Abstract
Small molecules have many important roles across the tree of life: they regulate processes from metabolism to transcription, they enable signaling within and between species, and they serve as the biochemical building blocks for cells. They also represent valuable phenotypic endpoints that are promising for use as biomarkers of disease states. In the context of engineering cell-based therapeutics, they hold particularly great promise for enabling finer control over the therapeutic cells and allowing them to be responsive to extracellular cues. The natural signaling and regulatory functions of small molecules can be harnessed and rewired to control cell activity and delivery of therapeutic payloads, potentially increasing efficacy while decreasing toxicity. To that end, this review considers small molecule-mediated regulation and signaling in bacteria. We first discuss some of the most prominent applications and aspirations for responsive cell-based therapeutics. We then describe the transport, signaling, and regulation associated with three classes of molecules that may be exploited in the engineering of therapeutic bacteria: amino acids, fatty acids, and quorum sensing signaling molecules. We also present examples of existing engineering efforts to generate cells that sense and respond to levels of different small molecules. Finally, we discuss future directions for how small molecule-mediated regulation could be harnessed for therapeutic applications, as well as some critical considerations for the ultimate success of such endeavors.
Keywords: small molecule, signaling, regulation, cellular therapeutics, microbiome, cancer, quorum sensing, bacteria
Graphical/Visual Abstract and Caption
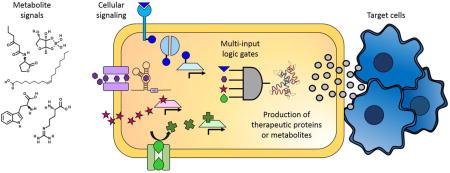
Small molecule signaling and regulation can be harnessed for the control of bacterial cellular therapeutics.
Introduction
Small molecules in biology are not just the biochemical intermediates and building blocks for cells: they also play critical regulatory roles across the tree of life. They can directly and quickly modulate the activity of metabolic enzymes, allowing cells to respond to environmental stimuli in timescales on the order of seconds1, 2. Some of the most basic and fundamental control mechanisms, found repeatedly across species and kingdoms, involve direct feedback inhibition of metabolic pathways via the binding of small molecule metabolic intermediates (metabolites) to enzymes3, 4. Small molecules also bind to and modulate transcription factors, which in turn control transcriptional responses yielding similar feedback regulation. Regulation by small molecules can also mediate large-scale transcriptional responses beyond just transcriptional feedback control. These responses can have fairly straightforward and easily understood results (e.g., catabolite repression5, 6) or extremely complex results; for example, retinoic acid is a small molecule that has been shown to influence the differentiation of stem cells down specific lineages7, 8.
Cellular engineering techniques have the potential to harness the tremendous breadth of small-molecule signaling and regulation found in nature to create cell-based therapeutics. Perhaps the most widely-known current cell-based therapy, stem cells have been heralded for their promise in regenerative medicine, and some treatments have even reached clinical trials9. Beyond stem cells, there is increasing interest in engineering cells to be a class of “theranostics” that sense a disease state and, upon detection, deliver a drug, therapeutic protein, or other payload. Similarly, cells could be engineered to recognize and regulate the dose of a therapeutic by continuously administering a drug until they sense a biomarker of treatment efficacy. Both applications require recognition of specific cues – likely biomarkers endogenous to humans, and often small molecules. Thus, a deep and broad understanding of the ways in which cells sense external small molecule cues and use them to mediate cellular signaling and regulation is critical to the design of dynamically responding cell-based therapeutics.
To that end, this review considers small molecule-mediated regulation and signaling in bacteria. We first summarize some of the most prominent applications and aspirations for responsive cell-based therapeutics. Based on these likely applications, we then consider in turn three different types of regulation that may be exploited in the engineering of therapeutic bacteria: amino acids, fatty acids, and quorum sensing. For those classes of small molecules, we consider their transport, signaling, and regulation, and we present examples of existing engineering efforts to generate cells that sense and respond to their levels. Finally, we conclude with discussion of some of the ways that small molecule-mediated regulation could be harnessed for therapeutic applications, as well as some critical considerations for the ultimate success of such endeavors.
APPLICATIONS AND ASPIRATIONS FOR CELL-BASED THERAPEUTICS
Application: Gut therapeutics
Probiotics are, in essence, one of the most widely-used (and simplified) forms of cell-based gut therapeutics. Multiple diseases are believed to involve the gut microbiome in some form – for example, irritable bowel disease is characterized not only by inflammation, but also by a disrupted gut microbiome10, 11. The idea behind the probiotics approach is bacterial interference12: to seed the gut with bacteria that are known to be harmless or beneficial, and then hope that these bacteria can colonize the host sufficiently to alter the microbiome and establish a more healthy community composition. The same scientific principle of bacterial interference is the basis for fecal microbiota transplants, which entail transplanting fecal samples from a healthy donor into a recipient. This approach has proven surprisingly effective in the treatment of Clostridium dificile infection, particularly in cases where the patient relapsed after initial antibiotic treatments13.
In each of these therapies, beneficial microbes are selected and delivered, but not genetically manipulated. To create responsive or advanced cell-based therapeutics, microbes must be engineered to synthesize and deliver a therapeutic protein or small molecule, preferably on-demand and in response to a specific signal.
There are numerous examples of engineered probiotics that are currently in development; a few representative examples follow. Lactococcus lactis has been engineered to secrete therapeutics for diabetes14, 15, irritable bowel disease16, and oral mucositis (ulcerative lesions that are a common side effect of chemotherapy)17. Additionally, Lactobacillus gasseri and Escherichia coli have been engineered to deliver therapeutics for diabetes and obesity, respectively18, 19. While this list is by no means comprehensive (and a more detailed treatment is provided by Mimee et al.20), it provides a sense of scope in terms of the diseases targeted and microbes used in recent efforts.
This approach still faces numerous challenges, though. Success, whether in bacterial interference or delivery of other therapeutic agents, requires stable engraftment (or at least survival of the delivery bacteria to its target niche in the gastrointestinal tract). Improvements in the delivery vehicles for probiotics can be used to help address this challenge21. In engineering microbes that sense and respond to specific disease states, identification of clinically relevant biomarkers and maintenance of circuit stability across many generations of the bacteria are additional significant challenges. Of course, biocontainment, safety, and regulation of these therapeutics are also critical concerns as well.
Application: Cancer therapeutics
Bacteria are also excellent candidate vehicles for the treatment of cancer. Work in this area began increasing exponentially at the turn of the century22, although the idea has been around for decades: it was noticed a long time ago that cancer patients sometimes had reductions in tumor sizes when suffering from bacterial infections23, and published bacterial cancer therapy clinical trials date as far back as the 1960’s24. Since tumors are essentially immunoprivileged environments25, bacteria that can localize to these tumors have the potential to expand rapidly and thus potentially deliver drugs or prodrugs to kill cancer cells.
Salmonella species have been explored extensively for their potential to treat cancer. Salmonella is a facultative anaerobe that, when injected systemically, attaches to the walls of tumor vasculature and can subsequently invade the tumor more completely26 via mechanisms ranging from the aforementioned immunoprivilege to chemotaxis towards molecules produced by tumors22. Just infection and proliferation of Salmonella itself can be sufficient to cause tumor regression (with multiple examples provided by Forbes22), but the effectiveness of these cells can be improved via engineering. For example, Salmonella strains that are auxotrophic for leucine and arginine, which are produced by dying tumor tissue, show increased tumor specificity regardless of any payload22, 27, 28. Strains that were auxotrophic for purines (also produced by necrotic tumor tissue) have been used to transform prodrugs into their active forms selectively in the tumor29, 30. Cytotoxic agents, cytokines, and tumor-specific antigens and antibodies have also been integrated into Salmonella delivery vectors, as described elsewhere22.
Clostridium species have also been the focus of significant efforts in bacterial tumor targeting. Clostridium species are obligate anaerobes, such that bacterial spores must be injected because live cells cannot survive in normoxic physiological environments. Over fifty years ago these spores were shown to selectively germinate in anoxic tumor regions31–33, making for a powerfully specific delivery agent since such anoxic regions do not normally occur elsewhere in the body. In follow-up clinical trials oncolysis was demonstrated, although tumor control and eradication were not demonstrated purely due to the growth of the bacteria24, 34, 35. Nonetheless, this has been the springboard for broader use of Clostridia to deliver therapeutics34.
Other bacterial cancer therapeutics exist and will be noted in later sections. The examples above suffice to demonstrate the broad potential for, and significant effort invested in, the use of bacteria to target cancer cells. As biomarkers for specific cancers and subtypes continue to be identified, the opportunity for on-demand, precision medicine will call for the development and engineering of increasingly sophisticated bacterial delivery vectors.
Aspiration: Finding biomarkers that identify diseased states
The ideal cell-based therapeutic would provide highly specific, selective delivery only to the targeted organ and/or only when the patient suffers from some pathological condition. A prerequisite to enabling this functionality is the existence of biomarkers that specifically indicate a given pathological condition. For all but the most straightforward examples (e.g., phenylketonuria, an inborn error of metabolism that causes decreased metabolism of phenylalanine36) the discovery of such biomarkers for individual pathologies is a grand challenge. Phenotypic endpoint indicators of disease are desirable for such biomarkers, to avoid complications due to multigenic, multiallelic diseases and penetrance that can arise from DNA-based biomarkers. Metabolites are a promising example of such a downstream readout. Significant effort has been put into discovering metabolite signatures of individual diseases, with recent advances in analytical chemistry instrumentation providing the impetus for continued effort and hope that unique biomarker signatures can be identified. We will briefly consider some of the ongoing biomarker identification efforts in the two general application areas discussed above.
