Abstract
Dihydrodipicolinate synthase (DHDPS) is critical to the production of lysine through the diaminopimelate (DAP) pathway. Elucidation of the function, regulation and structure of this key class I aldolase has been the focus of considerable study in recent years, given that the dapA gene encoding DHDPS has been found to be essential to bacteria and plants. Allosteric inhibition by lysine is observed for DHDPS from plants and some bacterial species, the latter requiring a histidine or glutamate at position 56 (Escherichia coli numbering) over a basic amino acid. Structurally, two DHDPS monomers form the active site, which binds pyruvate and (S)-aspartate β-semialdehyde, with most dimers further dimerising to form a tetrameric arrangement around a solvent-filled centre cavity. The architecture and behaviour of these dimer-of-dimers is explored in detail, including biophysical studies utilising analytical ultracentrifugation, small-angle X-ray scattering and macromolecular crystallography that show bacterial DHDPS tetramers adopt a head-to-head quaternary structure, compared to the back-to-back arrangement observed for plant DHDPS enzymes. Finally, the potential role of pyruvate in providing substrate-mediated stabilisation of DHDPS is considered.
Keywords: Allostery, Antibiotic, Crystal, Herbicide, SAXS, Sedimentation
Lysine biosynthesis
Lysine is an essential amino acid in animals, including humans, but can be synthesised de novo in bacteria, lower eukaryotes and plants for utilisation in protein and peptidoglycan cell wall syntheses (Velasco et al. 2002). Unlike the other naturally occurring amino acids, lysine is the only one known to have two distinct biosynthetic pathways (Torruella et al. 2009). The α-aminoadipate (AAA) pathway is part of the glutamate biosynthetic family and is thought to be present almost exclusively in fungi and euglenoids (Miyazaki et al. 2004; Xu et al. 2006). The diaminopimelate (DAP) pathway belongs to the aspartate biosynthetic family and is understood to be present in bacteria, plants and lower fungi (Hudson et al. 2005; Velasco et al. 2002). Here, we will review our current knowledge of the enzyme catalysing the first committed step in the DAP pathway, namely dihydrodipicolinate synthase (DHDPS) (EC 4.3.3.7). DHDPS has been of interest to biophysicists in recent times, given the enzyme’s diverse molecular evolution, particularly at the quaternary structure level, and its essentiality to bacteria and plants, vindicating its potential as a novel antibiotic and herbicide target.
DAP pathway
The DAP pathway is responsible for not only the production of the end-product (S)-lysine in bacteria, plants and lower fungi, but also the penultimate product meso-2,6-DAP (meso-DAP), which is a critical component of the crosslinking network in the bacterial cell wall (Atkinson et al. 2012a; Dogovski et al. 2009, 2012; Hutton et al. 2007; Soares da Costa et al. 2015).
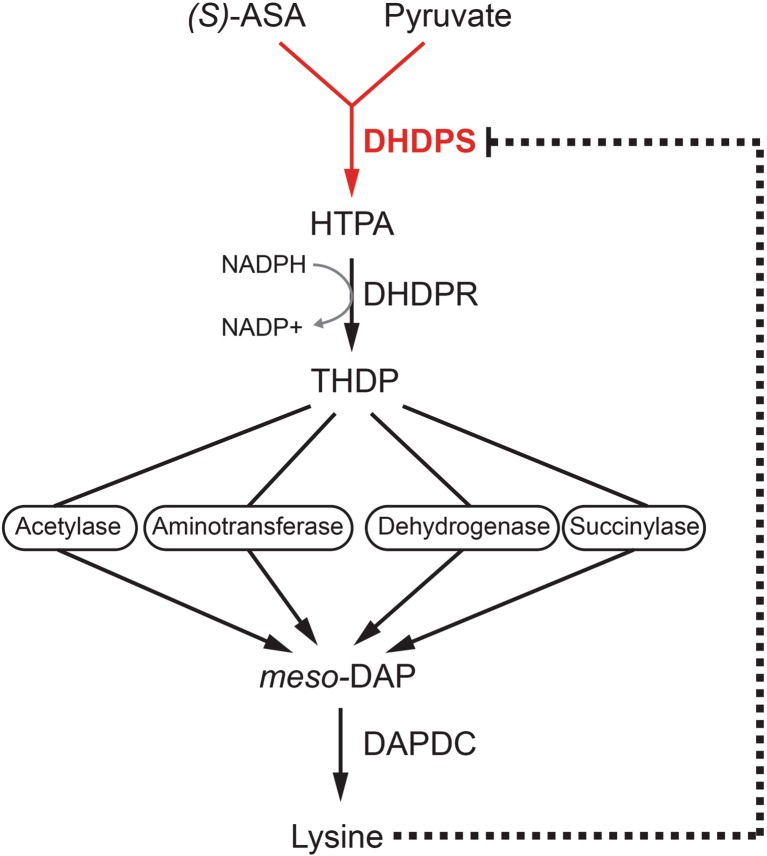
