Abstract
The production of blood cells is dependent on the activity of a rare stem cell population that normally resides in the bone marrow (BM) of the organism. These hematopoietic stem cells (HSCs) have the ability to both self-renew and differentiate, ensuring this lifelong hematopoiesis. Determining the regulation of HSC functions should thus provide critical insight to advancing regenerative medicine. Until quite recently, HSCs were primarily studied using in vitro studies and transplantations into immunodeficient hosts. Indeed, the definition of a bona fide HSC is its ability to reconstitute lymphopenic hosts. In this review, we discuss the development of novel HSC-specific genetic reporter systems that enable the prospective identification of HSCs and study of their functions in the absence of transplantation. Coupled with additional technological advances, these studies are now defining the fundamental properties of HSCs in vivo. Furthermore, complex cellular and molecular mechanisms that regulate HSC dormancy, self-renewal, and differentiation are being identified and further dissected. These novel reporter systems represent a major technological advance for the stem cell field and allow new questions to be addressed.
Keywords: Hematopoietic stem cell, hematopoiesis, in vivo model, lineage tracing, clonal analysis, label retention
Introduction
Adult hematopoietic stem cells (HSCs) have the ability to both self-renew and differentiate, reconstituting a majority of the lineages of the hematopoietic system and ensuring life-long hematopoiesis. These properties are essential for the success of bone marrow (BM) transplantations both in the clinic and laboratory. Over the years, numerous challenges have impeded the study of HSCs including their paucity in the BM as well as differences in the definition of an HSC at the phenotypic level as assessed by cell surface markers and at the functional level as measured by transplantation assays [1–7]. Moreover, variable outcomes have been observed upon transplantation of single cells that are phenotypically defined as HSCs [8–10]. Some cells were found to generate robust and balanced reconstitution of major lineages in both primary and secondary hosts while others had deficiencies in the reconstitution of one or more lineages in primary and/or secondary hosts. Studies are now focused on trying to establish phenotypic definitions of these functionally distinct cells as well as dissect the molecular mechanisms that underlie their biology.
In this review, we refer to the fraction of BM cells that give rise to the major hematopoietic lineages (myeloid, lymphoid, megakaryocytic, and erythroid) during serial transplantation as HSCs. Specifically, cells that give (1) reconstitution of only primary and not secondary hosts or (2) the full range of lineage reconstitution of primary hosts followed by loss of one or more lineages in secondary hosts would not be considered functional HSCs despite their matching the phenotypic criteria. Much of our knowledge of HSC function comes from transplantation, which is a major stress for the cell and the recipient animal [11, 12]. It involves the extraction of HSCs from their native environment, ex vivo manipulation, and introduction into immune-compromised hosts that have often undergone total body irradiation, a process that destroys the BM architecture [13]. While transplantation demonstrates the magnitude of what an HSC can do, this procedure does not reveal what an HSC actually does at the steady state. Although transplantation is an in vivo procedure, here we use the term “in vivo” to distinguish studies of endogenous HSCs in the absence of transplantation. Defining the fundamental characteristics of HSC in vivo and dissecting the molecular and cellular mechanisms that regulate these functions are essential for a better understanding of HSC biology and advancing regenerative medicine.
Strategies for studying HSCs in vivo
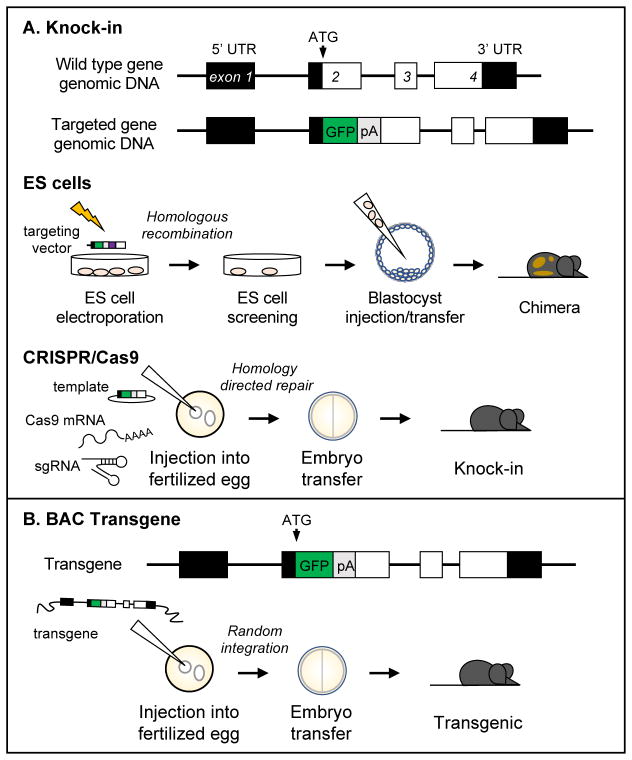
The main strategy employed to study HSCs in vivo has been the generation of HSC-specific genetic reporter systems using transgene (Tg) or “knock-in” (KI) approaches (Fig. 1A, B). These systems use the promoter of a gene of interest to drive expression of a reporter cassette, e.g. green fluorescent protein (GFP). In theory, the expression patterns of the gene of interest and reporter cassette should be similar, but experimentally this is not always the case. Multiple factors contribute to this discrepancy including the type of genetic system used, the specific manner in which the reporter cassette is cloned, and the stability of the reporter protein, which can be highly variable. To establish a KI mouse line, a reporter cassette is “knocked-in” to a targeted genomic locus (Fig. 1A), which largely preserves the global context of the genomic locus, specifically regulatory elements including enhancers as well as the 5′ untranslated region (UTR), and is thought to prevent “leaky” expression of the reporter cassette. Previously KI animals were generated by homologous recombination between a targeting vector and the genomic DNA of murine embryonic stem (ES) cells (Fig. 1A) [14, 15]. Correctly targeted ES cells are then injected into a blastocyst, which gives rise to a chimeric KI mouse. The F1 progeny from these founder chimeric animals must be screened to ensure germline contribution from the targeted ES cells. The discovery of clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (Cas) [16, 17] has greatly reduced the time needed to make KI animals as it bypasses the use of ES cells. In this case, mRNA encoding Cas9 nuclease, a single guide RNA (sgRNA) and a donor template encoding the reporter cassette are injected into a fertilized egg (Fig. 1A). The sgRNA encodes a short complementary sequence upstream of a protospacer-adjacent motif signal and provides target specificity for DNA binding by Cas9, which can induce double-stranded breaks (DSB) in the DNA. Genome-editing takes advantage of the endogenous DNA repair pathways used to resolve these DSBs, including the more frequent but imprecise non-homologous end joining and the precise homology directed repair (HDR) pathways [18, 19]. During the generation of KI animals, a donor template with sequence homology to the targeted locus is precisely inserted within genomic DNA by HDR [20, 21]. Despite the efficiency and versatility of CRISPR/Cas, any animal generated by this method should be carefully screened for off-target effects including differential mutation of the other allele. Furthermore, the introduction of a reporter cassette at a given genomic locus, regardless of the technology used, may disrupt or abrogate expression of the targeted endogenous gene. Thus, the phenotype of KI animals must be analyzed carefully for potential effects arising from the generated haploinsufficiency.