The identification of circulating or in situ metabolite biomarkers for cancer has long been a prominent emphasis in the field of metabolomics, which is the systems-scale analysis of metabolites in biological systems. A number of reviews provide in-depth and broad summaries of the different metabolites and metabolite combinations that have been implicated as biomarkers for different types of cancer37, 38. Extensive efforts have been undertaken in almost every type of cancer, from breast39 to prostate40 to lung41. A few general conclusions can be drawn from the trends observed in these efforts. First, two classes of molecules that are frequently identified as biomarkers for cancer are amino acids (and their derivative molecules) and lipids (fatty acids, carnitines, etc.). This makes sense, as dysfunctional metabolism is viewed as a hallmark of all cancers42, and so one would expect there to be commonalities in the metabolic mechanisms by which rapid cellular proliferation can be maintained. This leads to the second important conclusion, which is that due to the high degree of overlap in perturbed metabolite levels across cancer types, it is extremely difficult to find single metabolite biomarkers that are unique for a given cancer. The most promising (though quite difficult) option is to find a panel of a small number of metabolites which, when combined, provide a molecular diagnosis for a specific type of cancer. For the purposes of designing a responsive cell-based therapeutic, this would necessarily entail the construction of a complex biosensor network in the cell.
The prevalence of metabolomics applications to studying gastroenterological diseases, such as inflammatory bowel disease (IBD), has increased significantly in recent years, though it still lags behind the extensive study of cancer metabolomics. Nonetheless, metabolomics has been used to study human IBD in a number of contexts and in a wide variety of sample types, including urine, serum/plasma, fecal extracts, and colon tissue43. It has been shown that the sample type can have a significant impact on the metabolomic results and biological inferences drawn: for example, urine samples were shown to exhibit more biological variability and greater sensitivity to environmental cues (like diet) than plasma samples44, though fecal extracts and urine are expected to provide more direct phenotypic endpoint indicators of IBD (and gut-resident microbes would not easily be able to sense serum metabolite levels). This tradeoff between relevance and expected utility must be considered when determining which metabolites should be used to trigger production of therapeutic by a bacterial cell. Recent reviews, including one by Storr et al., provide a more detailed review of metabolomics studies of IBD and of the metabolites identified as significant in different tissue types43. Perhaps of most relevance to our discussion here is that differences in amino acid and fatty acid metabolism seem to be one signature of IBD45. These classes of molecules thus are of utmost importance in considering how metabolite biomarker-responsive bacterial cell therapeutics might be designed and implemented.
Aspiration: Sensing and responding to biomarkers
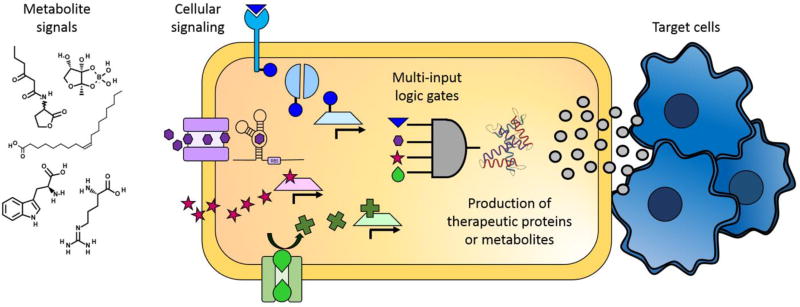
Once relevant biomarkers for a pathological condition are known, one must still create a cell capable of sensing these biomarkers and responding only in their presence (Figure 1). While the idea of simple in vitro evolution of biosensors for specific metabolites (e.g., aptamers)46 is appealing and exciting, in vitro evolved molecules may not exhibit the same sensitivity or specificity when used in vivo. In the context of cell-based therapeutics, this means that the natural repertoire of metabolite-responsive molecules and signaling networks is a useful starting point for engineering control of drug or biologic production in cell-based therapeutics.
Figure 1.
Overview of the ways that metabolite signals can be harnessed to create cellular therapeutics. Extracellular metabolites, including amino acids, fatty acids, and quorum sensing molecules, can trigger changes in cellular activity. Some metabolites enter the cytoplasm directly through the membrane or through specific transporters, and other metabolites bind to proteins either extracellularly or in the periplasm and trigger signaling cascades. Metabolites can effect changes in cells by changing the conformation of enzymes, mRNA, or transcription factors. Ultimately, to create effective cellular therapeutics, cells will have to sense and respond to multiple metabolite markers by enacting layered logic gates to tightly and specifically control the production of therapeutic proteins or metabolites. Finally, the therapeutic molecules must be released so that they can have the desired effect on the target cells.
Fortunately, the breadth of choices available for control of metabolite-responsive action in cells is significant. A recent bioinformatics analysis identified dozens of distinct ligand-responsive transcription factors that could be used to control cellular production of a target molecule in response to intracellular or extracellular levels of specific metabolites47. These sensors span diverse chemical classes including lipids, amino acids, carbohydrates, and steroids, each of which are likely sources of disease-indicating biomarkers.
Much of the study and review of metabolite-responsive transcription factors has been in the context of metabolic engineering – the manipulation of cellular metabolism to produce high levels of a specific target metabolite where the cell would normally produce low levels (or none) of the metabolite. In particular, excellent reviews of genetically-encoded biosensors48, and specifically transcription factor based biosensors49, provide perspective not only on the breadth of different sensors that have been identified or developed, but also on the various methods that can be used to develop or improve such sensors. Liu et al.50 also provide a detailed treatment of metabolite-responsive transcription factors that can be exploited for the responsive cell-based therapeutic goals described in this review. The insights and progress from the field of metabolic engineering, while in a different context, are quite relevant to the development of cell-based therapeutic approaches – instead of engineering cells to produce industrially relevant compounds, they can be engineered to produce drugs by changing the genes or pathways being controlled.
SMALL MOLECULE REGULATION IN BACTERIA RELEVANT TO RESPONSIVE CELL-BASED THERAPEUTICS
As described earlier, amino acids and fatty acids are two commonly-identified (and well-studied) classes of metabolite biomarkers that might be used to drive selective payload delivery of therapeutic cells. We now consider each of these in greater detail. We will also discuss another key class of small molecules that has proven useful in engineering inducible and controlled activity of bacterial cells: quorum sensing signaling molecules. For all three classes of molecules, we describe the relevant transport and signaling mechanisms that are critical to engineering an effective small molecule-responsive cell, and present examples of how these pathways have previously been exploited. Often the most common applications exploiting these regulatory and signaling pathways are in the context of metabolic engineering, and thus much of the discussion will be in that vein.
Amino acids
Transport
Extensive reviews on bacterial amino acid transport proteins in general are available elsewhere, including one with considerations for the biotechnological applications and implications of these proteins51. We will briefly describe some of the major transport mechanisms of these proteins.
Most bacterial amino acid transporters are major facilitator superfamily (MFS) transporters. MFS transporters undergo conformational changes that allow them to transport their substrates across biological membranes, and they can be divided into three main groups based on the transport mode: uniporters that transport a single substrate, symporters that transport a substrate in association with a coupling ion, and antiporters that transport a substrate and a co-substrate in opposite directions52. They rely on energy from concentration or electrochemical gradients of their coupling ions or co-substrates to transport molecules against the gradient. Some MFS transporters are specific for individual amino acids, while others may be for more general classes of molecules (e.g., hydrophobic or charged).
Of the bacterial amino acid transporters that are not in the MFS superfamily, quite a few are in the ATP binding cassette (ABC) superfamily of transporters, which is characterized by two integral membrane proteins, an extracytoplasmic binding protein, and ATPase activity that drives transport of amino acids into the cell. These transporters can be broken down into two classes: polar amino acid transporters (the larger of the two classes), and hydrophobic amino acid transporters53. While some of these transporters are constitutively expressed, some are only conditionally expressed (for example, under nitrogen limitation by σ54, the nitrogen assimilation factor54, 55).
Signaling and regulation
The presence or absence of amino acids in cells has major effects on transcription. Different effects are seen if the change in amino acids levels is general (e.g., nitrogen limitation) or specific to a small number of amino acids. When there is general amino acid starvation, ribosomes stall because they do not have sufficient amino acids (or more specifically, charged transfer RNA) to allow translation. This prompts ribosome-associated RelA to produce guanosine pentaphosphate (ppGpp), a global regulator alarmone that activates the stringent response56. The stringent response is a switch from a primarily anabolic mode devoted to cell division and the generation of building blocks for biomass production to a catabolic mode focused more on survival, using strategies such as cessation of DNA replication and re-assimilation of unused nucleotides and fatty acids back into central metabolism57.
