmsds of 0.27–0.37 Å to the monomers of the native structure. For both the 15-min and 12-h soaks, electron density appeared near all C2 carbons of the TPP thiazole rings. The shorter (15-min) soak generated omit density that is consistent with a lactyl-TPP intermediate state (SI Appendix, Fig. S6A), whereas the longer soak (12 h) yielded omit density that was consistent with an acetyl-TPP intermediate state (SI Appendix, Fig. S6B). The proposal that these density features are due to pyruvate reacting with TPP is supported by the experimental observation that MtPFOR that has been crystallized retains its ability to oxidize pyruvate (SI Appendix, Fig. S7).
As described above, a lactyl-TPP intermediate is formed upon nucleophilic attack of the C2 carbon of TPP on the carbonyl carbon of pyruvate (Fig. 1D). In the active sites with lactyl-TPP bound, the carboxylate moiety of lactyl-TPP forms hydrogen bonds with the side chains of residues Thr29, Arg112, and Asn1000 from domains I and VI. The hydroxyl moiety of lactyl-TPP forms a hydrogen bond with N4′ of TPP (Fig. 3A), consistent with findings of others related to the importance of the N4′ in proton transfer reactions of OFORs (27). Although the pyruvate-binding residues in MtPFOR and DaPFOR are conserved, namely Thr29/31, Arg112/114, and Asn1000/996, the distances between the conserved residues and the lactyl-TPP intermediate are quite different (Fig. 3 A and C). The source of the difference appears to be in the refinement of the adduct structures and not in the positioning of the protein side chains. In particular, the aromatic thiazolium ring of TPP in DaPFOR was refined as nonplanar, and the TPP-lactyl bond was long (1.9 Å) as was the bond between the two carbons of the lactyl moiety (1.8 Å). In contrast, these distances are both 1.6 Å in this structure of MtPFOR; furthermore, the thiazolium ring of TPP in MtPFOR is planar. The net result is that the lactyl moiety in MtPFOR is closer to the N4′ of TPP and Arg112/114 and farther from Thr29/31 and Asn1000/996 than it is in DaPFOR (Fig. 3 A and C). Notably, the long bond length and nonplanarity of the intermediate as refined in the DaPFOR structure is inconsistent with EPR spectroscopic and computational studies on the HE-TPP radical in MtPFOR (22, 31). In contrast, the geometry observed here is supported by these experimental and computational studies.
The electron density in the MtPFOR structure obtained following a 12-h soak of crystals with pyruvate suggests that decarboxylation of the TPP adduct has occurred (SI Appendix, Fig. S6B). Although the crystallographic data cannot distinguish the redox or protonation state of the TPP adduct observed in this structure, we assume that the HE-TPP radical state would have decayed during the 12-h soaking time of the experiment at room temperature [at 25 °C the t1/2 of decay of the radical in the absence of CoA is 4 min (21)] and thus refer to this adduct as acetyl-TPP. The acetyl group of the acetyl-TPP adduct forms a hydrogen bond with N4′ of TPP and is close to Arg112 (Fig. 3B). Similar to the lactyl-TPP intermediate in MtPFOR, the aromatic thiazolium ring is planar, and the carbon–carbon bond for the acetyl-TPP adduct is refined to 1.5 Å. In contrast, a HE-TPP radical intermediate was proposed to have been captured in a structure of DaPFOR (16). This adduct as refined contained a nonplanar thiazolium ring and a long (1.8 Å) carbon–carbon bond between the HE and the C2 of TPP (Fig. 3D and SI Appendix, Fig. S6D). Again, the structure observed here is a better fit to the experimental spectroscopic data and computational results (22, 31).
The Structure of MtPFOR with CoA Bound Reveals the Enigmatic CoA-Binding Site.