Figure 1. Use of KI and Tg approaches to generate HSC-specific genetic reporter animals.
Both strategies use regulatory elements of a gene of interest to drive expression of a reporter cassette. This cassette can be inserted 5′ (as depicted above, immediately after the ATG start codon) or 3′ of the coding sequence. (A) KI animals can be generated by targeting of ES cells or using the CRISPR/Cas9 system. To modify ES cells, a targeting vector that encodes reporter and antibiotic resistance cassettes flanked by arms of homologous sequence both directly 5′ and 3′ of the targeted locus is electroporated into ES cells. These flanking arms facilitate homologous recombination between the vector insert and ES cell genomic DNA, and ES cells are selected for the appropriate antibiotic resistance and screened for targeting vector incorporation. Correctly targeted ES cells are injected into a blastocyst of a different genetic background, which is transferred to a pseudopregnant female. ES cell contribution to the resulting pup is visualized by the different coat colors associated with the genetic backgrounds of the ES cells and blastocyst. Chimeric animals must be further bred to confirm germline contribution from the ES cells. Alternatively, KI animals can be generated by the direct injection of mRNA encoding the Cas9 nuclease, an sgRNA that directs DNA binding of Cas9, and a donor template vector encoding the reporter cassette into a fertilized egg. DSB mediated by Cas9 can be resolved by HDR, leading to incorporation of the reporter cassette at the desired genomic locus. The zygote is transferred to a pseudopregnant female, and the resulting progeny are screened for the correct genotype. (B) BAC Tg animals are made in a similar manner to KI animals made using CRISPR/Cas9. A reporter cassette is introduced into a BAC clone that encodes the locus of interest by recombineering. The targeted BAC DNA becomes randomly incorporated into the genomic DNA of a fertilized egg, which is then transferred to a pseudopregnant female mouse. Resulting offspring are genotyped for the Tg.
While different types of Tgs have been developed, several recently described HSC reporter systems were established using bacterial artificial chromosomes (BACs). BACs are plasmids based on bacterial F element that can encode large DNA inserts from 100–350 kilobases and as such have been used for the generation of genomic DNA libraries [22]. In the generation of BAC transgenic animals, a BAC clone that contains the genomic locus of interest is first selected and a reporter cassette is inserted in the locus by recombineering [23, 24]. The targeted BAC is injected into the pronucleus of a fertilized egg where it randomly inserts into the genomic DNA, and this early stage embryo is subsequently transferred to a pseudo-pregnant female mouse (Fig. 1B). The genomic DNA insert of a BAC is relatively large and encodes most if not all of the regulatory elements of the desired locus, leading to a similar expression pattern between the reporter cassette and endogenous gene. However, aberrant reporter cassette expression may arise from transgene copy number, position effects due to the precise genomic integration site, or the absence of distal gene regulatory elements like enhancers encoded by the BAC clone [25]. Therefore, multiple transgenic founder lines should be analyzed in order to assess the fidelity of Tg expression.
The development of HSC-specific genetic reporter systems requires a gene with an expression profile sufficiently restricted the HSC compartment. The selection of such an ideal candidate gene critically depends on robust analyses of the gene expression profile of functional HSCs as well as other hematopoietic cell types in the BM. Transcriptional profiling of single HSCs was only recently described [26, 27], and consequently most HSC gene expression profiles were derived from the bulk population. Considering the functional heterogeneity of HSCs observed upon transplantation [8–10] and the limits to which the transcriptional profile dictates cell function, the identification of differentially expressed genes that reliably distinguish HSCs from more differentiated cells presents a major challenge. As a result, many candidate genes have been tested and nearly all of the recently described HSC-specific transgenic reporter systems have been based on different genes.
Recent HSC-specific genetic reporter systems
Due to the persistence of inconsistent and often imprecise phenotypic definitions of HSCs in the literature, initial descriptions of HSC reporter systems showed nearly uniform labeling of the fraction of BM cells that are Lineage− Sca1+ cKit+ (LSK), which contains HSCs as well as populations of more differentiated progenitors [28, 29]. However, recently described constitutive and inducible genetic reporter systems using at a minimum LSK with the differential expression of SLAM family markers CD48− CD150+ [4] show improved specificity for HSCs. Each system presents a unique combination of advantages and caveats that must be considered in the interpretation of results and design of future studies as summarized in Table 1 [30–37]. Notably, only two genes used to drive reporter expression have established roles in the hematopoietic system. Tek encodes the Tie2 protein, which is a ligand for the angiopoietin receptor, and Tie2/angiopoietin signaling has been shown to regulate HSC quiescence [38]. von Willebrand factor (vWF, gene symbol Vwf) is critical for platelet adhesion and hemostasis [39], and defects in vWF lead to the coagulation disorder von Willebrand disease. Most of the other reporter systems use genes that are preferentially expressed by HSCs but have unresolved functions.
Table 1.