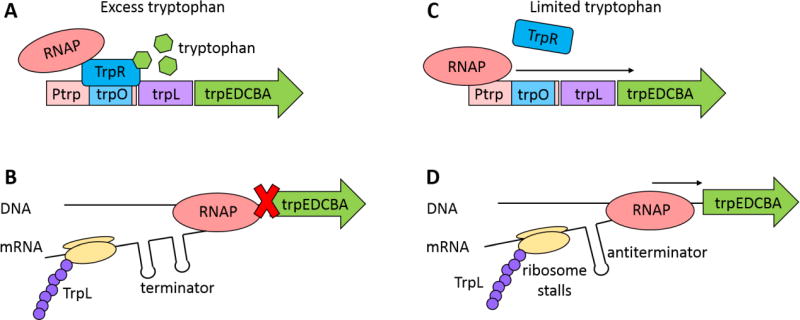
Bacteria have a variety of mechanisms to respond to changes in specific amino acid levels, and multiple regulatory networks or mechanisms are often activated simultaneously. For example, the tryptophan biosynthesis genes are controlled both by tryptophan-responsive transcription factors and through transcriptional attenuation. When the cell has sufficient tryptophan, the TrpR repressor prevents transcription of tryptophan biosynthesis genes (Figure 2A), and a transcriptional terminator upstream of the biosynthesis genes simultaneously helps to eliminate any leaky expression from the TrpR-controlled promoter (Figure 2B). In low tryptophan, the TrpR repressor is derepressed, turning on transcription of the trp operon (Figure 2C). Low tryptophan levels cause (via the lack of charged transfer RNAs) the ribosome to stall on tryptophan codons, which causes different mRNA folding kinetics in the still-forming transcript and thus a different hairpin structure to form that does not cause transcriptional termination (Figure 2D). B. subtilus has a slightly different method of transcriptional attenuation: in the presence of tryptophan, the trp RNA binding attenuation protein (TRAP) binds trp operon RNA as it is being transcribed, forming a transcriptional terminator that ceases transcription58, 59. Similarly, when tryptophan is in excess, cells induce expression of the tryptophanase operon, tna, to degrade tryptophan into other useful metabolic compounds. The tna operon is also regulated by transcriptional attenuation: at low to moderate tryptophan levels, a transcriptional terminator is formed that prevents transcription of the entire operon. When tryptophan is in excess, the transcriptional terminator is not formed, and the operon is transcribed60–62.
Figure 2.
Control of tryptophan biosynthesis genes in E. coli. Tryptophan biosynthesis is controlled through both transcriptional repression and transcriptional attenuation. (A) In the presence of tryptophan, the transcription factor TrpR binds to tryptophan, changes its conformation, and binds to the trp operator site to prevent binding of RNA polymerase (RNAP) to the promoter (Ptrp) and transcription of the trp operon. (B) Transcriptional attenuation is another layer of control in preventing unnecessary expression of tryptophan biosynthesis genes. If transcription is initiated in the presence of tryptophan, while translating the leader peptide TrpL, a transcriptional terminator is transcribed that causes RNAP dissociation from the DNA and prevents transcription of the trp operon. (C) In the absence of tryptophan, TrpR cannot bind to the trp operator site, and transcription of the trp operon proceeds. (D) In limited tryptophan, the ribosome stalls on tryptophan codons in the leader peptide because of a shortage of tryptophan-charged charged transfer RNAs. This stall alters messenger RNA (mRNA) folding kinetics such that an antiterminator forms, transcription continues, and tryptophan biosynthesis proteins are synthesized.
Cells also use riboswitches to control their transcriptional response to changes in amino acids63. When riboswitches for lysine and glycine bind those amino acids, transcript RNA folding is changed, which causes transcriptional attenuation either by creating a transcriptional terminator or by blocking the ribosomal binding site and preventing translation.
Implementation examples
To get a sense for how one could best exploit our understanding of natural signaling and regulation based on amino acid levels, it is instructive to look at existing synthetic biology and metabolic engineering efforts for amino acid production. Transcriptional regulators that respond to changes in lysine64, methionine65, and valine66 have been used to regulate reporter proteins to create biosensors for those amino acids that better enable high-throughput screens for altered amino acid metabolism.
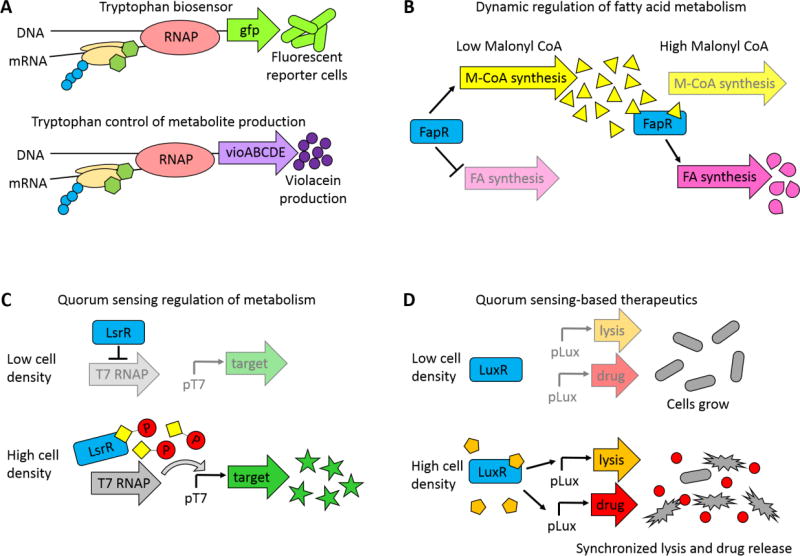
Beyond reporting on amino acid levels, amino acid biosensors can also be used to effect metabolic changes. Fang et al. increased the yields of the pigment violacein by replacing the tna operon, which normally degrades tryptophan, with the metabolic pathway for violacein that uses tryptophan as a precursor67, 68 (Figure 3A). Similarly, the lysine riboswitch described above was used to effect a larger-scale metabolic shift by controlling flux through the TCA cycle in E. coli and C. glutamicum69. Each of these cases demonstrates how natural mechanisms to regulate amino acid levels could potentially be used to directly control the production of a metabolite payload in a bacterial cell-based therapeutic.
Figure 3.
Examples of ways that metabolite signaling and regulation has been harnessed for metabolic engineering and therapeutic applications. (A) Tryptophan regulation of cell output. The genes of the tna operon, which naturally degrade tryptophan, were replaced with either GFP or the vioABCDE genes, which produce the purple pigment violacein from tryptophan. In the presence of tryptophan, tryoptophan binds to the ribosome to prevent premature transcriptional termination. Replacement of the naturally occurring genes with either reporter genes or metabolic pathways created a biosensor that reported on intracellular tryptophan levels and a dynamically controlled metabolic pathway that upregulated violacein synthesis in the presence of excess tryptophan. Adapted from Fang, et. al.68 (B) Dynamic regulation of fatty acid production to improve fatty acids yields. In the absence of malonyl-CoA (a precursor to fatty acids), the transcription factor FapR turns on the malonyl-CoA synthesis pathway and represses synthesis of fatty acids. When sufficient malonyl-CoA has accumulated, malonyl-CoA binds to FapR, and the FapR-malonyl-CoA complex then activates the fatty acid synthesis pathway. Adapted from Xu, et. al.70 (C) Cell-density dependent metabolite production. Target genes are expressed from a T7 promoter, which is only transcribed in the presence of T7 RNA polymerase (T7 RNAP). In low cell density, the transcription factor LsrR represses production of T7 RNAP, and the target genes are not expressed. Once the culture reaches a threshold cell density, phosphorylated autoinducer binds to LsrR, and LsrR no longer represses transcription of T7 RNAP. T7 RNAP then transcribes the target gene. Adapted from Tsao, et. al.71 (D) Cell-density dependent drug delivery. Genes for an anti-cancer drug and a lytic protein were placed under control of LuxR regulated promoters. In low cell density, these genes are not activated. Once the cells reach a threshold cell density, autoinducer binds to LuxR, turning on production of the drug and lytic protein. Synchronized lysis of cells delivers the drug to the surrounding environment, and a few unlysed bacteria enable reseeding and pulsatile treatment. Adapted from Din, et. al.72
Fatty acids
Transport
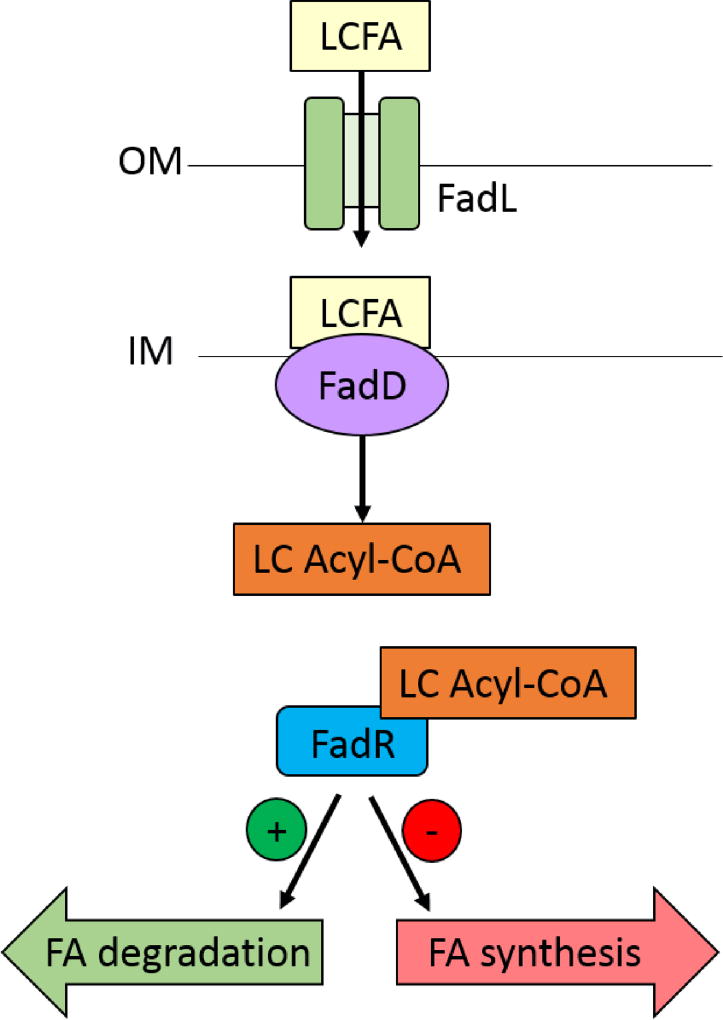
Transport of long chain fatty acids in E. coli happens via a very well-characterized mechanism73, shown in Figure 4. FadL, the only membrane-bound long-chain fatty acid transporter in E. coli, is an outer membrane protein. It binds long chain fatty acids with high affinity and specificity, with the length of the carbon tail providing a significant amount of the substrate specificity74. FadL then transports the fatty acid into the (slightly acidic) periplasm, where it gets protonated and partitions into and “flips” to the cytoplasmic face of the inner membrane. The acyl-CoA synthetase FadD partitions into the inner membrane while bound to ATP, and with broad substrate specificity it cleaves the protonated fatty acid to yield a long chain acyl-CoA that is released into the cytosol.
