Significance
The free radical theory of aging remains controversial. Accumulation of mitochondrial damage is commonly accepted as an age-related phenomenon associated with the inescapable side effects of oxidative metabolism. However, to date, molecular determinants of this phenomenon have not been identified. Previous evidence indicates that engineered mice deficient in the denitrosylase S-nitrosoglutathione reductase (GSNOR) show features of aging. Here, we show that due to epigenetic events, GSNOR expression declines with age, ultimately resulting in the accumulation of damaged mitochondria. By contrast, centenarians maintain high GSNOR expression. Collectively, these data suggest that GSNOR may act as a longevity protein countering defects in mitochondrial physiology that arise from age-related epigenetic deregulation.
Keywords: aging, S-nitrosylation, GSNOR, mitochondria, mitophagy
Abstract
S-nitrosylation, a prototypic redox-based posttranslational modification, is frequently dysregulated in disease. S-nitrosoglutathione reductase (GSNOR) regulates protein S-nitrosylation by functioning as a protein denitrosylase. Deficiency of GSNOR results in tumorigenesis and disrupts cellular homeostasis broadly, including metabolic, cardiovascular, and immune function. Here, we demonstrate that GSNOR expression decreases in primary cells undergoing senescence, as well as in mice and humans during their life span. In stark contrast, exceptionally long-lived individuals maintain GSNOR levels. We also show that GSNOR deficiency promotes mitochondrial nitrosative stress, including excessive S-nitrosylation of Drp1 and Parkin, thereby impairing mitochondrial dynamics and mitophagy. Our findings implicate GSNOR in mammalian longevity, suggest a molecular link between protein S-nitrosylation and mitochondria quality control in aging, and provide a redox-based perspective on aging with direct therapeutic implications.
The oxidative modification of cysteine residues by nitric oxide (NO) to form S-nitrosothiols (SNOs) is called S-nitrosylation, and provides the large part of ubiquitous NO bioactivity (1, 2). Physiological S-nitrosylation modulates the activity of proteins in all functional classes across phylogeny (3). By contrast, hyper–S-nitrosylation has been implicated in a wide range of pathophysiologies, including cell senescence and death (4–6). The steady-state concentration of protein SNOs (PSNOs) reflects manifold equilibria between protein and low-molecular-weight SNOs that are regulated by rates of S-nitrosylation and denitrosylation. S-nitrosylation is a function of NO synthesis, which is catalyzed by NO synthases (NOSs), as well as enzymes termed nitrosylases, which directly introduce NO into proteins (7), while denitrosylation is catalyzed by denitrosylases, including the enzyme S-nitrosoglutathione reductase (GSNOR). Although being formerly characterized as a class III alcohol dehydrogenase (ADH5) or glutathione-dependent formaldehyde dehydrogenase (FDH), GSNOR exhibits its highest catalytic efficiency and specificity for S-nitrosoglutathione (GSNO) reduction (8, 9). Through trans-nitrosylation, the exchange of an NO moiety between SNOs and free thiols, GSNO is in equilibrium with cellular PSNOs; therefore, GSNOR activity affects the extent of protein S-nitrosylation (10). Conditions in which NOSs are pathologically activated result in aberrantly enhanced S-nitrosylation, which is associated with cellular damage (11–13) and disease, such as neurodegeneration (14). Also, phenotypes of mice deficient in GSNOR (Gsnor−/−) (15–20), in which protein S-nitrosylation is greatly increased, overlap those found in experimental models of aging, supporting the idea that GSNOR preserves cellular function.
The free radical theory of aging postulates that oxidative stress, an inevitable consequence of oxygen consumption, results in mitochondrial injury and dysfunction. In fact, damaged mitochondria, with altered mitochondrial morphology, represent markers of senescent cells in aging tissues (21–23). Nitrosative stress has also been related to aging and linked to excessive S-nitrosylation of components of the mitochondrial respiratory complexes (24–26). However, the role of mitochondrial nitrosative stress in cellular senescence is not well understood. Here, we demonstrate that mammalian aging is regulated by epigenetic down-regulation of GSNOR and by the consequently enhanced S-nitrosylation of proteins controlling mitochondrial dynamics and mitophagy. This offers a unique perspective on the mitochondrial free radical theory of aging and may open new therapeutic approaches.
Results
GSNOR Expression Decreases During Aging and Cell Senescence.
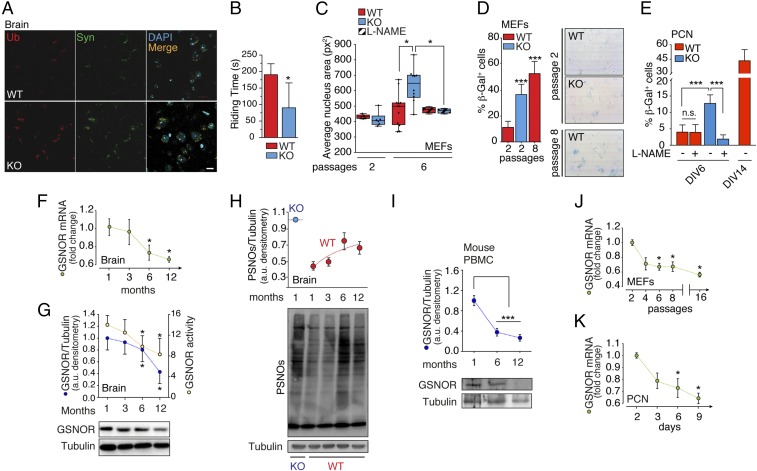
Based on the evidence that Gsnor−/− mice exhibit a number of phenotypes associated with aging (15–20), we decided to analyze the impact of GSNOR deficiency on the nervous system. To this end, we screened neocortical samples in 2-mo-old Gsnor−/− mice for accumulation of protein aggregates, with these commonly being accepted as markers of premature neurodegeneration. As shown in Fig. 1A, Gsnor−/− brains exhibited marked increases in ubiquitin- and α-synuclein–positive protein aggregates that correlated with accelerated declines in motor control coordination (observed at 2 mo) (Fig. 1B), with this also being a distinctive feature of aged brains. To investigate in further detail the mechanisms linking GSNOR to aging, we moved to cell systems and selected two different cell types: (i) the actively dividing mouse embryonic fibroblasts (MEFs), which were used as a model of replicative senescence, and (ii) primary cortical neurons (PCNs), which are postmitotic (terminally differentiated) cells, and thus used as a canonical model of aging in vitro. As also reported previously (27, 28), MEFs and PCNs preferentially expressed two different isoforms of NOS (i.e., NOS2 in MEFs and NOS1 in PCNs; SI Appendix, Fig. S1 A and B) and produced NO at detectable levels, which was reverted by incubation with the pan-NOS inhibitor L-NG-nitroarginine methyl ester (L-NAME) (SI Appendix, Fig. S1 C and D). When Gsnor−/− MEFs were maintained in culture, they showed senescence-related features, including an increase in nuclear size (Fig. 1C) and β-galactosidase activity (Fig. 1D) to an extent comparable to that observed in WT MEFs undergoing replicative senescence (i.e., after six or eight passages in culture). Similarly, β-galactosidase activity, evaluated in Gsnor−/− PCNs after 6 d in vitro, was significantly higher than in WT counterparts and, of note, reverted by L-NAME addition (Fig. 1E and SI Appendix, Fig. S2), thereby highlighting the physiological relevance of GSNOR in cell senescence. To establish any functional associations between GSNOR and aging, we measured GSNOR mRNA levels in brains of WT mice and ascertained that they decreased age-dependently (Fig. 1F). This phenomenon was associated with declines in GSNOR protein levels and enzymatic activity (Fig. 1G), and with an increase in PSNOs (Fig. 1H), which is a direct assessment of cellular denitrosylating capacity. Importantly, no significant changes in NOS levels were observed (SI Appendix, Fig. S3A), suggesting that the increase in PSNO levels reflects, in large part, the diminished denitrosylation. In line with this, PSNO levels of Gsnor−/− brain did not increase with age (SI Appendix, Fig. S3B), indicating that GSNOR deficiency induced steady-state high S-nitrosylation, which was maintained throughout life (at least under controlled laboratory conditions). Lastly, to evaluate whether the age-dependent down-regulation of GSNOR was specific to the brain or a molecular feature shared with other tissues, we collected peripheral blood mononuclear cells (PBMCs) from 1- to-12-mo-old WT mice and also observed in these cells an age-related decrease in GSNOR protein levels (Fig. 1I). In vitro assays, performed in murine primary cells, confirmed that GSNOR mRNA also declined during cell senescence, that is, in MEFs during passages in culture (Fig. 1J) and in PCNs maintained in vitro (Fig. 1K), indicating that down-regulation of GSNOR levels and excessive S-nitrosylation are biochemical features of age-related pathophysiology.