Comparison of recently generated HSC-specific fluorescent reporters
| Gene | Method | HSC expression | Expression in other BM populations | Labeled HSC observations | Ref. |
|---|---|---|---|---|---|
| Ctnnal1 (α-catulin) | Knock-in: GFP | 50% LSK CD48− CD150+ | Minor (<1%) fractions of cKit− or cKit+ SSChi | Localization of cells in BM sinusoids away from bone surface, no distinct niche for quiescent vs. proliferating HSCs | [30] |
| Fgd5 | Knock-in: mCherry, zsGreen | 80% LSK CD48− CD150+ (mCherry) | Small (5–10%) fractions of progenitors, endothelial cells | All reconstituting activity lies within the labeled population | [34] |
| Gprc5c | BAC Tg: EGFP | 28% LSK CD34− CD48− CD135− CD150+ | Small (<5%) fractions of progenitors | Enriched for dormant cells, improved outcomes upon transplantation | [31] |
| Hoxb5 | Knock-in: tri-mCherry | 22% LSK CD34− CD135− CD150+ | Small (5%) fractions of LSK CD34+ CD135− CD150+, unquantified fractions of Lin+ or Lin− cKit+ | Enriched for cells that have enhanced reconstitution of both primary and secondary hosts, proximity to VE-cadherin+ cells in BM | [33] |
| Pdzk1ip1 | BAC Tg: EGFP | 27% LSK CD48− CD150+ (EGFP) | 4% granulocytes and minor (<1%) fractions of progenitors (EGFP) | Enriched for cells that serially reconstitute (EGFP) | [37] |
| Tek (Tie2) | Tg: Green Lantern GFP | 5% LSK CD34− CD48− CD135− CD150+ | Minor fractions of other primitive populations, endothelial cells | Enriched for cells that have enhanced reconstitution and undergo mitophagy | [35] |
| Vwf | BAC Tg: EGFP, tdTomato | 60% LSK CD34− CD48− CD150+ | Nearly all MkP and platelets, 50% of endothelial cells | Increased contribution to platelet production | [32, 36] |
Some of the recent reporter systems like those based on Ctnnal1 (catenin (cadherin associated protein), alpha-like 1, also known as α-catulin) or Fgd5 (FYVE, RhoGEF and PH domain containing 5) genes label a majority of phenotypic HSCs [30, 34], distinguishing the bulk pool of HSCs from other hematopoietic populations. While resolving the functional heterogeneity of HSCs remains a challenge when phenotypic HSCs are labeled with high efficiency, as occurs in Fgd5 reporter animals, these systems may be useful to confirm HSC identity in visualization studies. Indeed, Acar et al. generated the Ctnnal1 reporter system to investigate the organization and distribution of HSCs throughout the BM. This system is insufficiently specific as a single marker of HSCs, but when combined with cKit expression, HSCs were identified at a frequency of 1/3.5 Ctnnal1-labeled cKit+ cells. 3-dimensional rendering of the distribution of Ctnnal1-labeled cKit+ cells in the context of the BM architecture demonstrated their preferential location away from the bone surface and in the diaphysis. Despite not all Ctnnal1-labeled cKit+ cells showing bona fide HSC activity, these cells were predominantly observed proximal to blood vessel sinusoids, which suggests the perisinusoidal nature of their microenvironment. The BM microenvironment is a complex network of myriad cell types, and reporter specificity is paramount for any study of HSCs, but in particular for visualization. Thus, reporter systems that label phenotypic HSCs with high efficiency still require additional genetic systems or markers to further refine the HSC population for further functional studies.
Other novel reporter systems mark subsets of HSCs that are associated with specific functions revealed in transplantation assays. Transgenic systems based on Vwf distinguish a subset of HSCs that show increased contribution to platelets upon transplantation [32, 36]. Transplantation of single Vwf-labeled HSCs demonstrated specific and stable combinations of lineages reconstituted [32]. Platelets were the only lineage invariably reconstituted and unipotential reconstitution was only associated with platelet production [32]. The function of Vwf-labeled HSCs was primarily analyzed in transplantation settings, whereas the in vivo functions of these cells remain largely unresolved. Two recently described reporter systems prospectively identify a fraction of HSCs with the capacity for balanced lineage output in serial transplantations. Hoxb5 (homeobox B5) is a member of the Antennapedia homeobox family and is a transcription factor with known roles in the gut [40]. The function of Hoxb5 in hematopoietic cells is not known, but it is preferentially expressed in HSCs. Hoxb5-tri-mCherry KI animals showed specific expression in HSCs with little expression in other hematopoietic populations, and limiting dilution analyses identified functional HSCs at a high frequency [33]. Superior reconstitution was seen in both the primary and secondary recipients, suggesting that this marker refines the pool of functional HSCs. Finally, we recently generated Pdzk1ip1-GFP transgenic animals and found reporter expression in the subset of HSCs with the phenotype of the least differentiated cells [37]. Pdzk1ip1-labeled HSCs displayed superior capacity for serial transplantation that, unlike cells labeled by Hoxb5 reporter, was revealed only in secondary recipients, demonstrating that this system labels the fraction of HSCs with durable self-renewal. Importantly, these reporter systems provide the tools to dissect the molecular mechanisms from these subsets of HSCs from the rest of the phenotypically defined population.
Lineage tracing of HSCs
Lineage tracing, also called fate mapping, is a technique that traces the appearance of heritable labels in different populations in order to study precursor to progeny relationships [41] and has been frequently used within the hematopoietic system to understand the developmental pathways of various hematopoietic cells. Genetic lineage tracing systems rely on Cre/Lox technology to irreversibly label cells, avoiding the need for ex vivo manipulation and transplantation. However, fate mapping of HSCs has been complicated as the available reporter systems were either constitutively expressed, often beginning within the embryo, or lacked sufficient specificity for HSCs [42–45]. Inducible systems based on tamoxifen-dependent Cre recombinase provide temporal control of the initial labeling event, and combined with sufficient reporter specificity, enable the tracing and quantification of output of a designated population (Fig. 2A). Inducible lineage tracing systems that provide temporal control of the initial labeling event and were recently generated for HSCs (Table 2). Using an inducible Tie2-Cre recombinase, Busch et al. [46] achieved very specific labeling of a minor fraction of HSCs: at the earliest time point, 1–3 weeks following induction with tamoxifen, HSC labeling was <1% with very little labeling detected in other populations shown. Pooled analysis from different time points showed an average of 1% HSC labeling, suggesting that the labeling of this population remained relatively constant. The appearance of labeled progeny was remarkably slow over time and the frequency of labeled cells was quite low, never reaching that of HSCs. Mathematical models estimating rates of cell differentiation and proliferation from each population based on these data support the conclusion that HSCs make relatively rare inputs to the populations and that hematopoiesis is largely driven by more differentiated short-term HSCs.