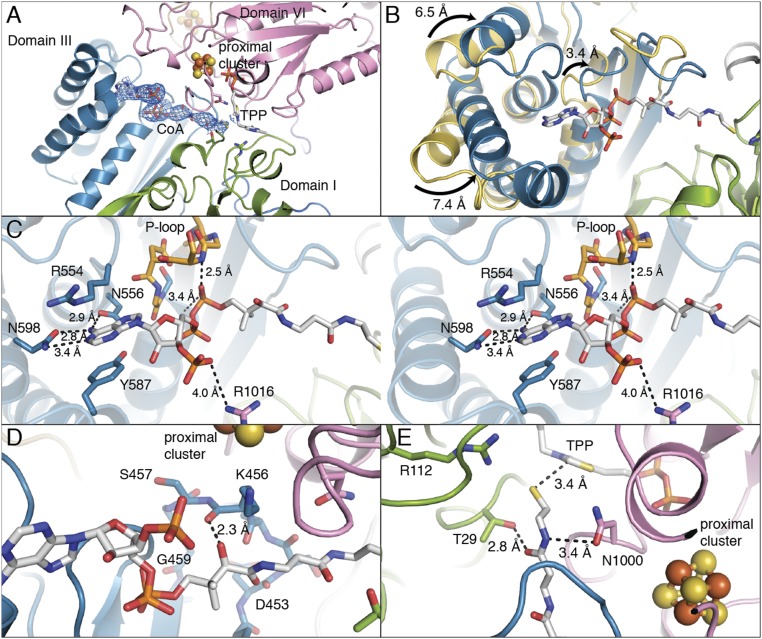
Since soaking crystals with CoA causes crystals to dissolve, we solved the CoA-bound MtPFOR structure to 3.3-Å resolution through cocrystallization (SI Appendix, Tables S1 and S2). Cocrystallization results in a reduction of the length of the c-axis of the crystal by approximately half and a reduction in the number of molecules in the ASU from six to three. Two of the three MtPFOR monomers form one homodimer in the ASU and the other MtPFOR monomer forms a dimer by crystallographic symmetry. The overall architecture of MtPFOR with CoA bound is similar to that in the native structure of MtPFOR: each monomer has a Cα rmsd of 0.25–0.37 Å. The 3′-phosophoadenosine diphosphate moiety of CoA is anchored by residues of domain III as the pantothenate reaches across this domain to position the cysteamine moiety of CoA into the active site, which is formed between domains I and VI (Fig. 4A). Although CoA is bound in all monomers, the best density is observed in chain C; thus, only CoA–protein interactions in chain C will be described. When CoA binds, domain III moves into an alternate conformation by swinging up to 7.4 Å toward the active site relative to the native structure (Fig. 4B), shortening the distance between TPP and CoA-binding residues 421GLGSDG426 (described below) from 21.6 to 17.7 Å. Additionally, these conformational changes in domain III appear to be required for CoA binding as residues are not positioned to bind CoA in the CoA-free structure (see below).
Fig. 4.
Domain III of MtPFOR is a CoA-binding domain. (A) The pyrophosphate and adenine end of CoA (in sticks) binds to domain III (teal), and the CoA molecule extends into the active site, which is composed of domains I (green) and VI (pink). The 2Fo − Fc composite-omit electron density for CoA is shown in blue mesh that is contoured to 1.0 σ. (B) Structural superposition of domain III from MtPFOR with (teal) and without (yellow) CoA bound. The overall Cα rmsd of domain III (residues 415–630) is 4.9 Å with parts of the domain moving up to 7.4 Å. (C) Stereo views of domain III of MtPFOR with CoA bound illustrate residues anchoring adenosine moiety and pyrophosphate of CoA. Domain III of MtPFOR with CoA bound is colored teal; CoA and CoA binding residues of MtPFOR are shown as sticks. Residues 421–426 form P-loop (orange), which binds the pyrophosphate moiety of CoA by the backbone of P-loop and the dipole of the subsequent alpha helix. Asn556 and Asn598 form hydrogen bonds with adenine of CoA. Arg554 and Tyr587 bind the adenine by cation–pi interaction and pi–pi stacking interactions. Arg1016 forms a charge–charge interaction with 3′-phospho group of CoA. (D) The carboxyl group on the pantothenate moiety of CoA forms a hydrogen bond with the carbonyl backbone of Lys456. (E) Thr29 and Asn1000, which form hydrogen bonds with lactyl-TPP intermediates (Fig. 3A), also form hydrogen bonds with the cysteamine moiety of CoA.
