Significance
The budding yeast Saccharomyces cerevisiae is one of the most commonly used model organisms in studying DNA damage and repair in eukaryotes. In this study, we adapted the recently invented excision repair-sequencing method to investigate the mode of in vivo repair of the two major UV-induced DNA lesions, the cyclobutane pyrimidine dimer and the (6-4) pyrimidine–pyrimidone photoproduct, and to generate the dynamic repair maps of the these two lesions at single-nucleotide resolution across the entire yeast genome. These data revealed a polarity of repair within genes and other effects of transcription and the time of repair on the repair pattern. Our findings should aid in research on carcinogenesis by UV and chemical carcinogens.
Keywords: XR-seq, DNA damage, excision repair, UV, yeast
Abstract
We have adapted the eXcision Repair-sequencing (XR-seq) method to generate single-nucleotide resolution dynamic repair maps of UV-induced cyclobutane pyrimidine dimers and (6-4) pyrimidine–pyrimidone photoproducts in the Saccharomyces cerevisiae genome. We find that these photoproducts are removed from the genome primarily by incisions 13–18 nucleotides 5′ and 6–7 nucleotides 3′ to the UV damage that generate 21- to 27-nt-long excision products. Analyses of the excision repair kinetics both in single genes and at the genome-wide level reveal strong transcription-coupled repair of the transcribed strand at early time points followed by predominantly nontranscribed strand repair at later stages. We have also characterized the excision repair level as a function of the transcription level. The availability of high-resolution and dynamic repair maps should aid in future repair and mutagenesis studies in this model organism.
Nucleotide excision repair removes a variety of bulky adducts from the genome, including DNA lesions induced by UV radiation, chemical carcinogens, and chemotherapeutic agents (1, 2). The basic mechanism of the repair reaction has been investigated in some detail both in prokaryotes and in eukaryotes (3–7). The repair reaction entails dual incisions bracketing the lesions to remove the damage mainly in oligomers 12–13 nt in length in prokaryotes and 24–32 nt in length in eukaryotes (8–11) followed by filling in the excision gap by DNA polymerase and ligation.
The core repair reaction is impacted by multiple factors in vivo, including chromatin structure, transcription, regulatory protein binding to DNA, and posttranscriptional modification of histones as well as DNA modification and compaction (12–14). Recently, a number of high-resolution methods have been employed to investigate the effects of these factors on nucleotide excision repair in vivo both at the single-gene level (15) and genome-wide (16–21). These methods have measured in vivo repair either by determining the disappearance of damage from the genome (subtractive/indirect method) or by capturing and quantifying the excised oligonucleotides (direct method). While the subtractive method has been successfully employed to address some fundamental questions regarding excision repair, it relies, by its own nature, on subtracting two large numbers from one another, which limits its sensitivity and hence general utility. In contrast, the direct method has virtually zero background and thus has certain advantages in measuring damage formation and repair as well as in studying damage-induced mutagenesis, carcinogenesis, and chemotherapy.
The yeast Saccharomyces cerevisiae has been the most commonly used eukaryotic model organism in studying replication, recombination, and repair and has been extensively used in studying UV-induced DNA damage, repair, and mutagenesis (22–25). Here, we have adapted the eXcision Repair-sequencing (XR-seq) method recently developed in our laboratory (16–18) to investigate the dual-incision mechanism in S. cerevisiae in vivo and to generate dynamic repair maps of UV-induced cyclobutane pyrimidine dimers (CPDs) and (6-4) pyrimidine–pyrimidone photoproducts [(6-4)PPs] at single-nucleotide resolution in S. cerevisiae genome. Our data reveal a dual-incision mode in yeast that differs from that in humans and show the sequential repair of the transcribed strand (TS) and nontranscribed strand (NTS) in transcriptionally active genes, and we report the association between transcription rate and excision repair level.
Results
Adaptation of XR-seq for Yeast.
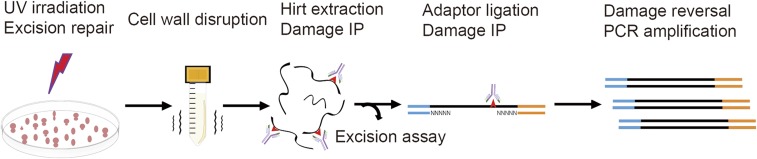
In the original XR-seq method for mapping nucleotide excision repair in human cells, the excised oligomers carrying the damage are separated from genomic DNA by gentle cell lysis, and then they are coimmunoprecipitated with the repair factor TFIIH (16). To perform XR-seq in bacteria, the method was modified by replacing the coimmunoprecipitation with TFIIH with direct immunoprecipitation with a UV-damaged DNA-specific antibody (26). To adapt XR-seq to analyze genome-wide excision repair in yeast cells, we compared three different strategies to isolate the excised oligomers and found that the optimal method is to use bead beating to disrupt the cell wall followed by a modified Hirt extraction procedure (Fig. S1). The scheme of in vivo excision assay and XR-seq library construction in yeast is shown in Fig. 1. Briefly, yeast cells in suspension were irradiated with 120 J/m2 UV (254 nm) and allowed to carry out excision repair for certain time periods. At different time points, cells were harvested, and the cell walls were disrupted by vigorous bead beating. The excised oligomers were then isolated and purified by a modified Hirt extraction procedure and immunoprecipitation with damage-specific antibodies. Then, the excised oligomers were used for either 3′ end radiolabeling or XR-seq library construction. For visual analysis of the excision products, the excised oligomers were 3′ end-labeled with 32P-Cordycepin and resolved on an 11% denaturing sequencing gel. For XR-seq library construction, the excised oligomers were ligated with adaptors and reimmunoprecipitated with damage-specific antibodies. After damage reversal by an appropriate photolyase, the ligated and purified excised oligomers were then amplified by PCR and subjected to next-generation sequencing. Sequences were then analyzed and aligned to the yeast genome, and genomic maps of repair were generated.
Fig. 1.
Scheme of excision assay and XR-seq library construction in yeast. Cells are treated with UV (254 nm) and allowed to carry out excision repair for certain periods of time. The cell walls are then disrupted by bead beating. The excised oligomers are isolated by Hirt extraction and DNA damage immunoprecipitation (IP). The isolated excision products can be analyzed either by excision assay or by XR-seq method. For XR-seq library construction, the excised oligomers are ligated to adaptors and then purified by a second DNA damage IP. The adaptor-containing oligomers are amplified by PCR after the DNA damage reversal by photoreactivation.
In Vivo Dual-Incision Pattern in Yeast.
