Abstract
Researchers in the field of structural biology, especially X-ray crystallography and protein nuclear magnetic resonance, are interested in knowing as much as possible about the state of their target protein in solution. Not only is this knowledge relevant to studies of biological function, it also facilitates determination of a protein structure using homogeneous monodisperse protein samples. A researcher faced with a new protein to study will have many questions even after that protein has been purified. Analytical ultracentrifugation (AUC) can provide all of this information readily from a small sample in a non-destructive way, without the need for labeling, enabling structure determination experiments without any wasting time and material on uncharacterized samples. In this article, I use examples to illustrate how AUC can contribute to protein structural analysis. Integrating information from a variety of biophysical experimental methods, such as X-ray crystallography, small angle X-ray scattering, electrospray ionization-mass spectrometry, AUC allows a more complete understanding of the structure and function of biomacromolecules.
Keywords: Protein, AUC, Crystallography, Interaction
Many studies of protein structure and function begin with the preparation of purified samples on the scale of a few milligrams. Even having achieved the non-trivial task of producing such as sample, a researcher faced with a new protein to study will have many questions regarding its form in solution. Is there any aggregation in the prepared protein solution? Is the protein monodisperse? Is the protein monomeric or mulitmeric? If the protein shows significant self-association, what are the thermodynamic binding constants? If the protein is a hetero-oligomer, what is the stoichiometry? When you want to crystallize a protein complex, does it form stably in solution? Analytical ultracentrifugation (AUC), a method used for the quantitative analysis of macromolecules in solution, can provide all of this information readily from a small sample in a non-destructive way, without the need for labeling, thereby allowing structure determination experiments without wasting any time and material on uncharacterized samples. AUC itself is a well-established method, but software developments in the present century have hugely improved the ease and accuracy of this analytical technique. The use of sedimentation velocity (SV-AUC) has in particular significantly expanded the applications of AUC, and new computational methods for the AUC analysis are being actively developed by several groups (Balbo et al. 2005; Philo 2006; Brown et al. 2007, 2009; Correia and Stafford 2009; Behlke and Ristau 2010; Brookes et al. 2010). SV-AUC data contain a huge amount of information on the hydrodynamic properties of the target proteins, and computational methods are focused on developing methods on how to extract useful information from the raw data. SEDFIT, for example, can calculate diffusion-deconvoluted sedimentation coefficient distributions from direct boundary modeling of SV-AUC experimental data (Schuck 2000; Schuck et al. 2002). High-resolution sedimentation coefficient distribution profiles give important insights into the hydrodynamic properties of the target proteins. In this article, I provide examples to illustrate how AUC can contribute to protein structural analysis. Integrating information from a variety biophysical experimental methods, such as X-ray crystallography, small angle x-ray scattering (SAXS), electrospray ionization (ESI)-mass spectrometry, AUC allows a more complete understanding of the structure and function of biomacromolecules.
