Significance
Cellulose is synthesized at the plasma membrane by cellulose synthase complexes (CSCs). The ability to deliver CSCs to discrete sites at the cell surface is critical for cellulose synthesis. We discovered that the de novo secretion of CSCs is mediated by cooperation among a recently identified cellulose synthase interacting protein, exocyst complex, and a plant-specific protein PATROL1 in Arabidopsis thaliana. We present a timeline of events for exocytosis of CSCs. CSCs represent unique cargo proteins that are not conserved in the mammalian or yeast system. Plants offer unique opportunities to characterize the function and regulation of exocytosis that may provide insight into the evolution of exocytosis in eukaryotes.
Keywords: exocytosis, exocyst complex, cellulose synthase complex, plasma membrane, microtubule
Abstract
Cellulose synthesis occurs exclusively at the plasma membrane by cellulose synthase complexes (CSCs). Therefore, delivery of CSCs to discrete sites at the plasma membrane is critical for cellulose synthesis. Despite their significance, the delivery of CSCs is poorly understood. Here we used proteomics approaches, functional genetics, and live cell imaging to show that the de novo secretion of CSCs is mediated by cooperation among cellulose synthase interactive 1 (CSI1), the plant-specific protein PATROL1, and exocyst complex in Arabidopsis thaliana. We propose that CSI1 plays a role in marking the docking site, which allows CSCs-containing vesicles access to the plasma membrane through its interaction with microtubules. PATROL1 assists in exocytosis by its interaction with multiple components, including CSI1, CSCs, and exocyst subunits. Both PATROL1 and the exocyst complex determine the rate of delivery of CSCs to the plasma membrane. By monitoring the exocyst complex, PATROL1, CSI1, and CSCs dynamics in real time, we present a timeline of events for exocytosis of CSCs. Our findings provide unique insights into the evolution of exocytosis in eukaryotes.
The invention of internal membranes has a profound change in cellular organization. The complex cellular compartmentalization is the driving force for the structural and functional complexity in eukaryotic cells. During the evolution of this course, eukaryotic cells develop a sophisticated system to regulate protein trafficking between organelles and the plasma membrane (PM). Exocytosis is a cellular process in which membrane-bound intracellular vesicles transiently fuse with the cell membrane and their content is secreted into the extracellular space. Exocytosis is best studied in synaptic vesicles that involve docking of synaptic vesicles at active zones, priming steps that render the vesicles competent to fuse with the PM and the final process of fusion. Studies in several eukaryotic systems, including mammals, Drosophila, and plants, have revealed that the exocyst complex is the central hub of multiple membrane trafficking processes (1–6). Yeast, mammals, and plants appeared to have conserved factors involved in vesicle trafficking to the cell surface, including the exocyst complex, SNARE proteins, and Rab GTPases. There is a wealth of knowledge about the role and regulation of the exocyst complex in yeast and mammals (7–10). However, most factors undergo gene duplications, as exemplified by 36 exocyst subunits, 65 SNARE proteins, and 57 Rab GTPases in Arabidopsis. Therefore, it is difficult to apply the existing information of exocyst complex-mediated vesicle trafficking in yeast and mammals to plants.
Over the past decades, a significant number of studies revealed that Arabidopsis is an excellent model plant for studying the exocytotic network (2, 6, 11–16). Plant cells use exocytosis to secrete the cell wall components, which may provide important insights for remodeling the extracellular matrix in other organisms. The ability to deliver newly synthesized proteins to discrete sites at the cell surface is critical for plant cells to grow anisotropically. It is particularly important in the case of cellulose synthase (CESA) proteins, which must be localized to the PM to synthesize cellulose microfibrils. The regulation of steady-state levels of CESA proteins depends not only on transcription/translation, but also on the rates of endocytosis and exocytosis. Recent studies showed that CESA is a cargo protein of the clathrin-mediated endocytosis (CME). The abundance of cellulose synthase complexes (CSCs) is affected in several CME-deficient mutants, including the medium subunit of CME adaptor protein 2 (AP2) complex and members in the TPLATE complex (TPC)/TSET (17–20). The newly endocytosed CSCs are subsequently trafficked to the trans-Golgi network/early endosome (TGN/EE), where they could be recycled back to the PM (19). It also has been reported that under abiotic stress, massive internalization of PM-localized CSCs occurs through AP2/CME. The internalized CSCs are temporarily stored in the small CESA compartments/microtubule-associated cellulose synthase compartments (SmaCC/MASCs); and are recycled to the PM after release of stress. Importantly, the formation of SmaCC/MASCs also requires the cellulose synthase interactive 1 (CSI1), a protein previously proved to be a linker between the cortical microtubules and CSCs (21–23).
Little is known about the de novo secretion of CSCs to the PM. Presumably, delivery of cargo to the PM requires the exocyst complex, an evolutionally conserved complex present in yeast, mammals, and plants. The exocyst complex is comprised of eight subunits: Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70, and Exo84. While null mutants of individual subunits led to severe growth and secretion defect in yeast and early lethality in mice and flies, null mutants in several exocyst subunits in Arabidopsis resulted in mild root hair phenotype or pollen tube growth defects (2, 4, 15, 24–26). Unlike its yeast and mammalian counterparts that have a single copy of each of the eight subunits, exocyst subunits in plants undergo gene duplications, exemplified by 2 isoforms of Sec3, Sec5, and Sec15, 3 isoforms of Exo84, and 23 isoforms of Exo70 in Arabidopsis (27). The possibility of a large number of distinct exocyst complexes in plants is compatible with the versatility and plasticity of plant life. The exocyst complex has been speculated to mediate the insertion of CSCs to the PM (19). However, how exocyst complex-mediated exocytosis is involved in cellulose synthesis is lacking any mechanistic details.
Here we report that in Arabidopsis, the de novo secretion of CSCs to the PM is mediated by cooperation among CSI1, the exocyst complex, and a recently identified protein, PATROL1 (PTL1). Previous studies have shown that PATROL1 is involved in the transportation of the plant H+-ATPase, AHA1, to and from the PM in guard cells and subsidiary cells (28, 29). In our present study, both the exocyst complex and PTL1 copurified with CSI1 and CESA6. Mutations of either exocyst subunits or PATROL1 affect the delivery of CSCs to the PM. Our results suggest that the exocyst complex and PTL1 are required for tethering/fusion of CSCs to the target membrane, while the CSI1 proteins serve as landmarks on the microtubules to define the position of secretion. By monitoring the exocyst complex, PTL1, CSI1, and CSC dynamics in real time, we observed a temporal sequence of de novo secretion of CSCs.
Results
Multiple Approaches Were Used to Identify Proteins Involved in CSC Trafficking.
Because cellulose synthesis occurs exclusively at the PM, the trafficking of CESA to the PM is an important mechanism for controlling the steady-state levels of CESA. To identify proteins involved in trafficking of CESA, multiple approaches were used. In a yeast two-hybrid method, the central domain (amino acids residues 323–863) of CESA6 was used as a bait to screen an Arabidopsis seedling library. A total of 25 nonduplicated positive clones were identified from 95.87 million yeast transformants (22). Among putative CESA interacting clones, one clone encodes Sec5B (Fig. S1 and Dataset S1). In an affinity purification method, the alpaca-based GFP Nano-Traps were used to isolate YFP-CESA6 and its associated proteins by an ultrasensitive high-throughput nano-LC/MS system. CSI1 protein appeared as top hit among the interactive candidates and multiple primary CESA proteins were copurified with CESA6, verifying the feasibility of the affinity-purification approach. Several exocyst subunits were copurified with YFP-CESA6 (Fig. S1 and Dataset S2). The putative interactions between exocyst subunits and CESA open up the possibility of probing the trafficking of CESA through exocyst complex-mediated exocytosis.
To further dissect the trafficking of CSCs, we also performed coimmunoprecipitation (co-IP) to identify CSI1 interacting proteins. Two different tags were generated for co-IP experiments. In addition to a GFP-tag (30), we used a GS tag that combines two IgG-binding domains of protein G with a streptavidin-binding peptide, separated by low-temperature active rhinovirus 3 C protease cleavage sites (Fig. S2A). To mimic the complex formation in vivo, we used the native promoter to drive the expression of a CSI1-GS construct. The CSI1-GS construct was introduced into csi1-3 and fully complemented the cell-expansion defect in csi1-3, indicating that the GS tag does not interfere with CSI1 function (Fig. S2B). The CSI1-associated proteins were enriched through two-step affinity purification before identification by the ultrasensitive high-throughput nano-LC/MS system (Fig. S2C). Similar to CESA, multiple exocyst subunits were copurified with CSI1 (Fig. S1 and Dataset S2), further indicating the potential involvement of the exocyst complex in trafficking of CSCs.