Yeast has all of the orthologs of human excision repair genes (22), and hence it is expected to have a dual-incision pattern similar to humans. Indeed, in a seminal study Guzder et al. (10) purified the six core yeast excision repair proteins and reconstituted the excision reaction in vitro. They observed excised fragments in the size range of 24–27 nt. It was assumed that these fragments were generated by asymmetric dual incision as in humans, but the actual incision sites were not analyzed.
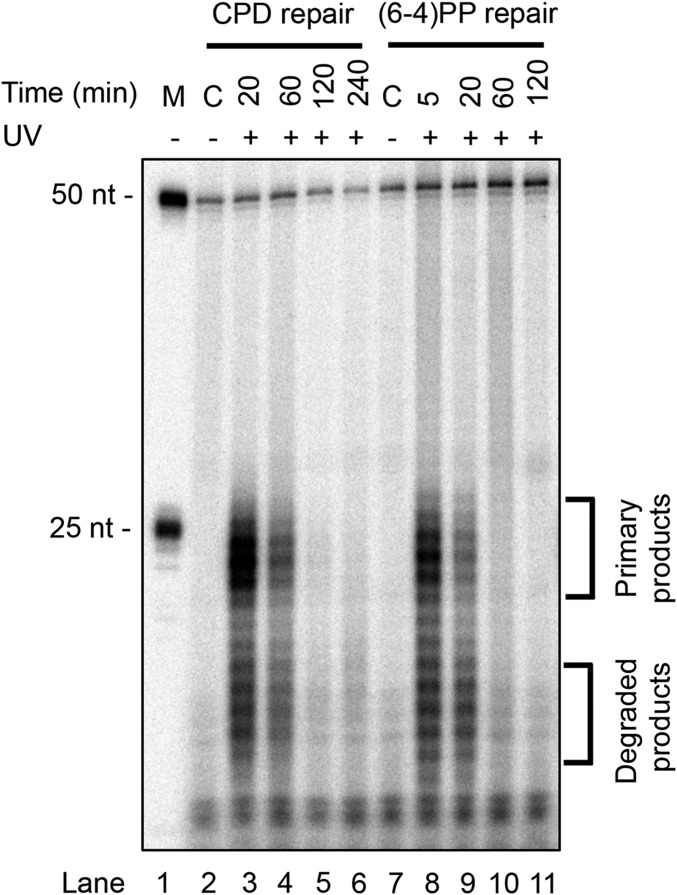
Here, with the availability of the in vivo excision assay and XR-seq, we revisited the yeast dual-incision pattern. We note, however, that in the in vivo excision assay, the fragments represent a snapshot of those generated shortly before cell lysis. We have found that in all organisms tested (human, mouse, Arabidopsis thaliana, Escherichia coli, and yeast) the excised fragments are rather rapidly degraded, and therefore the samples taken earliest represent the primary excision products (11, 16, 27). Fig. 2 shows a time course of the in vivo excision assay for CPDs and (6-4)PPs in wild-type yeast cells. As apparent, two size classes are observed for both lesions with the longer group representing the primary excision products. Of special note, while the 20-min time point for CPD is dominated by the longer species, in the case of (6-4)PP the longer species dominate at 5 min postirradiation, and by 20 min they become less abundant than the degraded products. This phenomenon, which has been observed in human cells as well (16), is due to the fact that (6-4)PPs are excised more rapidly than CPDs, and hence they are degraded more rapidly. Regardless of the properties of the in vivo excision assay, it appears that both CPDs and (6-4)PPs are excised mainly in the form of 21- to 27-nt-long oligomers, in general agreement with Guzder et al. (10).
Fig. 2.
Excised oligomers containing UV photoproducts from yeast cells. Cells were irradiated with 120 J/m2 UV (254 nm) and incubated for the indicated times. The cell walls were then disrupted, and excised oligomers were isolated by Hirt extraction. Excision products containing UV photoproducts were then purified by G-50 filtration column and immunoprecipitated either with anti-CPD antibody or with anti-(6-4)PP antibody. The purified excision products were then 3′ end-labeled with 32P-Cordycepin and resolved on an 11% denaturing sequencing gel. The 50-mer (0.2 fmol) was used as an internal control in each sample. Brackets indicate the locations of primary excision products and degraded excision products.
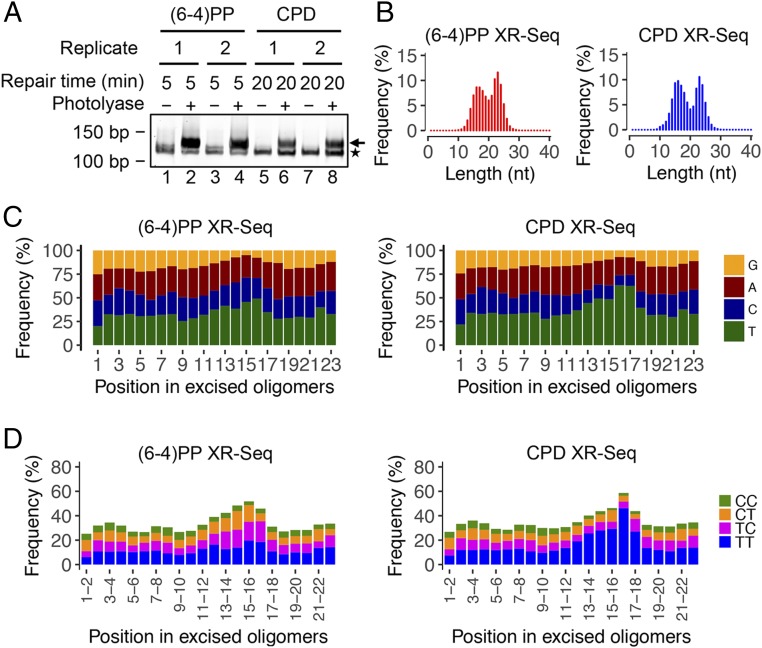
Next we proceeded to analyze the excision products by XR-seq both to identify the incision sites and to generate excised fragment libraries for repair mapping. Fig. 3A shows the libraries generated by PCR amplification of (6-4)PP or CPD-containing oligomers. Sequencing of the PCR products again yielded two populations of sizes for both lesions, consistent with what was observed in the in vivo excision assay (Fig. 3B and Fig. S2). Analyses of thymine frequencies for reads ranging from 15 to 27 mers show thymine enrichment 7–8 nt from the 3′ end for all reads in CPD XR-seq, which suggests that all of the excised oligomers are degraded from 5′ to 3′ (Fig. S3). As seen in Fig. 3 C and D, plotting both single- and double-nucleotide frequencies for the predominant 23-nt-long oligomers reveals that both photoproducts are located primarily 15 nucleotides away from the 5′ end and 6 nucleotides away from the 3′ end. The same pattern can be seen in the dipyrimidine frequencies for the 23 mers obtained from both (6-4)PP and CPD XR-seq at different time points (Fig. S4). Although some longer fragments up to 28 nucleotides are present, the vast majority are in the range of 21–27 nt in length. Thus, in yeast the photoproducts are removed from the genome primarily by incisions 13–18 nt 5′ and 6–7 nt 3′ to the UV damage, generating mostly 21- to 27-nt-long excision products. Models for the dual-incision modes for E. coli (8, 27), yeast, and humans (9, 28) are shown in Fig. 4.