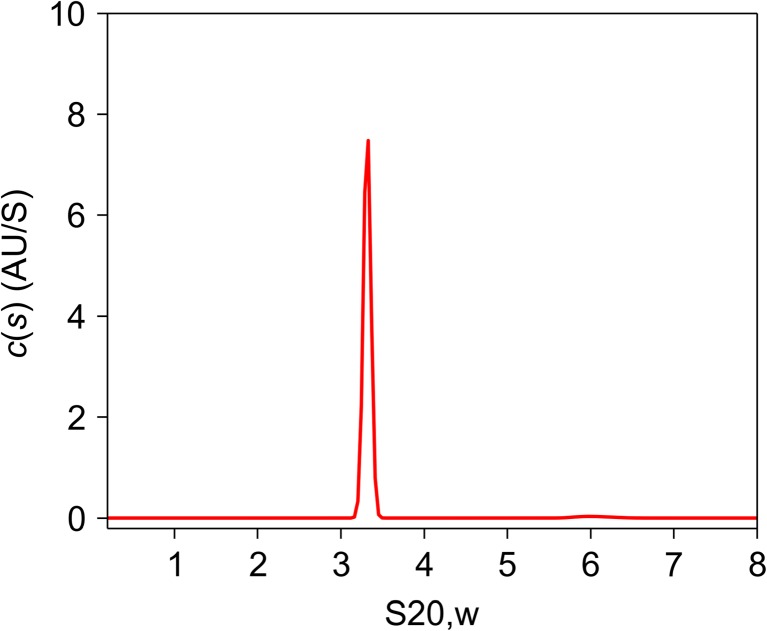
Figure 1 shows a typical protein sedimentation coefficient distribution profile, c(s), analyzed by the SEDFIT software tool. The protein in question is a human transcription factor, TFIIE (Itoh et al. 2005). Transcription of protein-encoding genes in eukaryotes requires the formation of a transcription preinitiation complex, which is composed of promoter DNA, RNA polymerase II, and five general transcription factors (Orphanides et al. 1996; Roeder 1996). TFIIE is one of the five factors. TFIIE is incorporated last, to complete the transcription preinitiation complex, activate it, and induce promoter melting. It has been suggested that human TFIIE consists of two alpha [57 kDa, estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)] and two beta (34 kDa; SDS-PAGE) subunits with a molecular mass of about 180 kDa as estimated by size exclusion chromatography (SEC) (Ohkuma et al. 1990). Human TFIIE was believed to be a hetero-tetramer from this result. We applied AUC velocity experiments to characterize recombinant human TFIIE, containing alpha subunits (49 kDa) and 6His-tagged beta subunits (35 kDa). The c(s) distribution (Fig. 1) has a quite sharp single peak with an S20,w value of 3.3 S. This c(s) distribution means that the protein complex is monodisperse and that there is no sample aggregation in the solution. TFIIE is well-behaved, forming a single, stable structure. In such cases, the c(s) distribution can be converted into a molecular mass distribution (Schuck et al. 2002). The molecular mass of the TFIIE was determined to be 81 (± 5) kDa, which means that the complex must be an alpha–beta hetero-dimer rather than the hetero-tetramer that was deduced from gel filtration experiments. The AUC results agree well with that of ESI-mass spectrometry (84 kDa). Why did TFIIE show such a large molecular mass in the SEC experiments? AUC analysis revealed another important hydrodynamic parameter, the frictional ratio (f/fo), which represents the degree of deviation due to hydration, rugosity, asymmetry, and expansion of the molecule from a minimum possible value of 1.0 for a hard, incompressible, unhydrated sphere. Typical values for globular proteins lie in the range of 1.05 ~ 1.30. TFIIE has the frictional ratio of 2.1, suggesting that it is a very highly asymmetric or expanded ellipsoid, with an axial ratio of around 15. This shape can explain the anomalously fast elution profile of TFEII in SEC. The low-resolution structure of TFIIE in solution was also analyzed by SAXS. The shape of TFIIE in solution was reconstructed from the experimental SAXS data using simulated annealing, and the reconstruction indicated that a dummy atom model for TFIIE has a highly extended rod-like structure. This explains why the molecular mass of TFIIE was estimated to be 180 kDa by SEC, whereas the results from the AUC and ESI-mass spectrometry studies show it to be about 84 kDa. Molecular shape has a huge impact on the mass estimated by SEC, since the method requires calibration with globular proteins, such as gamma-globulin and BSA (bovine serum albumin). If the molecular shape of a protein is far from globular, it may easily elute anomalously quickly from a gel-filtration column because long proteins do not easily enter into the SEC column gel matrix compared with spherical proteins of the same mass. An extended rod-like structure, such as TFIIE, may easily appear to be much larger than its true size in a SEC experiment.
Fig. 1.