Figure 4.
Fatty acid transport, signaling, and regulation in E. coli. The outer membrane protein FadL recognizes and transports long chain fatty acids (LCFA) into the periplasm. The slight acidity of the periplasm causes the carboxyl group of the LCFA to become protonated, which triggers it to “flip” to the cytoplasmic face of the inner membrane. The acyl-CoA synthetase FadD then localizes to the inner membrane, where it cleaves the charged fatty acid to yield a long chain acyl-CoA. The transcription factor FadR binds the LC Acyl-CoA, which makes FadR unable to bind to its operator site. Fatty acid degradation pathways are upregulated and fatty acid synthesis pathways are downregulated. Figure adapted from Dirusso and Black73.
Signaling and regulation
Transport of fatty acids into E. coli is ultimately regulated on a transcriptional level via the transcription factor FadR. Normally, FadR is bound to promoters such that it serves as a repressor of the fatty acid degradation pathway and the fatty acid transport pathway, and as an activator of fatty acid synthesis pathways75. However, when FadR binds to the long chain acyl-CoAs that are produced by FadD, it can no longer bind to its cognate promoters, causing fatty acid synthesis to be turned off, fatty acid degradation to be turned on, and fatty acid transport to be induced (Figure 4). This of course then serves as a positive feedback loop to accelerate the transport of fatty acids into the cell.
In Bacilus subtilus, regulation of fatty acid synthesis and degradation is more modular75. FadR again controls degradation of long-chain fatty acids by repressing fatty acid degradation pathways in the absence of long chain acyl-CoA but does not control fatty acid biosynthesis. Instead, another transcription factor, FapR, acts as a transcriptional repressor for fatty acid and phospholipid biosynthesis. FapR is regulated by malonyl-CoA; when malonyl-CoA binds to FapR, it can no longer bind its cognate promoters, causing fatty acid biosynthesis to be induced. It is worth noting that in contrast to the broad acyl-CoA specificity of FadR, FapR is specific to malonyl-CoA, an essential intermediate in fatty acid biosynthesis, yielding a tightly-regulated pool of malonyl-CoA via feedback inhibition.
Implementation examples
Since fatty acids are precursor molecules to many high-value compounds (e.g., pharmaceuticals and biofuels), fatty acid production has been the focus of significant metabolic engineering efforts. In those efforts, natural mechanisms for the regulation of fatty acid biosynthesis have been harnessed in numerous ways. For example, Xu et al. generated a malonyl-CoA-dependent biosensor in E. coli by using the B. subtilus FapR protein76. They constructed a hybrid lac-inducible, malonyl-CoA-inducible promoter containing the lac operator and the cognate cis-regulatory element for FapR, fapO, and they used it in a sensor they suggest can enable generation of strains that increase production of malonyl-CoA (a rate-limiting precursor in fatty acid chain elongation reactions).
In separate work, Xu et al. also found that FapR could act as an activator under certain circumstances77. In designing synthetic promoters, when fapO was added to an orthogonal promoter in E. coli as in the aforementioned work, FapR functioned as a repressor when it was expressed from the same construct. However, when fapO was added to a native E. coli promoter, they found that FapR could also act as an activator. As a result, the addition of malonyl-CoA to the cells in turn decreased, rather than increased, expression from the new promoter. They then integrated these two promoters with distinct functions into a metabolic switch that enabled dynamic regulation of fatty acid biosynthesis and efficiently redirected carbon flux towards that pathway (Figure 3B).
Another group used native FadR as the basis for a fatty acid/acyl-CoA biosensor as part of a dynamic sensor-regulator system47. They sought to improve production of fatty acid ethyl esters by using FadR to dynamically control the supply of precursors to the fatty acid biosynthesis pathway. Their approach overcame previously encountered toxicity issues associated with metabolic intermediate accumulation, and they engineered synthetic promoters with the FadR binding site that increased the dynamic range of transcriptional response to fatty acid levels compared to native E. coli promoters. This promoter was then used to drive transcription of specific modules of the fatty acid ethyl ester biosynthesis pathway to optimize supply and utilization of pathway intermediates, leading to enhanced yield.
Quorum Sensing
Another set of small molecules with great potential for use in cell-based therapeutics are the quorum-sensing signaling molecules. In quorum sensing, cells sense and respond to changes in population density by activating different genetic programs. Cells produce small molecules, called autoinducers, that regulate these transcriptional programs. Recognition of and response to a specific autoinducer could be used to create targeted antibiotics, in which the therapeutic cells sense and target harmful bacteria. Alternatively, cell-based therapeutics could use quorum sensing networks to coordinate cell response, such that cells only release their payload when the cells reach a critical population density; this would enable targeted treatment in areas where bacterial cells accumulate and help to minimize off-target drug release. Similarly, treatment specificity could be increased by requiring two different types of bacteria to be present to trigger payload delivery.
Transport
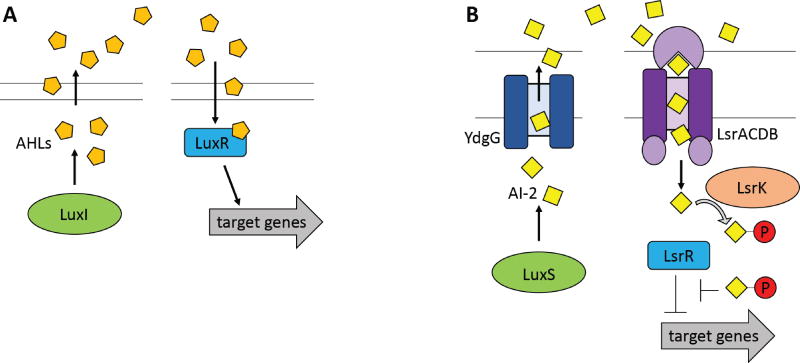
Transport of quorum-sensing signaling molecules varies by species, and in some cases it is much simpler than the transport mechanisms described in previous sections. For example, multiple gram-negative bacteria use an N-acyl-homoserine lactone (AHL) molecule as a quorum-sensing signaling molecule. AHLs have homoserine lactone rings with N-acyl chains that vary in length and side chain modification. The widely studied Vibrio fischeri AHL freely diffuses into and out of the cell, without use of transporters in either direction (see Figure 5A). In other organisms (e.g., Pseudomonas aeruginosa) that use AHLs with longer N-acyl side chains, the AHL can partition into the membrane and is more effectively transported out of a cell through active efflux systems78, 79. The size of the N-acyl side chain also affects AHL diffusion rates in aqueous environments, meaning that species with smaller AHLs (higher diffusion rates) can communicate across longer distances80.
Figure 5.
Quorum sensing signaling and regulation. (A) AHL-mediated quorum-sensing networks. LuxI is an AHL synthase that produces acyl-homoserine lactones (AHLs) that can freely diffuse through the membrane. When the AHL concentration reaches a critical threshold, LuxR family proteins bind AHL to alter the transcription of target genes. (B) The AI-2 mediated quorum sensing network in E. coli. LuxS produces the quorum sensing molecule AI-2. The transporter YdgG facilitates AI-2 export. When the AI-2 concentration has reached a threshold level, the ABC transporter LsrACDB imports AI-2 into the cytoplasm, where it is phosphorylated by LsrK. The phosphorylated AI-2 molecule binds to LsrR, prompting derepression of target genes. Figure adapted from (A) Miller & Bassler85 and (B) Li et al.88.