Fig. 3.
Analyses of dsDNA libraries, length distribution, and nucleotide frequencies of the excised oligomers in XR-seq. (A) dsDNA libraries for XR-seq analyzed by 10% native polyacrylamide gel electrophoresis. One percent of ligation products was amplified by 20 cycles of PCR. The arrow on the right indicates the PCR products with inserts. The asterisk on the right indicates adaptor-only PCR products. (B) Length distribution profiles of excised oligomers containing (6-4)PP or CPD from XR-seq. The total read number for each XR-seq was ∼9 million. The repair incubation times for (6-4)PP and CPD XR-seq were 5 and 20 min, respectively. (C) Single-nucleotide frequencies for 23 mers obtained by (6-4)PP and CPD XR-seq. (D) Dipyrimidine frequencies for the 23 mers from (6-4)PP and CPD XR-seq. The enrichment of thymine cytosine (TC) frequency for (6-4)PP XR-seq and that of thymine thymine (TT) frequency for CPD XR-seq are at a position 6 nt from the 3′ end of the excised oligomers.
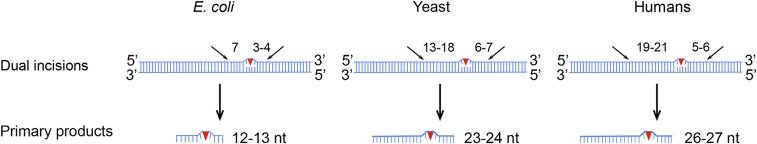
Fig. 4.
Nucleotide excision repair modes for E. coli, yeast, and humans. In E. coli, dual incisions occur 7 nt 5′ and 3–4 nt 3′ to the UV photoproduct. In yeast cells, the photoproducts are excised by dual incisions 13–18 nt 5′ and 6–7 nt 3′ to the damage. In humans, the dual-incision sites are at 19–21 nt 5′ and 5–6 nt 3′ to the UV damage. In yeast and humans, there is considerable variability in incision sites, principally at the 5′ incision site, giving rise to fragments ranging from 21 to 28 in yeast and 24 to 32 in humans. Shown in the figure are the sizes of predominant excision products for E. coli, yeast, and humans, which are 12–13 nt, 23–24 nt, and 26–27 nt, respectively.
Excision Repair Kinetics of Individual Genes.
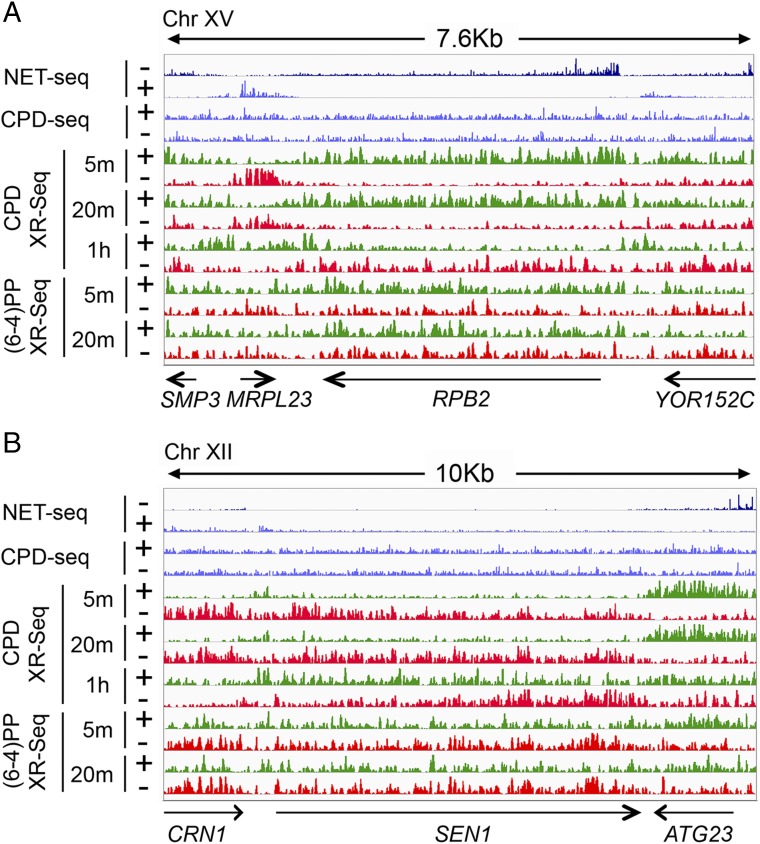
We aligned the XR-seq reads to the yeast genome and thus generated genome-wide dynamic repair maps of both CPDs and (6-4)PPs at single-nucleotide resolution. Correlation analyses of the two biological replicates at different time points show high reproducibility (minimum Pearson r = 0.96, Fig. S5). To compare our XR-seq signals with gene expression levels, we integrated published native elongating transcript sequencing (NET-seq) and RNA-seq data (29). We also integrated the published CPD-seq data (19) to compare the initial CPD damage formation levels with our CPD repair results. Our repair maps enable us to visualize repair signals at both individual gene and genome-wide levels (Fig. 5 and Fig. S6). We illustrate gene-level repair kinetics in Fig. 5A with RPB2, an actively transcribed representative gene. The repair of (6-4)PPs in RPB2 reveals a small preference for the TS at the two time points. The preferential TS repair is relatively small because of the highly efficient recognition of (6-4)PPs by the core excision repair system (3, 30). In contrast, the preferential repair of CPDs in the TS compared with the NTS is much greater at both 5- and 20-min time points because CPDs in the NTS are poorly recognized by the core repair system. Interestingly, at the 1-h time point this preferential repair trend is reversed. This seemingly paradoxical result follows from the nature of the XR-seq assays: the excised oligonucleotides are degraded rapidly to smaller species that are not included in library preparation. As a consequence, the XR-seq data show a snapshot of the oligomers excised within a few minutes before harvesting cells for library preparation. Thus, within the first 20 min the TS repair is nearly complete and then the excised oligomers are degraded such that by 1 h postirradiation the only lesions remaining are in the NTS, giving the appearance of preferential repair of the NTS. It has been independently reported that repair of the RPB2 TS is nearly complete after 20 min (31), and at 1 h overall UV damage repair is ∼80% complete (32). This reversal of the repair pattern is the result of unrepaired damage asymmetry due to the TS repair completion, thus freeing the core repair machinery to work on the NTS.