Fig. 1.
GSNOR decrease is associated with age and cell senescence. (A) Immunofluorescence analyses of brain cortexes from 2-mo-old WT and Gsnor−/− (KO) mice incubated with antiubiquitin (Ub, red) and anti–α-synuclein (Syn, green) antibodies. DAPI was used to stain nuclei. Ub spot count per field = 30.67 ± 4.17 (WT); 62.67 ± 8.67 (KO); average Syn spot count per field = 33.67 ± 6.96 (WT); 64.67 ± 6.96 (KO). Values represent mean ± SEM of n = 3 fields containing ∼35 cells per field. (Scale bar: 10 μm.) (B) Rotarod test. Values are expressed as latency to fall off the rod (riding time) and represent mean ± SEM of 2-mo old WT (n = 5) and KO (n = 5) mice, respectively, analyzed at three different times. *P < 0.05 (Student’s t test). (C) Nuclear size evaluated in WT and KO MEFs after two and six passages in culture in the presence or lack of 500 μM L-NAME added every 48 h. Size is expressed as squared pixels (px2). Values represent mean ± SEM of n ≥ 6 independent experiments. *P < 0.05 (Student’s t test). β-Galactosidase (β-Gal) activity of WT and KO MEFs (D) and PCNs (E) is shown. Values represent mean ± SEM of n = 3 independent experiments performed in duplicate. ***P < 0.001 (Student’s t test). DIV, days in vitro. (F) Real-time PCR analysis of GSNOR expression in brain from WT mice 1 to 12 mo of age. Actin and the ribosomal protein L34 were used as internal standards. Values represent mean ± SEM of n = 5 animals per experimental point analyzed in triplicate. *P < 0.05 (Student’s t test). (G) GSNOR activity and protein levels in brain homogenates from WT mice from 1 to12 mo of age. Results of GSNOR immunoreactive band densitometry (relative to tubulin) and enzymatic activity (expressed as nanomoles of NADH × milligrams of protein−1 × min−1) are shown and represent mean ± SEM of n ≥ 3 animals per experimental point analyzed in duplicate. *P < 0.05 (Student’s t test). (H) Amount of PSNOs in the brain from WT and KO mice from 1 to 12 mo of age. Results of PSNO densitometry (relative to tubulin) are shown. Values represent mean ± SD of n = 3 animals for each experimental point. (I) Western blot analysis of GSNOR in PBMCs from WT mice 1 to 12 mo of age. Results of band densitometry (relative to tubulin) are shown. Values represent mean ± SD of n = 3 animals for each experimental point. ***P < 0.001 (Student’s t test). Real-time PCR analysis of GSNOR expression in MEFs (J) and PCNs (K) is shown. L34 was selected as an internal standard, and values are expressed as fold change relative to cells maintained in culture for two passages (MEFs) or 2 d (PCNs). Values represent mean ± SEM of n = 3 independent experiments performed in triplicate. *P < 0.05 (Student’s t test).
GSNOR Expression Is Regulated by Ten-Eleven Translocation 1 Protein and Associated in Vivo and in Vitro with Promoter Methylation.
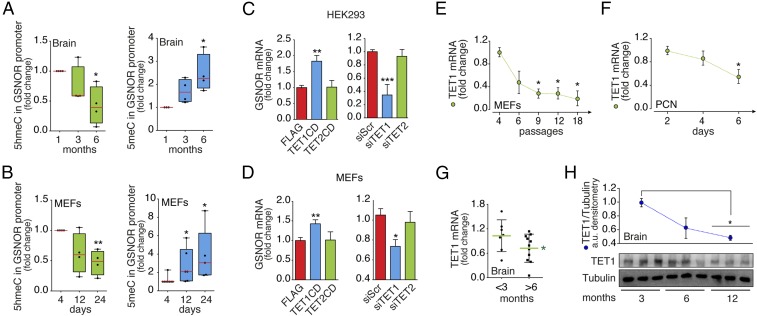
These observations prompted us to explore whether GSNOR underwent epigenetic regulation, thus possibly explaining its silencing during aging/cell senescence. Indeed, in silico analyses of the Adh5 (Gsnor) promoter revealed the presence of CpG islands surrounding the transcription start sequence (SI Appendix, Fig. S4A), with these being a predictive marker of epigenetic silencing by cytosine methylation. Methylated cytosine can be demethylated via a sequence of oxidation reactions catalyzed by ten-eleven translocation (TET) proteins (29). Oxidation of 5-methylcytosine (5meC) to 5-hydroxymethylcytosine (5hmeC) is the first reaction catalyzed by TETs to restore cytosine levels; therefore, 5hmeC is a molecular signature of active cytosine demethylation. We thus evaluated the abundance of 5meC and 5hmeC specifically in Gsnor promoters of brain specimens and in MEFs from WT mice. Our results revealed a concomitant decrease of 5hmeC and an increase in 5meC over time (i.e., with age) (Fig. 2 A and B), suggesting that GSNOR expression might be regulated epigenetically. To test this hypothesis, we manipulated the activity/expression of TET1 and TET2 (the TET isoforms mostly expressed in the adult phase) by either overexpressing their catalytic domains or, alternatively, knocking down their mRNA by small interference RNA (siRNA) in MEFs and human embryonic kidney (HEK293) cells (SI Appendix, Fig. S4 B and C). In line with its level of expression and binding preference for CpG-rich promoter regions (30), TET1, but not its cognate partner TET2, was found to modulate Gsnor transcription (Fig. 2 C and D). Notably, TET1 expression in MEFs, PCNs, and brains of WT mice showed an age-related decrease (Fig. 2 E–G), which, in brains, was concomitant with a decline in protein levels (Fig. 2H), confirming an age-related down-regulation of the entire TET1/GSNOR axis. These results were confirmed by in silico meta-analyses of RNA-sequencing studies performed on three distinct mouse strains other than C57BL/6J (the background strain of Gsnor−/− mice), demonstrating that both GSNOR and TET1 were markedly underexpressed in brains from old mice (SI Appendix, Fig. S5). Interestingly, TET1 decreased age-dependently during replicative senescence in Gsnor−/− MEFs with a trend comparable to its WT counterpart (SI Appendix, Fig. S4D), confirming that decline of TET1 is independent of excessive S-nitrosylation. Moreover, the evidence that DNA methyl transferases and TET1 expression levels did not significantly change between WT and Gsnor−/− cells and brains (SI Appendix, Fig. S4 E and F) suggested that, seemingly, alterations in GSNOR levels and activity had no visible effect on DNA methylation machinery.
Fig. 2.
Age-dependent GSNOR expression is regulated by DNA methylation and controlled by TET1. Real-time PCR analyses of (5hmeC, green) and (5meC, blue) levels in the Gsnor promoter in the mouse brain (A) and MEFs (B) are shown. Values are expressed as fold change relative to 1-mo-old WT brains or MEFs maintained in culture for four passages, and represent mean ± SEM of n ≥ 4 independent experiments performed in quadruplicate. *P < 0.05; **P < 0.01 (Student’s t test). Real-time PCR analyses of GSNOR expression in HEK293 cells (C) and WT MEFs (D) transfected with either catalytic domains or siRNA for TET1 and TET2 are shown. Values represent mean ± SEM of n = 3 independent experiments performed in duplicate. *P < 0.05; **P < 0.01; ***P < 0.001 (Student’s t test). Real-time PCR analyses of TET1 expression in WT MEFs (E) and PCNs (F) maintained in culture for up to 18 passages or 6 d, respectively, as well as in brains obtained from WT mice of different ages, are shown. (G) Values are normalized on actin, and represent mean ± SEM of n = 3 independent experiments (MEFs and PCNs) and n ≥ 6 animals (mouse brain) performed in triplicate. *P < 0.05 (Student’s t test). (H, Bottom) Western blot analysis of TET1 in brain homogenates from WT mice in a range from 3 to 12 mo of age. (H, Top) Results of band densitometry (relative to tubulin) are shown. Values represent mean ± SD of n = 3 animals per age. *P < 0.05 (Student’s t test). CD, catalytic domain.
GSNOR Impacts Mitochondrial Function and Regulates Mitochondrial Shape by Modulating Dynamin-Related Protein 1 S-Nitrosylation.