Residues 421GLGSDG426 of domain III, which form the P-loop (phosphate-binding loop), move 3.4 Å to engage in both hydrogen bonding and electrostatic interactions with the pyrophosphate moiety of CoA (Fig. 4 B and C). The 3′-phospho group of CoA is further stabilized by charge–charge interactions with Arg1016 from domain VI (Fig. 4C). Interestingly, the KD for dephospho-CoA is increased by 102-fold compared with CoA (21), suggesting that the charge–charge interaction between Arg1016 and the 3′-phospho group of CoA is important. It is the only interaction made by a nondomain III residue to the pyrophosphate moiety of CoA. Without the conformational change of domain III that accompanies CoA binding, Arg1016 would be too far to interact with the 3′-phospho group (∼6.3 Å), suggesting that this interaction could aid in more than just CoA binding; it might also stabilize the alternative conformation of domain III. Completing the interactions from the protein to the nucleotide end of CoA, the adenine moiety hydrogen bonds with the side chains of Asn556 and Asn598 and appears to make cation–pi and pi–pi interactions with Arg554 and Tyr587. Notably, the majority of these CoA-binding residues adopt different positions in the native CoA-free structure (SI Appendix, Fig. S8).
The pantothenate moiety of CoA extends past residues 453–459 of domain III, forming one hydrogen bond to Lys456, a lysine residue that is directly below the proximal [4Fe-4S] cluster (Fig. 4D). At the business end of the CoA, the cysteamine moiety is positioned to form hydrogen bonds with active-site residues Thr29 and Asn1000 (Fig. 4E). These residues were also found to bind the carboxyl group of the lactyl-TPP intermediate (Fig. 3A).
As a result of CoA binding, the MtPFOR active site is no longer solvent-exposed (SI Appendix, Fig. S9). The swinging motion of domain III partially seals the active site, and CoA fills the remaining space. Notably, domain VII of DaPFOR is located where CoA is expected to bind and where domain III would be expected to move upon CoA binding, based on the MtPFOR structure (SI Appendix, Fig. S10). This observation could explain why it was not possible to obtain a CoA-bound crystal structure of the DaPFOR enzyme. It also could explain the observation that a variant of DaPFOR without domain VII displays improved activity compared with the full-length DaPFOR (33).
Sequence Alignment Supports Domain III as a Universal CoA-Binding Module for CoA-Dependent OFORs.
The alignment of domain III sequences from 43 different CoA-dependent OFORs, which include PFOR, OGOR, VOR, and IOR, reveal three highly conserved motifs (SI Appendix, Tables S3 and S4). The amino acid sequences near the conserved site of structurally characterized OFORs are shown in Fig. 5. The first motif corresponds to residues 421–426, which form the P-loop in MtPFOR. In this region, Gly426 is absolutely conserved, whereas residues 421, 423, and 424 are conserved glycine or small amino acids (alanine and serine). The second and the third motifs are composed of residues that surround Asn556 and Asn598 in MtPFOR, respectively. Asn556, the side chain amides of which form hydrogen bonds with the adenine moiety of CoA, is found in a Rx1–4N motif, and Asn598, which also forms hydrogen bonds with the adenine moiety of CoA with its side chain, is found in a F/Yx7–11N motif. R and F/Y in two motifs correspond to Arg554 and Tyr587 in MtPFOR that form cation–pi and pi–pi interactions with adenine and are found in 58 and 72% of aligned sequences.
Fig. 5.
Sequence alignment of CoA-dependent OFORs shows that CoA-binding motifs are highly conserved across different OFORs. The conserved motifs of structurally characterized CoA-dependent OFORs are shown. Conserved glycine sites of P-loop are highlighted in yellow, and the conserved adenine-binding asparagine sites are highlighted in blue. Positively charged arginine residues that may form a cation–pi interaction with adenine of CoA are highlighted in magenta. Aromatic residues that may form pi–pi interactions with adenine of CoA are highlighted in green. The complete sequence alignment and UniProt IDs are shown in SI Appendix, Tables S3 and S4.