Fig. 5.
Excision repair kinetics of individual genes in yeast cells. (A) Distribution of XR-seq signals at each time point, in each strand, along a 7.6-kb region of chromosome XV. For (6-4)PP XR-seq, the normalized read counts for the two strands over the RPB2 gene at 5 and 20 min are 58/37 (TS/NTS) and 55/52 (TS/NTS), respectively. Arrows at the bottom indicate positions and directions of four genes including the highly transcribed RPB2 gene. (B) Distribution of XR-seq signals at each time point, in each strand, along a 10-kb region of chromosome XII. Arrows at the bottom indicate positions and directions of three genes including the SEN1 gene. The NET-seq. (29) and CPD-seq. (19) signals are plotted in blue. The “+” denotes plus-strand (5′ to 3′ direction) and “−” means minus-strand (3′ to 5′ direction). The y axis, the normalized read counts, is in a scale of 1–20 for NET-seq and CPD-seq and 1–50 for XR-seq.
In addition to the sequential repair of TS and NTS in actively transcribed genes, we found transcription-coupled repair exhibits a 5′ to 3′ gradient within genes, with fast repair at the 5′ region and slow repair at the 3′ end region, which is consistent with a previous study that measured repair by the subtractive method (33). Fig. 5B shows a screenshot of repair profiles of the SEN1 gene (6.7 kb), which codes for an important transcription-coupled repair protein Sen1 (34). Transcription-coupled repair of CPDs at the transcription start site (TSS) region of SEN1 peaks at 5 min, and then it decreases at 20 min and reaches its lowest level at the 1-h time point. At the transcription end site (TES) region, the trend is the opposite: transcription-coupled repair of CPDs is low at early time points and gradually increases up to 1 h. In agreement with the sequential repair pattern in RPB2, there is strong preference of NTS repair of SEN1 only at the 1-h time point. The (6-4)PP repair kinetics of SEN1 also show (as seen with RPB2) a small trend of preferential TS repair within 20 min (Fig. 5B).
Genome-Wide Analyses of Excision Repair Kinetics.
Among the many factors affecting repair efficiency, transcription plays a unique role: in organisms ranging from E. coli to humans transcription is known to strongly stimulate the repair of the TS (7) by essentially employing RNA polymerase II (RNAP II) as a highly specific damage recognition protein (35). In yeast, stalling of RNAP II at a damage site in the TS has been shown to be essential for efficient transcription-coupled repair (31).
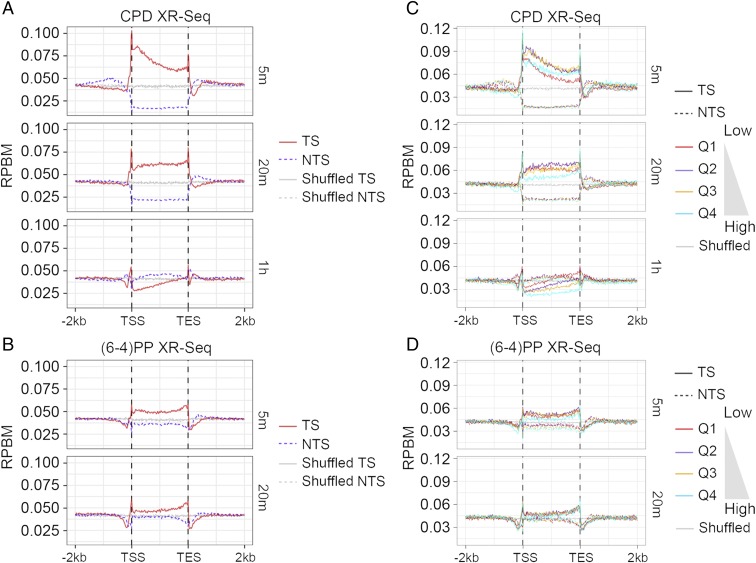
With this general view, we analyzed the strand-specific CPD repair profiles across all genes (Fig. 6A). The analysis normalizes the length of all genes to a constant so that the average repair levels between the TSS and TES may be plotted. Also plotted are average repair levels for the 2 kb upstream and downstream of all genes. The analysis in Fig. 6A shows that CPD repair of the TS in the gene body is highest at 5 min, when there is over fivefold more repair of the TS than the NTS near the TSS. At 20-min and 1-h time points, TS repair gradually decreases starting from the 5′ end. This genome-wide TS repair pattern, in which TS repair sequentially occurs in the 5′ to 3′ direction, is in agreement with the pattern shown in SEN1 (Fig. 5B). In contrast, the NTS repair exhibits the lowest relative repair level at 5 min and gradually increases at 20-min and 1-h time points. As expected, there is preferential repair of the NTS at the 1-h time point, consistent with our observation in RPB2 (Fig. 5A).
Fig. 6.
Average repair profiles for CPD and (6-4)PP. (A) CPD and (B) (6-4)PP repair profiles for TS and NTS over 6,291 yeast genes (>300 bp). TS is shown by the red solid line, and NTS is denoted by the purple dashed line. The gray solid and dashed lines represent the TS and NTS repair observed from randomly shuffled windows, respectively. (C) CPD and (D) (6-4)PP repair profiles over the 6,291 yeast genes (>300 bp) separated into expression quartiles with Q1 being the lowest expression quartile and Q4 the highest. Gray lines represent the repair observed from randomly shuffled windows.