In addition to exocyst subunits, we identified PTL1 among the top hits using both GFP-tagged and GS-tagged CSI1. In a reciprocal co-IP, CSI1 was copurified with PTL1, validating the association of CSI1 and PTL1 (Fig. S1 and Dataset S2). The CSI1–PTL1 interaction was further validated by in vitro pull-down assays. GST-tagged PTL1 was able to pull down His-CSI1, while no detectable His-CSI1 was pulled down by purified GST. We also determined that direct interaction occurs between PTL1 and the C2 domain of CSI1 (Fig. S3A). In the co-IP, Sec10, a core component of the exocyst complex, was copurified with PTL1. The direct interaction between Sec10 and PTL1 was verified by in vitro pull-down assays (Figs. S1 and S3B). No direct interaction between PTL1 and truncated fragments of two exocyst subunits, Sec5B and Sec6, were detected in the pull-down assays (Fig. S3C). The negative interaction between PTL1 and Sec5B or Sec6 is consistent with the results that GFP–PTL1 copurified with Sec10, but not Sec5B or Sec6. However, we used the truncations of Sec5B and Sec6 for in vitro pull-down assays as we failed to express full-length proteins of Sec5B and Sec6 in Escherichia coli. Therefore, we cannot rule out the possibility that PTL1 interacts with full-length Sec5B and Sec6 or other regions of Sec5B and Sec6.
PTL1 Is Functionally Associated with the Exocyst Complex.
Several subunits of the exocyst complex have been shown to label punctate both at the PM and within the cytoplasm (31–33). PTL1 has also been shown to label punctate in Arabidopsis guard cells and subsidiary cells (29). However, these studies used the constitutive 35S promoter that is known to cause high levels of gene expression. Therefore, the fluorescent protein (FP) fusion proteins might not faithfully report the trafficking and localization of native proteins. To avoid artifacts generated by overexpression, we constructed various FP fusion proteins for PTL1 using the native promoter. All of the FP–PTL1 constructs were transformed in to a T-DNA knockout line for PATROL1 (patrol1-2, SALK_018676C) (Fig. S4 A and B), which led to mild growth retardation compared with WT plants (Fig. 1A and Fig. S4 E and F). FP–PTL1 fully complemented the patrol1-2 mutant phenotype, suggesting that fluorescently tagged protein is functional (Fig. 1A). In contrast to a previous finding that endosomal localization of PTL1 mirrors Munc13-1–YFP in mouse brain cells (28), there were almost no overlaps between GFP–PTL1 and various endosomal markers (34), including ARA7, RabC1, RabG3F, Rha1, RabA1E, and RabA1G (Fig. S5). The discrepancy of localization may be due to the 35S promoter that is known to cause high levels of gene expression. Alternatively, PTL1 may have differential subcellular localization in different cell types.
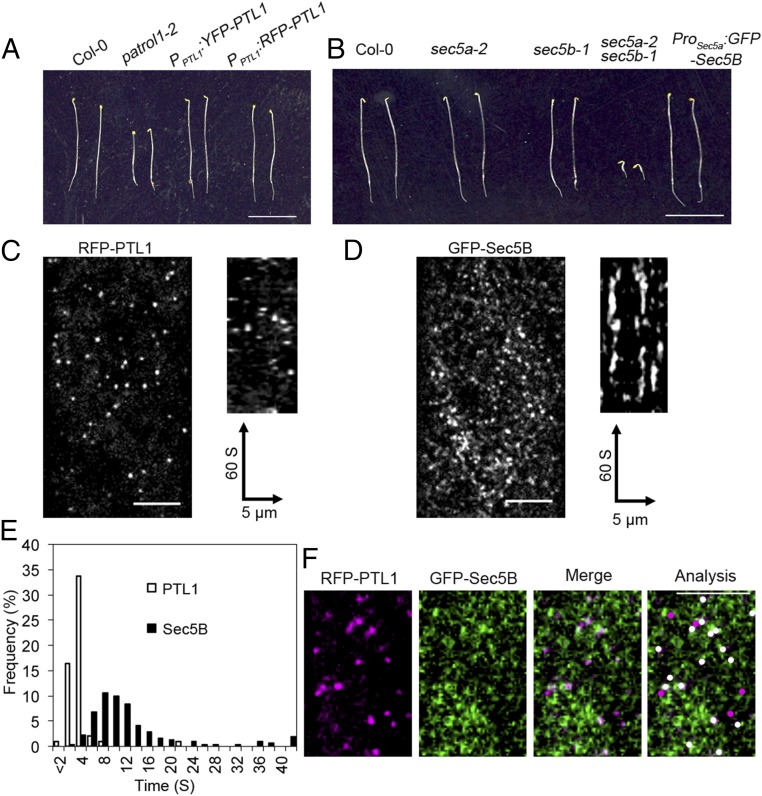
Fig. 1.
Visualization of PTL1 and Sec5B in living cells. (A) Introducing either YFP–PTL1 or RFP–PTL1 under the PTL1 native promoter restores etiolated hypocotyl growth in the patrol1-2 mutants. Seedlings were grown on 1/2 MS medium in dark for 4 d before imaging. (Scale bar, 1 cm.) (B) The Sec5A and Sec5B genes in Arabidopsis have redundant functions. Single mutants of either gene (sec5a-2 and sec5b-1) don’t show growth defect, but the double mutant sec5a-2 sec5b-1 is seedling lethal. Introduction of GFP–Sec5B under the native Sec5A promoter complemented the growth defect of double mutants. Seedlings were grown on 1/2 MS medium in dark for 4 d before imaging. (Scale bar, 1 cm.) (C and D) Single-frame images (Left) and kymographs (Right) showing the subcellular localizations of RFP–PTL1 (C) and GFP–Sec5B (D) in living cells (Left). Images were obtained from the epidermal cells of the Arabidopsis etiolated hypocotyls. Kymographs were derived from a 60-s movie with 1-s interval for both C and D (Right). (Scale bar, 5 μm.) (E) Histogram showing the distribution of particle lifetimes of PTL1 and Sec5B measured in kymographs. n = 298 particles from seven cells for PTL1; n = 312 particles from nine cells for Sec5B. (F) PTL1 particles colocalize with Sec5B on the PM. Shown are single images obtained from RFP–PTL1 and GFP–Sec5B simultaneously, and a merged image generated by combining both signals. In the analysis panel, white dots represent colocalized RFP–PTL1 and GFP–Sec5B. (Scale bar, 5 μm.)
The Arabidopsis genome encodes two isoforms of Sec10 genes, namely Sec10A and Sec10B. They are tandem duplications in the genome. Analysis of the T-DNA insertion mutant in Sec10A and Sec10B revealed no morphological defect. Because of the tight genetic linkage, it’s virtually impossible to obtain sec10a sec10b-null mutant (35). Therefore, we turned to genetically amenable sec5a sec5b for functional studies and localization analysis. Single mutants of Sec5A and Sec5B showed no morphological difference to WT plants, while mutation of both Sec5A and Sec5B led to seedling lethality (Fig. 1B and Fig. S4 A, C, G, and H). A GFP–Sec5B construct was transformed into the sec5a-2/sec5a-2 sec5b-1/+ line and GFP–Sec5B transgenic lines in the sec5a-2 sec5b-1 double mutants were identified by PCR genotyping. The reliability of FP–Sec5B fusions was demonstrated by mutant complementation (Fig. 1B). In the hypocotyl cells of 2.5-d-old etiolated Arabidopsis seedlings, both RFP–PTL1 and GFP–Sec5B appeared as distinctive puncta that closely associated with the PM (Fig. 1 C and D and Movies S1 and S2). Both RFP–PTL1 and GFP–Sec5B exhibited an appearing and disappearing behavior, with a transient pausing at the PM (Movies S1 and S2). Quantification results showed resident lifetimes were 2–4 s for PTL1 particles, while GFP–Sec5B remained at the PM for 8–12 s (Fig. 1E). To examine the correlation between PTL1 and the exocyst complex, we generated a transgenic line expressing both RFP–PTL1 and GFP–Sec5B. In the colocalization analysis, RFP–PTL1 and GFP–Sec5B was tracked temporally. Two particles were functionally colocalized if they were spatiotemporally colocalized for at least two frames of live-cell imaging. We analyzed 791 RFP–PTL1 particles from 16 cells in 7 seedlings and 85.8 ± 5.38% of RFP–PTL1 colocalized with GFP–Sec5B (Fig. 1F).