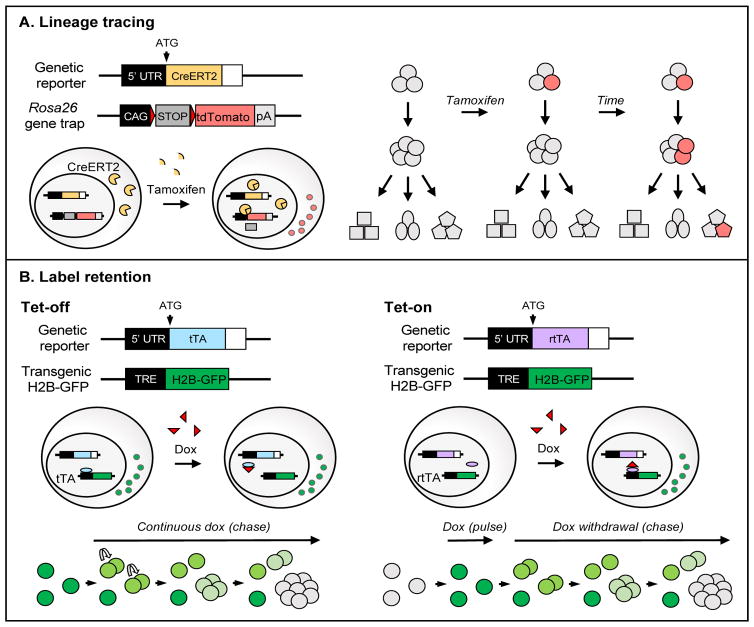
Figure 2. Studying HSC functions in vivo using genetic reporter systems.
HSC-specific reporters can be combined with other inducible genetic mouse strains to study in vivo functions of HSCs. (A) In vivo HSC contribution to hematopoiesis can be studied by lineage tracing. In the absence of tamoxifen, the CreERT2 recombinase is retained in the cytoplasm and a LoxP-flanked Stop cassette encoding repeating polyadenylation signals prevents expression of the fluorescent reporter (e.g. tdTomato) in cells that express the HSC reporter. Tamoxifen treatment enables CreERT2 translocation to the nucleus where it mediates excision of the Stop cassette. This results in irreversible expression of tdTomato, and any daughter cell that arises from the labeled cell will also express tdTomato. The contribution of the labeled cell to the production of other cell types is estimated by measuring the fraction of tdTomato+ cells over time. (B) Dormant HSCs can be identified in vivo by the ability to stably retain human histone H2B-GFP fluorescent label. A TRE regulates the expression of H2B-GFP, and this system can function as a “Tet-off” or “Tet-on” system. In Tet-off systems, H2B-GFP is induced in cells that express the HSC-specific tTA. In the presence of the tetracycline analog doxycycline (dox) the de novo synthesis of H2B-GFP is inhibited. The H2B-GFP protein is highly stable but becomes diluted upon proliferation of labeled cells over time. Cells that retain the fluorescent label over a long period of continuous dox treatment are considered stable label retaining cells. In a Tet-on system, H2B-GFP expression is induced in cells that express the HSC-specific reverse tetracycline transactivator (rtTA) only upon treatment with dox. Once complete labeling of the population of interest is confirmed, dox is removed and cells are assessed for label retention over time.
Table 2.
Recently generated HSC-specific inducible Cre recombinase mouse strains
| Gene | Method | HSC Labeling | Initial labeling in other BM populations | Labeled HSC observations | Ref. |
|---|---|---|---|---|---|
| Fgd5 | Knock-in: CreERT2 | 60% LSK CD150+ (tdTomato) | Small (5–10%) fractions of progenitors, endothelial cells | Not reported | [34] |
| Pdzk1ip1 | BAC Tg: CreERT2 | 32% LSK CD48− CD150+ (tdTomato) | Small (<5%) fractions of progenitors | Major contribution to steady-state hematopoiesis (CreERT2) | [37] |
| Tek (Tie2) | Knock-in: Mer-Cre-Mer | Minor fractions (<1%) LSK CD48− CD150+ (YFP) 1–3 weeks after induction | Very minor fractions (<0.1%) of progenitors, endothelial cells not reported | Minimal contributions to steady-state hematopoiesis | [46] |
| Vwf | Knock-in: CreERT2-P2A-EGFP | 12% LSK CD34− CD48− CD150+ (tdTomato) 2–3 weeks after induction | Not reported | Preferential contribution to steady-state production of platelets | [32] |
We recently demonstrated efficient labeling of HSCs in Pdzk1ip1-CreERT2 Rosa26-tdTomato reporter animals, with an average 32% of phenotypic HSCs tdTomato+ following tamoxifen treatment. We found that the labeling of phenotypic HSCs increased over time and that this labeling steadily spread to differentiated progenitors and mature cells, nearly but not quite reaching that observed in HSCs at the corresponding time point. The difference observed in labeling of HSCs and downstream populations can be explained by HSCs that did not differentiate, which would be a minor fraction of the population and may correspond to the recently described stable label retaining HSCs [47]. Our data fit a two-subset model of the HSC population in which labeled HSCs fully self-renew with the fraction of their labeled progeny steadily growing over time, and this initial input from HSCs to the downstream progeny becomes amplified with each step of differentiation. These results support the conclusion that HSCs make a major contribution to hematopoiesis at the steady state. Although different, we believe our results are generally compatible with those of Busch et al. [46] and that the discrepancies are related to the specific expression pattern inherent to each system leading to a major difference in labeling kinetics associated with each system. HSC labeling induced in Tie2-Mer-Cre-Mer animals is specific but inefficient, which precludes the study of dynamics within individual animals over time. As a result, mature cell labeling was estimated relative to an average HSC labeling pooled from all time points rather than initial HSC labeling. The low efficiency of HSC labeling approaches the level of clonal analysis, while the comparatively high dose of tamoxifen, which is thought to induce HSC proliferation [48], and the delay in the onset of lineage tracing from the initial treatment may select for more quiescent cells. By comparison our system allowed a more efficient and rapid labeling of HSCs. This increased efficiency permitted both the initial measurement of labeling and continuous analysis of animals over time, but it was also associated with labeling in small fractions of downstream progenitors. This background labeling cannot completely account for the major labeling observed in downstream populations and was considered in our mathematical modeling. Although it does not address the activity of distinct HSC clones, we believe our study captures the in vivo output of the population of HSCs.