We then performed the same analysis with (6-4)PP repair across all genes and found preferential repair of the TS within 20 min after UV irradiation (Fig. 6B), which is consistent with the repair pattern seen in the RPB2 and SEN1 genes (Fig. 5). Similar to CPD repair, preferential TS repair of (6-4)PP is observed from 5 to 20 min, albeit at lower magnitude than CPD repair observed genome-wide. This genome-wide observation is in qualitative agreement with a previous report showing the transcription-coupled repair of (6-4)PPs in the yeast URA3 gene (30). As in the case for genome-wide CPD repair (Fig. 6A), the NTS repair of (6-4)PP (Fig. 6B) is relatively low at 5 min, but increases at the 20-min time point, indicating the sequential repair pattern from the TS to the NTS as repair is completed in the TS. Interestingly, the TS repair peaks at the TES region for both CPD and (6-4)PP repair, which may be due to the high density of RNAP II and/or AT-rich sequences near the TES (36, 37).
Effect of Transcription Rate on Excision Repair.
Since the discovery of transcription-coupled repair, it has been widely assumed that transcription-coupled repair is positively correlated with transcription level (12). Indeed, genome-wide there is high correlation between the levels of transcription-coupled repair and transcription in human cells (16, 38), and this correlation has also been seen in the lacZ gene in E. coli (26). To examine the effect of the transcription rate on excision repair in yeast cells, we sorted all of the genes into quartiles based on their expression levels (29) and plotted the strand-specific average repair profiles for each quartile (Fig. 6 C and D). Surprisingly, for the CPD repair shown in Fig. 6C, while at 5 min the TS repair level increases with expression level from quartiles 1–3 (Q1, Q2, and Q3), the TS repair level of the highest expression quartile 4 (Q4) is lower than that of Q2 and Q3, and it becomes the lowest at both 20-min and 1-h time points. The relative TS repair level of Q1 turns out to be the highest at the 1-h time point when NTS repair is dominant. Similar trends were observed for (6-4)PP repair (Fig. 6D). These results indicate a positive association between transcription and repair, but lower repair in the highest quartile transcription group Q4. A high transcription rate may interfere with repair; alternatively, many damage sites in the Q4 group may have been repaired well before the 5-min time point.
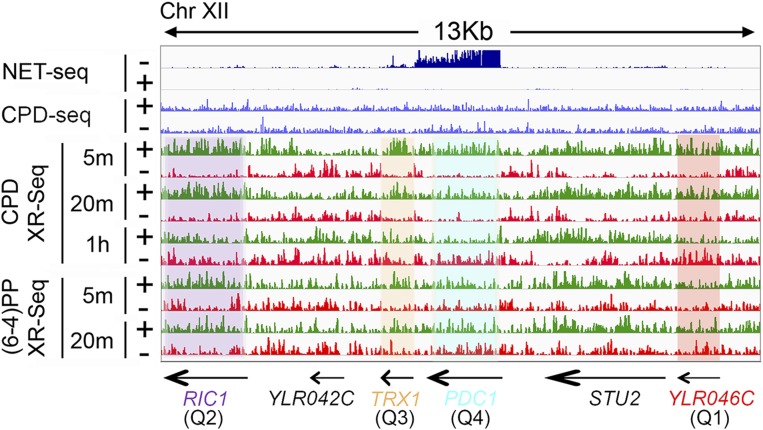
The lower repair associated with Q4 is further illustrated in Fig. 7 with four individual genes that exhibit different expression levels representing the four quartiles. These genes are located along a 13-kb region of chromosome XII. For CPD repair, the TS repair level in the most highly expressed gene, PDC1 (Q4), is lower than that in the TRX1 and RIC1 genes from Q3 and Q2, respectively. The TS repair level in the YLR046C gene, which belongs to the lowest Q1, is similar to that of the most highly expressed PDC1 gene. For (6-4)PP repair, this trend can also be observed in individual genes. The normalized read counts [reads per kilobase per million mapped reads (RPKM)] for both CPD and (6-4)PP repair at different time points in these four individual genes are listed in Table S1. Moreover, the correlation analyses between XR-seq and NET-seq in Figs. S7 and S8 further illustrate trends in transcription and repair. Especially at the earliest time point (5 min), there is an overall increase in the repair level of the TS as a function of transcription, but after a point, increased transcription is associated with decreased repair. Of note, the overall TS repair level decrease for highly transcribed genes (Fig. S7) appears to be principally caused by a subset of genes rather than all genes.
Fig. 7.
NET-seq, CPD-seq, and XR-seq patterns of representative genes. The repair profiles of PDC1, TRX1, RIC1, and YLR046C genes representing the four quartiles defined in Fig. 6 are highlighted in blue, yellow, purple, and red, respectively. Arrows at the bottom indicate positions and directions of six genes including the highly transcribed PDC1 gene. The y-axis scale is 1–20 for CPD-seq and 1–50 for both NET-seq and XR-seq.
In light of the above results and the unique features of transcribed DNA, we were interested to see if the transcription level influences damage formation by UV irradiation. In Fig. S9 we plot CPD damage levels for each gene (19) as a function of expression level computed from NET-seq datasets (29). Interestingly, with the CPD-seq dataset (19) we found that there is a slight negative correlation between transcription level and damage formation. The decrease in damage formation was observed in both strands and may account for the slight decrease in NTS repair of the highly expressed genes (Fig. S7 and S8). However, the decrease in damage does not parallel the increase and then decrease in TS repair; therefore, the repair impairment on TS of the highly expressed genes cannot be explained by the decrease in damage formation.
Overall, our results are consistent with a positive association between transcription and excision repair rates at low and intermediate levels of transcription. In the most strongly transcribed genes, the high transcription rate may have interfered with repair as it has been proposed for E. coli (Discussion). However, the very rapid excision repair followed by the complete degradation of the excised oligomers within 5 min may have prevented the capture of earlier repair events.
Discussion
There have been numerous genetic, biochemical, and structural studies on repair of UV damage in yeast at the single-gene and structural levels (15, 34, 39–41). A recent important study based on mapping T4 UV endonuclease incision at CPD sites followed by genome-wide mapping of the incision sites reported differential UV damage formation and inferred repair (19). This method, named CPD-seq, has been very useful in defining certain genomic parameters that influence damage formation and repair. However, because in this method “repair” is defined by subtracting two large numbers from one another, it has low sensitivity for detecting all of the factors that affect repair. A high-resolution repair map that directly measures damage formation and repair is expected to provide a more accurate view of the dynamics of damage formation and excision repair of UV-induced DNA damage that includes both CPDs and (6-4)PPs. Here, we have adapted the XR-seq method, originally developed for mammalian cells (16), to analyze dynamic repair events in yeast, and we have generated single-nucleotide resolution dynamic repair maps of the two major UV photoproducts over the course of 1 h following UV irradiation.