Distribution of sedimentation coefficients c(s20,w) for the general transcription factor TFIIE. Calculated c(s20,w) is plotted against the sedimentation coefficient s20,w. Experiments were conducted at an initial protein concentration of 1.37 mg/mL in 20 mM sodium phosphate (pH 7.9), 500 mM NaCl, 10 mM 2-mercaptoethanol, 10% glycerol. Data were collected at a rotor speed of 40,000 rpm and at time intervals of 5 min. The calculated values of the weight-average s20,w and frictional ratio (f/fo) are 3.3 S and 2.1, respectively
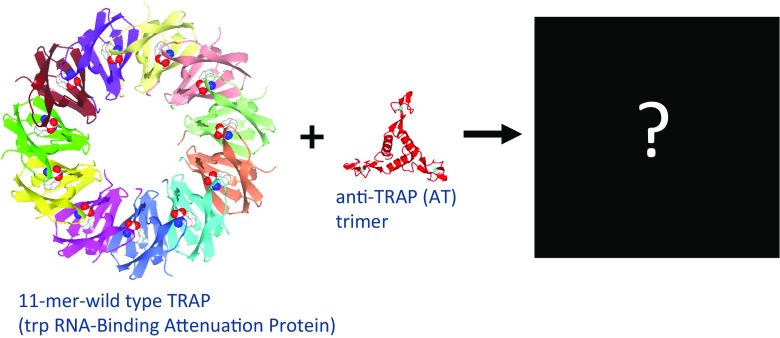
AUC velocity experiments analyzed by the SEDFIT software tool allow very sensitive detection of different molecular mass species—for example, to quantify protein–protein interactions and observe protein complex formation. We used AUC to find the size and shape of the complex formed between TRAP (trp RNA-binding attenuation protein, where Trp is tryptophan) and a regulator called Anti-TRAP (AT) (Watanabe et al. 2009). TRAP plays a central role in the highly intricate regulation of transcription and translation of the trp operon in several species of Bacillus, and this system has provided important insights into several mechanisms of gene regulation (Babitzke et al. 1994, 1995; Babitzke 1997, 2004; Gollnick et al. 2005). The protein forms an 11-mer ring that binds 11 molecules of tryptophan at symmetry-related sites, and the X-ray crystal structure of the TRAP–Trp–RNA complex has been solved for the Bacillus stearothermohilus protein (Chen et al. 1999). With tryptophan bound, TRAP can bind trp mRNA and induce transcription termination. In Bacillus subtilis, when levels of charged tRNA fall, the protein AT is expressed, which blocks RNA binding to Trp-bound TRAP and relieves expression of the trp operon. AT forms a stable trimer, four copies of which can further associate into a dodecamer form. Several TRAP–AT complex models have been proposed, but there is as yet no explanation for the ability of AT to compete with RNA binding to TRAP. The nature of the interaction has proved elusive (Fig. 2). We addressed this issue with a variety of biophysical methods, such as AUC, ESI-mass spectrometry, and protein crystallography.
Fig. 2.
Schematic drawing of 11-mer trp RNA-binding attenuation protein (TRAP) and trimer anti-TRAP (AT) interaction. Wild-type Bacillus stearothermophilus TRAP (PDB ID 1QAW; Chen et al. 1999), Bacillus subtilis anti-TRAP (PDB ID 2BX9; Shevtsov et al. 2005)
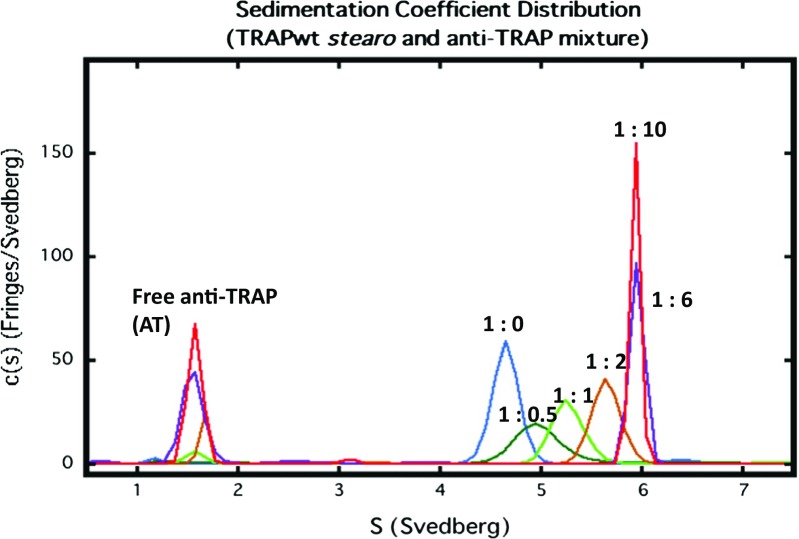
In the sedimentation velocity c(s) analysis, protein–protein interaction can be detected through the emergence of new peaks at higher concentrations, shifts in the ratios of the peak areas, and/or shifts in the peak positions (Dam et al. 2005). Figure 3 shows the sedimentation velocity analysis of mixtures of wild-type B. stearothemophilus 11-mer TRAP and AT. The TRAP concentration was fixed at 0.5 mg/ml, and varying amounts of AT were added. The data obtained from six experiments are overlaid. The molar ratios (by monomers) of TRAP-AT are 1:0 (blue), 1:0.5 (green), 1:1 (light green), 1:2 (brown), 1:6 (purple), 1:10 (red). When AT was added the c(s) distribution peak position was shifted. Shifting peak positions indicate the rapid chemical interconversion of species during the sedimentation. The reaction causes the sedimenting system to assume an average sedimentation rate in between those of the reacting species, shifting according to their relative population. We estimated that there were several species of the TRAP–AT complex and that these caused broadening of the c(s) distribution. The c(s) peak, however, did not show a further shift when excess AT was added (Fig. 3). We found that the TRAP ring was saturated with AT at a 1:6 stoichiometry and that the TRAP–AT complex was stable to excess AT.