Another widely studied family of quorum sensing molecules is autoinducer-2 (AI-2), which has a completely different structure that contains a cyclic ether and is often complexed with a borate81, 82. Unlike AHLs, which have a wide variety of highly species-specific structures, a small number of unique AI-2 molecules is produced and sensed by a wide variety of bacteria. However, transport of AI-2 varies by species: for example, E. coli imports AI-2 through the ABC transporter LsrACDB83 (see Figure 5B), while in Vibrio harveyi, AI-2 binds to the two-component sensor-kinase LuxPQ to effect a phosphorylation cascade84.
Signaling and regulation
A variety of signaling and regulation modalities are observed in the diverse quorum-sensing mechanisms across bacterial species. In the AHL family of quorum-sensing molecules in gram-negative bacteria, AHL synthases (generally homologs of the Vibrio fischeri protein LuxI) produce AHLs, AHLs diffuse into neighboring cells, and upon reaching a critical concentration in neighboring cells, AHLs bind to LuxR family transcriptional regulators85 (see Figure 5A). Notably, these LuxR proteins often recognize a specific type of AHL, such that quorum sensing only occurs between cells of the same species. However, some species can recognize a variety of AHL molecules, allowing for interspecies communication86.
When AI-2 molecules enter the cell, they trigger a phosphorylation-mediated positive feedback loop. AI-2 is produced indirectly by the S-ribosylhomocysteine lyase LuxS82. In E. coli and Salmonella, the inner membrane transporter YdgG facilitates its export to the periplasm87 for sending the AI-2 signal; for receiving the signal, the ABC transporter LsrACDB mediates import of AI-2, which is then phosphorylated by LsrK into an active form88 (see Figure 5B). Production of LsrACDB is regulated both by AI-2 and by cAMP/CRP, which is a global regulator in E. coli. Consistent with the name of the molecule, this regulation allows for a positive feedback autoinduction, such that when some AI-2 enters, it turns on expression of more AI-2 importers, thus causing more AI-2 to enter. In other species, such as Vibrio harveyi, AI-2 and some other autoinducers (AI1, CAI1) are imported into the periplasm, where they work through two-component sensor kinase proteins. For example, in Vibrio species, the autoinducer sensor LuxPQ autophosphorylates and controls transfer of phosphorylation to transcriptional regulators that control expression of target pathways89. Some of the transcription that is regulated includes noncoding sRNAs, which in turn can disrupt or activate translation of target proteins90–94.
Ultimately, the cellular activities controlled by quorum-sensing molecules are diverse and vary significantly by species. These activities can include toxin production95, biofilm formation96, bioluminescence97, protein secretion98, motility99, and cell division100. In the same way, a variety of cellular readouts and outputs could easily be engineered into these regulatory systems for therapeutic purposes.
Implementation examples
One obvious class of applications of quorum-sensing regulatory networks is to coordinate cell outputs, similar to the overall functional purpose of natural quorum-sensing systems. For example, Danino et al.101 used quorum sensing components to develop synchronized genetic clock oscillators and then used microfluidics to characterize the spatiotemporal dynamics of the system. (Similar quorum sensing circuits have also been made in eukaryotes, including the yeast Saccharomyces cerevisiae, using mating-type pheromones as signaling molecules102, though eukaryotic applications of quorum sensing are beyond the scope of this review.)
Another application of these regulatory networks is in the self-regulation of protein induction, which has been primarily explored in the context of metabolic engineering, but could readily be applied to cellular therapeutics. Tsao et al. made a system in E. coli that responds to AI-2 such that T7 RNA polymerase is only expressed when AI-2 is sensed71. A protein to be overexpressed was placed under the control of a T7 promoter, yielding a protein expression system that would only be induced at a critical cell density (Figure 3C). Gupta et al. improved an E. coli metabolic engineering system by tuning the cell density that turns on the quorum sensing cascade, which was accomplished by changing the level of protein that produces AHL103. By varying promoter and ribosomal binding site strength to lower levels of AHL synthase enzymes, they were able to cause gene expression to be turned off at higher cell densities, and used this to determine the optimal point at which to redirect metabolic flux towards their engineering target.
Quorum sensing regulatory pathways have already been applied to therapeutics. For example, quorum sensing pathways have been used to control when bacterial cells invade cancer cells104. (Of note, they needed to carefully tune expression levels to ensure that cell invasion only started when a critical density was reached; see Future Directions for more discussion of the importance of tight control vs. leakiness.) Since the bacteria already naturally localize to tumors, they will likely only be at the threshold density in these tumors, and so the bacteria would only invade cells at the tumor site (though there was no in vivo characterization of efficacy or toxicity in that work). Din et al. used AHL quorum sensing circuits to couple drug delivery to lysis based on cell density72 (Figure 3D). Genetically encoded payloads were delivered to tumor cells upon the accumulation of sufficient population density via simultaneous lysis of cells; the stochasticity of the process yielded a few unlysed bacteria which allowed for reseeding and subsequent retreatment of the tumor, ultimately yielding a pulsatile treatment regimen. Altogether, the approach allowed for enhanced specificity of drug delivery and minimization of toxicity effects from bacterial accumulation. The authors also demonstrated in vivo validation of their concept in mice. Swofford et al. incorporated an AHL-based quorum sensing circuit into (previously discussed) tumor hypoxia-targeting Salmonella, increasing the specificity of drug release.
A microbiome application has also been explored, using non-pathogenic E. coli cells to target pathogenic Pseudomonas aeruginosa in the respiratory and gastrointestinal tracts105. Saeidi et al. designed E. coli cells to recognize AHL secreted by P. aeruginosa and, upon recognition, produce proteins (pyocins) that kill P. aeruginosa and a lysis protein that enables complete secretion of the pyocin toxin.
Future Applications and Conclusion
As is evident from the above discussions, metabolite signaling pathways have been successfully harnessed for numerous metabolic engineering applications. These applications may be directly relevant to the development of cell-based therapeutics if the metabolite-sensing system is similar and the payload to be produced by the cells is a small-molecule drug; if not, the general approaches used to harness and engineer metabolite-sensing systems can be applied to other signaling pathways and other types of payloads.
For example, the strategy known as dynamic metabolic engineering offers a framework for how to design cells to respond to changes in their environment106, 107. The goal of dynamic metabolic engineering is to redirect flux from one pathway to another at a specific point during a fermentation so as to maximize the final titer or productivity of cells for a target metabolite product. This allows one to balance trade-offs between growth and production, overcome toxicity issues of intermediates and products, and even enhance the stability of the engineered cells. As indicated above, engineering of natural quorum sensing regulatory networks has already shown significant utility for controlling cell behavior in a dynamic metabolic engineering context103 and in therapeutic design72, 104, 105.
One of the most obvious ways that systems and synthetic biology can be applied in the development of the cell-based therapeutics considered here is in creating logic gates that respond to multiple metabolites. We have already discussed the fact that finding a single, unique biomarker for a given pathological condition is extremely difficult, if not impossible. To get a truly specific and selective responsive cell, multiple biomarker signals will need to be integrated into a single overall regulatory control network.
Hybrid promoters, which include cis-encoded sites for multiple cis- or trans-acting regulatory mechanisms, are a promising approach to achieve such combinatorial control. Hybrid promoters have been explored rather extensively in the context of commonly-used transcription factors like the lac repressor, tet repressor, and the activator/repressor araC108, 109, including optimization of location and order of different operator sites in the promoter. This can be expanded further to incorporate operator sites for metabolically relevant transcription factors or regulatory mechanisms. As previously discussed, combinatorial regulation has been explored with FapR76, 77 and with FadR47, and this approach is likely generalizable enough to be used modularly with many other metabolite-transcription factor pairs. It is particularly promising as a direct way to integrate quorum sensing and metabolite-regulated responses.
Of course, there are limitations on what kinds of combinatorial regulation can be effected by directly combining the transcription factor binding sites of multiple metabolite-responsive transcription factors. For example, the dominance of an activator or a repressor over another transcription factor may allow only an AND gate, a NAND gate, or some other specific gate to be constructed via a hybrid promoter. The design of more complex gates regardless of the epistasis of individual regulatory molecules will necessarily draw from recent advances in synthetic biology to construct arbitrary logic combinations110. This could also include other types of regulation as well, including more cis-regulatory mechanisms like aptamers or transcriptional attenuation approaches described previously.
Synthetic biology can also help in tuning response sensitivity and selectivity. The concentration of analyte that activates production of the target metabolite can be shifted by modifying the sequence of the operator site or polymerase binding domain, adding decoy operator sites, and decreasing the affinity of a transcriptional regulator for the analyte, reviewed more thoroughly in Ang, et. al.111 Response cooperativity can be improved through operator site optimization, transcriptional attenuators, and feedback loops111. Directed evolution of regulators can be used to improve substrate specificity and prevent activation by off-target molecules112, 113, though it suffices to say that protein engineering (whether rational or otherwise) remains a task requiring significant effort and resources for a given engineering goal.
Three important challenges lie ahead, though. First, as discussed previously, the design of therapeutics that sense and respond to biomarkers of disease will necessarily be limited by knowledge of biomarkers in the appropriate biofluid or niche. Second, even if these biomarkers are identified, there may not be well-characterized regulators or sensor mechanisms for the biomarker of interest. One way to combat this could be a systems approach to identify promoters that drive changes in transcription in response to the target biomarker. For example, Dahl et al. determined genes that were up- or down-regulated in response to the levels of a molecule of interest, and then used these genes’ promoters to characterize and control production of that molecule114.