Clonal analysis of hematopoiesis
The contributions of individual HSC clones to the overall maintenance of hematopoiesis are poorly defined. To this end, new genetic approaches have recently been developed including a barcoding approach based on transposase insertion sites mediated by a tetracycline-regulated Sleeping Beauty system [49, 50]. An initial study assessed the distribution of cloned transposition sites across various hematopoietic populations upon pan-hematopoietic induction of transposase over time [49]. Minimal overlap was observed in the integration sites detected in HSCs compared to downstream multipotent progenitors (MPPs) and granulocytes, leading to the conclusion that the bulk of hematopoiesis proceeds independently of HSCs. A second report using this same system analyzed the distribution of integration sites across hematopoietic populations with particular focus on the megakaryocytic lineage [50]. The extent of overlap in sites detected exclusively in HSCs and megakaryocytes but not progenitor populations suggested an early divergence of megakaryocytes. While this is consistent with other reports suggesting a close relation between HSC and megakaryocytes [32, 36, 51, 52], results generated by this system should be interpreted cautiously. Transposase induction occurs across all hematopoietic populations, so whether tagged cells arise de novo or from differentiation is unclear. Hierarchical relationships thus can be inferred but not demonstrated. Additionally, false positives from cloning of integration sites and secondary transposition events due to the persistence of transposase in cells were not formally excluded. These technical caveats could contribute to the relative lack of HSC output observed in the first study, and notably a different method was used to retrieve integration sites in the second study [50]. Furthermore, the results that HSCs make little contribution to steady state hematopoiesis contrasts with recent clonal analysis of hematopoiesis using the HUe system [53], a transgenic system that contains multiple copies of the Brainbow2.1 cassette [54], which encodes LoxP-flanked green, yellow, red, and cyan fluorescent protein cassettes in tandem. The high copy number of tandem cassettes enables the generation of a large range (>103) of distinct fluorescent color labels based on the pattern of recombination upon induction of Mx1-Cre with polyinosinic:polycytidilic acid (polyI:C). With the caveat that polyI:C is known to activate HSC proliferation through type I interferon [55], pan-hematopoietic labeling of HUe animals resulted in the generation of a number of fluorescent clones with different sizes and hematopoietic activity. Importantly, these clones contained HSCs that showed similar capacities for proliferation and differentiation upon transplantation as displayed in the original donor in vivo.
The recently described Polylox system expresses a cassette containing 10 LoxP sites from the Rosa26 locus [56]. Cre-mediated recombination of this locus has the potential to generate more than 1.8×106 different “barcodes” based on patterns involving up to 10 different recombination events although experimentally derived barcodes showed at most 6 recombination events. Using inducible Tie2-Cre recombination [46], this system was recently used to investigate the in vivo output of HSC clones during fetal and adult hematopoiesis. In experiments of Polylox labeling in the embryo at embryonic day 9.5, nearly all HSCs in the adult bone marrow contained barcodes demonstrating the high efficiency of Tie2-mediated recombination in the embryo. Furthermore, barcode overlap between populations demonstrated output to mature lineages from embryonically marked HSCs. The observed distribution of barcodes among populations from the embryonic labeling experiments suggest that the adult HSC pool is composed of many different embryonically derived clones with different sizes and mostly multilineage or oligolineage potential. Nevertheless, major overlap was observed in the barcode sequences obtained between experiments, indicating that some barcodes have a high probability of being generated thus complicating the estimation of clone size. Analysis of the output of adult HSC clones showed little overlap in the barcodes observed in HSCs and progenitors compared to mature cells, confounding further analysis. This was likely related to barcoding that was observed in more differentiated short-term HSCs, MPPs, and restricted progenitors upon induction. Furthermore, depending on their frequency within a given population and the extent to which the sample size captures the population, some barcodes are likely to be missed, leading to underestimation of the lineage potential of clones. Thus, the in vivo contribution of adult HSC clones to multi-lineage hematopoiesis is still unresolved.
In vivo HSC dormancy and self-renewal
Inducible genetic systems for label retention assays have provided insight into the effects of proliferative history on HSC function. The originally described system comprises two transgenes, the first encoding human histone H2B-GFP fusion protein under the control of a tetracycline responsive promoter element (TRE) and the second a tetracycline transactivator (tTA) cassette driven by the T cell acute lymphocytic leukemia 1 gene (Tal1 also known as Scl) [57, 58]. In double transgenic animals, H2B-GFP is expressed and incorporated in bulk hematopoietic stem and progenitor cells (HSPCs), but upon chase with the tetracycline analog doxycycline (dox) H2B-GFP expression is inhibited (Fig. 2B). Cells progressively lose GFP expression as a function of their proliferation, whereas cells that retain the GFP label over time are identified as quiescent. Studies identified a population of HSCs that stably retained the H2B-GFP label for almost one year. Label retaining HSCs contained most of the serial reconstituting capacity compared to those that did not retain label [57, 58]. Although quiescent, these cells could be activated to proliferate upon challenge to immune stress mediated by the granulocyte colony stimulating factor cytokine or cytotoxic stress upon exposure to 5-fluorouracil or bromodeoxyuridine [58]. The functions of the pool of label retaining HSCs were further studied in a modified version of the H2B-GFP system that uses a human CD34-tTA transgene to regulate H2B-GFP expression [47, 59]. In this system, only a small fraction of HSCs was found to stably retain label after 22 months chase with dox, and although not all label-retaining HSCs were able to engraft, these cells enriched for repopulating activity upon serial transplantation. Notably, a loss of HSC self-renewal was estimated to occur after four rounds of cell division based on one-half dilutions in the fluorescent intensity of GFP label [47]. These studies demonstrate the existence of a rare subset of dormant HSCs that is held in reserve even with age, but further work is needed to understand how the dynamics of HSC quiescence and proliferation are regulated in vivo.
Building on these studies of HSC label retention, recent work from Cabezas-Wallscheid et al. coupled label retention assays with single-cell RNA-Sequencing (scRNA-Seq) to generate transcriptional profiles of single label-retaining HSCs, nonlabel-retaining HSCs with an otherwise identical phenotype and downstream progenitors [31]. Their analyses suggest that all HSCs irrespective of label retention are generally quiescent while the nonlabel-retaining (active) HSCs had higher expression of genes involved in biosynthesis activity and priming for cell cycle and label-retaining (dormant) HSCs displayed a signature of low biosynthetic activity and high retinoic acid (RA) signaling. To further study dormant HSCs, the gene Gprc5c (G protein coupled receptor, family C, group 5, member C), which was observed to be enriched in label retaining HSCs, was used to generate a transgenic reporter. Although the overlap between the transcriptional profiles and delayed kinetics in cell cycle entry of Gprc5c-labeled cells upon stimulation were shown, label retention capacity of Gprc5c-labeled HSCs was not confirmed. Moreover, Gprc5c-labeled HSCs displayed enhanced reconstitution kinetics, but did not robustly separate HSCs with superior reconstitution capacity as observed in other systems [33, 37]. Gprc5c-labeled HSCs were increased following treatment with all trans-retinoic acid (ATRA), consistent with Gprc5c being a RA inducible gene [60]. Furthermore, in vivo induction of RA signaling was associated with increased retention of HSCs in a quiescent, G0 state even upon stimulation with polyI:C. In the absence of RA signaling mediated by vitamin A, a reduction in Gprc5c-labeled HSCs and an overall decrease in hematopoietic cell numbers was observed. These results demonstrate the in vivo role of RA signaling in hematopoiesis. Future studies should confirm the effects of RA signaling on maintenance of HSCs with superior self-renewal identified by other reporters as previous studies in human HSCs showed the opposite effect of RA signaling [61]. Furthermore, this study provides new molecular definitions of HSC functionally “dormant” and “active” states and raises general questions about how we define and measure HSC quiescence.