Discussion
The structures presented in this work provide snapshots of CoA-dependent OFOR chemistry at different stages of the reaction cycle. They facilitate the identification of consensus motifs for CoA binding in this superfamily and allow for a consideration of the molecular basis by which CoA binding accelerates turnover. For MtPFOR, the HE-TPP radical intermediate decay kinetics have been determined both in the absence and the presence of CoA or CoA analogs (21). Binding of CoA increases the rate of the electron transfer from the HE-TPP radical into the enzyme-bound [4Fe-4S] clusters by 105-fold (21). There are a number of possible explanations for this dramatic rate acceleration, including the following: that CoA binding releases bound water molecules, leading to a favorable entropic effect; that CoA binding alters the redox potentials of one or more of the [4Fe-4S] clusters, favoring cluster reduction; and/or that CoA binding promotes electron transfer to the [4Fe-4S] clusters through charge repulsion between the negatively charged CoA thiolate and the HE-TPP radical (21, 31).
The entropic argument was largely disfavored when it was discovered that desulfo-CoA does not increase the rate of HE-TPP radical decay, despite the fact that the dissociation constants for desulfo-CoA and CoA are only 10-fold different (21). Thus, desulfo-CoA can bind, albeit more weakly, but cannot accelerate electron transfer. In fact, the rate with desulfo-CoA is slower than the baseline rate in the absence of CoA (21). Since one would expect a similar release of water molecules for CoA binding and desulfo-CoA binding, entropic factors seemed less likely to be the determining factor. Instead, these observations emphasize the importance of the thiol group of CoA.
Our structures are consistent with the experimental observation that desulfo-CoA can bind to MtPFOR as only a small percentage of the CoA–protein interactions involve the thiol moiety (Fig. 4). The modestly increased dissociation constant for desulfo-CoA may be due to the loss of a favorable electrostatic interaction between the CoA thiolate and positively charged active-site residue Arg112 (Fig. 4E). On the other hand, our structural data do not favor the idea that CoA binding accelerates electron transfer through altering the redox potential of a [4Fe-4S] cluster. CoA does make contact with residues that are adjacent to the proximal [4Fe-4S] cluster. Lys456, for example, is one of the closer positively charged residues to this cluster, and the Lys456 backbone carbonyl hydrogen bonds to a carbonyl of CoA (Fig. 4D). However, there is no reason to believe that desulfo-CoA would interact any differently with Lys456, and certainly not differently enough to explain a 106 difference of electron transfer rate. Additionally, the solvent accessibility of the proximal cluster does not change between CoA-free (65 Å2) and CoA-bound states (61 Å2). Thus, these structural data cannot rule out that a change in redox potential is the cause of electron transfer rate enhancement, but they do not provide support either.
The third possibility—that the rate of acceleration of HE-TPP radical oxidation is due to a negatively charged CoA thiolate coming into proximity of the TPP adduct—is supported by the structure. We find that CoA binding does indeed place the thiol moiety directly adjacent to TPP; the distance between the S of CoA and C2 of TPP is a very close 3.4 Å. Such a short distance should lead to significant charge–charge repulsion. Additionally, the solvent accessibility of the HE-TPP radical would be expected to decrease upon CoA binding. Without CoA, the active site is open and solvent accessible, and with CoA bound, the active site is closed. In this closed active site, the negatively charged thiolate of CoA is sequestered, a condition that should enhance the effect of charge–charge repulsion, favoring oxidization of a TPP intermediate. For these reasons, we favor a mechanism in which a thiolate form of CoA drives HE-TPP radical oxidation through charge–charge repulsion, which would be followed by, or simultaneous with, nucleophilic attack on the TPP adduct, forming acetyl-CoA. EPR spectroscopy and computational studies additionally support a thiolate instead of a thiol radical as the form of the CoA that reacts with HE-TPP (21, 31). Also, facile removal of the acetyl group from TPP by CoA should shift the equilibrium toward product formation (14). Kinetics of other OFORs need to be established to determine whether this rate acceleration due to CoA binding is a common feature. Sequence alignments do suggest that CoA-binding interactions will be conserved. Taken together with the active-site similarities, it seems likely that CoA-dependent OFORs will employ molecular mechanisms comparable to that of MtPFOR even if the rate of acceleration upon CoA binding is not as dramatic in all cases.