Fig. 3.
Sedimentation velocity analysis of mixtures of wild-type Bacillus stearothemophilus 11-mer TRAP and AT. The TRAP concentration was fixed at 0. 5 mg/ml, and varying amounts of AT were added. The data obtained from six experiments are overlaid. The molar ratios (by monomers) of TRAP-AT are 1:0 (blue), 1:0.5 (green), 1:1 (light green), 1:2 (brown), 1:6 (purple), 1:10 (red)
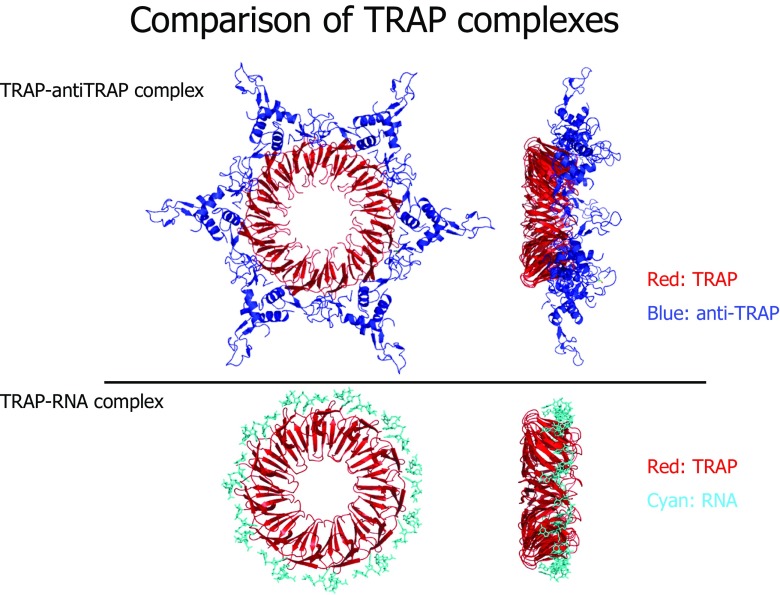
Using the c(s) analysis, we were able to crystallize the saturated TRAP–AT complex. This complex formed crystals that diffracted to 3.2 Å and allowed a clear molecular replacement solution to be determined in space group R32 with two AT trimers and four TRAP subunits in the asymmetric unit. Three AT trimers and six TRAP subunits were found in the asymmetric unit of a P6 crystal form. The structures reveal that the wild-type TRAP has formed 12-mer rings (Fig. 4). The model of the 12-mer TRAP–AT complex is radically different from those previously proposed, but immediately explains the ability of AT to block RNA binding to TRAP since AT contacts the RNA-binding residues directly. One AT trimer binds to two neighboring TRAP subunits within a ring, with the majority of the contacts being between one AT chain and one TRAP subunit. The contacts between TRAP and AT include both hydrophobic interactions and salt bridges. Phe-32 of TRAP, a residue close to bound RNA (Antson et al. 1999), sits in an apolar pocket formed by Pro-13, Ala-28 and Ile-35 of AT. The model immediately explains the known importance of Lys-37 and Arg-58 residues to TRAP-AT binding (Valbuzzi et al. 2002) and how AT sterically blocks RNA binding to TRAP. Phe-32 appears from the model to be a key residue because two copies contact each AT trimer, and replacing it with alanine abolishes binding to AT.