The transition of HSCs from quiescence to a more proliferative state is associated with a switch in metabolism away from a glycolytic state to oxidative phosphorylation, activation of phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin pathways and generation of reactive oxygen species as recently reviewed [62, 63]. Moreover, defects in cellular metabolism and autophagy lead to exhaustion of the pool of HSCs [64–66]. Ito et al [35] recently set out to address the mechanisms that regulate HSC self-renewal using a previously described Tie2-GFP transgenic reporter [67]. Importantly, this reporter was generated in a completely independent manner from the inducible Tie2-Cre system previously discussed, and the overlap between HSC populations labeled by each system is not known. About 5% of phenotypic HSCs were Tie2-GFP+, and functional assays demonstrated an enrichment for cells that could undergo serial transplantation in reporter-labeled HSCs. However, this reporter did not capture all functional HSCs as serial reconstitution was observed in 40% of recipients of GFP- HSCs. Expression analyses of Tie2-GFP+ and GFP− HSCs was performed and signatures for peroxisome proliferator-activated receptor (PPAR) signaling, fatty acid metabolism, and mitochondrial autophagy (mitophagy) were detected in reporter-labeled HSCs. Tie2-GFP+ HSCs were found to undergo mitophagy and this was induced by PPAR signaling. Using in vivo paired daughter cell assays, the capacity for self-renewal and differentiation of HSCs was measured. While Tie2-GFP+ HSCs were found to preferentially undergo self-renewal and provided reconstitution in transplantation assays, the ability to reconstitute was strongly decreased upon knock-down of Parkin, a key intermediate of mitophagy in Tie2-GFP HSCs. Collectively, these results indicate a role for mitophagy in HSCs, providing a link between this process and self-renewal in Tie2-GFP+ HSCs.
Concluding remarks
In vitro colony formation and transplantation assays have previously been the basis and will continue to be valuable tools for the study of HSCs. Indeed, the functionality of labeled cells must be validated by transplantation assays in the development of HSC-specific reporter animals. Nevertheless, these reporter systems are a major technological advance that permit characterization of the fundamental properties of HSCs in vivo. Specifically, adult HSCs make a major contribution to hematopoiesis at the steady state [68]. That HSCs do contribute to steady state hematopoiesis is supported by recent clonal analysis that found hematopoietic clones with distinct fluorescent labels that are composed of phenotypic HSCs, progenitors and mature cells [53]. The extent to which HSCs contribute to hematopoiesis is also demonstrated by label retaining assays in which the majority of HSCs progressively lost label, while <5% of the population remained dormant and stably retained the label [47]. Importantly, the major contribution to steady state hematopoiesis established after transplantation was also recently confirmed in human HSCs [69]
In vivo clonal analysis of the contribution of fetal HSCs suggests that adult hematopoiesis comprises the activity of many distinct clones with different sizes [56, 70]. While recent analyses suggest an active contribution of adult HSCs to megakaryopoiesis [50], the clonal contribution of adult HSCs to the full spectrum of lineages during steady state hematopoiesis remains poorly understood, requiring further studies that incorporate alternative systems and approaches [53, 70]. HSC self-renewal and differentiation were recently shown to be affected by basic processes that regulate mitochondria, including fission/fusion and turnover [35, 71]. Although the mechanisms by which this occurs are unclear, the impact of these processes on cell fate has been shown in other types of stem cells [72] suggesting they are general mechanisms to preserve stem cell fitness. As new interactions between the metabolic state and HSC output in vivo are being defined [31], better resolution of the overwhelmingly complex metabolic pathways of HSCs especially at the epigenetic level is warranted. A number of metabolic intermediates feed into the pathways regulating the epigenetic status of a cell, which could affect its overall gene expression profile and function or prime specific functional states in daughter cells following cell division [73]. Indeed, this was recently shown for ascorbate acid (vitamin C) in HSCs, which promotes the activity of the Tet (Ten-Eleven-translocation) family of proteins that regulate DNA demethylation [74, 75]. Treatment with ascorbate acid reversed the defects associated with in vivo loss of TET2 function, including DNA hypermethylation, aberrant proliferation of HSPCs and the development of leukemia.
New strategies that facilitate in vivo study of HSCs, including human HSCs, continue to be developed [76]. In addition to the generation of entirely novel genetic reporter systems, such strategies employ previously established reporter systems in new ways [77]. Such recently described genetic reporter systems have begun to provide insight to HSCs biology in the absence of perturbation. The application of new single cell -omics approaches [78–80] to such systems promises to reveal fundamental properties of individual HSCs at high resolution. In light of the studies documenting detrimental clonal hematopoiesis associated with age and disease [81–83], understanding the general dynamics and molecular mechanisms that regulate HSC function at the single cell level in vivo is imperative for not just stem cell aficionados but for the basic fields of biology and medicine.
HIGHLIGHTS.