Both PTL1 and Subunits of Exocyst Complex Are Required for the Delivery of CSCs to the PM.
Consistent with a previously identified patrol1 mutant (patrol1-1) that harbored a point mutation, which led to a premature stop codon, patrol1-2 showed mild growth retardation (28). Hypocotyls in etiolated seedlings of patrol1-2 were shorter than WT plants, a phenotype often observed in cellulose-deficient mutants (Fig. 1A). Consistent with the predicted role of PTL1 in cellulose synthesis, patrol1-2 mutants showed a 12.6% reduction in the crystalline cellulose content (Fig. S6A).
As we cannot gather enough materials for cellulose content analysis because of seedling lethality of sec5a-2 sec5b-1, we generated a pollen-rescued sec6-3 mutant in which a pollen-specific transmission defect was complemented by expression of the WT Sec6 coding sequence driven by the pollen-specific Lat52 promoter (Fig. S6B). Homozygous pLat52::Sec6 sec6-3 mutants were viable and propagable, which has enabled us to use functional genetics to investigate how defects in exocytosis interfere with cellulose synthesis. pLat52::Sec6 sec6-3 transgenic lines exhibited growth defect in etiolated seedlings as well as light-grown plants, and a reduction in crystalline cellulose content (Fig. S6 A and C). Cellulose deficiency and reduced growth is consistent with the hypothesis that the integrity of the exocyst complex is important for cellulose biosynthesis.
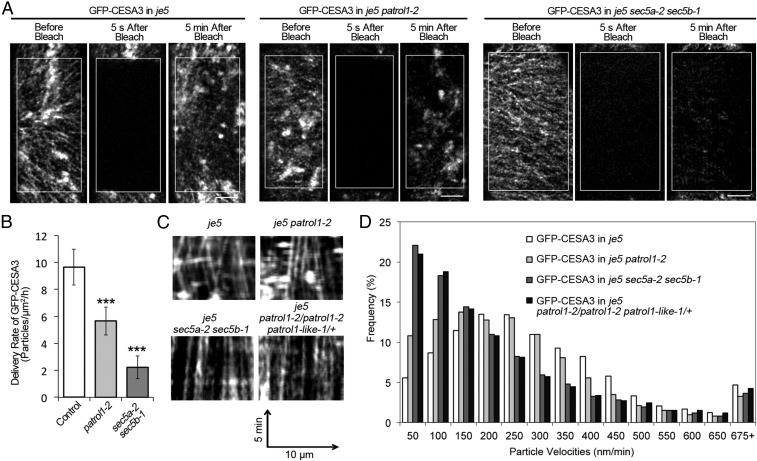
To better understand the role of PTL1 and exocyst complex in the cellulose synthesis, transgenic lines expressing either GFP–CESA3 or YFP–CESA6 were crossed into patrol1-2, sec5a-2 sec5b-1 and pLat52::Sec6 sec6-3, respectively. In case of analysis of GFP–CESA3 in sec5a-2 sec5b-1, segregating populations from parental sec5a-2/sec5a-2 sec5b-1/+ lines were imaged before PCR genotyping to confirm the double homozygous background. To evaluate the delivery efficiency of CSCs, a fluorescence recovery after photobleaching (FRAP) assay was performed. Newly delivered particles within the photobleached area were monitored after photobleaching. Only those particles that showed a transient stationary motion before moving with constant velocities were considered as newly delivered CSCs (17, 36). The delivery rate of CESAs in patrol1-2 mutants and in both exocyst mutants was significantly lower than that in WT (Fig. 2 A and B and Fig. S6 E and F).
Fig. 2.
Both PTL1 and subunits of exocyst are required for the delivery of CSCs to the PM. (A) Single-frame images displaying the PM CSCs before, 5 s and 5 min after photobleaching. FRAP assays were performed in the control GFP–CESA3 in je5 lines, the patrol1-2 mutants, and the sec5a-2 sec5b-1 double mutants. White boxes mark the photobleached area. (Scale bar, 5 μm.) (B) Quantifications of the delivery rate of CSCs from the FRAP assays descripted in A. n = 18 Regions of interest (ROIs) from nine cells for the GFP-CESA3 control line; n = 28 ROIs from 18 cells in the patrol1-2 mutant line (GFP–CESA3 in je5 patrol1-2), n = 22 ROIs from nine cells in the sec5a-2 sec5b-1 double-mutant line (GFP–CESA3 in je5 sec5a-2 sec5b-1). Error bars represent SD. ***P < 0.0001. (C and D) The velocity of PM CSCs is significantly reduced in both exocyst mutants and PATROL1 PATROL1-like double mutants. (C) Kymographs derived from 5-min movies with 5-s intervals. (D) Histograms displaying the distribution of CSC particle velocities. Average velocities in control line, patrol1-2, sec5a-2 sec5b-1, and patrol1-2/patrol1-2 patrol1-like-1/+ mutants are 302.89 ± 201.41 nm/min, 256.96 ± 195.75 nm/min, 218.67 ± 216.92 nm/min, and 225.66 ± 217.19 nm/min, respectively. n = 6,890 particles from six cells for the control line (GFP–CESA3 in je5), n = 8,976 particles from six cells for the patrol1-2 mutant line (GFP–CESA3 in je5 patrol1-2), n = 8,158 particles from 11 cells for the sec5a-2 sec5b-1 double mutant line (GFP-CESA3 in je5 sec5a-2 sec5b-1), n = 8,692 particles from nine cells for the PATROL1 PATROL1-like double mutants (GFP–CESA3 in je5 patrol1-2/patrol1-2 patrol1-like-1/+).
During the analysis of delivery rate of CSC, we noticed that a significant number of particles showed reduced mobility in sec5a-2 sec5b-1 and pLat52::Sec6 sec6-3. Analysis showed that the velocity of CSCs was significantly reduced. More than 20% of CSCs remains stationary in sec5a-2 sec5b-1 compared with 5% in WT (Fig. 2 C and D). However, in patrol1-2 mutants, the average motility of the PM-localized CSCs was slightly lower than WT (Fig. 2 C and D). The mild phenotype of patrol1-2 may be attributed to functional redundancy as PATROL1 has 5 PATROL1-like genes in Arabidopsis. We crossed patrol1-2 to its closest homolog patrol1-like-1 and double mutants were embryo lethal (Fig. S7 and Dataset S3). Similar to sec5a-2 sec5b-1, patrol1-2/patrol1-2 patrol1-like-1/+ had stunt dark-grown seedlings and mature plants (Fig. S7). The average velocity of CSCs was reduced to 226 ± 217 nm/min compared with 302.89 ± 201.41 nm/min in WT. A similar percentage of CSCs remained stationary in patrol1-2/patrol1-2 patrol1-like-1/+ compared with that of sec5a-2 sec5b-1 (Fig. 2 C and D and Movie S3). We hypothesize that stationary CSCs represent faulty CSCs that were unable to complete fusion/scission. Moreover, in both PATROL1/PATROL1-LIKE and the exocyst complex mutants, the density of PM CSCs was significantly higher than that in WT seedlings (Fig. S8). Taken together, these results suggest that both PTL1 and the exocyst complex are required for the secretion of CSCs to the PM.
CSI1 Serves as a Landmark in Exocyst Complex-Mediated CSC Secretion.