Finally, and perhaps most importantly, is the challenge of maintaining tight control in the design of cells that sense and respond to environmental cues. One wants to make sure that the payload is delivered only in response to specific signals indicating diseased cells or tissue, so as to minimize the release of potentially harmful products near healthy cells. As discussed above, in developing bacterial cells that can invade tumors, Anderson et al. found that even baseline expression from a theoretically tightly-repressed, non-leaky promoter was sufficient to cause unwanted cell invasion104. In metabolic engineering space, this is a challenge recently referred to as “precision metabolic engineering”115, where one needs to completely switch cell states – for example, establishing an “off” state that is truly and completely off. This is expected to be of greatest concern for the synthesis of small molecules, as even extremely low-level leaky expression of metabolic enzymes that produce non-degradable products can cause amplification of that baseline that can prevent the original engineering goal from being achieved116. In short, while it may be tempting to view very low baseline levels and high dynamic response range during fluorescence-based characterization of promoters or circuits as sufficient, it is imperative to assess whether the construct behaves sufficiently with the real output of interest117.
These challenges, though, are not insurmountable with the appropriate systems and synthetic biology toolboxes. With the high potential reward for so many different kinds of cell-based therapeutics, these challenges will undoubtedly be overcome either on an ad hoc basis or through the development of new techniques or technologies. In the interim, they need careful consideration as we slowly move towards a new era of responsive, cell-based therapeutics.
Acknowledgments
The authors acknowledge the National Science Foundation (MCB-1254382) and the National Institutes of Health (R35-GM119701) for funding support. MPM was supported by an NSF graduate research fellowship (DGE-1148903).
Footnotes
Monica P. McNerney, No conflicts of interest
Mark P. Styczynski, No conflicts of interest
Contributor Information
Monica P. McNerney, School of Chemical & Biomolecular Engineering, Georgia Institute of Technology, monica.mcnerney@gatech.edu
Mark P. Styczynski, School of Chemical & Biomolecular Engineering, Georgia Institute of Technology, Mark.styczynski@chbe.gatech.edu
References
- 1.Link H, Kochanowski K, Sauer U. Systematic identification of allosteric protein-metabolite interactions that control enzyme activity in vivo. Nature biotechnology. 2013;31:357–61. doi: 10.1038/nbt.2489. [DOI] [PubMed] [Google Scholar]
- 2.van Eunen K, Kiewiet JA, Westerhoff HV, Bakker BM. Testing biochemistry revisited: how in vivo metabolism can be understood from in vitro enzyme kinetics. PLoS Comput Biol. 2012;8:e1002483. doi: 10.1371/journal.pcbi.1002483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Machado D, Herrgard MJ, Rocha I. Modeling the Contribution of Allosteric Regulation for Flux Control in the Central Carbon Metabolism of E. coli. Front Bioeng Biotechnol. 2015;3:154. doi: 10.3389/fbioe.2015.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braus GH. Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: a model system for the regulation of a eukaryotic biosynthetic pathway. Microbiol Rev. 1991;55:349–70. doi: 10.1128/mr.55.3.349-370.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hofman-Bang J. Nitrogen catabolite repression in Saccharomyces cerevisiae. Mol Biotechnol. 1999;12:35–73. doi: 10.1385/MB:12:1:35. [DOI] [PubMed] [Google Scholar]
- 6.Kayikci O, Nielsen J. Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res. 2015;15 doi: 10.1093/femsyr/fov068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bibel M, Richter J, Schrenk K, et al. Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nat Neurosci. 2004;7:1003–9. doi: 10.1038/nn1301. [DOI] [PubMed] [Google Scholar]
- 8.Guan K, Chang H, Rolletschek A, Wobus AM. Embryonic stem cell-derived neurogenesis. Retinoic acid induction and lineage selection of neuronal cells. Cell Tissue Res. 2001;305:171–6. doi: 10.1007/s004410100416. [DOI] [PubMed] [Google Scholar]
- 9.Sanz-Ruiz R, Casado Plasencia A, L RB, et al. Rationale and Design of a Clinical Trial to Evaluate the Safety and Efficacy of Intracoronary Infusion of Allogeneic Human CARdiac stEm Cells in Patients with Acute Myocardial Infarction and Left Ventricular Dysfunction: The Randomized Multicenter Double-Blind Controlled CAREMI Trial. Circ Res. 2017 doi: 10.1161/CIRCRESAHA.117.310651. [DOI] [PubMed] [Google Scholar]
- 10.Jonkers D, Stockbrugger R. Probiotics and inflammatory bowel disease. Journal of the Royal Society of Medicine. 2003;96:167–71. [PMC free article] [PubMed] [Google Scholar]
- 11.Sheil B, Shanahan F, O'Mahony L. Probiotic effects on inflammatory bowel disease. The Journal of nutrition. 2007;137:819s–24s. doi: 10.1093/jn/137.3.819S. [DOI] [PubMed] [Google Scholar]
- 12.Falagas ME, Rafailidis PI, Makris GC. Bacterial interference for the prevention and treatment of infections. Int J Antimicrob Agents. 2008;31:518–22. doi: 10.1016/j.ijantimicag.2008.01.024. [DOI] [PubMed] [Google Scholar]
- 13.Drekonja D, Reich J, Gezahegn S, et al. Fecal Microbiota Transplantation for Clostridium difficile Infection: A Systematic Review. Ann Intern Med. 2015;162:630–8. doi: 10.7326/M14-2693. [DOI] [PubMed] [Google Scholar]
- 14.Takiishi T, Korf H, Van Belle TL, et al. Reversal of autoimmune diabetes by restoration of antigen-specific tolerance using genetically modified Lactococcus lactis in mice. The Journal of clinical investigation. 2012;122:1717–25. doi: 10.1172/JCI60530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robert S, Gysemans C, Takiishi T, et al. Oral delivery of glutamic acid decarboxylase (GAD)-65 and IL10 by Lactococcus lactis reverses diabetes in recent-onset NOD mice. Diabetes. 2014;63:2876–87. doi: 10.2337/db13-1236. [DOI] [PubMed] [Google Scholar]
- 16.Souza BM, Preisser TM, Pereira VB, et al. Lactococcus lactis carrying the pValac eukaryotic expression vector coding for IL-4 reduces chemically-induced intestinal inflammation by increasing the levels of IL-10-producing regulatory cells. Microb Cell Fact. 2016;15:150. doi: 10.1186/s12934-016-0548-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caluwaerts S, Vandenbroucke K, Steidler L, et al. AG013, a mouth rinse formulation of Lactococcus lactis secreting human Trefoil Factor 1, provides a safe and efficacious therapeutic tool for treating oral mucositis. Oral oncology. 2010;46:564–70. doi: 10.1016/j.oraloncology.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Duan FF, Liu JH, March JC. Engineered commensal bacteria reprogram intestinal cells into glucose-responsive insulin-secreting cells for the treatment of diabetes. Diabetes. 2015;64:1794–803. doi: 10.2337/db14-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Z, Guo L, Zhang Y, et al. Incorporation of therapeutically modified bacteria into gut microbiota inhibits obesity. The Journal of clinical investigation. 2014;124:3391–406. doi: 10.1172/JCI72517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mimee M, Citorik RJ, Lu TK. Microbiome therapeutics - Advances and challenges. Advanced drug delivery reviews. 2016;105:44–54. doi: 10.1016/j.addr.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anselmo AC, McHugh KJ, Webster J, Langer R, Jaklenec A. Layer-by-Layer Encapsulation of Probiotics for Delivery to the Microbiome. Adv Mater. 2016;28:9486–90. doi: 10.1002/adma.201603270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forbes NS. Engineering the perfect (bacterial) cancer therapy. Nature Reviews Cancer. 2010;10:784–93. doi: 10.1038/nrc2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsung K, Norton JA. Lessons from Coley's Toxin. Surg Oncol. 2006;15:25–8. doi: 10.1016/j.suronc.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 24.Carey RW, Holland JF, Whang HY, Neter E, Bryant B. Clostridial oncolysis in man. European journal of cancer. 1967;3:37–42. [Google Scholar]
- 25.Streilein JW. Unraveling immune privilege. Science. 1995;270:1158–9. doi: 10.1126/science.270.5239.1158. [DOI] [PubMed] [Google Scholar]
- 26.Forbes NS, Munn LL, Fukumura D, Jain RK. Sparse initial entrapment of systemically injected Salmonella typhimurium leads to heterogeneous accumulation within tumors. Cancer research. 2003;63:5188–93. [PubMed] [Google Scholar]
- 27.Zhao M, Yang M, Li XM, et al. Tumor-targeting bacterial therapy with amino acid auxotrophs of GFP-expressing Salmonella typhimurium. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:755–60. doi: 10.1073/pnas.0408422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao M, Yang M, Ma H, et al. Targeted therapy with a Salmonella typhimurium leucine-arginine auxotroph cures orthotopic human breast tumors in nude mice. Cancer research. 2006;66:7647–52. doi: 10.1158/0008-5472.CAN-06-0716. [DOI] [PubMed] [Google Scholar]
- 29.King I, Bermudes D, Lin S, et al. Tumor-targeted Salmonella expressing cytosine deaminase as an anticancer agent. Hum Gene Ther. 2002;13:1225–33. doi: 10.1089/104303402320139005. [DOI] [PubMed] [Google Scholar]