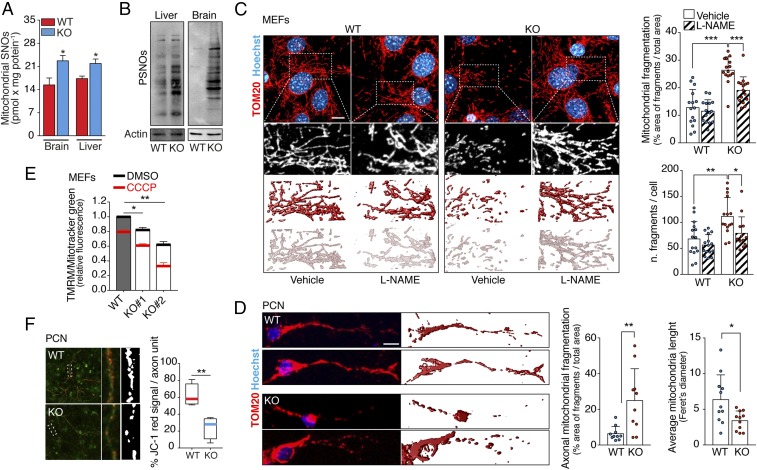
The mitochondrial free radical theory of aging states that oxidative stress, generated as a byproduct of cellular respiration, negatively impacts mitochondrial function, causing damage, and ultimately leading to aging. S-nitrosylation of mitochondrial respiratory complexes (particularly complexes I, III, and IV) can also inhibit their activity (24, 31, 32), potentially inducing cellular injury. In agreement, we observed that the extent of protein S-nitrosylation in mitochondria purified from the liver and brain of Gsnor−/− mice increased compared with the WT mice (Fig. 3A), recapitulating the difference observed in whole-liver and -brain extracts (Fig. 3B). Moreover, Gsnor−/− MEFs showed a fragmented mitochondrial network (Fig. 3C and SI Appendix, Fig. S6A), which was rescued by L-NAME incubations. A similar phenotype was observed in PCNs (Fig. 3D) and neuronal cells (SI Appendix, Fig. S6B), with both showing a higher relative extent of mitochondrial fragmentation compared with MEFs. This condition is frequently associated with mitochondrial dysfunction and is predictive of cell senescence (33–35). Accordingly, we observed that the mitochondrial transmembrane potential (Δψm) of Gsnor−/− cells was also markedly reduced (Fig. 3 E and F) and that ATP concentration was slightly decreased versus WT cells (SI Appendix, Fig. S7A).
Fig. 3.
GSNOR deficiency results in the accumulation of S-nitrosylated proteins and affects mitochondrial homeostasis. (A) S-nitrosothiols (SNOs) of the mitochondria-enriched fraction of brain and liver obtained from WT and Gsnor−/− (KO) mice at 2 mo of age evaluated by the Seville–Griess assay. (B) PSNO amount in the brain and liver from WT and KO mice at 2 mo of age. Actin was used as a loading control. Three-dimensional reconstruction of the mitochondrial network of WT and KO MEFs (C, Top) and PCNs (D, Top) is revealed by confocal fluorescence microscopy upon incubation with an antibody against the mitochondrial protein TOM20. A total of seven to 11 z-stacks (0.3-μm size) were merged. (C, Bottom and D, Bottom) Three-dimensional rendering of TOM20 signal is shown. Hoechst 33342 dye was used to visualize nuclei. (C) In MEFs, values are expressed as both the percentage area of fragments within the total mitochondrial area and the number of fragments per cell. (Scale bar: 10 μm.) (D) In PCNs, the analysis was performed in the axonal region, and Feret’s diameter is reported as an estimation of average mitochondrial length. (Scale bar: 5 μm.) The data shown represent mean ± SD of at least n ≥ 15 cells per experimental point. *P < 0.05; **P < 0.01; ***P < 0.001 (Student’s t test). (E) Δψm in WT and KO MEFs was evaluated by flow cytometry analysis of tetramethylrhodamine methyl ester (TMRM) fluorescence. CCCP (20 μM) was used as a positive control of mitochondrial depolarization. Values are normalized on the total mitochondrial mass (obtained upon incubation with MitoTracker Green) and expressed as the TMRM fluorescence fold change. Values represent mean ± SD of n = 3 independent experiments performed in triplicate. *P < 0.05; **P < 0.01. (F) Δψm in WT and KO PCNs was evaluated upon JC-1 staining. Values are expressed as a percentage area of red signal (polarized mitochondria) within the green signal area (axon). The red signal was thresholded, and the resulting area is shown in white. Values represent mean ± SD of n ≥ 15 axons counted deriving from three independent experiments. **P < 0.01.
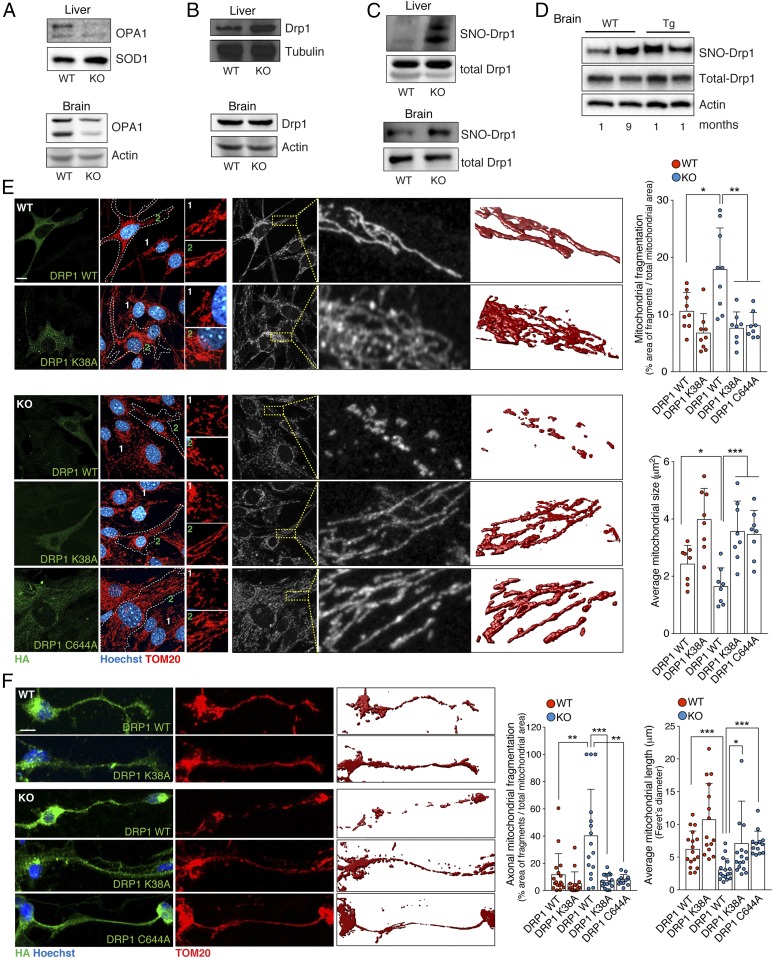
Mitochondrial dynamics depend on continuous fusion and fission events, which are indispensable for cell homeostasis (36). Both processes are regulated by large GTPases. Among them, optic atrophy 1 (OPA1) is required to fuse the inner membranes of adjacent mitochondria, while dynamin-related protein 1 (Drp1) regulates mitochondrial fission (31, 36). Western blot analyses performed on protein extracts from Gsnor−/− liver and brain showed a significant decrease in the levels of OPA1 with respect to the WT genotype (Fig. 4A), which was accompanied by no significant change in the amount of Drp1 (Fig. 4B). However, it has been previously shown that Drp1 GTPase activity can be positively modulated by S-nitrosylation of Cys644 (31, 37). Pull-downs from livers and brains, performed with biotin switch assays for PSNOs, confirmed that S-nitrosylated Drp1 (SNO-Drp1) was abundant in the extracts of Gsnor−/− tissues (Fig. 4C). Interestingly, SNO-Drp1 seemed to increase in old WT brains, reaching values comparable to those in Tg2576 mice (Fig. 4D), which represents a common in vivo experimental model of Alzheimer disease (AD) already reported to show high levels of SNO-Drp1 (37). Ectopic expression of the nonnitrosylable C644A mutant of Drp1, which was refractory to nitrosylation with GSNOR deficiency (SI Appendix, Fig. S7B), restored the mitochondrial network in Gsnor−/− MEFs and PCNs (Fig. 4 E and F). This mutant recapitulated the effects induced by the K38A variant of Drp1, which is unable to bind GTP (38) and correctly promote mitochondrial fission (Fig. 4 E and F). Similar effects were also obtained by overexpressing OPA1, used to force mitochondrial fusion and counterbalance excessive fragmentation, or, alternatively, by incubating Gsnor−/− MEFs with the Drp1 inhibitor mitochondrial division inhibitor 1 (Mdivi-1) (SI Appendix, Fig. S7C), although this compound was recently demonstrated to exert additional (Drp1-independent) effects as well (39). Notably, OPA1 levels in Gsnor−/− MEFs were restored by L-NAME treatment, but not by the expression of C644A Drp1 (SI Appendix, Fig. S7D); this suggests that OPA1 decrease is dependent on excessive S-nitrosylation induced by GSNOR deficiency, but unrelated to Drp1 S-nitrosylation state. Taken together, our data indicate that GSNOR-mediated increases in S-nitrosylation contribute to mitochondrial dysfunction and trigger dysregulation of mitochondrial dynamics, a well-established signature of accelerated cell senescence and aging (34).