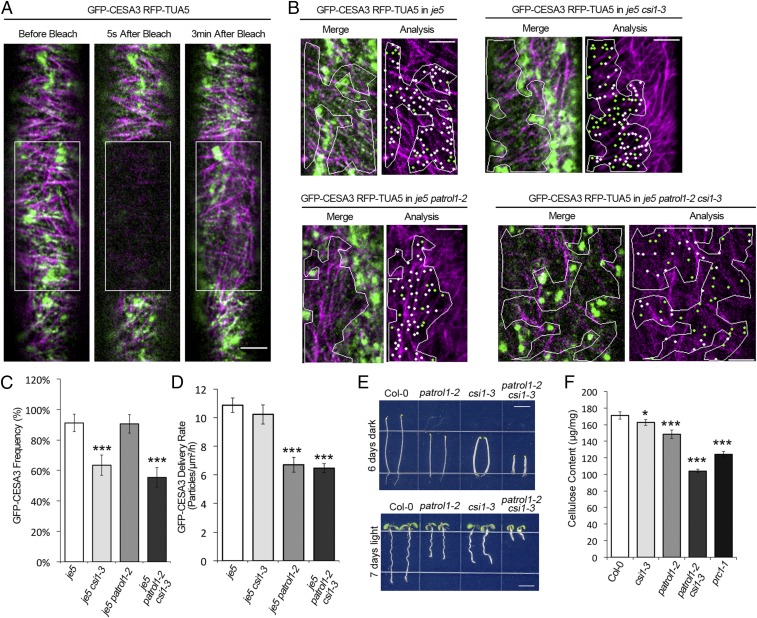
It has been shown that the delivery of CSCs to the PM is directed by cortical microtubules (37); and that CSI1 serves as a linker between CSCs and cortical microtubules (23). Based on the observations that CSI1 closely associated with PTL1 and the exocyst complex, we hypothesized that CSI1 might function as a landmark, which positions the tethering of the exocyst complex and CSCs along the cortical microtubules. To test this hypothesis, a transgenic line coexpressing GFP–CESA3 and mCherry-TUA5 was crossed into csi1-3. FRAP experiments were performed to capture nascent delivery of CSCs and the position of delivery event was analyzed for coincidence with microtubules. Quantifications showed that in control (GFP–CESA3 mCherry-TUA5 in je5) plants, over 90% of the nascent CSCs were delivered to the cortical microtubules. However, in csi1-3, only 63% of newly delivered CSCs were positioned to microtubules, indicating that CSI1 is required for directing CSCs to microtubules. In contrast, the tethering of CSCs along the cortical microtubules was not affected in patrol1-2 mutants (Fig. 3 B and C). Furthermore, the delivery rate of CSCs was not affected in csi1-3 mutants, but it was significantly reduced in patrol1-2 mutants (Fig. 3D).
Fig. 3.
CSI1 serves as a landmark for the delivery of CSCs. (A) To visualize the cortical microtubule-directed delivery of nascent CSCs, a FRAP assay was performed in dual-labeled lines containing both GFP–CESA3 and mCherry-TUA5. Single-frame images were obtained from two fluorescent channels simultaneously at 5 s before, 5 s after, and 3 min after photobleaching. Boxes mark the photobleached area. (Scale bar, 5 μm.) (B) Representative merge images displaying the coincidence between newly delivered CSCs and microtubules. FRAP assays were performed in control (GFP–CESA3 mCherry-TUA5 in je5), csi1-3, patrol1-2, and patrol1-2 csi1-3 mutants, respectively. For analysis, ROI that exclude the underlying intracellular compartments were marked by white outlines. White and green dots mark CSC particles that were delivered to microtubule-occupied regions and microtubule-free regions, respectively. (Scale bars, 5 μm.) (C) CSI1 is required for position the delivery of CSCs to microtubules. The frequency of coincidence between CSC delivery site and microtubule in control, csi1-3, patrol1-2, and patrol1-2 csi1-3 mutants from B was quantified, respectively. n = 383 particles from seven cells for control cells (GFP–CESA3 mCherry-TUA5 in je5), n = 286 particles from five cells for csi1-3, n = 203 particles from six cells for patrol1-2, n = 126 particles from five cells for patrol1-2 csi1-3, respectively. Error bars represent SD. ***P < 0.0001. (D) CSI1 does not affect the overall delivery rate of CSC. The delivery rate of CSCs was measured from observations in B. n = 538 particles from 10 cells for control cells (GFP–CESA3 mCherry-TUA5 in je5); n = 347 particles from 10 cells for csi1-3, n = 353 particles from 10 cells for patrol1-2, n = 387 particles from nine cells for patrol1-2 csi1-3, respectively. Error bars represent SD. ***P < 0.0001. (E) Morphology of patrol1-2 csi1-3 mutants shows genetic interactions between PTL1 and CSI1. Seedlings were grown on 1/2 MS medium in dark for 6 d (Upper) or on 1/2 MS medium plus 1% sucrose under light for 7 d (Lower) before imaging. (Scale bars, 1 cm.) (F) The graph compares the crystalline cellulose content of 4-d-old etiolated seedlings of Col-0, csi1-3, patrol1-2, csi1-3 patrol1-2, and prc1-1. Error bars represent SD. *P < 0.01, ***P < 0.0001.
Because PTL1 and CSI1 influence the trafficking of CSCs to the PM in different ways, patrol1-2 csi1-3 mutants were generated. In patrol-2 csi1-3 mutants, both the delivery rate and the percentage of newly delivered CSCs to the microtubules were reduced, but it showed no enhancement of either phenotype over one of the single mutants (Fig. 3 C and D and Dataset S4). However, patrol-2 csi1-3 mutants exhibited enhanced growth defects compared with one of the single mutants and showed a further reduction of the crystalline cellulose content in etiolated seedlings (Fig. 3 E and F, Fig. S4 E and F, and Dataset S4). Taken together, the above data suggest that CSI1 and PTL1 play distinct roles in the CSCs delivery process. Specifically, CSI1 is required for providing a landmark that allows CSC-containing vesicles access to the cortical microtubules, whereas PTL1 is involved in late step of delivery of CSCs.
Temporal Sequence of CSCs Delivery Event.
To capture the sequence of the delivery event, we generated several dual-labeled transgenic lines coexpressing RFP–CSI1 and GFP–CESA6, RFP–PTL1 and YFP–CESA6, and GFP–SEC5B and RFP–CESA3, respectively. A FRAP assay was performed and both fluorescent channels were monitored in real time. We observed that RFP–CSI1 appeared about 44 s prior (41.88 ± 11.38 s after photobleaching, n = 16) to that of GFP–CESA3 (85.93 ± 19.34 s after photobleaching, n = 16) (Fig. 4 A and B). In all events, the appearance of a CSI1 particle was followed by the coappearance of a Golgi-tethered CESA. After static motion about 1–2 min, both RFP–CSI1 and GFP–CESA3 particles displayed linear motility (Fig. 4A), indicating cellulose synthesis after a successful tethering and fusion. The tethering of CESA to the PM was accompanied by the appearance of both PTL1 and Sec5B. PTL1 proteins transiently associated with CSCs (1–2 s) at the new delivery site, while the GFP–Sec5B remained with CSCs much longer (∼10 s) before steady movement of CSCs (Fig. 4 E and F).
Fig. 4.
CSI1, PTL1, and the exocyst sequentially regulate the delivery of CSCs. (A) CSI1 shows a strong bias to appear before the delivery of CSCs. To visualize the delivery sequence of CSI1 and CSCs, a FRAP assay was performed in a dual-labeled line expressing RFP–CSI1 and GFP–CESA6. Kymographs were generated from a 10-min movie (5-s interval) to visualize the dynamics of particles over time. Shown is a representative kymograph displaying a CSC delivery event, in which CSC was delivered to an existing RFP–CSI1. The delivery of CSC is accompanied by pausing of the Golgi apparatus. A cartoon representation of the delivery event is shown on the Right. (B) Quantification of the time required for the delivery of CSCs and CSI1 in A. n = 16 particles for both GFP–CESA6 and RFP–CSI1. Red bar represents average time. P < 0.0001. (C) In the patrol1-2 mutants, CSI1 still appears before the delivery of CSCs. The same FRAP assay, as describe in A, was performed in the patrol1-2 mutants expressing RFP–CSI1 and YFP–CESA6. Shown is a representative delivery event and the corresponding cartoon representation. (D) Quantification of the delivery time of CSI1 and CSCs in the patrol1-2 mutants. n = 16 particles for both YFP–CESA6 and RFP–CSI1. Red bar represents average time. P < 0.001. (E) PTL1 shows a strong bias to appear at the same time as CSCs. FRAP assays were performed in dual-labeled lines of RFP–PTL1 and YFP–CESA6. Kymographs were derived from 6-min movies (1-s intervals). Arrowheads point to PTL1 particles that appeared at the same time as delivery of CSCs to the PM. n = 105 observations from eight cells. A cartoon representation of the delivery event is shown on the Right. (F) Exocyst shows a strong bias to appear at the same time as CSCs. FRAP assays were performed in dual-labeled lines of GFP–Sec5B and RFP–CESA3. Kymographs were derived from 5-min movies (2-s intervals). Shown are representative kymographs where an exocyst particle labeled by GFP–Sec5B coappeared with a CSC particle labeled by RFP–CESA3. n = 98 observations from six cells. A cartoon representation of the delivery event is shown on the Right.