It is also interesting to compare this CoA-dependent mechanism with the putative mechanism for the one known CoA-independent enzyme, OOR. We have recently proposed that MtOOR operates via a bait-and-switch mechanism in which positively charged residues in the active site (the “bait”) attract and activate the dicarboxylic acid substrate oxalate for nucleophilic attack by the TPP (13). Oxalate-TPP adduct formation would be followed by the “switch,” in which a loop in the active site (“switch loop”) flips 180° to put a negatively charged Asp residue (Asp116α) directly toward the TPP-substrate adduct to drive decarboxylation and electron transfer (SI Appendix, Fig. S11). In this way, Asp116α and the switch loop act in a manner similar to the role that we propose above for CoA, driving electron transfer through charge repulsion.
The similarities between MtPFOR and MtOOR do not end there. Like MtPFOR, MtOOR has a domain III that undergoes a conformational change (Fig. 6) (13). In PFOR, domain III rearranges to bind CoA and swings into position the thiolate of CoA directly adjacent to the HE-TPP radical intermediate state. Presumably, domain III will return to an “out” position as acetyl-CoA is released. In OOR, domain III swings out to facilitate decarboxylation of oxalate and release of CO2 from the active site, and a loop called the “plug loop” becomes disordered, opening up the active site. In one crystal structure of MtOOR, domain III movement is blocked by lattice contacts, and an oxalate-bound form of TPP is trapped. Thus, domain III movement appears key to both reactions, but the domain direction and timing of the movement are not the same.
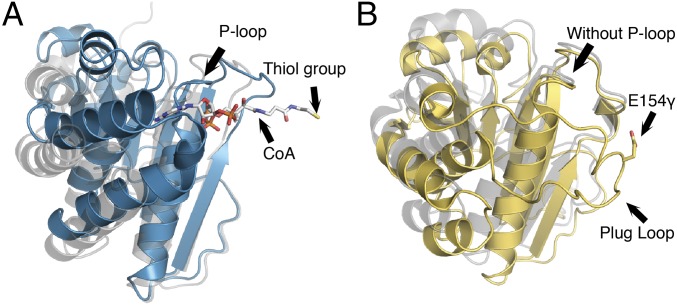
Fig. 6.
Domain III comparison for MtPFOR and MtOOR. (A) Domain III of MtPFOR with CoA bound (teal) and without CoA bound (gray). In PFOR, domain III anchors pyrophosphate and adenine of CoA and swings toward the active site, initiating the second electron transfer step in the PFOR reaction. (B) Domain III of MtOOR swung-in toward the active site [yellow; PDB ID code 5C4I (12)] and swung-out away from the active site [gray; PDB ID code 5EXE (13)]. In OOR, domain III contains a plug loop housing Glu154γ. The plug-loop movement facilitates oxalate decarboxylation. 19γARGVVM24γ in OOR spatially corresponds to the location of the P-loop in PFOR, but the motif adapts a different geometry that does not have sufficient space to bind a phosphate.
The overall structure of each domain III is very similar (Fig. 6), but domain III of MtOOR does not have any of the CoA-binding motifs of MtPFOR, and MtPFOR does not have enough residues to form a loop similar to OOR’s plug loop. Thus, each domain III displays modifications required for their specialized functions. We expect that MtOOR evolved from a CoA-dependent OFOR. PFOR is likely an ancient enzyme, responsible for funneling into cellular metabolism the acetyl-CoA produced through the Wood–Ljungdahl pathway from CO2, a carbon source thought to be abundant in the early atmosphere. OOR likely evolved later as a means for microbes to utilize oxalate as a carbon source. The CoA-binding motif would have been lost over time, but the domain movement feature was kept and repurposed. OOR seems to be a family outlier, the only CoA-independent OFOR, but the similarities in mechanisms are striking.