Fig. 4.
Comparison of TRAP complexes. Wild-type TRAP-AT complex (PDB ID 2ZP8); TRAP–RNA complex (PDB ID 1C9S) (Antson et al. 1999)
Why did the 11-mer wild-type TRAP crystallize as the 12-mer TRAP in the TRAP–AT complex? The ESI-mass spectrometry experiment confirmed that a small proportion of wild-type TRAP exists in a 12-mer ring form in solution. A minor population of a 12-mer form of wild-type B. subtilis TRAP has previously been suggested, based on the results of mass spectrometry studies (McCammon et al. 2004). The ESI-mass spectrometry results clearly show that not only does the 11-mer TRAP possibly bind five AT trimers but also that the 12-mer TRAP may bind six AT timers. Crystallization preferentially selects the 12-mer form, even though most of the TRAP is present as an 11-mer form, and the crystal structure confirms that wild-type TRAP can make a 12-mer ring. The result remains surprising, however, given the known thermostability of the 11-mer TRAP (Heddle et al. 2006). AT trimers are just large enough to contact each neighbor around the 12-mer TRAP ring, but the wild-type 11-mer TRAP is too small to accommodate more than five trimers and could not therefore give a similar crystal form. Increasing the diameter of the TRAP ring has little effect on its intrinsic affinity for the AT trimer, but it does increase the binding capacity to six trimers, giving the overall complex suitable symmetry for crystallization. In conclusion, our crystal structures, AUC analysis, and mass spectrometry results reveal that individual AT trimers may bind around the TRAP ring. Further work is required to determine whether a single AT trimer bound to TRAP is sufficient to relieve transcription termination, and whether a 12-mer form of TRAP could function equally well in vivo. More generally, our results show the importance of using a variety of biophysical methods and solution conditions in studies of protein complex.
Acknowledgments
I would like to thank my colleagues at Yokohama City University for their support during my stay, when the main results of this article were obtained.
Compliance with ethical standards
Conflicts of interest
Satoru Unzai declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by the author.
Footnotes
This article is part of a Special Issue on ‘Biomolecules to Bio-nanomachines—Fumio Arisaka 70th Birthday’ edited by Damien Hall, Junichi Takagi and Haruki Nakamura.
References
- Antson AA, Dodson EJ, Dodson G, et al. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature. 1999;401:235–242. doi: 10.1038/45730. [DOI] [PubMed] [Google Scholar]
- Babitzke P (1997) Regulation of tryptophan biosynthesis: Trp-ing the TRAP or how Bacillus subtilis reinvented the wheel. Mol Microbiol 26:1–9 [DOI] [PubMed]
- Babitzke P (2004) Regulation of transcription attenuation and translation initiation by allosteric control of an RNA-binding protein: the Bacillus subtilis TRAP protein. Curr Opin Microbiol 7:132–139. 10.1016/j.mib.2004.02.003 [DOI] [PubMed]
- Babitzke P, Bear DG, Yanofsky C (1995) TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a toroid-shaped molecule that binds transcripts containing GAG or UAG repeats separated by two nucleotides. Proc Natl Acad Sci USA 92:7916–7920 [DOI] [PMC free article] [PubMed]
- Babitzke P, Stults JT, Shire SJ, Yanofsky C (1994) TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a multisubunit complex that appears to recognize G/UAG repeats in the trpEDCFBA and trpG transcripts. J Biol Chem 269:16597–16604 [PubMed]
- Balbo A, Minor KH, Velikovsky CA, et al. Studying multiprotein complexes by multisignal sedimentation velocity analytical ultracentrifugation. Proc Natl Acad Sci USA. 2005;102:81–86. doi: 10.1073/pnas.0408399102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlke J, Ristau O. Enhanced resolution of sedimentation coefficient distribution profiles by extrapolation to infinite time. Eur Biophys J. 2010;39:449–455. doi: 10.1007/s00249-009-0425-1. [DOI] [PubMed] [Google Scholar]