Genetic reporter systems enable prospective identification of bona fide hematopoietic stem cells (HSCs)
Most adult HSCs make a major contribution to steady state hematopoiesis
Minor fraction of stable label-retaining HSCs persists even with age
New roles for mitochondria and metabolic state in regulating HSC function in vivo
Acknowledgments
Work on HSCs from the Reizis laboratory is supported by NIH grant AG049074.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Adolfsson J, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121(2):295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Christensen JL, I, Weissman L. Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proc Natl Acad Sci U S A. 2001;98(25):14541–6. doi: 10.1073/pnas.261562798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodell MA, et al. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183(4):1797–806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiel MJ, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121(7):1109–21. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 5.Osawa M, et al. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273(5272):242–5. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 6.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241(4861):58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 7.Benz C, et al. Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell Stem Cell. 2012;10(3):273–83. doi: 10.1016/j.stem.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Dykstra B, et al. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1(2):218–29. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 9.Muller-Sieburg CE, et al. Deterministic regulation of hematopoietic stem cell self-renewal and differentiation. Blood. 2002;100(4):1302–9. [PubMed] [Google Scholar]
- 10.Yamamoto R, et al. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell. 2013;154(5):1112–26. doi: 10.1016/j.cell.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Harrison DE, Astle CM, Delaittre JA. Loss of proliferative capacity in immunohemopoietic stem cells caused by serial transplantation rather than aging. J Exp Med. 1978;147(5):1526–31. doi: 10.1084/jem.147.5.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu H, et al. Hematopoietic stem cell exhaustion impacted by p18 INK4C and p21 Cip1/Waf1 in opposite manners. Blood. 2006;107(3):1200–6. doi: 10.1182/blood-2005-02-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao X, et al. Irradiation induces bone injury by damaging bone marrow microenvironment for stem cells. Proc Natl Acad Sci U S A. 2011;108(4):1609–14. doi: 10.1073/pnas.1015350108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller U. Ten years of gene targeting: targeted mouse mutants, from vector design to phenotype analysis. Mech Dev. 1999;82(1–2):3–21. doi: 10.1016/s0925-4773(99)00021-0. [DOI] [PubMed] [Google Scholar]
- 15.Hall B, Limaye A, Kulkarni AB. Overview: generation of gene knockout mice. Curr Protoc Cell Biol. 2009;Chapter 19(Unit 19):12 19 12 1–17. doi: 10.1002/0471143030.cb1912s44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327(5962):167–70. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 17.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482(7385):331–8. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 18.Gong C, et al. Mechanism of nonhomologous end-joining in mycobacteria: a low-fidelity repair system driven by Ku, ligase D and ligase C. Nat Struct Mol Biol. 2005;12(4):304–12. doi: 10.1038/nsmb915. [DOI] [PubMed] [Google Scholar]
- 19.Overballe-Petersen S, et al. Bacterial natural transformation by highly fragmented and damaged DNA. Proc Natl Acad Sci U S A. 2013;110(49):19860–5. doi: 10.1073/pnas.1315278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh P, Schimenti JC, Bolcun-Filas E. A mouse geneticist’s practical guide to CRISPR applications. Genetics. 2015;199(1):1–15. doi: 10.1534/genetics.114.169771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H, et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154(6):1370–9. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osoegawa K, et al. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 2000;10(1):116–28. [PMC free article] [PubMed] [Google Scholar]
- 23.Heintz N. BAC to the future: the use of bac transgenic mice for neuroscience research. Nat Rev Neurosci. 2001;2(12):861–70. doi: 10.1038/35104049. [DOI] [PubMed] [Google Scholar]
- 24.Sharan SK, et al. Recombineering: a homologous recombination-based method of genetic engineering. Nat Protoc. 2009;4(2):206–23. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Keuren ML, et al. Generating transgenic mice from bacterial artificial chromosomes: transgenesis efficiency, integration and expression outcomes. Transgenic Res. 2009;18(5):769–85. doi: 10.1007/s11248-009-9271-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kowalczyk MS, et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015;25(12):1860–72. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nestorowa S, et al. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood. 2016;128(8):e20–31. doi: 10.1182/blood-2016-05-716480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hills D, et al. Hoxb4-YFP reporter mouse model: a novel tool for tracking HSC development and studying the role of Hoxb4 in hematopoiesis. Blood. 2011;117(13):3521–8. doi: 10.1182/blood-2009-12-253989. [DOI] [PubMed] [Google Scholar]
- 29.Gothert JR, et al. In vivo fate-tracing studies using the Scl stem cell enhancer: embryonic hematopoietic stem cells significantly contribute to adult hematopoiesis. Blood. 2005;105(7):2724–32. doi: 10.1182/blood-2004-08-3037. [DOI] [PubMed] [Google Scholar]
- 30.Acar M, et al. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature. 2015;526(7571):126–30. doi: 10.1038/nature15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cabezas-Wallscheid N, et al. Vitamin A-Retinoic Acid Signaling Regulates Hematopoietic Stem Cell Dormancy. Cell. 2017;169(5):807–823e19. doi: 10.1016/j.cell.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 32.Carrelha J, et al. Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells. Nature. 2018;554(7690):106–111. doi: 10.1038/nature25455. [DOI] [PubMed] [Google Scholar]
- 33.Chen JY, et al. Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature. 2016;530(7589):223–7. doi: 10.1038/nature16943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gazit R, et al. Fgd5 identifies hematopoietic stem cells in the murine bone marrow. J Exp Med. 2014;211(7):1315–31. doi: 10.1084/jem.20130428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ito K, et al. Self-renewal of a purified Tie2+ hematopoietic stem cell population relies on mitochondrial clearance. Science. 2016;354(6316):1156–1160. doi: 10.1126/science.aaf5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanjuan-Pla A, et al. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature. 2013;502(7470):232–6. doi: 10.1038/nature12495. [DOI] [PubMed] [Google Scholar]
- 37.Sawai CM, et al. Hematopoietic Stem Cells Are the Major Source of Multilineage Hematopoiesis in Adult Animals. Immunity. 2016;45(3):597–609. doi: 10.1016/j.immuni.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arai F, et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118(2):149–61. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 39.Denis C, et al. A mouse model of severe von Willebrand disease: defects in hemostasis and thrombosis. Proc Natl Acad Sci U S A. 1998;95(16):9524–9. doi: 10.1073/pnas.95.16.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu M, et al. HOXB5 expression is spatially and temporarily regulated in human embryonic gut during neural crest cell colonization and differentiation of enteric neuroblasts. Dev Dyn. 2003;228(1):1–10. doi: 10.1002/dvdy.10350. [DOI] [PubMed] [Google Scholar]