- 30.Pawelek JM, Low KB, Bermudes D. Tumor-targeted Salmonella as a novel anticancer vector. Cancer research. 1997;57:4537–44. [PubMed] [Google Scholar]
- 31.Engelbart K, Gericke D. Oncolysis by Clostridia .V. Transplanted Tumors of Hamster. Cancer research. 1964;24:239-&. [PubMed] [Google Scholar]
- 32.Mose JR, Mose G. Oncolysis by Clostridia .I. Activity of Clostridium Butyricum (M-55) + Other Nonpathogenic Clostridia against Ehrlich Carcinoma. Cancer research. 1964;24:212-&. [PubMed] [Google Scholar]
- 33.Thiele EH, Boxer GE, Arison RN. Oncolysis by Clostridia .3. Effects of Clostridia + Chemotherapeutic Agents on Rodent Tumors. Cancer research. 1964;24:222-&. [PubMed] [Google Scholar]
- 34.Brown JM, Wilson WR. Exploiting tumour hypoxia in cancer treatment. Nature reviews Cancer. 2004;4:437–47. doi: 10.1038/nrc1367. [DOI] [PubMed] [Google Scholar]
- 35.Heppner F, Mose JR. The liquefaction (oncolysis) of malignant gliomas by a non pathogenic Clostridium. Acta Neurochir (Wien) 1978;42:123–5. doi: 10.1007/BF01406639. [DOI] [PubMed] [Google Scholar]
- 36.Mak CM, Lee HC, Chan AY, Lam CW. Inborn errors of metabolism and expanded newborn screening: review and update. Crit Rev Clin Lab Sci. 2013;50:142–62. doi: 10.3109/10408363.2013.847896. [DOI] [PubMed] [Google Scholar]
- 37.Patel S, Ahmed S. Emerging field of metabolomics: big promise for cancer biomarker identification and drug discovery. Journal of pharmaceutical and biomedical analysis. 2015;107:63–74. doi: 10.1016/j.jpba.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 38.Armitage EG, Barbas C. Metabolomics in cancer biomarker discovery: current trends and future perspectives. Journal of pharmaceutical and biomedical analysis. 2014;87:1–11. doi: 10.1016/j.jpba.2013.08.041. [DOI] [PubMed] [Google Scholar]
- 39.Hart CD, Tenori L, Luchinat C, Di Leo A. Metabolomics in Breast Cancer: Current Status and Perspectives. Adv Exp Med Biol. 2016;882:217–34. doi: 10.1007/978-3-319-22909-6_9. [DOI] [PubMed] [Google Scholar]
- 40.Lima AR, Bastos Mde L, Carvalho M, Guedes de Pinho P. Biomarker Discovery in Human Prostate Cancer: an Update in Metabolomics Studies. Transl Oncol. 2016;9:357–70. doi: 10.1016/j.tranon.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vargas AJ, Harris CC. Biomarker development in the precision medicine era: lung cancer as a case study. Nature Reviews Cancer. 2016;16:525–37. doi: 10.1038/nrc.2016.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 43.Storr M, Vogel HJ, Schicho R. Metabolomics: is it useful for inflammatory bowel diseases? Curr Opin Gastroen. 2013;29:378–83. doi: 10.1097/MOG.0b013e328361f488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lenz EM, Bright J, Wilson ID, Morgan SR, Nash AFP. A H-1 NMR-based metabonomic study of urine and plasma samples obtained from healthy human subjects. J Pharmaceut Biomed. 2003;33:1103–15. doi: 10.1016/s0731-7085(03)00410-2. [DOI] [PubMed] [Google Scholar]
- 45.Jansson J, Willing B, Lucio M, et al. Metabolomics reveals metabolic biomarkers of Crohn's disease. PLoS One. 2009;4:e6386. doi: 10.1371/journal.pone.0006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruscito A, DeRosa MC. Small-Molecule Binding Aptamers: Selection Strategies, Characterization, and Applications. Front Chem. 2016;4:14. doi: 10.3389/fchem.2016.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang F, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nature Biotechnology. 2012;30:354–U166. doi: 10.1038/nbt.2149. [DOI] [PubMed] [Google Scholar]
- 48.Michener JK, Thodey K, Liang JC, Smolke CD. Applications of genetically-encoded biosensors for the construction and control of biosynthetic pathways. Metabolic engineering. 2012;14:212–22. doi: 10.1016/j.ymben.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahr R, Frunzke J. Transcription factor-based biosensors in biotechnology: current state and future prospects. Applied microbiology and biotechnology. 2016;100:79–90. doi: 10.1007/s00253-015-7090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu D, Evans T, Zhang FZ. Applications and advances of metabolite biosensors for metabolic engineering. Metabolic engineering. 2015;31:35–43. doi: 10.1016/j.ymben.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 51.Burkovski A, Kramer R. Bacterial amino acid transport proteins: occurrence, functions, and significance for biotechnological applications. Applied microbiology and biotechnology. 2002;58:265–74. doi: 10.1007/s00253-001-0869-4. [DOI] [PubMed] [Google Scholar]
- 52.Quistgaard EM, Low C, Guettou F, Nordlund P. Understanding transport by the major facilitator superfamily (MFS): structures pave the way. Nature reviews Molecular cell biology. 2016;17:123–32. doi: 10.1038/nrm.2015.25. [DOI] [PubMed] [Google Scholar]
- 53.Hosie AH, Poole PS. Bacterial ABC transporters of amino acids. Research in microbiology. 2001;152:259–70. doi: 10.1016/s0923-2508(01)01197-4. [DOI] [PubMed] [Google Scholar]
- 54.Hirschman J, Wong PK, Sei K, Keener J, Kustu S. Products of Nitrogen Regulatory Genes Ntra and Ntrc of Enteric Bacteria Activate Glna Transcription Invitro - Evidence That the Ntra Product Is a Sigma-Factor. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:7525–9. doi: 10.1073/pnas.82.22.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao K, Liu M, Burgess RR. Promoter and regulon analysis of nitrogen assimilation factor, sigma54, reveal alternative strategy for E. coli MG1655 flagellar biosynthesis. Nucleic acids research. 2010;38:1273–83. doi: 10.1093/nar/gkp1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH. Dissection of the mechanism for the stringent factor RelA. Molecular cell. 2002;10:779–88. doi: 10.1016/s1097-2765(02)00656-1. [DOI] [PubMed] [Google Scholar]
- 57.Traxler MF, Summers SM, Nguyen HT, et al. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–48. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yanofsky C, Konan KV, Sarsero JP. Some novel transcription attenuation mechanisms used by bacteria. Biochimie. 1996;78:1017–24. doi: 10.1016/s0300-9084(97)86725-9. [DOI] [PubMed] [Google Scholar]
- 59.Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends in biochemical sciences. 2004;29:11–7. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 60.Gong F, Yanofsky C. Reproducing tna operon regulation in vitro in an S-30 system. Tryptophan induction inhibits cleavage of TnaC peptidyl-tRNA. The Journal of biological chemistry. 2001;276:1974–83. doi: 10.1074/jbc.M008892200. [DOI] [PubMed] [Google Scholar]
- 61.Stewart V, Landick R, Yanofsky C. Rho-dependent transcription termination in the tryptophanase operon leader region of Escherichia coli K-12. Journal of bacteriology. 1986;166:217–23. doi: 10.1128/jb.166.1.217-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stewart V, Yanofsky C. Evidence for transcription antitermination control of tryptophanase operon expression in Escherichia coli K-12. Journal of bacteriology. 1985;164:731–40. doi: 10.1128/jb.164.2.731-740.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Serganov A, Patel DJ. Amino acid recognition and gene regulation by riboswitches. Biochimica et biophysica acta. 2009;1789:592–611. doi: 10.1016/j.bbagrm.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Binder S, Schendzielorz G, Stabler N, et al. A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome biology. 2012;13:R40. doi: 10.1186/gb-2012-13-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mustafi N, Grunberger A, Kohlheyer D, Bott M, Frunzke J. The development and application of a single-cell biosensor for the detection of l-methionine and branched-chain amino acids. Metabolic engineering. 2012;14:449–57. doi: 10.1016/j.ymben.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 66.Mustafi N, Grunberger A, Mahr R, et al. Application of a genetically encoded biosensor for live cell imaging of L-valine production in pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum strains. PLoS One. 2014;9:e85731. doi: 10.1371/journal.pone.0085731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fang M-Y, Zhang C, Yang S, et al. High crude violacein production from glucose by Escherichia coli engineered with interactive control of tryptophan pathway and violacein biosynthetic pathway. Microbial Cell Factories. 2015;14 doi: 10.1186/s12934-015-0192-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fang M, Wang T, Zhang C, et al. Intermediate-sensor assisted push-pull strategy and its application in heterologous deoxyviolacein production in Escherichia coli. Metabolic engineering. 2016;33:41–51. doi: 10.1016/j.ymben.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 69.Zhou LB, Zeng AP. Exploring lysine riboswitch for metabolic flux control and improvement of L-lysine synthesis in Corynebacterium glutamicum. ACS Synth Biol. 2015;4:729–34. doi: 10.1021/sb500332c. [DOI] [PubMed] [Google Scholar]