- Brookes E, Cao W, Demeler B. A two-dimensional spectrum analysis for sedimentation velocity experiments of mixtures with heterogeneity in molecular weight and shape. Eur Biophys J. 2010;39:405–414. doi: 10.1007/s00249-009-0413-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PH, Balbo A, Schuck P. On the analysis of sedimentation velocity in the study of protein complexes. Eur Biophys J. 2009;38:1079–1099. doi: 10.1007/s00249-009-0514-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PH, Balbo A, Schuck P. Using prior knowledge in the determination of macromolecular size-distributions by analytical ultracentrifugation. Biomacromolecules. 2007;8:2011–2024. doi: 10.1021/bm070193j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XP, Antson AA, Yang M, et al. Regulatory features of the trp operon and the crystal structure of the trp RNA-binding attenuation protein from Bacillus Stearothermophilus. J Mol Biol. 1999;289:1003–1016. doi: 10.1006/jmbi.1999.2834. [DOI] [PubMed] [Google Scholar]
- Correia JJ, Stafford WF. Extracting equilibrium constants from kinetically limited reacting systems. Meth Enzymol. 2009;455:419–446. doi: 10.1016/S0076-6879(08)04215-8. [DOI] [PubMed] [Google Scholar]
- Dam J, Velikovsky CA, Mariuzza RA et al (2005) Sedimentation velocity analysis of heterogeneous protein–protein interactions: Lamm equation modeling and sedimentation coefficient distributions c(s). Biophysj 89:619–634. 10.1529/biophysj.105.059568 [DOI] [PMC free article] [PubMed]
- Gollnick P, Babitzke P, Antson A, Yanofsky C (2005) Complexity in regulation of tryptophan biosynthesis in Bacillus subtilis. Annu Rev Genet 39:47–68. 10.1146/annurev.genet.39.073003.093745 [DOI] [PubMed]
- Heddle JG, Yokoyama T, Yamashita I, et al. Rounding up: engineering 12-membered rings from the cyclic 11-mer TRAP. Structure. 2006;14:925–933. doi: 10.1016/j.str.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Unzai S, Sato M, et al. Investigation of molecular size of transcription factor TFIIE in solution. Proteins. 2005;61:633–641. doi: 10.1002/prot.20647. [DOI] [PubMed] [Google Scholar]
- McCammon MG, Hernández H, Sobott F, Robinson CV. Tandem mass spectrometry defines the stoichiometry and quaternary structural arrangement of tryptophan molecules in the multiprotein complex TRAP. J Am Chem Soc. 2004;126:5950–5951. doi: 10.1021/ja0317170. [DOI] [PubMed] [Google Scholar]
- Ohkuma Y, Sumimoto H, Horikoshi M, Roeder RG. Factors involved in specific transcription by mammalian RNA polymerase II: purification and characterization of general transcription factor TFIIE. Proc Natl Acad Sci USA. 1990;87:9163–9167. doi: 10.1073/pnas.87.23.9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. doi: 10.1016/S0968-0004(96)10050-5. [DOI] [PubMed] [Google Scholar]
- Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P, Perugini MA, Gonzales NR, et al. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys J. 2002;82:1096–1111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov MB, Chen Y, Gollnick P, Antson AA. Crystal structure of Bacillus Subtilis anti-TRAP protein, an antagonist of TRAP/RNA interaction. Proc Natl Acad Sci USA. 2005;102:17600–17605. doi: 10.1073/pnas.0508728102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valbuzzi A, Gollnick P, Babitzke P, Yanofsky C. The anti-trp RNA-binding attenuation protein (anti-TRAP), AT, recognizes the tryptophan-activated RNA binding domain of the TRAP regulatory protein. J Biol Chem. 2002;277:10608–10613. doi: 10.1074/jbc.M111813200. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Heddle JG, Kikuchi K, et al. The nature of the TRAP-anti-TRAP complex. Proc Natl Acad Sci USA. 2009;106:2176–2181. doi: 10.1073/pnas.0801032106. [DOI] [PMC free article] [PubMed] [Google Scholar]