Given the direct interaction between PTL1 and CSI1, we then examined the in vivo correlation between PTL1 and CSI1 in transgenic plants coexpressing RFP–PTL1 and GFP–CSI1 (30). We observed that over 70% of RFP–PTL1 colocalized with GFP–CSI1 (70.53 ± 5.8%, n = 556 from nine cells) (Fig. S9). Considering the short resident lifetimes of PTL1 at the PM, this could indicate a transient association between PTL1 and CSI1. We therefore monitored the temporal behavior of the colocalized particles. We found that in all cases (147 events in 6 cells), PTL1 preferentially docked on an existing CSI1 particle before disappearing from the PM (Fig. 5 A and C and Movie S4).
Fig. 5.
Further dissection of the timeline for the appearance of CSI1, PTL1, and exocyst. (A) Representative time-lapse images showing that CSI1 has a strong bias to appear before as PTL1, and PTL1 transiently associated with CSI1 for ∼4 s before it disappeared. n = 147 observations from six cells. (Scale bars, 2 μm.) (B) Exocyst shows a strong bias to appear before as PTL1. Shown are representative time-lapse images from dual-labeled line expressing GFP–Sec5B and RFP–PTL1. (Scale bars, 5 μm.) (C) A timeline displaying the estimated appearing/disappearing sequences of CSCs, CSI1, exocyst, and PTL1. (D) A hypothetical model shows the cooperation among CSI1, PTL1, and exocyst complex during the delivery of CSC to the PM.
To further dissect the roles of CSI1 and PTL1 in the delivery of CSCs, we evaluated the delivery of CSI1 and CSCs in the patrol1-2 mutants. In the absence of PTL1, CSI1 still appeared before CSCs. However, the delivery of CSI1 (56.06 ± 19.26 s after photobleaching, n = 16) and CSCs (153.25 ± 4.76 s after photobleaching, n = 16) were both significantly delayed. Moreover, the time lag between delivery of CSI1 and CSCs was also prolonged by ∼60 s compared to that of WT (Fig. 4 C and D). These results further support the codependency of CSI1 and PTL1’s function in the secretion of CSC-containing vesicles.
To further investigate temporal behavior of PTL1 and the exocyst complex, the dynamic relationship between functional associated RFP–PTL1 and GFP–Sec5B was examined. GFP–Sec5B showed a strong bias to appear 1 ∼ 2 s before PTL1 on the PM (Fig. 5 B and C and Movie S5). Moreover, the GFP–Sec5B dynamics were not affected in the patrol1-2 mutants. However, the lifetimes of PTL1 (9.20 ± 6.53 s) were significantly prolonged in sec5a-2 sec5b-1 (Fig. S10 and Movies S6 and S7). These data indicate that PTL1 may function in a later step than the exocyst complex in the CSC delivery process.
Discussion
The Cooperative and Distinct Function of CSI1, PTL1, and the Exocyst Complex at the PM.
In this study, we used multiple approaches to identify proteins involved in delivery of CSCs to the PM. Three independent affinity-purification experiments with CESA6 or CSI1 as bait identified multiple subunits of the exocyst complex. The interaction between CESA6 and exocyst complex was supported by a yeast two-hybrid assay. In addition to the exocyst complex, PTL1 was copurified with CSI1. The direct interaction between PTL1 and CSI1 was validated by reverse-affinity purification and in vitro pull-down experiments. Several lines of evidence suggest that CSI1, PTL1, and the exocyst complex cooperate in the process of de novo secretion of CSCs. Both PTL1 and the exocyst complex were required for efficient delivery of CSCs to the PM. Lesions in PTL1 and multiple subunits of the exocyst complex led to reduced delivery rate of CSCs. In contrast, mutation of CSI1 did not affect the delivery rate of CSCs, but the delivery of CSCs along microtubules was significantly affected. We propose that CSI1 plays a role in marking the docking site, which allows CSC-containing vesicles access to the membrane through its interaction with microtubules.
PTL1 and the exocyst complex had shared as well unique features in their function and phenotypes. The lethality in sec5a-2 sec5b-1 and patrol1-2 patrol1-like-1 suggests that PTL1 and the exocyst complex are essential in plant cells. PTL1 colocalized with the exocyst complex in vivo. However, the resident lifetimes of PTL1 were significantly shorter compared with that of the exocyst complex. The density of PTL1 was also significantly lower compared with that of the exocyst complex, suggesting that PTL1 may serve a specific population of vesicles. Lesions in both PTL1 and exocyst subunits resulted in reduced delivery rate of CSCs and increased density of CSCs. The increase of PM density of CSCs suggests that a feedback mechanism may be in place when delivery is deficient. Alternatively, exocyst complex-independent delivery may compensate for the delivery deficiency. Moreover, abolishment of the exocyst complex function caused a prolonged resident time of PTL1, indicating that the function of PTL1 relies on successful assembly of the exocyst complex. This is consistent with the observation that the exocyst complex showed a strong bias to appear before PTL1. We hypothesize that severe deficiencies of CSC delivery in sec5a-2 sec5b-1 and patrol1-2/patrol1-2 patrol1/+ mutants lead to the excessive faulty CSCs at the PM and that eventually affects overall efficiency of cellulose synthesis, exemplified by reduced motility of CSCs. Our data support an evolutionally conserved role of the exocyst complex in physically tethering vesicles to the PM. Because PTL1 interacted with multiple molecular components, such as CSI1, Sec10, and PTL1’s temporal dynamics during secretion of CSCs, we propose that PTL1 plays a role in assisting tethering and priming for vesicle fusion. Our data also suggest that a distinct set of exocyst subunits (Sec3A, Sec5B, Sec6, Sec8, Sec10, Sec15B, Exo84B, Exo70A1, and Exo70B1) may be involved in secretion of CSCs. While function of other exocyst subunits in secretion of CSCs awaits validation, the duplication and specialization of the exocyst complex is compatible with the sessile lifestyle of plants that allows plants to rapidly adapt to environment and modulate growth.
Function of Microtubule-Dependent Positioning of CSC Delivery.
In the evolution of plants and animals, actin and microtubules have switched places. In plants, actin filaments provide tracks for organelle movement, while cortical microtubules regulate cell expansion. Conversely, in animals interphase microtubule arrays direct organelle trafficking, while actin filaments dominate the cortex and regulate cell motility (38). The role of the cytoskeleton in exocytosis is far more complex than ensuring the access of vesicles to secretory sites. Many studies in mammalian cells and yeast demonstrated that interphase microtubule arrays serve as major tracks for vesicle transport from the cell interior to the cell cortex, while F-actin and myosin motors directly involve in vesicle transport and fusion during the late phases of the secretory process (9). It’s unclear whether plants adopt the similar mechanism in the regulation of exocytosis through the cytoskeleton. CSCs represent unique cargo proteins that are not conserved in the mammalian or yeast systems in which exocytosis has been most rigorously investigated. Previous work showed that actin and microtubules had different roles in trafficking of CESA. Microtubules play key role in positioning the delivery of CSCs to the PM, whereas actin is important for cell-wide distribution of CSCs (37). Results from our study support the hypothesis that microtubule-dependent positioning of CSCs delivery via CSI1. Loss of CSI1 resulted in significant reduction in CSCs-microtubule tethering, whereas loss of PTL1 did not have the same effect on CSC-microtubule tethering. We propose that CSI1 marks the docking site along microtubules at the PM, which ensures association of CSCs with cortical microtubules as they spin out newly synthesized cellulose microfibrils. The temporal sequence suggests that PTL1 preferentially docks on an existing CSI1 particle. Through interaction with Sec10, PTL1 may assist vesicle fusion by transiently interacting with CSI1 and the exocyst complex.