This adaptation of XR-seq has enabled us to make the following conclusions regarding excision repair of UV damage in yeast. First, we were able to find that, while the excised fragment sizes are similar in humans and yeast, the dual-incision sites in yeast are unique with the incisions mode 13–18 nt 5′ and 6–7 nt 3′ to the UV photoproducts compared with 19–21 nt 5′ and 5–6 nt 3′ to the damage in humans. The similarity in excision products is consistent with yeast and humans having homologous repair proteins and contrasts with the smaller 12- to 13-mer products made by E. coli using fewer, nonhomologous repair proteins and a different reaction mechanism (Fig. 4). We also find that TS repair progresses in the 5′ to 3′ direction within genes in agreement with previous reports that measured repair by subtractive methods (19, 30). This may result from a greater density of RNAP II near the promoter than in the gene body, which increases the likelihood of a lesion near the 5′ end blocking transcription and undergoing transcription-coupled repair at the early time point.
Finally, as has been reported in organisms ranging from E. coli to yeast to humans (7, 12), we find in our XR-seq analyses in yeast that the TS is also preferentially repaired over the NTS. Interestingly, as in E. coli and humans, we observe a positive correlation between transcription rate and the magnitude of transcription-coupled repair. We observed a discrepancy among the most highly expressed genes, which may be due to rapid repair occurring before our earliest time point or may reflect interference of excision repair by high transcription rate. The topic of high transcription rate and excision repair has been of interest especially in relation to the phenomenon “mutation frequency decline” (MFD) observed in E. coli. MFD refers to the loss of certain suppressor mutations observed when irradiated cells are held in minimal media before plating (42). The suppressor mutations are thought to result from unrepaired UV damage in the template strand of anticodon-encoding loops. It was suggested that the decline in suppressor mutations associated with a stringent response condition is associated with going from complete medium [strong tRNA transcription (hindered repair)] to stringent response conditions [weaker tRNA transcription (enhanced repair)] (42, 43), and there is experimental support for this model (44, 45). Conceptually, strong transcription could interfere with excision repair when trailing RNAPs “catch up” to and interfere with a leading RNAP that has been blocked by DNA damage and is engaging in repair. Interference of this sort is not observed in human cells (16–18) presumably because high-density RNAP II accumulation in a given gene is rare or nonexistent due to the large genome size and relatively slow growth rate compared with yeast and bacteria. Indeed, it has been reported that the average RNAP II density in higher eukaryotes is lower than in yeast (46).
In conclusion, by adapting the XR-seq method to yeast we have obtained data that have enabled us to determine the unique dual-incision pattern in yeast to generate single-nucleotide and genome-wide repair maps for CPD and (6-4)PP in the yeast genome and to examine the relationships between transcription by RNAP II and excision repair. We believe these findings should aid in future repair and mutagenesis studies in this widely used model organism.
Materials and Methods
Yeast Strain.
The yeast strain used in this study is Y452 (MATα, ura3-52, his3–1, leu2-3, leu2-112, cir°), which is wild type with respect to repair. Yeast cells were cultured in YPD (1% yeast extract, 2% peptone, and 2% dextrose) medium at 30 °C in a shaking incubator at 250 × g.
Antibodies and Oligonucleotides.
The rabbit anti-mouse IgG (ab46540) was purchased from Abcam. The anti-CPD (NMDND001) and anti-(6-4)PP (NMDND002) antibodies were obtained from Cosmo Bio. All of the oligonucleotides used for adaptor ligation and PCR amplification were the same as described previously (16).
UV Irradiation and Repair Incubation.
Yeast cells were grown to late log phase (OD600 ∼ 1.0), washed twice with ice-cold ddH2O, resuspended in 2% dextrose, and irradiated in ice-cold 150-mm tissue culture dishes with 120 J/m2 of UV (254 nm). Following irradiation, one-tenth volume of a stock solution (10% yeast extract, 20% peptone) was immediately added to the cells, which were then incubated at 30 °C in the dark to allow excision repair. At different times of the repair incubation (5 min, 20 min, and 1 h), 10-mL aliquots were rapidly cooled in ice water to stop repair and then processed for excision assay and XR-seq.
Cell-Wall Disruption and in Vivo Excision Assay.
To isolate the excised oligomers carrying UV photoproducts in vivo, we first tested three different strategies to disrupt yeast cell walls and lyse cell membranes: (i) Cells were pelleted and resuspended in 300 µL STES (500 mM NaCl, 200 mM Tris–Cl, 10 mM EDTA, and 0.1% SDS), 300 µL phenol:chloroform:isoamyl alcohol (25:24:1), and 600 µL acid-washed glass beads. (ii) The same strategy as the aforementioned method except STET (500 mM NaCl, 200 mM Tris–Cl, 10 mM EDTA, and 1% Triton X-100) was used instead of STES. (iii) Cells were pelleted and resuspended in 600 µL TEP (150 mM Tris–Cl, 30 mM EDTA, and 10 µL Proteinase K) and 600 µL acid-washed glass beads. All of the samples in procedures from i to iii were bead-beat on a Mini-Beadbeater-16 (Bio Spec) for 2.5 min at 4 °C. For samples containing STES or STET buffer, the cell lysates were then centrifuged, and the aqueous supernatants were incubated with 5 µL RNase A at 37 °C for 1 h and then with 5 µL Proteinase K at 55 °C for 1 h. Samples were then extracted with phenol:chloroform:isoamyl alcohol (25:24:1) and precipitated with ethanol. The precipitated small DNA molecules were further purified by a G-50 filtration column (GE Healthcare) and subjected to immunoprecipitation with anti-(6-4)PP antibody as described previously (16). For samples containing TEP buffer, 66 µL of 10% SDS was mixed with the disrupted cells, and 165 μL of 5 M NaCl was mixed in after incubation at room temperature for 10 min. Following overnight incubation at 4 °C, the supernatant fraction was separated by centrifugation and incubated with 10 µL RNase A at 37 °C for 1 h and then with 5 µL Proteinase K at 55 °C for 30 min. The excised oligomers were then sequentially purified by phenol–chloroform extraction, ethanol precipitation, G-50 column filtration, and DNA damage-specific immunoprecipitation. This latter approach approximates the Hirt lysis method used previously (27). For direct excision assay, the excised oligomers isolated by each of the three different methods were 3′ end-labeled with 32P-Cordycepin and resolved on an 11% denaturing sequencing gel.
XR-seq Library Preparation and Sequencing.