Four different routes of the DAP pathway have been discovered (Dogovski et al. 2012). They all share the same reactions at the beginning and end of the pathway, but differ in the intermediate steps depending on the species (Dogovski et al. 2012) (Fig. 1). All DAP sub-pathways commence with the condensation reaction between pyruvate and (S)-aspartate β-semialdehyde [(S)-ASA)] to form (4S)-4-hydroxy-2,3,4,5-tetrahydro-(2S)-dipicolinic acid (HTPA) (Dogovski et al. 2009, 2012; Soares da Costa et al. 2015). This is the first committed and rate-limiting step of the DAP pathway catalysed by DHDPS (Dogovski et al. 2009, 2012; Soares da Costa et al. 2015) (Fig. 1). HTPA is then reduced by dihydrodipicolinate reductase in an NADPH-dependent reaction to yield 2,3,4,5-tetrahydrodipicolinate (THDP) (Dommaraju et al. 2011; Girish et al. 2008) (Fig. 1). From this point on, the DAP pathway diverges into four sub-pathways, namely the succinylase, acetylase, dehydrogenase and aminotransferase pathways (Dogovski et al. 2009, 2012; Hutton et al. 2007) (Fig. 1). These four alternative sub-pathways all converge for the final step of the DAP pathway involving the diaminopimelate decarboxylase (DAPDC)-catalysed decarboxylation of meso-DAP to produce lysine (Peverelli et al. 2016; Ray et al. 2002) (Fig. 1). Lysine also regulates flux through the pathway by binding allosterically to DHDPS and inhibiting the enzyme from plants and some bacterial species, which will be discussed in the section entitled Allosteric regulation.
Fig. 1.
Diaminopimelate (DAP) biosynthesis pathway of bacteria and plants. The pathway commences with the condensation of pyruvate with (S)-aspartate β-semialdehyde [(S)-ASA] to form the heterocyclic product, (4S)-4-hydroxy-2,3,4,5-tetrahydro-(2S)-dipicolinic acid (HTPA) catalysed by dihydrodipicolinate synthase (DHDPS). HTPA is subsequently reduced by dihydrodipicolinate reductase (DHDPR) to form 2,3,4,5-tetrahydrodipicolinate (THDP), which is then converted via one of four sub-pathways depending on the species to meso-2,6-DAP (meso-DAP). Lysine is then formed by the decarboxylation of meso-DAP by diaminopimelate decarboxylase (DAPDC). The pathway is regulated by feedback inhibition by lysine, which binds allosterically to DHDPS
DHDPS
Gene and protein nomenclature
dapA gene
DHDPS is the product of the dapA gene (Dogovski et al. 2009, 2012). The gene was initially mapped in Escherichia coli in 1971 (Bukhari and Taylor 1971) and first cloned in 1986 (Richaud et al. 1986). In contrast to other enzymes in the DAP pathway, the expression of dapA in E. coli was not found to be regulated by cellular free lysine levels or any other stimuli (Butour et al. 1974). In the last three decades, the dapA gene has been cloned and sequenced from a variety of other bacterial (Atkinson et al. 2011, 2012a; Chen et al. 1993; Cremer et al. 1988; Devenish et al. 2009; Dommaraju et al. 2010; Evans et al. 2011; García-Rodríguez et al. 2000; Girish et al. 2008; Gunji et al. 2004; Kaur et al. 2011; Pisabarro et al. 1993; Siddiqui et al. 2013; Skovpen and Palmer 2013; Wolterink-van Loo et al. 2008; Wubben et al. 2010) and plant species (Atkinson et al. 2011, 2014; Frisch et al. 1991; Ghislain et al. 1995; Kaneko et al. 1990; Silk et al. 1994; Vauterin and Jacobs 1994). Typically, bacteria have a single dapA gene that is 800–900 bp, whereas plants have two annotated dapA genes consisting of ~ 1000 bp each. Gene duplication of dapA and other biosynthetic pathway genes in plants is prevalent as a means to enhance metabolic flux in vivo (Panchy et al. 2016). Sequence similarity at the amino acid level is high among plant enzymes, whereas bacterial enzymes appear to be more divergent (Blickling et al. 1997a; Cremer et al. 1988; Gunji et al. 2004; Kaneko et al. 1990; Mirwaldt et al. 1995; Pisabarro et al. 1993).