It’s unclear how PTL1 fulfills its role in vesicle fusion. Protein sequence analysis revealed that PTL1 harbors a MUN domain that is homologous to Munc13 proteins in animals (28). Munc13 protein plays a key role in mediating the transition from the Munc18-1/closed syntaxin-1 complex to the Munc18-1/SNARE complex assembly (39, 40). Munc13 is also implicated to have a role in vesicle docking (41). However, the MUN domain from PTL1 and Munc13 shared only ∼8% amino acid sequence similarity (28). Different from Munc13, PTL1 does not possess any additional domains, such as calmodulin-binding domain, C1 domain, and C2 domains. We could not rule out the possibility that PTL1 adopts a similar 3D structure despite the low sequence similarity with Munc13. PTL1 had a plant-specific DUF810 domain, which is present in a variety of dicots, monocots, conifers, and the moss Physcomitrella patens, but not in animal or yeast, indicating that PTL1 protein might have plant-specific function in vesicle trafficking. It has not been reported that mammalian Munc13 directly interacts with the exocyst complex. Nevertheless, lesion in PTL1 did affect AHA1 internalization in guard cells, suggesting PTL1’s role in translocation of AHA1 (33). It remains to be determined whether the PTL1-mediated translocation of AHA1 depends on the exocyst complex. Given the universal expression of PTL1 and lethality of patrol1-2 patrol1-like-1, it is possible that the PTL1 and its partners are involved in the trafficking of many proteins, including CSCs and AHA1. Additional genetic and proteomic studies are desired to identify other protein cargos of the PTL1-invloved secretory pathway.
A Temporal Sequence During de Novo Secretion of CSCs.
Here we present a timeline of events for exocytosis of CSCs. CSI1 appeared at the PM over 40 s before the appearance of CSCs. It’s unlikely that CSI1 plays a role in vesicle delivery and fusion because CSC delivery rate was not affected in csi1-3–null mutants. Two-color temporal analysis shows that the exocyst complex associated with CSC-containing vesicles for about 8–12 s and disappeared, presumably at the moment of fusion. PTL1 appeared after the exocyst complex and associated with CSC-containing vesicles for only 2–4 s. CSI1 remained its association with CSCs after fusion and displayed typical directional motility as CSCs move through the PM during cellulose synthesis. The recruitment of the exocyst complex is independent of PTL1 because patrol1-2 did not affect GFP–Sec5B localization, nor did it affect resident lifetimes of GFP–Sec5B. In contrast, we observed a substantial increase in resident lifetimes of YFP–PTL1 in sec5a-2 sec5b-1.
In this study, CESA represents a protein that is regulated by exocyst complex-mediated exocytosis in plants. We provide a sneak peek of the highly regulated process of CSC delivery mediated by CSI1, PTL1, and exocyst. Plants offer exciting opportunities to characterize the function and regulation of exocytosis that may provide insight into the evolution of exocytosis in eukaryotes.
Materials and Methods
Plant Material and Growth Conditions.
Arabidopsis thaliana seeds were sterilized using 30% bleach, stratified at 4 °C for 3 d, and then plated on Murashige and Skoog (MS) plates [1/2 × MS salts, 0.8% agar, 0.05% monohydrate 2-(N-Morpholino) ethanesulfonic acid, pH 5.7]. For etiolated seedlings, plates were put in darkness at 22 °C and grown for a specified number of days. For soil-grown plants, seedlings were germinated and grown under light on MS plates containing 1% sucrose for several days and then transferred to pots and grown in a growth chamber at 22 °C under a 16-h light and 8-h dark cycle.
Transgenic Lines.
Seeds of all of the T-DNA mutants [patrol1-2 (SALK_018676C), sec5a-2 (GABI_731C01), sec5b-1 (SALK_001525), sec6-3 (SAIL_1231_H05)] were obtained from the Arabidopsis Biological Resource Center (ABRC). PCR reactions were performed to identify single plants for the T-DNA insertion. Primers used for T-DNA genotyping of T-DNA mutant alleles are listed in Dataset S5. To generate the PPTL1::YFP/RFP-PTL1 patrol1-2 transgenic line, a DNA fragment containing 2-kb PATROL1 promoter and the full-length cDNA clone of PATROL1 were amplified using primers listed in Dataset S5 and cloned into the PCR8/GW/TOPO vector (Life Technologies). After sequencing, both fragments were cloned to the pH7WYF2/pH7WRF2 vectors (42). The verified constructs were introduced into patrol1-2 using Agrobacterium tumefaciens-mediated transformation (43). An RFP-PATROL1–expressing A. thaliana transgenic line was crossed with one expressing GFP–Sec5B, GFP–CSI1 (30), and YFP–CESA6 (44), respectively, to generate dual labeled transgenic lines.
To generate the CSI1–GS transgenic line, a DNA fragment containing a 3-kb CSI1 promoter was cloned into pENTR 5-TOPO vector (ThermoFisher Scientific) using primers listed in Dataset S5. The CSI1–GS construct was generated by multisite Gateway via three DNA fragments from pENTR 5-TOPO CSI1 promoter, pEN-R2-GSrhinotage-L3 (Vlaams Instituut voor Biotechnologie, VIB), and pDONR-CSI1 (22) into the destination vector pKCTAP (VIB). The verified constructs were introduced into csi1-6 using A. tumefaciens-mediated transformation.
To generate the pSec5A::GFP-Sec5B transgenic line in sec5a-2 sec5b-1 double-mutant background, a DNA fragment containing 2-kb Sec5A promoter and the full-length cDNA of Sec5B were amplified using primers listed in Dataset S5 and cloned into the pGEM-T and pDONR/Zeo vectors (ThermoFisher Scientific), respectively. After sequencing, both fragments were cloned to the pH7WGF2 (VIB) vector and then transferred into Arabidopsis with a background of sec5a-2 (homozygous) sec5b-1 (heterozygous). To generate the pLat52::GFP-Sec6 transgenic line in sec6-3 mutant background, a DNA fragment containing 600-bp Lat52 promoter was amplified using primers listed in Dataset S5 and cloned into the pGEM-T vector. After sequencing, the Lat52 promoter fragment was cloned to the pH7WGF2 vector. The cDNA fragment of Sec6 (PENTR-SEC6, ABRC ID G09918) were then cloned into the above vector and then transferred into Arabidopsis with a background of sec6-3.
A GFP–CESA3-expressing A. thaliana transgenic line (45) was crossed into the patrol1-2 homozygous and sec5a-2 (homozygous) 5b-1 (heterozygous) background, respectively. F2 homozygous for patrol1-2, sec5a-2 and sec5b-1 were identified by PCR genotyping; F2 homozygous for je5 was genotyped by PCR followed by Hph1 digestion (Dataset S5). An YFP–CESA6-expressing A. thaliana transgenic line was crossed into the pLat52::Sec6 sec6-3 transgenic background to generate YFP–CESA6 in prc1-1 pLat52::Sec6 sec6-3. F2 homozygous for prc1-1 was genotyped by PCR followed by HpyCH4V digestion; homozygous for sec6-3 was identified by PCR genotyping.
Yeast Two-Hybrid Assay.
The central domain of CESA6 (amino acids 323–863) was fused to GAL4 DNA binding domain in bait vector pB66 and used for yeast two-hybrid screen against prey library A. thaliana RP1 (22). Approximately 95.8 million interactions were tested and 91 clones were sequenced.
Affinity Purification.