Excised oligomers, purified by G-50 column filtration and immunoprecipitated with anti-CPD or anti-(6-4)PP antibody, were ligated to adaptors as described previously (16). The ligated products were then subjected to a second round of DNA damage-specific immunoprecipitation. The CPDs and (6-4)PPs were then repaired by photoreactivation using CPD photolyase or (6-4)PP photolyase as described (16). One percent of the unrepaired and repaired ligation products were used in sets of PCR amplification reactions to determine the minimum number of PCR cycles needed for the final XR-seq library preparation. The number of PCR cycles used for CPD and (6-4)PP XR-seq in this study ranged from 12 to 14. Libraries were gel-purified, pooled, and sequenced using the Illumina HiSeq 2500 platform.
Data Processing and Visualization.
Sequencing reads from two biological replicates were adaptor-trimmed using BBDuk from the BBMap package (https://sourceforge.net/projects/bbmap/). Duplicate reads were removed by a FASTX-Toolkit. Reads longer than 50 bp were excluded from further analysis. The processed reads were aligned to the sacCer3 yeast genome by using bowtie with the arguments: –nomaqround -m 4 -v 1 -k 1 -y –strata –best -p 4 –seed = 123. The length distribution and nucleotide(s) frequencies were obtained by using custom scripts. Sequencing data for RNA-seq and NET-seq were downloaded from previously published datasets in the Gene Expression Omnibus (GEO) database (accession no. GSE68484), and CPD-seq datasets were obtained from the GEO database (accession no. GSE79977). Genome-wide distribution of the XR-seq, NET-seq, RNA-seq, and CPD-seq reads were normalized and viewed in Integrative Genomics Viewer. Pearson’s correlation coefficient between the two biological replicates was calculated and plotted by using R. The sequencing data for XR-seq in this study were deposited in the GEO database, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE110621).
Average Repair Profile Analysis.
The gene list was retrieved from The Saccharomyces Genome Database. Genes shorter than 300 bp were excluded from the analysis. Each gene was divided into 100 equal windows, and the repair signal for each window was counted and normalized to RPBM (reads per base per million mapped reads). Up- and downstream flanking regions were divided into 20-bp windows (n = 100), and XR-seq reads were counted and normalized to RPBM. The expression score for each gene was performed using the NET-seq dataset. The raw reads were retrieved from the Sequence Read Archive database (SRX1016113). The adaptor was trimmed using cutadapt, and reads were aligned to the yeast genome by using bowtie followed by deduplication of the mapped reads. Read counts were normalized to RPKM.
Genic Read Counts.
Each XR-seq dataset was subsampled to 6 million aligned reads. Aligned reads obtained from the XR-seq, NET-seq, and CPD-seq datasets were counted for each yeast gene using bedtools (47). The read count values were adjusted with respect to read depth and gene length by RPKM normalization. The correlation between CPD-seq and NET-seq was analyzed by using the 0-h CPD-seq sample (SRX1683881) normalized by the control (no UV, SRX1683880). In the scatter plots, the genes containing no mapped read for any of the datasets and the genes shorter than 300 bp were excluded from the analysis.
Supplementary Material
Acknowledgments
We thank Dr. Shisheng Li (Louisiana State University) for providing the yeast strain and useful comments and Dr. Brian D. Strahl (University of North Carolina at Chapel Hill) for cell culture support. This work was supported by NIH Grants GM118102 and ES027255.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE110621).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801687115/-/DCSupplemental.
References
- 1.Sancar A. Mechanisms of DNA repair by photolyase and excision nuclease (Nobel Lecture) Angew Chem Int Ed Engl. 2016;55:8502–8527. doi: 10.1002/anie.201601524. [DOI] [PubMed] [Google Scholar]
- 2.Wood RD. Nucleotide excision repair in mammalian cells. J Biol Chem. 1997;272:23465–23468. doi: 10.1074/jbc.272.38.23465. [DOI] [PubMed] [Google Scholar]
- 3.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 4.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 5.Goosen N, Moolenaar GF. Repair of UV damage in bacteria. DNA Repair (Amst) 2008;7:353–379. doi: 10.1016/j.dnarep.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Naegeli H, Sugasawa K. The xeroderma pigmentosum pathway: decision tree analysis of DNA quality. DNA Repair (Amst) 2011;10:673–683. doi: 10.1016/j.dnarep.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 7.Hu J, Selby CP, Adar S, Adebali O, Sancar A. Molecular mechanisms and genomic maps of DNA excision repair in Escherichia coli and humans. J Biol Chem. 2017;292:15588–15597. doi: 10.1074/jbc.R117.807453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sancar A, Rupp WD. A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell. 1983;33:249–260. doi: 10.1016/0092-8674(83)90354-9. [DOI] [PubMed] [Google Scholar]
- 9.Huang JC, Svoboda DL, Reardon JT, Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc Natl Acad Sci USA. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guzder SN, Habraken Y, Sung P, Prakash L, Prakash S. Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J Biol Chem. 1995;270:12973–12976. doi: 10.1074/jbc.270.22.12973. [DOI] [PubMed] [Google Scholar]
- 11.Canturk F, et al. Nucleotide excision repair by dual incisions in plants. Proc Natl Acad Sci USA. 2016;113:4706–4710. doi: 10.1073/pnas.1604097113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 13.Gong F, Kwon Y, Smerdon MJ. Nucleotide excision repair in chromatin and the right of entry. DNA Repair (Amst) 2005;4:884–896. doi: 10.1016/j.dnarep.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Mao P, Wyrick JJ, Roberts SA, Smerdon MJ. UV-induced DNA damage and mutagenesis in chromatin. Photochem Photobiol. 2017;93:216–228. doi: 10.1111/php.12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W, Li S. Facilitators and repressors of transcription-coupled DNA repair in Saccharomyces cerevisiae. Photochem Photobiol. 2017;93:259–267. doi: 10.1111/php.12655. [DOI] [PubMed] [Google Scholar]