Fig. 4.
Gsnor−/− cells exhibit increased fragmentation due to S-nitrosylation of Drp1. Representative Western blots of OPA1 (A) and Drp1 (B) in the brain and liver from 2-mo-old WT and GSNOR-KO mice are shown. Actin, SOD1, or tubulin was used as a loading control. (C) Western blot analysis of SNO-Drp1 performed on a pool of PSNOs obtained from liver and brain lysates of WT and KO mice. As a control, Western blot analyses of total Drp1 were performed on the lysates using the same anti-Drp1 antibody. (D) Western blot analysis of SNO-Drp1 performed on a pool of PSNOs obtained from brain lysates of 1- and 9-mo-old WT mice and 1-mo-old Tg2576 mice. Actin was used as a loading control. Three-dimensional reconstructions of the mitochondrial network of WT and KO MEFs (E) and PCNs (F) are revealed by confocal fluorescence microscopy as described in Fig. 3C. (Scale bars: 5 μm.) Data shown represent mean ± SD of at least n ≥ 8 cells (MEFs) and n ≥ 15 axons (PCNs) per experimental point. *P < 0.05; **P < 0.01; ***P < 0.001 (Student’s t test).
GSNOR Sustains Mitophagy and Modulates Parkin S-Nitrosylation.
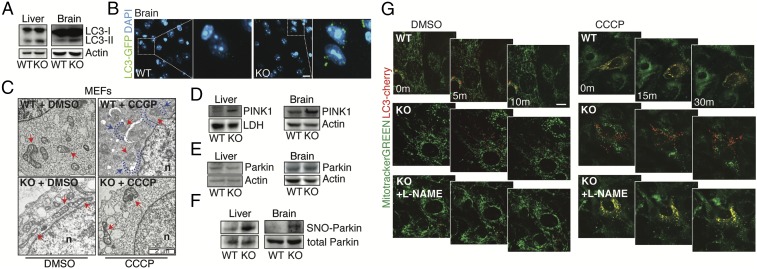
An increased fission rate of mitochondria is usually a prerequisite for their turnover by autophagy, the so-called mitophagy. In this context, loss of Δψm (a condition that we observed in Gsnor−/− cells) represents the first step in mitochondrial sequestration and delivery to lysosomes for degradation. An autophagy-inhibitory role for NO was previously ascribed to S-nitrosylation in tumor cell lines subjected to high NO flux due to treatment with NO donors or NOS overexpression (40, 41). However, an analysis of skeletal muscle from Gsnor−/− mice indicated that mitophagy, rather than autophagy, seemed to be impaired (19). We set out to find whether autophagy and/or mitophagy was modulated in GSNOR-deficient brain and liver. Western blot analysis of the autophagic marker microtubule-associated protein light chain 3 (LC3) indicated that the autophagy-proficient (autophagosome-bound) isoform of the protein (LC3-II) was increased in both the liver and brain of Gsnor−/− mice (Fig. 5A). Immunofluorescence detection of LC3 in brain slices of mice expressing GFP-LC3 confirmed the increases of LC3 in Gsnor−/− (Lc3+/GFP:Adh5−/−) background (Fig. 5B). However, bulk autophagic flux remained unchanged in cells. WT or Gsnor−/− MEFs did not show any significant differences in the amount of LC3+ puncta, either before or after pharmacological inhibition of autophagy by chloroquine (SI Appendix, Fig. S8 A and B). It is noteworthy that the sole change observed was a trend toward increases in p62 levels, with this possibly being a marker of impaired autophagy (42). However, this phenomenon was not detected when autophagy was induced by rapamycin or starvation (SI Appendix, Fig. S8 A and B). By contrast, transmission electron micrographs proved that the majority of Gsnor−/− mitochondria displayed severely damaged cristae (Fig. 5C) but were rarely localized inside autophagosomes, indicating that they were not properly recognized by mitophagy. Incubations with carbonyl cyanide m-chlorophenylhydrazone (CCCP), an uncoupling molecule used to induce mitophagy in vitro, further worsened the already compromised mitochondrial ultrastructure of Gsnor−/− cells, although, again, mitochondria were essentially excluded from autophagic vesicles (Fig. 5C), suggesting that Gsnor−/− cells are deficient in targeting damaged mitochondria for removal by mitophagy.
Fig. 5.
Gsnor−/− cells show defects in mitophagy linked to S-nitrosylation. (A) Representative Western blot of LC3 in brain and liver homogenates obtained from 2-mo old WT and Gsnor−/− (KO) mice. Actin was used as a loading control. (B, Left) Brain slices (i.e., cerebral cortex) obtained from Lc3+/GFP:Adh5+/+ (2) and Lc3+/GFP:Adh5−/− (KO) mice analyzed by fluorescence microscopy to visualize autophagosomes. DAPI was used to visualize nuclei. (B, Right) Digital magnification is shown. (Scale bar: 10 μm.) (C) Transmission electron microscopy images of WT and KO MEFs treated for 6 h with CCCP or vehicle (DMSO). Nuclei (n) and mitochondria (red arrows) are indicated, along with internal double-membraned structures surrounding mitochondria (phagophores, blue dotted lines and arrows). Representative Western blots of PINK1 (D) and Parkin (E) in brain and liver total extracts obtained from 2-mo old WT and KO mice are shown. LDH or actin was used as a loading control. (F) Western blot analysis of SNO-Parkin performed on PSNOs obtained from liver and brain lysates of WT and KO mice. Western blot analysis of total Parkin is shown as a control. (G) Representative frames captured upon live-imaging fluorescence microscopy (Movies S1–S6) of mitochondrial networks in WT and KO MEFs expressing the fluorescent protein LC3-cherry (red) and stained with MitoTracker Green (green) to visualize autophagosomes and mitochondria, respectively. MEFs were pretreated with 0.5 mM L-NAME (where indicated) or with the vehicle solution for 1 wk with administration every 48 h. CCCP was added before image acquisition to induce mitophagy. m, minute(s). (Scale bar: 10 μm.)
With mitochondria depolarization, the PTEN-induced putative kinase 1 (PINK1) accumulates on the outer mitochondrial membrane (43), where it recruits and activates the E3 ligase Parkin and other autophagic receptors to mitochondria (44) to induce mitophagy. Western blot analyses showed that PINK1 accumulated in the brain and liver of Gsnor−/− mice (Fig. 5D), confirming that Gsnor−/− cells had low Δψm, and that PINK1 was responsive to it. However, total levels of Parkin did not change in Gsnor−/− versus WT cells (Fig. 5E). It has been reported that Parkin undergoes S-nitrosylation, resulting in a loss-of-function modification that compromises Parkin activity (45, 46). S-nitrosylated Parkin (SNO-Parkin) accumulated in the liver and brain of Gsnor−/− mice (Fig. 5F), suggesting that GSNOR deficiency, by inducing excessive S-nitrosylation, suppresses Parkin activity. To demonstrate a causal relationship between S-nitrosylation and mitophagy, live-imaging fluorescence microscopy analyses were performed. WT and Gsnor−/− MEFs were treated for three passages in culture with repeated additions of L-NAME every 48 h, and mitophagy was then induced by CCCP. As expected, mitochondria of WT cells changed from elongated to fragmented, and were properly internalized by autophagosomes upon CCCP administration (Fig. 5G and Movies S1 and S2). By contrast, Gsnor−/− cells showed fragmented mitochondria, which, in the presence of CCCP, were not (or only rarely) engulfed by autophagic vesicles, although these were produced and accumulated inside the cytoplasm (Fig. 5G and Movies S3 and S4). Remarkably, L-NAME was able, per se, to significantly restore a more elongated mitochondrial shape and a correct mitophagy upon challenge with CCCP (Fig. 5G and Movies S5 and S6). Similar experiments were also done in GSNOR-downregulating HEK293 cells in which, as a denitrosylating agent, we used the thiol-reductant DTT. Also in this case, mitophagy was restored (SI Appendix, Fig. S8C and Movies S7–S11), further confirming that defects in mitophagy are related to enhanced S-nitrosylation.
TET1 Down-Regulation Recapitulates Mitochondrial Phenotype and Senescence Induced by GSNOR Deficiency.