Transgenic plants of YFP–CESA6, GFP–CSI1, YFP–PTL1, or CSI1–GS were grown on plate [1/2 × MS salts, 0.8% agar, 0.05% monohydrate 2-(N-Morpholino) ethanesulfonic acid, 1% sucrose, pH 5.7] for 7 d (22 °C, 16-h-light and 8-h-dark cycle). Ten grams of seedlings from each genetic background were harvested from plate and homogenized in liquid nitrogen. The ground material was resuspended in 10 mL lysis buffer [50 mM Tris pH7.5, 150 mM NaCl, 0.5% Triton X-100, two tablets of protease inhibitor mixture (cOmplete and PhosSTOP by Roche)] and incubated for 30 min at 4 °C. The ground material/lysis buffer mixture was then subjected to a primary spin at 2,100 × g for 5 min (4 °C); and a secondary spin at 20,200 × g for 20 min (4 °C). The supernatant was incubated with 15-μL GFP-TRAP (GFP-TRAP_A, Chromotek) or IgG (IgG Sepharose 6 Fast Flow; GE Healthcare) beads at 4 °C for 2 h. After incubation, the beads were collected and rinsed for five times with wash buffer [50 mM Tris pH 7.5, 150 mM NaCl, 0.2% Triton X-100, two tablets of protease inhibitor mixture (cOmplete and PhosSTOP by Roche)] and one final wash with trypsin buffer (50 mM ammonium bicarbonate, pH 8.0). Proteomics analyses were performed as described below.
Proteomics Analyses.
Protein samples were treated with 1 μg trypsin overnight at 37 °C in 100-μL trypsin buffer supplemented with 10% acetonitrile. The samples were placed on top of a vortex mixer for continuous agitation to keep beads in suspension. After trypsin digestion, supernatants were collected and pelleted with a speedvac concentrator and redissolved in 20 μL 0.1% formic acid. A MudPIT approach was employed to analyze these trypsin-treated samples. A two-dimension nanoAcquity UPLC (Waters) and an Orbitrap Fusion MS (ThermoFisher Scientific) were configured together to perform online 2D-nano LC/MS/MS analysis. The 2D-nanoLC was operated with a 2D-dilution method that was configured with nanoAcquity UPLC. Two mobile phases for the first dimension LC fractionation were 20 mM ammonium formate (pH 10) and acetonitrile, respectively. Online fractionation was achieved by 5-min elution off a NanoEase trap column (PN# 186003682; Waters) using stepwise-increased concentration of acetonitrile. A total of five fractions were generated with 13%, 18%, 21.5%, 27%, and 50% of acetonitrile, respectively. A final flushing step used 80% acetonitrile to clean up the trap column. Each and every fraction was then analyzed online using a second dimension LC gradient. The second dimension nano-UPLC method was described previously (46).
Orbitrap Fusion MS method was based on a data-dependent acquisition survey. The acquisition time was set from 1 to 70 min. A nano electrospray ionization source was used with spray voltage at 2,600 V, sweep gas at 0, and ion transfer tube temperature at 275 °C. An orbitrap mass analyzer was used for MS1 scan with resolution set at 60,000. MS mass range was 300–1,800 m/z. The AGC target for each scan was set at 500,000 with maximal ion injection time set at 100 ms.
For the MS2 scan, an Orbitrap mass analyzer was used using an auto/normal mode with resolution set at 30,000. Only precursor ions with intensity 50,000 or higher were selected for the MS2 scan. The sequence of individual MS2 scanning was from most-intense to least-intense precursor ions using a top-speed mode under time control of 4-s higher energy CID (HCD) was used for fragmentation activation with 30% normalized activation energy. Quadrupole was used for precursor isolation with a 2 m/z isolation window. MS2 mass range was set auto/normal with the first mass set at 110 m/z. Maximal injection time was 100 ms with the AGC target set at 20,000. Ions were injected for all available parallelizable time. A 5-s exclusion window was applied to all abundant ions to avoid repetitive MS2 scanning on the same precursor ions using 10-ppm error tolerance. Only charge states from 2 to 6 were allowed for MS2 scan, but undetermined charge states were also included. All MS2 spectra were recorded in the centroid mode.
The raw MS files were processed and analyzed using Proteome Discoverer v2.1 (ThermoFisher Scientific). A Sequest HT search engine was used to match all MS data to the Arabidopsis TAIR10 protein database supplemented with common contaminant proteins, such as keratins. The search parameters were as follows: trypsin with two missed cleavage, minimal peptide length for six amino acids, MS1 mass tolerance 20 ppm, MS2 mass tolerance 0.05 Da, Gln → pyro-Glu (N-term Q), oxidation (M), N-terminal acetylation as variable modification. Only proteins with 1% false-discovery rate cut-off were considered in the final result.
Reverse Transcription and PCR Analysis.
RNA was purified from 25 mg of rosette leaves from Col-0, patrol1-2, sec5a-2, sec5b-1, sec5a-2 sec5b-1, and pLat52::Sec6 sec6-3 using the Direct-zol RNA MiniPrep Kit (Zymo Research). The corresponding cDNA was derived via reverse-transcriptase synthesis reaction using the RevertAid Reverse Transcriptase (Thermo Scientific). cDNA from each genotype was subjected to PCR using primer pairs that were specific to the coding sequence of PATROL1, Sec5A, Sec5B, Sec6, and ACTIN2 (Dataset S5). Thirty-five temperature cycles were applied in all reactions and the PCR products were analyzed by gel electrophoresis.
Cellulose Content Measurement.
Etiolated homozygous seedlings of 4-d-old were collected from MS plates. Crystalline cellulose was measured using the Updegraff method (47). Data were collected from six technical replicates for each tissue sample.
Protein Purification.
Full-length coding sequence of the genes of interest were cloned in to either the YG201 vector, which contains a His tag, or the pGEX-KG vector, which contains a GST tag, and expressed in BL21 E. coli. Protein expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside at 15 °C for 20 h. Protein purification was performed as described previously (17).
In Vitro Pull-Down Assay.
GST resin-bound proteins were washed three times in interaction buffer (20 mM Hepes, pH 7.4, 1 mM EDTA, 5 mM MgCl2, 1 mM DTT, and 0.1% Triton X-100) for equilibration. Aliquots of ∼10 μg of equilibrated GST-bound proteins were incubated with ∼10 μg of soluble His-tagged proteins in a total volume of 0.5 mL of interaction buffer for 2 h at 4 °C on a rocker. The resin was then washed 10 times with interaction buffer, and resuspended in SDS loading buffer, boiled for 5 min, and subjected to SDS/PAGE and Western blotting for analysis. For Western blots, His-tagged proteins were detected on film using a horseradish peroxidase-conjugated His antibody and SuperSignal West Femto substrate (ThermoFisher Scientific).
Live Cell Imaging.
See SI Materials and Methods for details of live cell imaging.
Supplementary Material
Acknowledgments
We thank Ryan Gutierrez and David Ehrhardt for providing the mCherry-TUA5 GV3101 agrobacteria strain; Herman Höfte for providing GFP–CESA3 and GFP–CESA6 transgenic seeds; Chris Somerville for providing YFP–CESA6 transgenic seeds; and Niko Geldner for providing mCherry-ARA7, -RabA1E, -RabA1G, -RabC1, -RabG3F, and -Rha1. Y.G. and X.Z. were supported by National Science Foundation Grant 1121375. S.L. and X.X. were supported by The Center for LignoCellulose Structure and Formation, an Energy Frontier Research Center funded by the Department of Energy, Office of Science, Basic Energy Sciences under Award DE-SC0001090.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800182115/-/DCSupplemental.