- 70.Xu P, Li L, Zhang F, Stephanopoulos G, Koffas M. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:11299–304. doi: 10.1073/pnas.1406401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsao C-Y, Hooshangi S, Wu H-C, Valdes JJ, Bentley WE. Autonomous induction of recombinant proteins by minimally rewiring native quorum sensing regulon of E. coli. Metabolic engineering. 2010;12:291–7. doi: 10.1016/j.ymben.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 72.Din MO, Danino T, Prindle A, et al. Synchronized cycles of bacterial lysis for in vivo delivery. Nature. 2016;536:81–5. doi: 10.1038/nature18930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dirusso CC, Black PN. Bacterial long chain fatty acid transport: gateway to a fatty acid-responsive signaling system. The Journal of biological chemistry. 2004;279:49563–6. doi: 10.1074/jbc.R400026200. [DOI] [PubMed] [Google Scholar]
- 74.Black PN. Characterization of FadL-specific fatty acid binding in Escherichia coli. Biochim Biophys Acta. 1990;1046:97–105. doi: 10.1016/0005-2760(90)90099-j. [DOI] [PubMed] [Google Scholar]
- 75.Fujita Y, Matsuoka H, Hirooka K. Regulation of fatty acid metabolism in bacteria. Mol Microbiol. 2007;66:829–39. doi: 10.1111/j.1365-2958.2007.05947.x. [DOI] [PubMed] [Google Scholar]
- 76.Xu P, Wang W, Li L, Bhan N, Zhang F, Koffas MA. Design and kinetic analysis of a hybrid promoter-regulator system for malonyl-CoA sensing in Escherichia coli. ACS chemical biology. 2014;9:451–8. doi: 10.1021/cb400623m. [DOI] [PubMed] [Google Scholar]
- 77.Xu P, Li L, Zhang F, Stephanopoulos G, Koffas M. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:11299–304. doi: 10.1073/pnas.1406401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pearson JP, Van Delden C, Iglewski BH. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. Journal of bacteriology. 1999;181:1203–10. doi: 10.1128/jb.181.4.1203-1210.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smith RS, Iglewski BH. Pseudomonas aeruginosa quorum sensing as a potential antimicrobial target. Journal of Clinical Investigation. 2003;112:1460–5. doi: 10.1172/JCI20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Decho AW, Frey RL, Ferry JL. Chemical challenges to bacterial AHL signaling in the environment. Chemical reviews. 2011;111:86–99. doi: 10.1021/cr100311q. [DOI] [PubMed] [Google Scholar]
- 81.Federle MJ. Autoinducer-2-based chemical communication in bacteria: complexities of interspecies signaling. Contrib Microbiol. 2009;16:18–32. doi: 10.1159/000219371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tavender TJ, Halliday NM, Hardie KR, Winzer K. LuxS-independent formation of AI-2 from ribulose-5-phosphate. BMC Microbiol. 2008;8:98. doi: 10.1186/1471-2180-8-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li J, Attila C, Wang L, Wood TK, Valdes JJ, Bentley WE. Quorum sensing in Escherichia coli is signaled by AI-2/LsrR: effects on small RNA and biofilm architecture. Journal of bacteriology. 2007;189:6011–20. doi: 10.1128/JB.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Neiditch MB, Federle MJ, Miller ST, Bassler BL, Hughson FM. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol Cell. 2005;18:507–18. doi: 10.1016/j.molcel.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 85.Miller MB, Bassler BL. Quorum sensing in bacteria. Annual review of microbiology. 2001;55:165–99. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 86.Bassler BL, Greenberg EP, Stevens AM. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. Journal of bacteriology. 1997;179:4043–5. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Herzberg M, Kaye IK, Peti W, Wood TK. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. Journal of bacteriology. 2006;188:587–98. doi: 10.1128/JB.188.2.587-598.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li J, Attila C, Wang L, Wood TK, Valdes JJ, Bentley WE. Quorum sensing in Escherichia coli is signaled by AI-2/LsrR: Effects on small RNA and Biofilm architecture. J Bacteriol. 2007;189:6011–20. doi: 10.1128/JB.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pereira CS, Thompson JA, Xavier KB. AI-2-mediated signalling in bacteria. FEMS Microbiol Rev. 2013;37:156–81. doi: 10.1111/j.1574-6976.2012.00345.x. [DOI] [PubMed] [Google Scholar]
- 90.Bardill JP, Zhao X, Hammer BK. The Vibrio cholerae quorum sensing response is mediated by Hfq-dependent sRNA/mRNA base pairing interactions. Mol Microbiol. 2011;80:1381–94. doi: 10.1111/j.1365-2958.2011.07655.x. [DOI] [PubMed] [Google Scholar]
- 91.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 92.Tu KC, Bassler BL. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes & development. 2007;21:221–33. doi: 10.1101/gad.1502407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hammer BK, Bassler BL. Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A. 2007;104:11145–9. doi: 10.1073/pnas.0703860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bardill JP, Hammer BK. Non-coding sRNAs regulate virulence in the bacterial pathogen Vibrio cholerae. RNA biology. 2012;9:392–401. doi: 10.4161/rna.19975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harbor perspectives in medicine. 2012;2 doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–4. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 97.Bassler BL, Wright M, Silverman MR. Sequence and function of LuxO, a negative regulator of luminescence in Vibrio harveyi. Mol Microbiol. 1994;12:403–12. doi: 10.1111/j.1365-2958.1994.tb01029.x. [DOI] [PubMed] [Google Scholar]
- 98.Henke JM, Bassler BL. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. Journal of bacteriology. 2004;186:3794–805. doi: 10.1128/JB.186.12.3794-3805.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kohler T, Curty LK, Barja F, van Delden C, Pechere JC. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. Journal of bacteriology. 2000;182:5990–6. doi: 10.1128/jb.182.21.5990-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Withers HL, Nordstrom K. Quorum-sensing acts at initiation of chromosomal replication in Escherichia coli. Proc Natl Acad Sci U S A. 1998;95:15694–9. doi: 10.1073/pnas.95.26.15694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Danino T, Mondragon-Palomino O, Tsimring L, Hasty J. A synchronized quorum of genetic clocks. Nature. 2010;463:326–30. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Williams TC, Nielsen LK, Vickers CE. Engineered quorum sensing using pheromone-mediated cell-to-cell communication in Saccharomyces cerevisiae. ACS Synth Biol. 2013;2:136–49. doi: 10.1021/sb300110b. [DOI] [PubMed] [Google Scholar]
- 103.Gupta A, Reizman IM, Reisch CR, Prather KL. Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat Biotechnol. 2017;35:273–9. doi: 10.1038/nbt.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. Journal of molecular biology. 2006;355:619–27. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- 105.Saeidi N, Wong CK, Lo TM, et al. Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Molecular systems biology. 2011;7:521. doi: 10.1038/msb.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Venayak N, Anesiadis N, Cluett WR, Mahadevan R. Engineering metabolism through dynamic control. Current Opinion in Biotechnology. 2015;34:142–52. doi: 10.1016/j.copbio.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 107.Brockman IM, Prather KL. Dynamic metabolic engineering: New strategies for developing responsive cell factories. Biotechnology journal. 2015;10:1360–9. doi: 10.1002/biot.201400422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I-1-I-2 regulatory elements. Nucleic Acids Research. 1997;25:1203–10. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cox RS, 3rd, Surette MG, Elowitz MB. Programming gene expression with combinatorial promoters. Molecular systems biology. 2007;3:145. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nielsen AA, Der BS, Shin J, et al. Genetic circuit design automation. Science. 2016;352:aac7341. doi: 10.1126/science.aac7341. [DOI] [PubMed] [Google Scholar]
- 111.Ang J, Harris E, Hussey BJ, Kil R, McMillen DR. Tuning response curves for synthetic biology. ACS Synth Biol. 2013;2:547–67. doi: 10.1021/sb4000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cobb RE, Sun N, Zhao H. Directed evolution as a powerful synthetic biology tool. Methods. 2013;60:81–90. doi: 10.1016/j.ymeth.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Khan S, Brocklehurst KR, Jones GW, Morby AP. The functional analysis of directed amino-acid alterations in ZntR from Escherichia coli. Biochemical and biophysical research communications. 2002;299:438–45. doi: 10.1016/s0006-291x(02)02660-8. [DOI] [PubMed] [Google Scholar]
- 114.Dahl RH, Zhang F, Alonso-Gutierrez J, et al. Engineering dynamic pathway regulation using stress-response promoters. Nature Biotechnology. 2013;31:1039-+. doi: 10.1038/nbt.2689. [DOI] [PubMed] [Google Scholar]
- 115.McNerney MP, Watstein DM, Styczynski MP. Precision metabolic engineering: The design of responsive, selective, and controllable metabolic systems. Metabolic engineering. 2015;31:123–31. doi: 10.1016/j.ymben.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Watstein DM, McNerney MP, Styczynski MP. Precise metabolic engineering of carotenoid biosynthesis in Escherichia coli towards a low-cost biosensor. Metabolic engineering. 2015;31:171–80. doi: 10.1016/j.ymben.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McNerney MP, Styczynski MP. Precise control of lycopene production to enable a fast-responding, minimal-equipment biosensor. Metabolic engineering. 2017 doi: 10.1016/j.ymben.2017.07.004. In revision. [DOI] [PMC free article] [PubMed] [Google Scholar]