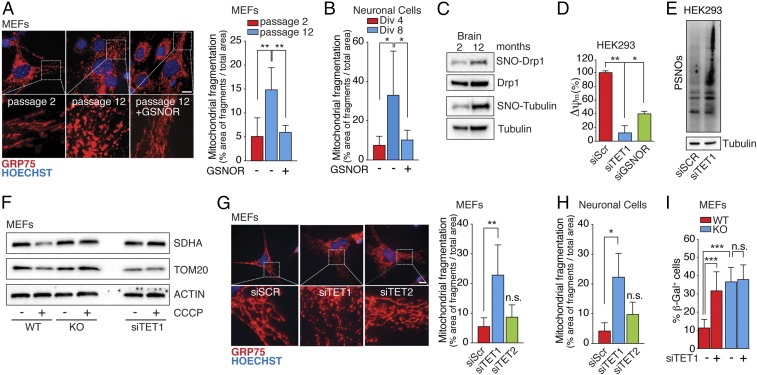
Finally, to put our findings into a more physiological context, we assessed mitochondrial fragmentation patterns in WT cells. Mitochondria of MEFs obtained from WT mice showed increased fragmentation during the passages in culture (Fig. 6A), which paralleled the decline in GSNOR (Fig. 1 J and K). Similarly, neuronal cells obtained from WT mice showed gradual loss of their mitochondrial network over time (12 d of growth in vitro) (Fig. 6B and SI Appendix, Fig. S9A). Notably, ectopic expression of GSNOR restored the mitochondrial reticulum in both cell types (Fig. 6 A and B). Taken together, our data suggest that loss of GSNOR activity, and consequent nitrosative stress, may be a causal factor in the loss of mitochondrial integrity observed during cell senescence. This assumption was also supported by evidence obtained in brain lysates of 12-mo-old WT mice that displayed hypernitrosylation of Drp1 (Fig. 6C), but not of the deacetylase SIRT1 (SI Appendix, Fig. S10), which has been recently demonstrated to be inhibited by S-nitrosylation, leading to apoptosis (47). Following this line of reasoning, we tested whether TET1 down-regulation would recapitulate nitrosative stress-mediated senescence. Our results indeed showed that siRNA for TET1 (siTET1), which decreases GSNOR expression (Fig. 2 C and D), displayed a marked decrease in Δψm even more accentuated than that produced by siGSNOR, (Fig. 6D) as well as a concomitant increase in PSNOs (Fig. 6E). In addition, CCCP treatment of WT MEFs following TET1 silencing did not reduce mitochondrial protein levels (Fig. 6F), with such a reduction representing a measure of their degradative rate. This finding confirms that mitophagy was negatively affected by TET1 down-regulation. The siTET1 also induced mitochondrial fragmentation in WT MEFs and neuronal cells, whereas no significant change was observed upon TET2 silencing (Fig. 6 G and H and SI Appendix, Fig. S9B). Also, β-galactosidase staining showed that signs of senescence in siTET1 cells were similar to those observed in Gsnor−/− cells (Fig. 6I). In sum, our results strongly indicate that TET1 and GSNOR regulate mitochondrial structure, function, and survival, and that hypoexpression of either entity results in a switch from a juvenile phenotype to a senescent phenotype.
Fig. 6.
GSNOR and TET1 manipulation impact on mitochondrial homeostasis, dynamics, and cell senescence. Fluorescence microscopy analyses of mitochondrial networks performed upon incubation with an antibody against Grp75 in WT MEFs maintained in culture for two or 12 passages (A, Top) or in WT neuronal cells after 4 or 8 d in vitro (Div) (B) are shown. Transfection with the WT form of GSNOR (+GSNOR) or an empty vector as a control is shown. Hoechst 33342 dye was used to stain nuclei in blue. (A, Bottom) Digital magnifications are shown. Values are expressed as a percentage area of fragments within the total mitochondrial area. Data shown represent mean ± SD of at least n ≥ 8 cells per experimental point. *P < 0.05; **P < 0.01 (Student’s t test). (Scale bar: 10 μm.) (C) Western blot analysis of SNO-Drp1 performed on a pool of PSNOs obtained from the brain of WT mice aged 2 and 12 mo upon biotin-switch assay. As a control, Western blot analysis of total Drp1 was performed. Tubulin is also shown as an indicator that excessive nitrosylation occurs with age. (D) Δψm in HEK293 cells transfected with siRNA against TET1 or GSNOR. siScr cells were used as a control. Values are expressed as a percentage of siScr cells and represent mean ± SEM of n = 3 independent experiments performed in duplicate. *P < 0.05; **P < 0.01 (Student’s t test). (E) Amounts of PSNOs in siScr and siTET1 HEK293 cells measured by biotin-switch assay. Tubulin was selected as a loading control. (F) Western blot analysis of the mitochondrial proteins SDHA and TOM20 in GSNOR KO and WT MEFs (with or deficient in TET1); CCCP was added for 8 h to induce mitophagy. Actin was selected as a loading control. Fluorescence microscopic images of mitochondrial networks performed in WT MEFs (G, Top) and WT neuronal cells (H) transfected with siRNA against TET1 or TET2 are shown; the antibody is against Grp75, and siScr cells were used as a control. Hoechst 33342 dye was used to stain nuclei in blue. (G, Bottom) Digital magnifications are shown. Values are expressed as a percentage area of fragments within the total mitochondrial area. Data shown represent mean ± SD of at least n ≥ 8 cells per experimental point. n.s., not significant. *P < 0.05; **P < 0.01 (Student’s t test). (Scale bar: 10 μm.) (I) β-Galactosidase (β-Gal) activity of WT and KO MEFs with or without TET1 knockdown. Values represent mean ± SD of n = 3 independent experiments performed in triplicate. ***P < 0.001 (Student’s t test).
GSNOR and TET1 Are Down-Regulated in Aging Humans but Not in Centenarians.
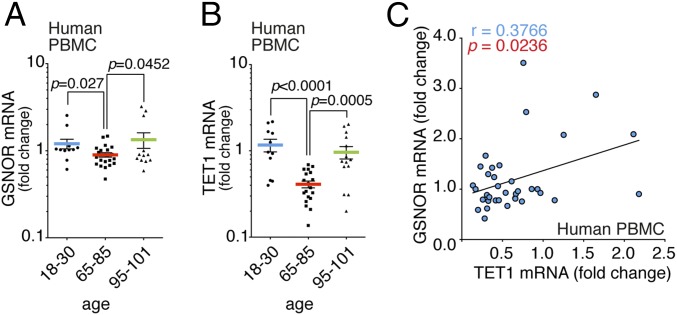
Results shown so far point toward down-regulation of the TET1/GSNOR axis during aging, regulating mitochondrial morphology and function. As a corollary, we speculated that S-nitrosylation profoundly impacts cell senescence and, thus, GSNOR serves as a “longevity gene.” In support of this hypothesis, we noted that ADH5 (GSNOR-coding gene) is localized in 4q23-25, a chromosomal locus tied to exceptional human longevity (48). Primed by this evidence, we measured GSNOR mRNA levels in PBMCs from healthy humans of different ages and compared results with those of exceptionally long-lived individuals (>95 y of age). The results shown in Fig. 7A indicate that GSNOR mRNA was reduced with age, whereas, remarkably, levels were unaffected in long-lived individuals. Real-time PCR analyses of TET1 showed an even more marked trend (Fig. 7B), with centenarians maintaining TET1 mRNA levels similar to young individuals. Furthermore, we confirmed that a statistically significant correlation between TET1 and GSNOR expression was found in humans (Fig. 7C), thus supporting the idea that both genes are functionally related.
Fig. 7.
TET1 and GSNOR are hypoexpressed in elderly, but not in exceptionally long-lived, humans. Real-time PCR assays of GSNOR (A) and TET1 expression (B) in PBMCs obtained from humans of different ages, including long-lived individuals (95–101 y of age), are shown. Values are expressed as fold change relative to a young group (18–30 y of age) after normalizing to two different internal standards (actin and L34), and they represent mean ± SEM of n ≥ 12 individuals per experimental point analyzed in triplicate. P values are shown in the graph (Student’s t test). (C) Correlation between the paired mRNA levels of GSNOR and TET1 analyzed by real-time PCR in PBMCs from humans of different ages (as shown in A and B). The linear regression and Pearson correlation coefficient were calculated using GraphPad Prism 6.0 (GraphPad Software, Inc.). R2 = 0.1418, P = 0.0236.
Discussion
We have discovered that the protein denitrosylase GSNOR is down-regulated in multiple models of aging and cell senescence as a consequence of the hypermethylation of its promoter by TET1, and that levels are maintained in exceptionally long-lived humans. We further show that GSNOR deficiency impairs mitochondrial function as a result of the nitrosative stress that is characteristic of cellular senescence and disorders of aging. Thus, our work may provide a new perspective on the “free radical theory of aging,” whereby excessive S-nitrosylation of key mitochondrial proteins contributes to aging and related disorders.