In summary, this work has provided a view of CoA binding in the OFOR family, a family of ancient microbial enzymes that are key to both redox chemistry and one-carbon chemistry. These data on the prototypic OFOR show that CoA binds to domain III and that CoA binding and domain III movement drive electron transfer from the TPP, which is coordinated by domains I and IV, into the domain V [4Fe-4S] clusters. Ultimately, external ferredoxins accept these low-potential electrons, which can be used for cellular processes such as nitrogen fixation (6) and sulfur reduction (34). Although here we have described the PFOR chemistry in terms of pyruvate oxidation, it is important to remember that the reaction is reversible, providing a means to build longer chain-carbon molecules through the reductive addition of CO2. OFORs are in fact present in three of the six known autotrophic pathways of CO2 fixation and thus contribute in a substantial fashion to global one-carbon metabolism. We still do not know if all OFORs are fully reversible. Reversibility of OOR has not been demonstrated, for example, nor do we fully understand the conditions that affect directionally for other OFORs. However, these crystallographic snapshots greatly enhance a molecular understanding of OFORs, which will facilitate further characterization and engineering efforts of CO2 fixation pathways.
Materials and Methods
M. thermoacetica growth, enzyme isolation, and protein quantification were performed as described previously (19). MtPFOR was crystallized by the sitting drop crystallization method in an anoxic atmosphere. The structure of MtPFOR was solved using the structure of DaPFOR [Protein Data Bank (PDB) ID code 2C42 (17)] as a molecular replacement model. Detailed protocols can be found in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Marcus I. Gibson and Dr. Steven O. Mansoorabadi for insightful discussion. This work was supported in part by National Institutes of Health (NIH) Grants GM069857 (to C.L.D.) and GM39451 (to S.W.R.). C.L.D. is a Howard Hughes Medical Institute Investigator. This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences of the NIH (Grant P41 GM103403). The Pilatus 6M detector on the 24-ID-C beamline is funded by NIH Office of Research Infrastructure Programs High-End Instrumentation Grant S10 RR029205. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes: Native MtPFOR, 6CIN; MtPFOR with lactyl-TPP bound, 6CIO; MtPFOR with acetyl-TPP bound, 6CIP; and MtPFOR with coenzyme A bound, 6CIQ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722329115/-/DCSupplemental.
References
- 1.Fuchs G. Alternative pathways of carbon dioxide fixation: Insights into the early evolution of life? Annu Rev Microbiol. 2011;65:631–658. doi: 10.1146/annurev-micro-090110-102801. [DOI] [PubMed] [Google Scholar]
- 2.Kletzin A, Adams MW. Molecular and phylogenetic characterization of pyruvate and 2-ketoisovalerate ferredoxin oxidoreductases from Pyrococcus furiosus and pyruvate ferredoxin oxidoreductase from Thermotoga maritima. J Bacteriol. 1996;178:248–257. doi: 10.1128/jb.178.1.248-257.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ragsdale SW. Pyruvate ferredoxin oxidoreductase and its radical intermediate. Chem Rev. 2003;103:2333–2346. doi: 10.1021/cr020423e. [DOI] [PubMed] [Google Scholar]
- 4.Gibson MI, Chen PY-T, Drennan CL. A structural phylogeny for understanding 2-oxoacid oxidoreductase function. Curr Opin Struct Biol. 2016;41:54–61. doi: 10.1016/j.sbi.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ragsdale SW, Pierce E. Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation. Biochim Biophys Acta. 2008;1784:1873–1898. doi: 10.1016/j.bbapap.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wahl RC, Orme-Johnson WH. Clostridial pyruvate oxidoreductase and the pyruvate-oxidizing enzyme specific to nitrogen fixation in Klebsiella pneumoniae are similar enzymes. J Biol Chem. 1987;262:10489–10496. [PubMed] [Google Scholar]