References
- 1.Friedrich GA, Hildebrand JD, Soriano P. The secretory protein Sec8 is required for paraxial mesoderm formation in the mouse. Dev Biol. 1997;192:364–374. doi: 10.1006/dbio.1997.8727. [DOI] [PubMed] [Google Scholar]
- 2.Hála M, et al. An exocyst complex functions in plant cell growth in Arabidopsis and tobacco. Plant Cell. 2008;20:1330–1345. doi: 10.1105/tpc.108.059105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu SC, et al. The mammalian brain rsec6/8 complex. Neuron. 1996;17:1209–1219. doi: 10.1016/s0896-6273(00)80251-2. [DOI] [PubMed] [Google Scholar]
- 4.Murthy M, Garza D, Scheller RH, Schwarz TL. Mutations in the exocyst component Sec5 disrupt neuronal membrane traffic, but neurotransmitter release persists. Neuron. 2003;37:433–447. doi: 10.1016/s0896-6273(03)00031-x. [DOI] [PubMed] [Google Scholar]
- 5.Murthy M, et al. Sec6 mutations and the Drosophila exocyst complex. J Cell Sci. 2005;118:1139–1150. doi: 10.1242/jcs.01644. [DOI] [PubMed] [Google Scholar]
- 6.Samuel MA, et al. Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell. 2009;21:2655–2671. doi: 10.1105/tpc.109.069740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennwald P, Rossi G. Spatial regulation of exocytosis and cell polarity: Yeast as a model for animal cells. FEBS Lett. 2007;581:2119–2124. doi: 10.1016/j.febslet.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He B, Guo W. The exocyst complex in polarized exocytosis. Curr Opin Cell Biol. 2009;21:537–542. doi: 10.1016/j.ceb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heider MR, Munson M. Exorcising the exocyst complex. Traffic. 2012;13:898–907. doi: 10.1111/j.1600-0854.2012.01353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polgar N, Fogelgren B. Regulation of cell polarity by exocyst-mediated trafficking. Cold Spring Harb Perspect Biol. 2017;10:a031401. doi: 10.1101/cshperspect.a031401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jürgens G, Geldner N. Protein secretion in plants: From the trans-Golgi network to the outer space. Traffic. 2002;3:605–613. doi: 10.1034/j.1600-0854.2002.30902.x. [DOI] [PubMed] [Google Scholar]
- 12.Lavy M, et al. A novel ROP/RAC effector links cell polarity, root-meristem maintenance, and vesicle trafficking. Curr Biol. 2007;17:947–952. doi: 10.1016/j.cub.2007.04.038. [DOI] [PubMed] [Google Scholar]
- 13.Li S, et al. EXO70A1-mediated vesicle trafficking is critical for tracheary element development in Arabidopsis. Plant Cell. 2013;25:1774–1786. doi: 10.1105/tpc.113.112144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rybak K, et al. Plant cytokinesis is orchestrated by the sequential action of the TRAPPII and exocyst tethering complexes. Dev Cell. 2014;29:607–620. doi: 10.1016/j.devcel.2014.04.029. [DOI] [PubMed] [Google Scholar]
- 15.Wen TJ, Hochholdinger F, Sauer M, Bruce W, Schnable PS. The roothairless1 gene of maize encodes a homolog of sec3, which is involved in polar exocytosis. Plant Physiol. 2005;138:1637–1643. doi: 10.1104/pp.105.062174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang C, et al. Endosidin2 targets conserved exocyst complex subunit EXO70 to inhibit exocytosis. Proc Natl Acad Sci USA. 2016;113:E41–E50. doi: 10.1073/pnas.1521248112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bashline L, Li S, Anderson CT, Lei L, Gu Y. The endocytosis of cellulose synthase in Arabidopsis is dependent on μ2, a clathrin-mediated endocytosis adaptin. Plant Physiol. 2013;163:150–160. doi: 10.1104/pp.113.221234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bashline L, Li S, Zhu X, Gu Y. The TWD40-2 protein and the AP2 complex cooperate in the clathrin-mediated endocytosis of cellulose synthase to regulate cellulose biosynthesis. Proc Natl Acad Sci USA. 2015;112:12870–12875. doi: 10.1073/pnas.1509292112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bashline L, Li S, Gu Y. The trafficking of the cellulose synthase complex in higher plants. Ann Bot. 2014;114:1059–1067. doi: 10.1093/aob/mcu040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirst J, et al. Characterization of TSET, an ancient and widespread membrane trafficking complex. eLife. 2014;3:e02866. doi: 10.7554/eLife.02866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lei L, et al. CELLULOSE SYNTHASE INTERACTIVE1 is required for fast recycling of cellulose synthase complexes to the plasma membrane in Arabidopsis. Plant Cell. 2015;27:2926–2940. doi: 10.1105/tpc.15.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu Y, et al. Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc Natl Acad Sci USA. 2010;107:12866–12871. doi: 10.1073/pnas.1007092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S, Lei L, Somerville CR, Gu Y. Cellulose synthase interactive protein 1 (CSI1) links microtubules and cellulose synthase complexes. Proc Natl Acad Sci USA. 2012;109:185–190. doi: 10.1073/pnas.1118560109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cole RA, Synek L, Zarsky V, Fowler JE. SEC8, a subunit of the putative Arabidopsis exocyst complex, facilitates pollen germination and competitive pollen tube growth. Plant Physiol. 2005;138:2005–2018. doi: 10.1104/pp.105.062273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizuno S, et al. Peri-implantation lethality in mice carrying megabase-scale deletion on 5qc3.3 is caused by Exoc1 null mutation. Sci Rep. 2015;5:13632. doi: 10.1038/srep13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, Guo W. The exocyst complex in exocytosis and cell migration. Protoplasma. 2012;249:587–597. doi: 10.1007/s00709-011-0330-1. [DOI] [PubMed] [Google Scholar]
- 28.Hashimoto-Sugimoto M, et al. A Munc13-like protein in Arabidopsis mediates H+-ATPase translocation that is essential for stomatal responses. Nat Commun. 2013;4:2215. doi: 10.1038/ncomms3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higaki T, Hashimoto-Sugimoto M, Akita K, Iba K, Hasezawa S. Dynamics and environmental responses of PATROL1 in Arabidopsis subsidiary cells. Plant Cell Physiol. 2014;55:773–780. doi: 10.1093/pcp/pct151. [DOI] [PubMed] [Google Scholar]
- 30.Lei L, Li S, Du J, Bashline L, Gu Y. Cellulose synthase INTERACTIVE3 regulates cellulose biosynthesis in both a microtubule-dependent and microtubule-independent manner in Arabidopsis. Plant Cell. 2013;25:4912–4923. doi: 10.1105/tpc.113.116715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding Y, et al. Exo70E2 is essential for exocyst subunit recruitment and EXPO formation in both plants and animals. Mol Biol Cell. 2014;25:412–426. doi: 10.1091/mbc.E13-10-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fendrych M, et al. Visualization of the exocyst complex dynamics at the plasma membrane of Arabidopsis thaliana. Mol Biol Cell. 2013;24:510–520. doi: 10.1091/mbc.E12-06-0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, et al. EXPO, an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in Arabidopsis and tobacco cells. Plant Cell. 2010;22:4009–4030. doi: 10.1105/tpc.110.080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geldner N, et al. Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 2009;59:169–178. doi: 10.1111/j.1365-313X.2009.03851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vukašinović N, et al. Dissecting a hidden gene duplication: The Arabidopsis thaliana SEC10 locus. PLoS One. 2014;9:e94077. doi: 10.1371/journal.pone.0094077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li S, et al. Cellulose synthase complexes act in a concerted fashion to synthesize highly aggregated cellulose in secondary cell walls of plants. Proc Natl Acad Sci USA. 2016;113:11348–11353. doi: 10.1073/pnas.1613273113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gutierrez R, Lindeboom JJ, Paredez AR, Emons AMC, Ehrhardt DW. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol. 2009;11:797–806. doi: 10.1038/ncb1886. [DOI] [PubMed] [Google Scholar]
- 38.Baskin TI, Gu Y. Making parallel lines meet: Transferring information from microtubules to extracellular matrix. Cell Adhes Migr. 2012;6:404–408. doi: 10.4161/cam.21121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma C, Li W, Xu Y, Rizo J. Munc13 mediates the transition from the closed syntaxin-Munc18 complex to the SNARE complex. Nat Struct Mol Biol. 2011;18:542–549. doi: 10.1038/nsmb.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma C, Su L, Seven AB, Xu Y, Rizo J. Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science. 2013;339:421–425. doi: 10.1126/science.1230473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Camacho M, et al. Heterodimerization of Munc13 C2A domain with RIM regulates synaptic vesicle docking and priming. Nat Commun. 2017;8:15293. doi: 10.1038/ncomms15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 43.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 44.Paradez A, Wright A, Ehrhardt DW. Microtubule cortical array organization and plant cell morphogenesis. Curr Opin Plant Biol. 2006;9:571–578. doi: 10.1016/j.pbi.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 45.Desprez T, et al. Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104:15572–15577. doi: 10.1073/pnas.0706569104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drakakaki G, et al. Isolation and proteomic analysis of the SYP61 compartment reveal its role in exocytic trafficking in Arabidopsis. Cell Res. 2012;22:413–424. doi: 10.1038/cr.2011.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Updegraff DM. Semimicro determination of cellulose in biological materials. Anal Biochem. 1969;32:420–424. doi: 10.1016/s0003-2697(69)80009-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.