Prior analyses on PBMCs obtained from a large cohort of European individuals, indicated an age-related decline of TETs (i.e., TET1, TET3) and 5hmeC (49). Similarly, a screening conducted in T cells suggested that TET1 down-regulation is a predictive marker of aging (50). Consistent with these findings, we have shown that TET1 levels decrease in senescent cells in vitro, as well as in mouse tissues and in human PBMCs from old, but not exceptionally long-lived, individuals. This decrease correlates with decreased GSNOR expression, in agreement with previous genome-wide analysis linking an ADH5/GSNOR-encompassing gene cluster to human longevity (48). Thus, we provide a mechanistic basis for TET effects in aging, connecting TET activity to dysregulation of S-nitrosylation.
TETs play pivotal roles in disease development and onset, including cancer (51) and aging (49, 50). They show different (and sometimes overlapping) functions, with TET1 targeting the largest number of loci, whereas TET2 and TET3 are particularly active in introns of highly expressed genes (30, 52). Results from wide-genome DNA methylation studies are not entirely clear. However, increasing evidence supports the idea that aging is associated with a general increase in 5meC and decrease in 5hmeC levels within the CpG islands located in gene promoter regions, with these serving as distinctive signs of gene expression silencing (53).
S-nitrosylation resulting from GSNOR deficiency has been strongly implicated in hepatocellular carcinoma (HCC) (18), so it is notable that TET1 down-regulation and decreases in 5hmeC are also markers of HCC progression (54). In this context, we recently demonstrated that HCC cells deficient in GSNOR show increased activity of mitochondrial complex II, also known as succinate dehydrogenase (SDH) (32). TETs belong to the large class of 2-oxoglutarate–dependent dioxygenases, which are reportedly inhibited by succinate and fumarate (55, 56), which are metabolites arising from the tricarboxylic acid cycle (56). Our preliminary data indicate that, perhaps due to increased SDH activity, fumarate accumulates over time in aging GSNOR−/− cells (57). Thus, excessive S-nitrosylation arising from TET1/GSNOR reduction may lead to fumarate accumulation and to a further inhibition of TET1 activity. The altered mitochondrial homeostasis and impairment of mitophagy observed in Gsnor−/− cells support this hypothesis. From this point of view, GSNOR deficiency can be seen as an age-accelerating condition wherein excessive S-nitrosylation of mitochondrial proteins impairs mitochondrial function.
Our finding that both Drp1 and Parkin are hypernitrosylated in Gsnor−/− mice is consistent with the accumulation of α-synuclein–positive and ubiquitin-positive protein aggregates in brain cortexes of young Gsnor−/− mice. As previously reported in AD models (37, 58), we found that with GSNOR deficiency, the mitochondrial fission-promoting activity of Drp1 is increased. While Drp1 nitrosylation in brains of normal old mice was equal to that found in mouse AD, Cho et al. (37) have shown that aged normal human brains do not, in fact, have elevated SNO-Drp1. We attribute the difference between mice and humans to the higher mass-specific metabolic rates in mice, which correlate with aging (59). Higher metabolic rates will increase reactive oxygen species (ROS) production, and thus, potentially, SNOs. Because the brain has a higher metabolic rate than other tissues, differences between mice and humans may be exaggerated.
Our results are in line with data (45) showing that excessive S-nitrosylation in aging impairs the E3 ubiquitin ligase activity of Parkin, and its capability to act as an enhancer of mitophagy (60, 61), but are in conflict with those recently reporting that Parkin S-nitrosylation is indispensable for mitophagy (62). It is worthwhile mentioning that, at variance with prior studies, we did not overexpress Parkin. Rather, we took advantage of Gsnor−/− mice and cells, which may represent more physiologically relevant models, and found that mitophagy, rather than bulk autophagy (40, 41), is severely compromised. Interestingly, it has recently been reported that PINK1 is S-nitrosylated at Cys-658, dampening its kinase activity (on substrate Parkin) and impairing mitophagy in cellular models of Parkinson’s disease (63). Mitochondrial insults simulating age-related stress induce PINK1 S-nitrosylation and affect mitochondrial removal; this phenomenon may thus contribute to mitophagy impairment of Gsnor−/− cells and animals.
It is important to stress that many mechanisms of mitophagy exist, with that mediated by PINK/Parkin exemplifying regulation by S-nitrosylation. However, excessive S-nitrosylation, as observed with GSNOR deficiency, may likely inhibit mitophagy through mechanisms other than PINK/Parkin. In addition, S-nitrosylation–independent mechanisms resulting from GSNOR down-regulation should be considered. For example, ROS, produced by oxidative metabolism, can lead to generation of reactive aldehydes (e.g., formaldehyde), and we cannot exclude the possibility that the FDH activity of GSNOR has a protective role. However, the evidence that L-NAME was able to almost completely reverse mitophagy defects strongly argues for a driving role of S-nitrosylation in this process. It has also been reported that GSNOR catalyzes the oxidation of retinol to generate retinal, which is essential for postnatal development of some tissues (64). Therefore, we cannot exclude that some effects of GSNOR deficiency are related to development defects rather than to aging or senescence. However, again, results obtained in aged animals and long-lived humans support the hypothesis that excessive S-nitrosylation plays a role in aging.
In conclusion, our results provide a unique perspective that may link inflammation, cancer, and degenerative disease with the free radical theory of aging. In this model, aging and related pathophysiology reflect a significant contribution from excessive S-nitrosylation (16, 18, 65, 66) that is regulated by GSNOR.
Materials and Methods
Cells.
HEK293 cells were obtained from Banca Biologica e Cell Factory (Istituto Tumori Genova) and grown in DMEM, 10% FBS, and antibiotics at 37 °C in an atmosphere of 5% CO2. Mouse PCNs and neuronal cells were obtained from cerebral cortices of WT and Gsnor−/− C57BL/6J mouse embryos at embryonic day (E) 14.5. Tissue was digested with trypsin 0.25%/EDTA (Sigma–Aldrich) at 37 °C for 7 min and minced until cell suspension became homogeneous. After three washes in PBS, viable cells were seeded at a concentration of 1 × 105 cells per milliliter on poly-d-lysine–coated coverslips (Sigma–Aldrich) in DMEM (Sigma–Aldrich) with 10% FBS, 2 mM glutamine, and 0.1 mg/mL gentamicin. After 1 h, DMEM was replaced with Neurobasal supplemented with B27, 2 mM glutamine, and 0.1 mg/mL gentamicin. Cell cultures were kept at 37 °C in a humidified atmosphere containing 5% CO2, with half of the growing medium replaced every 3 d. MEFs were prepared from WT and Gsnor−/− C57BL/6J mouse embryos at E13.5 as previously described (67). MEFs were cultured in DMEM with 10% FBS and used for experiments from the second to 16th passage in culture. All of the media and supplements were purchased from Gibco, Thermo Fisher Scientific.
Immunofluorescence Microscopy and Analyses.
Mice were anesthetized with Avertin (0.25 mg per g of body weight, i.p.) and perfused through the heart sequentially with PBS and then with 4% paraformaldehyde/PBS (Sigma–Aldrich) for 5 h at 4 °C. After perfusion-fixation, brains were immersion-fixed in the same fixative overnight at 4 °C and permeated sequentially with 10%, 20%, and 30% sucrose/PBS (Sigma–Aldrich) for 48 h at 4 °C for cryoprotection. Next, they were embedded in Tissue-Tek O.C.T. compound (Bio-Optica) and flash-frozen in liquid nitrogen-cooled isopentane (VWR); the coronal and sagittal cryosections (15 μm thick) were obtained with a Leica cryostat (−20 °C) and used for immunostaining, or collected at −80 °C.
HEK293, PCNs, neuronal cells, and MEFs were grown on coverslips; washed twice in PBS; and fixed with 4% paraformaldehyde for 10 min at room temperature.
Cells and slices were incubated with a permeabilization solution [PBS/Triton X-100, 0.4% (vol/vol)], blocked for 1 h with a blocking solution [PBS/normal goat serum, 10% (vol/vol)], and then incubated for 1 h (for cells) or overnight (for brain) with the following primary antibodies: anti–α-synuclein (Abcam), anti-Grp75 (Enzo Life Sciences), anti-p62 (MBL), anti-LC3 (Cell Signaling), anti-TOM20 (Santa Cruz Biotechnology), and anti-HA (Sigma–Aldrich). Cells and brains were then washed with PBS and incubated for 1 h with fluorophore-conjugated secondary antibodies (Thermo Fisher Scientific). Nuclei were stained with 1 μg/mL Hoechst 33342 dyes (for cells) or DAPI (for brains) (Thermo Fisher Scientific). Epifluorescence analyses were performed using a Delta Vision (Applied Precision) Olympus IX70 microscope. Confocal microscopy experiments were performed using LSM800 (on MEFs) and LSM700 (on PCNs) microscopes (ZEISS) equipped with ZEN imaging software. Brains were observed with a Leica TCS SP5 confocal microscope and Leica Application Suite (LAS) software (Leica Microsystems). Fluorescence images were adjusted for brightness, contrast, and color balance using Fiji (68) analysis software. For brain tissue, confocal digital single-plane (1-μm) images from z-stacks were generated.