- 7.Pierce E, Becker DF, Ragsdale SW. Identification and characterization of oxalate oxidoreductase, a novel thiamine pyrophosphate-dependent 2-oxoacid oxidoreductase that enables anaerobic growth on oxalate. J Biol Chem. 2010;285:40515–40524. doi: 10.1074/jbc.M110.155739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mai X, Adams MW. Characterization of a fourth type of 2-keto acid-oxidizing enzyme from a hyperthermophilic archaeon: 2-ketoglutarate ferredoxin oxidoreductase from Thermococcus litoralis. J Bacteriol. 1996;178:5890–5896. doi: 10.1128/jb.178.20.5890-5896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heider J, Mai X, Adams MW. Characterization of 2-ketoisovalerate ferredoxin oxidoreductase, a new and reversible coenzyme A-dependent enzyme involved in peptide fermentation by hyperthermophilic archaea. J Bacteriol. 1996;178:780–787. doi: 10.1128/jb.178.3.780-787.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tersteegen A, Linder D, Thauer RK, Hedderich R. Structures and functions of four anabolic 2-oxoacid oxidoreductases in Methanobacterium thermoautotrophicum. Eur J Biochem. 1997;244:862–868. doi: 10.1111/j.1432-1033.1997.00862.x. [DOI] [PubMed] [Google Scholar]
- 11.Mai X, Adams MW. Indolepyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. A new enzyme involved in peptide fermentation. J Biol Chem. 1994;269:16726–16732. [PubMed] [Google Scholar]
- 12.Gibson MI, et al. The structure of an oxalate oxidoreductase provides insight into microbial 2-oxoacid metabolism. Biochemistry. 2015;54:4112–4120. doi: 10.1021/acs.biochem.5b00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson MI, et al. One-carbon chemistry of oxalate oxidoreductase captured by X-ray crystallography. Proc Natl Acad Sci USA. 2016;113:320–325. doi: 10.1073/pnas.1518537113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pierce E, Mansoorabadi SO, Can M, Reed GH, Ragsdale SW. Properties of intermediates in the catalytic cycle of oxalate oxidoreductase and its suicide inactivation by pyruvate. Biochemistry. 2017;56:2824–2835. doi: 10.1021/acs.biochem.7b00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chabrière E, et al. Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate. Nat Struct Biol. 1999;6:182–190. doi: 10.1038/5870. [DOI] [PubMed] [Google Scholar]
- 16.Chabrière E, et al. Crystal structure of the free radical intermediate of pyruvate:ferredoxin oxidoreductase. Science. 2001;294:2559–2563. doi: 10.1126/science.1066198. [DOI] [PubMed] [Google Scholar]
- 17.Cavazza C, et al. Flexibility of thiamine diphosphate revealed by kinetic crystallographic studies of the reaction of pyruvate-ferredoxin oxidoreductase with pyruvate. Structure. 2006;14:217–224. doi: 10.1016/j.str.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 18.Yan Z, Maruyama A, Arakawa T, Fushinobu S, Wakagi T. Crystal structures of archaeal 2-oxoacid:ferredoxin oxidoreductases from Sulfolobus tokodaii. Sci Rep. 2016;6:33061. doi: 10.1038/srep33061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menon S, Ragsdale SW. Mechanism of the Clostridium thermoaceticum pyruvate:ferredoxin oxidoreductase: Evidence for the common catalytic intermediacy of the hydroxyethylthiamine pyropyrosphate radical. Biochemistry. 1997;36:8484–8494. doi: 10.1021/bi970403k. [DOI] [PubMed] [Google Scholar]