- 16.Hu J, Adar S, Selby CP, Lieb JD, Sancar A. Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 2015;29:948–960. doi: 10.1101/gad.261271.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adar S, Hu J, Lieb JD, Sancar A. Genome-wide kinetics of DNA excision repair in relation to chromatin state and mutagenesis. Proc Natl Acad Sci USA. 2016;113:E2124–E2133. doi: 10.1073/pnas.1603388113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu J, Adebali O, Adar S, Sancar A. Dynamic maps of UV damage formation and repair for the human genome. Proc Natl Acad Sci USA. 2017;114:6758–6763. doi: 10.1073/pnas.1706522114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao P, Smerdon MJ, Roberts SA, Wyrick JJ. Chromosomal landscape of UV damage formation and repair at single-nucleotide resolution. Proc Natl Acad Sci USA. 2016;113:9057–9062. doi: 10.1073/pnas.1606667113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W, et al. Human genome-wide repair map of DNA damage caused by the cigarette smoke carcinogen benzo[a]pyrene. Proc Natl Acad Sci USA. 2017;114:6752–6757. doi: 10.1073/pnas.1706021114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.García-Nieto PE, et al. Carcinogen susceptibility is regulated by genome architecture and predicts cancer mutagenesis. EMBO J. 2017;36:2829–2843. doi: 10.15252/embj.201796717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prakash S, Prakash L. Nucleotide excision repair in yeast. Mutat Res. 2000;451:13–24. doi: 10.1016/s0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- 23.Petes TD. Meiotic recombination hot spots and cold spots. Nat Rev Genet. 2001;2:360–369. doi: 10.1038/35072078. [DOI] [PubMed] [Google Scholar]
- 24.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 25.Nag R, Smerdon MJ. Altering the chromatin landscape for nucleotide excision repair. Mutat Res. 2009;682:13–20. doi: 10.1016/j.mrrev.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Adebali O, Sancar A, Selby CP. Mfd translocase is necessary and sufficient for transcription-coupled repair in Escherichia coli. J Biol Chem. 2017;292:18386–18391. doi: 10.1074/jbc.C117.818807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adebali O, Chiou YY, Hu J, Sancar A, Selby CP. Genome-wide transcription-coupled repair in Escherichia coli is mediated by the Mfd translocase. Proc Natl Acad Sci USA. 2017;114:E2116–E2125. doi: 10.1073/pnas.1700230114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu J, et al. Nucleotide excision repair in human cells: fate of the excised oligonucleotide carrying DNA damage in vivo. J Biol Chem. 2013;288:20918–20926. doi: 10.1074/jbc.M113.482257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harlen KM, et al. Comprehensive RNA polymerase II interactomes reveal distinct and varied roles for each phospho-CTD residue. Cell Reports. 2016;15:2147–2158. doi: 10.1016/j.celrep.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tijsterman M, de Pril R, Tasseron-de Jong JG, Brouwer J. RNA polymerase II transcription suppresses nucleosomal modulation of UV-induced (6-4) photoproduct and cyclobutane pyrimidine dimer repair in yeast. Mol Cell Biol. 1999;19:934–940. doi: 10.1128/mcb.19.1.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W, Selvam K, Ko T, Li S. Transcription bypass of DNA lesions enhances cell survival but attenuates transcription coupled DNA repair. Nucleic Acids Res. 2014;42:13242–13253. doi: 10.1093/nar/gku1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao P, Smerdon MJ. Yeast deubiquitinase Ubp3 interacts with the 26 S proteasome to facilitate Rad4 degradation. J Biol Chem. 2010;285:37542–37550. doi: 10.1074/jbc.M110.170175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andrade-Lima LC, Veloso A, Paulsen MT, Menck CF, Ljungman M. DNA repair and recovery of RNA synthesis following exposure to ultraviolet light are delayed in long genes. Nucleic Acids Res. 2015;43:2744–2756. doi: 10.1093/nar/gkv148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W, Selvam K, Rahman SA, Li S. Sen1, the yeast homolog of human senataxin, plays a more direct role than Rad26 in transcription coupled DNA repair. Nucleic Acids Res. 2016;44:6794–6802. doi: 10.1093/nar/gkw428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindsey-Boltz LA, Sancar A. RNA polymerase: the most specific damage recognition protein in cellular responses to DNA damage? Proc Natl Acad Sci USA. 2007;104:13213–13214. doi: 10.1073/pnas.0706316104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaughency P, Merran J, Corden JL. Genome-wide mapping of yeast RNA polymerase II termination. PLoS Genet. 2014;10:e1004632. doi: 10.1371/journal.pgen.1004632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baejen C, et al. Genome-wide analysis of RNA polymerase II termination at protein-coding genes. Mol Cell. 2017;66:38–49.e6. doi: 10.1016/j.molcel.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 38.Chiou YY, Hu J, Sancar A, Selby CP. RNA polymerase II is released from the DNA template during transcription-coupled repair in mammalian cells. J Biol Chem. 2018;293:2476–2486. doi: 10.1074/jbc.RA117.000971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li S, Smerdon MJ. Rpb4 and Rpb9 mediate subpathways of transcription-coupled DNA repair in Saccharomyces cerevisiae. EMBO J. 2002;21:5921–5929. doi: 10.1093/emboj/cdf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li W, Giles C, Li S. Insights into how Spt5 functions in transcription elongation and repressing transcription coupled DNA repair. Nucleic Acids Res. 2014;42:7069–7083. doi: 10.1093/nar/gku333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu J, et al. Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature. 2017;551:653–657. doi: 10.1038/nature24658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Witkin EM. Time, temperature, and protein synthesis: a study of ultraviolet-induced mutation in bacteria. Cold Spring Harb Symp Quant Biol. 1956;21:123–140. doi: 10.1101/sqb.1956.021.01.011. [DOI] [PubMed] [Google Scholar]
- 43.Selby CP, Sancar A. Mechanisms of transcription-repair coupling and mutation frequency decline. Microbiol Rev. 1994;58:317–329. doi: 10.1128/mr.58.3.317-329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li BH, Ebbert A, Bockrath R. Transcription-modulated repair in Escherichia coli evident with UV-induced mutation spectra in supF. J Mol Biol. 1999;294:35–48. doi: 10.1006/jmbi.1999.3265. [DOI] [PubMed] [Google Scholar]
- 45.Li S, Waters R. Induction and repair of cyclobutane pyrimidine dimers in the Escherichia coli tRNA gene tyrT: Fis protein affects dimer induction in the control region and suppresses preferential repair in the coding region of the transcribed strand, except in a short region near the transcription start site. J Mol Biol. 1997;271:31–46. doi: 10.1006/jmbi.1997.1154. [DOI] [PubMed] [Google Scholar]
- 46.Pérez-Ortín JE, Medina DA, Chávez S, Moreno J. What do you mean by transcription rate?: the conceptual difference between nascent transcription rate and mRNA synthesis rate is essential for the proper understanding of transcriptomic analyses. BioEssays. 2013;35:1056–1062. doi: 10.1002/bies.201300057. [DOI] [PubMed] [Google Scholar]
- 47.Quinlan AR. BEDTools: The Swiss-Army tool for genome feature analysis. Curr Protoc Bioinform. 2014;47:11.12.11–11.12.34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.