Counting of LC3 and p62 puncta was performed on at least 50 cells per experimental point using Fiji.
Average nucleus area was determined on MEFs after two and eight passages upon incubation with the cell-permeable nuclear staining dye Hoechst 33342. Acquisition and analysis were then performed on a ScanR screening station (Olympus). At least 1,000 cells per experimental point were analyzed.
Mitochondrial fragmentation was assessed by adapting the protocols reported by Barsoum et al. (69) and Marchi et al. (70) to our experimental settings. In particular, 3D reconstruction of MEFs and PCNs was achieved by Fiji analysis software (68), summing the TOM20-fluorescence signal z-stacks (seven to 11 planes, 0.3 μm). UCSF Chimera (71) was used for the 3D rendering of acquired z-stacks. Images were analyzed with Fiji by means of the following procedure: The same number of z-stacks for each image was merged, and a threshold was adjusted to isolate mitochondria signal from background. Thresholding algorithms used were “Otsu” for MEFs and “Triangle” for PCNs. The thresholding method was not modified between samples. Mitochondrial particles larger than 0.1 μm were analyzed. The total area of mitochondria, the area of the single particles, and Feret’s diameter were obtained from each field. Mitochondrial fragmentation (MF) was calculated with the following formula:
MEF and PCN fluorescence was acquired with oil-immersion 63× and 40× objectives, respectively. When analyzed, particles with an area of ≤1 (0.1 × 0.1 × 0.3-μm3 voxel size) in MEFs or ≤3 (0.156 × 0.156 × 0.5-μm3 voxel size) in PCNs were considered fragments. The analysis of mitochondria in PCNs was performed exclusively in the axon region. Due to the longitudinal disposition of the organelles along the axon, Feret’s diameter was used to estimate mitochondrial length, describing the longest distance between any two points along the particle analyzed.
Live-Imaging Confocal Microscopy.
HEK293 cells stably expressing GFP-LC3 and Mito-DsRed2 were grown in chamber slides and then transfected with siGNSOR (or siScr as a control) using Lipofectamine 2000 reagent, according to the manufacturer’s protocol (Thermo Fisher Scientific). Cells grown in different conditions (control setting or upon treatment with 20 μM CCCP, with or without 5 mM DTT) were placed in an incubator chamber for live imaging. Pictures were collected using a Leica TCS SP5 confocal microscope equipped with a 63× or 100× objective and LAS software. Images were captured every 30 s. MEFs (pretreated for 1 wk with a vehicle solution or with 0.5 mM L-NAME administered every 48 h) were grown in eight wells on a μ-Slide chambered coverslip (Ibidi) and transfected with a Cherry-LC3–expressing plasmid using Lipofectamine 3000 reagent (Thermo Fisher Scientific). Mitochondria were stained with 50 nM MitoTracker Green-FM. Cells were placed in the incubator chamber for live confocal imaging, and images were acquired with an Ultraview Vox Spinning Disk (PerkinElmer) equipped with a 63× objective. Videos were recorded with Volocity software. The time of acquisition was 30 min per experimental point, and images were captured every 30 s.
Supplementary Material
Acknowledgments
We thank Prof. Yi Zheng (University of North Carolina at Chapel Hill) for kindly providing TET1 catalytic domain (CD) and TET2CD coding plasmids, Prof. Stuart A. Lipton (Sanford–Burnham Medical Research Institute) for WT and C644A mutant of Drp1, Prof. Luca Scorrano (University of Padua) for WT OPA1, Vanda Turcanova for laboratory assistance, and Laila Fisher and Martin W. Bennett for secretarial and editorial work. This work has been supported by Danish Cancer Society Grant KBVU R146-A9414 (to G.F.) and Grant R146-A9364 (to F.C.); Associazione Italiano per la Ricerca sul Cancro (AIRC) Grant IG20719 (to G.F.), Grant IG2017 (to F.C.), and Start-Up Grant 20464 (to S. Cardaci); Ministero dell’Istruzione, dell’Università e della Ricerca (MIUR) Grants MIUR/Fondo per gli Investimenti della Ricerca di Base (FIRB) AUTOMED-RBAP11Z3YA and PRIN- 20157ATSLF_009 (to A.A.P.); NIH Grants P01HL075443 (Proj.3), R01GM099921, R01HL126900, and P01HL128192 (Proj.1) (to J.S.S.); and Bjarne Saxhof Foundation and NovoNordisk Grant 22544 (to F.C.). M.B. was supported by “Fondazione Umberto Veronesi” fellowship.
Footnotes
Conflict of interest statement: J.S.S. has received a commercial research grant from Novartis and has ownership interest (including patents) in Adamas and Lifehealth.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722452115/-/DCSupplemental.
References
- 1.Hess DT, Stamler JS. Regulation by S-nitrosylation of protein post-translational modification. J Biol Chem. 2012;287:4411–4418. doi: 10.1074/jbc.R111.285742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stamler JS, Lamas S, Fang FC. Nitrosylation. The prototypic redox-based signaling mechanism. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- 3.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: A physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 4.Iyer AKV, Rojanasakul Y, Azad N. Nitrosothiol signaling and protein nitrosation in cell death. Nitric Oxide. 2014;42:9–18. doi: 10.1016/j.niox.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamura T, Lipton SA. Redox modulation by S-nitrosylation contributes to protein misfolding, mitochondrial dynamics, and neuronal synaptic damage in neurodegenerative diseases. Cell Death Differ. 2011;18:1478–1486. doi: 10.1038/cdd.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura T, et al. Aberrant protein S-nitrosylation contributes to the pathophysiology of neurodegenerative diseases. Neurobiol Dis. 2015;84:99–108. doi: 10.1016/j.nbd.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seth D, et al. A multiplex enzymatic machinery for cellular protein S-nitrosylation. Mol Cell. 2018;69:451–464.e6. doi: 10.1016/j.molcel.2017.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu L, et al. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 9.Rizza S, Filomeni G. Chronicles of a reductase: Biochemistry, genetics and physio-pathological role of GSNOR. Free Radic Biol Med. 2017;110:19–30. doi: 10.1016/j.freeradbiomed.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Beigi F, et al. Dynamic denitrosylation via S-nitrosoglutathione reductase regulates cardiovascular function. Proc Natl Acad Sci USA. 2012;109:4314–4319. doi: 10.1073/pnas.1113319109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boullerne AI, Benjamins JA. Nitric oxide synthase expression and nitric oxide toxicity in oligodendrocytes. Antioxid Redox Signal. 2006;8:967–980. doi: 10.1089/ars.2006.8.967. [DOI] [PubMed] [Google Scholar]
- 12.Aquilano K, Baldelli S, Cardaci S, Rotilio G, Ciriolo MR. Nitric oxide is the primary mediator of cytotoxicity induced by GSH depletion in neuronal cells. J Cell Sci. 2011;124:1043–1054. doi: 10.1242/jcs.077149. [DOI] [PubMed] [Google Scholar]
- 13.Hara MR, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura T, et al. Aberrant protein S-nitrosylation in neurodegenerative diseases. Neuron. 2013;78:596–614. doi: 10.1016/j.neuron.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z, et al. Lymphocyte development requires S-nitrosoglutathione reductase. J Immunol. 2010;185:6664–6669. doi: 10.4049/jimmunol.1000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L, et al. Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell. 2004;116:617–628. doi: 10.1016/s0092-8674(04)00131-x. [DOI] [PubMed] [Google Scholar]
- 17.Wei W, Yang Z, Tang CH, Liu L. Targeted deletion of GSNOR in hepatocytes of mice causes nitrosative inactivation of O6-alkylguanine-DNA alkyltransferase and increased sensitivity to genotoxic diethylnitrosamine. Carcinogenesis. 2011;32:973–977. doi: 10.1093/carcin/bgr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei W, et al. S-nitrosylation from GSNOR deficiency impairs DNA repair and promotes hepatocarcinogenesis. Sci Transl Med. 2010;2:19ra13. doi: 10.1126/scitranslmed.3000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montagna C, et al. S-nitrosoglutathione reductase deficiency-induced S-nitrosylation results in neuromuscular dysfunction. Antioxid Redox Signal. 2014;21:570–587. doi: 10.1089/ars.2013.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao Y, et al. S-nitrosoglutathione reductase-dependent PPARγ denitrosylation participates in MSC-derived adipogenesis and osteogenesis. J Clin Invest. 2015;125:1679–1691. doi: 10.1172/JCI73780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimm A, Eckert A. Brain aging and neurodegeneration: From a mitochondrial point of view. J Neurochem. 2017;143:418–431. doi: 10.1111/jnc.14037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loeb LA, Wallace DC, Martin GM. The mitochondrial theory of aging and its relationship to reactive oxygen species damage and somatic mtDNA mutations. Proc Natl Acad Sci USA. 2005;102:18769–18770. doi: 10.1073/pnas.0509776102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kauppila TES, Kauppila JHK, Larsson NG. Mammalian mitochondria and aging: An update. Cell Metab. 2017;25:57–71. doi: 10.1016/j.cmet.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 24.Piantadosi CA. Regulation of mitochondrial processes by protein S-nitrosylation. Biochim Biophys Acta. 2012;1820:712–721. doi: 10.1016/j.bbagen.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clementi E, Brown GC, Feelisch M, Moncada S. Persistent inhibition of cell respiration by nitric oxide: Crucial role of S-nitrosylation of mitochondrial complex I and protective action of glutathione. Proc Natl Acad Sci USA. 1998;95:7631–7636. doi: 10.1073/pnas.95.13.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dahm CC, Moore K, Murphy MP. Persistent S-nitrosation of complex I and other mitochondrial membrane proteins by S-nitrosothiols but not nitric oxide or peroxynitrite: Implications for the interaction of nitric oxide with mitochondria. J Biol Chem. 2006;281:10056–10065. doi: 10.1074/jbc.M512203200. [DOI] [PubMed] [Google Scholar]