- 20.Furdui C, Ragsdale SW. The role of pyruvate ferredoxin oxidoreductase in pyruvate synthesis during autotrophic growth by the Wood-Ljungdahl pathway. J Biol Chem. 2000;275:28494–28499. doi: 10.1074/jbc.M003291200. [DOI] [PubMed] [Google Scholar]
- 21.Furdui C, Ragsdale SW. The roles of coenzyme A in the pyruvate:ferredoxin oxidoreductase reaction mechanism: Rate enhancement of electron transfer from a radical intermediate to an iron-sulfur cluster. Biochemistry. 2002;41:9921–9937. doi: 10.1021/bi0257641. [DOI] [PubMed] [Google Scholar]
- 22.Mansoorabadi SO, et al. EPR spectroscopic and computational characterization of the hydroxyethylidene-thiamine pyrophosphate radical intermediate of pyruvate:ferredoxin oxidoreductase. Biochemistry. 2006;45:7122–7131. doi: 10.1021/bi0602516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Astashkin AV, Seravalli J, Mansoorabadi SO, Reed GH, Ragsdale SW. Pulsed electron paramagnetic resonance experiments identify the paramagnetic intermediates in the pyruvate ferredoxin oxidoreductase catalytic cycle. J Am Chem Soc. 2006;128:3888–3889. doi: 10.1021/ja0585275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drake HL, Hu SI, Wood HG. Purification of five components from Clostridium thermoaceticum which catalyze synthesis of acetate from pyruvate and methyltetrahydrofolate. Properties of phosphotransacetylase. J Biol Chem. 1981;256:11137–11144. [PubMed] [Google Scholar]
- 25.Menon S, Ragsdale SW. Evidence that carbon monoxide is an obligatory intermediate in anaerobic acetyl-CoA synthesis. Biochemistry. 1996;35:12119–12125. doi: 10.1021/bi961014d. [DOI] [PubMed] [Google Scholar]
- 26.Buchanan BB, Arnon DI. A reverse Krebs cycle in photosynthesis: Consensus at last. Photosynth Res. 1990;24:47–53. [PubMed] [Google Scholar]
- 27.Kern D, et al. How thiamine diphosphate is activated in enzymes. Science. 1997;275:67–70. doi: 10.1126/science.275.5296.67. [DOI] [PubMed] [Google Scholar]
- 28.Breslow R. On the mechanism of thiamine action. IV. 1 Evidence from studies on model systems. J Am Chem Soc. 1958;80:3719–3726. [Google Scholar]
- 29.Cammack R, Kerscher L, Oesterhelt D. A stable free radical intermediate in the reaction of 2-oxoacid: Ferredoxin oxidoreductases of Halobacterium halobium. FEBS Lett. 1980;118:271–273. [Google Scholar]
- 30.Kerscher L, Oesterhelt D. The catalytic mechanism of 2-oxoacid:ferredoxin oxidoreductases from Halobacterium halobium. One-electron transfer at two distinct steps of the catalytic cycle. Eur J Biochem. 1981;116:595–600. doi: 10.1111/j.1432-1033.1981.tb05377.x. [DOI] [PubMed] [Google Scholar]
- 31.Reed GH, Ragsdale SW, Mansoorabadi SO. Radical reactions of thiamin pyrophosphate in 2-oxoacid oxidoreductases. Biochim Biophys Acta. 2012;1824:1291–1298. doi: 10.1016/j.bbapap.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Costelloe SJ, Ward JM, Dalby PA. Evolutionary analysis of the TPP-dependent enzyme family. J Mol Evol. 2008;66:36–49. doi: 10.1007/s00239-007-9056-2. [DOI] [PubMed] [Google Scholar]
- 33.Pieulle L, Magro V, Hatchikian EC. Isolation and analysis of the gene encoding the pyruvate-ferredoxin oxidoreductase of Desulfovibrio africanus, production of the recombinant enzyme in Escherichia coli, and effect of carboxy-terminal deletions on its stability. J Bacteriol. 1997;179:5684–5692. doi: 10.1128/jb.179.18.5684-5692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pieulle L, et al. Isolation and characterization of the pyruvate-ferredoxin oxidoreductase from the sulfate-reducing bacterium Desulfovibrio africanus. Biochim Biophys Acta. 1995;1250:49–59. doi: 10.1016/0167-4838(95)00029-t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.