- 41.Kretzschmar K, Watt FM. Lineage tracing. Cell. 2012;148(1–2):33–45. doi: 10.1016/j.cell.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 42.Alva JA, et al. VE-Cadherin-Cre-recombinase transgenic mouse: a tool for lineage analysis and gene deletion in endothelial cells. Dev Dyn. 2006;235(3):759–67. doi: 10.1002/dvdy.20643. [DOI] [PubMed] [Google Scholar]
- 43.Rybtsov S, et al. Hierarchical organization and early hematopoietic specification of the developing HSC lineage in the AGM region. J Exp Med. 2011;208(6):1305–15. doi: 10.1084/jem.20102419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samokhvalov IM, Samokhvalova NI, Nishikawa S. Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature. 2007;446(7139):1056–61. doi: 10.1038/nature05725. [DOI] [PubMed] [Google Scholar]
- 45.Zovein AC, et al. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3(6):625–36. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Busch K, et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518(7540):542–6. doi: 10.1038/nature14242. [DOI] [PubMed] [Google Scholar]
- 47.Bernitz JM, et al. Hematopoietic Stem Cells Count and Remember Self-Renewal Divisions. Cell. 2016;167(5):1296–1309e10. doi: 10.1016/j.cell.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanchez-Aguilera A, et al. Estrogen signaling selectively induces apoptosis of hematopoietic progenitors and myeloid neoplasms without harming steady-state hematopoiesis. Cell Stem Cell. 2014;15(6):791–804. doi: 10.1016/j.stem.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 49.Sun J, et al. Clonal dynamics of native haematopoiesis. Nature. 2014;514(7522):322–7. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodriguez-Fraticelli AE, et al. Clonal analysis of lineage fate in native haematopoiesis. Nature. 2018;553(7687):212–216. doi: 10.1038/nature25168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo G, et al. Mapping cellular hierarchy by single-cell analysis of the cell surface repertoire. Cell Stem Cell. 2013;13(4):492–505. doi: 10.1016/j.stem.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kent DG, et al. Prospective isolation and molecular characterization of hematopoietic stem cells with durable self-renewal potential. Blood. 2009;113(25):6342–50. doi: 10.1182/blood-2008-12-192054. [DOI] [PubMed] [Google Scholar]
- 53.Yu VW, et al. Epigenetic Memory Underlies Cell-Autonomous Heterogeneous Behavior of Hematopoietic Stem Cells. Cell. 2016;167(5):1310–1322e17. doi: 10.1016/j.cell.2016.10.045. [DOI] [PubMed] [Google Scholar]
- 54.Livet J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450(7166):56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 55.Essers MA, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458(7240):904–8. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- 56.Pei W, et al. Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature. 2017;548(7668):456–460. doi: 10.1038/nature23653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Foudi A, et al. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol. 2009;27(1):84–90. doi: 10.1038/nbt.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson A, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135(6):1118–29. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 59.Qiu J, et al. Divisional history and hematopoietic stem cell function during homeostasis. Stem Cell Reports. 2014;2(4):473–90. doi: 10.1016/j.stemcr.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robbins MJ, et al. Molecular cloning and characterization of two novel retinoic acid-inducible orphan G-protein-coupled receptors (GPRC5B and GPRC5C) Genomics. 2000;67(1):8–18. doi: 10.1006/geno.2000.6226. [DOI] [PubMed] [Google Scholar]
- 61.Ghiaur G, et al. Regulation of human hematopoietic stem cell self-renewal by the microenvironment’s control of retinoic acid signaling. Proc Natl Acad Sci U S A. 2013;110(40):16121–6. doi: 10.1073/pnas.1305937110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suda T, Takubo K, Semenza GL. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell. 2011;9(4):298–310. doi: 10.1016/j.stem.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 63.Kohli L, Passegue E. Surviving change: the metabolic journey of hematopoietic stem cells. Trends Cell Biol. 2014;24(8):479–87. doi: 10.1016/j.tcb.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ho TT, et al. Autophagy maintains the metabolism and function of young and old stem cells. Nature. 2017;543(7644):205–210. doi: 10.1038/nature21388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ito K, et al. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med. 2012;18(9):1350–8. doi: 10.1038/nm.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Warr MR, et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature. 2013;494(7437):323–7. doi: 10.1038/nature11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Motoike T, et al. Universal GFP reporter for the study of vascular development. Genesis. 2000;28(2):75–81. doi: 10.1002/1526-968x(200010)28:2<75::aid-gene50>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 68.Sawai CM, et al. Hematopoietic Stem Cells Are the Major Source of Multilineage Hematopoiesis in Adult Animals. Immunity. 2016;45(3):597–609. doi: 10.1016/j.immuni.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Biasco L, et al. In Vivo Tracking of Human Hematopoiesis Reveals Patterns of Clonal Dynamics during Early and Steady-State Reconstitution Phases. Cell Stem Cell. 2016;19(1):107–19. doi: 10.1016/j.stem.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ganuza M, et al. Lifelong haematopoiesis is established by hundreds of precursors throughout mammalian ontogeny. Nat Cell Biol. 2017;19(10):1153–1163. doi: 10.1038/ncb3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luchsinger LL, et al. Mitofusin 2 maintains haematopoietic stem cells with extensive lymphoid potential. Nature. 2016;529(7587):528–31. doi: 10.1038/nature16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen H, Chan DC. Mitochondrial Dynamics in Regulating the Unique Phenotypes of Cancer and Stem Cells. Cell Metab. 2017;26(1):39–48. doi: 10.1016/j.cmet.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ryall JG, et al. Metabolic Reprogramming of Stem Cell Epigenetics. Cell Stem Cell. 2015;17(6):651–62. doi: 10.1016/j.stem.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Agathocleous M, et al. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature. 2017;549(7673):476–481. doi: 10.1038/nature23876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cimmino L, et al. Restoration of TET2 Function Blocks Aberrant Self-Renewal and Leukemia Progression. Cell. 2017;170(6):1079–1095e20. doi: 10.1016/j.cell.2017.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rahmig S, et al. Improved Human Erythropoiesis and Platelet Formation in Humanized NSGW41 Mice. Stem Cell Reports. 2016;7(4):591–601. doi: 10.1016/j.stemcr.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Perez-Cunningham J, et al. Hematopoietic stem cell-specific GFP-expressing transgenic mice generated by genetic excision of a pan-hematopoietic reporter gene. Exp Hematol. 2016;44(8):755–764e1. doi: 10.1016/j.exphem.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Buenrostro JD, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523(7561):486–90. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Flyamer IM, et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature. 2017;544(7648):110–114. doi: 10.1038/nature21711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heath JR, Ribas A, Mischel PS. Single-cell analysis tools for drug discovery and development. Nat Rev Drug Discov. 2016;15(3):204–16. doi: 10.1038/nrd.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jaiswal S, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–98. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Steensma DP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xie M, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472–8. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]