- 27.Mehta DR, Ashkar AA, Mossman KL. The nitric oxide pathway provides innate antiviral protection in conjunction with the type I interferon pathway in fibroblasts. PLoS One. 2012;7:e31688. doi: 10.1371/journal.pone.0031688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komatsu T, Ireland DDC, Chen N, Reiss CS. Neuronal expression of NOS-1 is required for host recovery from viral encephalitis. Virology. 1999;258:389–395. doi: 10.1006/viro.1999.9734. [DOI] [PubMed] [Google Scholar]
- 29.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Y, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Giacomo G, Rizza S, Montagna C, Filomeni G. Established principles and emerging concepts on the interplay between mitochondrial physiology and S-(De)nitrosylation: Implications in cancer and neurodegeneration. Int J Cell Biol. 2012;2012:361872. doi: 10.1155/2012/361872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rizza S, et al. S-nitrosylation of the mitochondrial chaperone TRAP1 sensitizes hepatocellular carcinoma cells to inhibitors of succinate dehydrogenase. Cancer Res. 2016;76:4170–4182. doi: 10.1158/0008-5472.CAN-15-2637. [DOI] [PubMed] [Google Scholar]
- 33.Seo AY, et al. New insights into the role of mitochondria in aging: Mitochondrial dynamics and more. J Cell Sci. 2010;123:2533–2542. doi: 10.1242/jcs.070490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan DC. Mitochondria: Dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 35.Chan DC. Fusion and fission: Interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 36.Kasahara A, Scorrano L. Mitochondria: From cell death executioners to regulators of cell differentiation. Trends Cell Biol. 2014;24:761–770. doi: 10.1016/j.tcb.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 37.Cho D-HH, et al. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smirnova E, Shurland D-L, Ryazantsev SN, van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bordt EA, et al. The putative Drp1 inhibitor mdivi-1 is a reversible mitochondrial complex I inhibitor that modulates reactive oxygen species. Dev Cell. 2017;40:583–594.e6. doi: 10.1016/j.devcel.2017.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarkar S, et al. Complex inhibitory effects of nitric oxide on autophagy. Mol Cell. 2011;43:19–32. doi: 10.1016/j.molcel.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopez-Rivera E, et al. Inducible nitric oxide synthase drives mTOR pathway activation and proliferation of human melanoma by reversible nitrosylation of TSC2. Cancer Res. 2014;74:1067–1078. doi: 10.1158/0008-5472.CAN-13-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komatsu M, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 43.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lazarou M, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chung KKK, et al. S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 46.Yao D, et al. Nitrosative stress linked to sporadic Parkinson’s disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc Natl Acad Sci USA. 2004;101:10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shinozaki S, et al. Inflammatory stimuli induce inhibitory S-nitrosylation of the deacetylase SIRT1 to increase acetylation and activation of p53 and p65. Sci Signal. 2014;7:ra106. doi: 10.1126/scisignal.2005375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puca AA, et al. A genome-wide scan for linkage to human exceptional longevity identifies a locus on chromosome 4. Proc Natl Acad Sci USA. 2001;98:10505–10508. doi: 10.1073/pnas.181337598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valentini E, et al. Analysis of the machinery and intermediates of the 5hmC-mediated DNA demethylation pathway in aging on samples from the MARK-AGE study. Aging (Albany NY) 2016;8:1896–1922. doi: 10.18632/aging.101022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Truong TP, et al. Age-dependent decrease of DNA hydroxymethylation in human T cells. J Clin Exp Hematop. 2015;55:1–6. doi: 10.3960/jslrt.55.1. [DOI] [PubMed] [Google Scholar]
- 51.Huang Y, Rao A. Connections between TET proteins and aberrant DNA modification in cancer. Trends Genet. 2014;30:464–474. doi: 10.1016/j.tig.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Putiri EL, et al. Distinct and overlapping control of 5-methylcytosine and 5-hydroxymethylcytosine by the TET proteins in human cancer cells. Genome Biol. 2014;15:R81. doi: 10.1186/gb-2014-15-6-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu C, et al. Decrease of 5-hydroxymethylcytosine is associated with progression of hepatocellular carcinoma through downregulation of TET1. PLoS One. 2013;8:e62828. doi: 10.1371/journal.pone.0062828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laukka T, et al. Fumarate and succinate regulate expression of hypoxia-inducible genes via TET enzymes. J Biol Chem. 2016;291:4256–4265. doi: 10.1074/jbc.M115.688762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao M, et al. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng L, et al. Fumarate induces redox-dependent senescence by modifying glutathione metabolism. Nat Commun. 2015;6:6001. doi: 10.1038/ncomms7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X, et al. Impaired balance of mitochondrial fission and fusion in Alzheimer’s disease. J Neurosci. 2009;29:9090–9103. doi: 10.1523/JNEUROSCI.1357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Demetrius L. Of mice and men. When it comes to studying ageing and the means to slow it down, mice are not just small humans. EMBO Rep. 2005;6(Suppl 1):S39–S44. doi: 10.1038/sj.embor.7400422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Narendra D, Tanaka A, Suen D-F, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ozawa K, et al. S-nitrosylation regulates mitochondrial quality control via activation of parkin. Sci Rep. 2013;3:2202. doi: 10.1038/srep02202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oh CK, et al. S-nitrosylation of PINK1 attenuates PINK1/parkin-dependent mitophagy in hiPSC-based Parkinson’s disease models. Cell Rep. 2017;21:2171–2182. doi: 10.1016/j.celrep.2017.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Molotkov A, et al. Stimulation of retinoic acid production and growth by ubiquitously expressed alcohol dehydrogenase Adh3. Proc Natl Acad Sci USA. 2002;99:5337–5342. doi: 10.1073/pnas.082093299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rizza S, Montagna C, Di Giacomo G, Cirotti C, Filomeni G. S-nitrosation and ubiquitin-proteasome system interplay in neuromuscular disorders. Int J Cell Biol. 2014;2014:428764. doi: 10.1155/2014/428764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bellinger AM, et al. Hypernitrosylated ryanodine receptor calcium release channels are leaky in dystrophic muscle. Nat Med. 2009;15:325–330. doi: 10.1038/nm.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garfield AS. Derivation of primary mouse embryonic fibroblast (PMEF) cultures. In: Ward A, Tosh D, editors. Mouse Cell Culture: Methods and Protocols. Humana Press; Totowa, NJ: 2010. pp. 19–27. [DOI] [PubMed] [Google Scholar]
- 68.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barsoum MJ, et al. Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. EMBO J. 2006;25:3900–3911. doi: 10.1038/sj.emboj.7601253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marchi S, Bonora M, Patergnani S, Giorgi C, Pinton P. Methods to assess mitochondrial morphology in mammalian cells mounting autophagic or mitophagic responses. Methods Enzymol. 2017;588:171–186. doi: 10.1016/bs.mie.2016.09.080. [DOI] [PubMed] [Google Scholar]
- 71.Pettersen EF, et al. UCSF Chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.