Essentiality of dapA
Gene knockout studies have shown that dapA is essential in a number of bacterial species, including Salmonella typhimurium (Becker et al. 2006), Bacillus subtilis (Kobayashi et al. 2003), E. coli (Gerdes et al. 2003), Staphylococcus aureus (Forsyth et al. 2002) and Streptococcus pneumoniae (Dogovski et al. 2013). However, a recent study demonstrated that a dapA knockout in Pseudomonas aeruginosa results in no change in bacterial counts or virulence (Kaur et al. 2011). Interestingly, unlike other bacteria, there are four annotated dapA genes in the P. aeruginosa genome, of which two contain all the residues required for DHDPS function. Thus, the essentiality of these putative dapA genes for the survival of this bacterium remains to be elucidated.
The dapA gene has also been shown to be essential to plants. In Arabidopsis thaliana, double dapA gene knockouts results in lethality even after exogenous feeding with lysine (Jones-Held et al. 2012). The single-gene knockouts indicate that dapA1 contributes 30% towards the total DHDPS activity in A. thaliana, whereas dapA2 contributes 70% of the total activity (Jones-Held et al. 2012).
Due to the essentiality of the dapA gene to both bacteria and plants, and its absence in humans, DHDPS has been studied extensively as a target for the development of antibiotics and herbicides (Hutton et al. 2007; Mitsakos et al. 2008). However, the current status of anti-DHDPS inhibitors will not be discussed here.
DHDPS protein
DHDPS activity was first observed in 1965 from E. coli cell lysates (Yugari and Gilvarg 1965) and, five years later, the enzyme was purified to homogeneity (Shedlarski and Gilvarg 1970). Replacement of the substrates with closely related compounds resulted in a significant decrease in activity, suggesting that DHDPS specifically turns over pyruvate and (S)-ASA (Wolterink-van Loo et al. 2008).
Function and regulation
Aldolase family
DHDPS belongs to the class I aldolase sub-family of the (β/α)8-barrel proteins, whose members also include N-acetylneuraminate lyase, trans-o-hydroxybenzylidenepyruvate hydratase-aldolase, D-5-keto-4-deoxyglucarate dehydratase, trans-2′-carboxybenzalpyruvate hydratase aldolase and D-2-keto-3-deoxygluconate aldolases (Aghaie et al. 2008; Barbosa et al. 2000; Gefflaut et al. 1995; Izard et al. 1994; Lawrence et al. 1997; Soares da Costa et al. 2017; Theodossis et al. 2004). These enzymes catalyse different reactions on separate biochemical pathways, but they all have common structural features. It has been proposed that class I aldolases share a unifying step in their reaction pathway, namely the Schiff base formation between a strictly conserved lysine residue and the C2 carbon of the common α-keto acid moiety of the substrate (Fullerton et al. 2006; Gefflaut et al. 1995; Soares da Costa et al. 2015).
Reaction mechanism
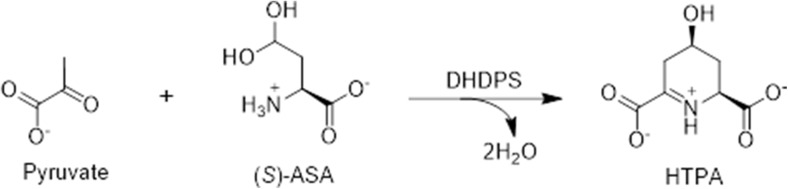
The DHDPS-catalysed reaction proceeds via a typical ping-pong mechanism, in which pyruvate binds to the active site, first forming a covalent enzyme–substrate intermediate, resulting in the release of a protonated water molecule (Blickling et al. 1997b; Dogovski et al. 2009, 2012; Laber et al. 1992). (S)-ASA then binds in the active site and condenses with the bound pyruvate intermediate to form the heterocyclic product, HTPA (Blickling et al. 1997b; Laber et al. 1992) (Fig. 2). Studies performed using isothermal titration calorimetry confirmed that (S)-ASA does not interact with DHDPS in the absence of a Schiff base with pyruvate (Muscroft-Taylor et al. 2010).
Fig. 2.
Schematic of the DHDPS-catalysed reaction. Shown is the condensation of pyruvate and (S)-ASA to form HTPA and water catalysed by DHDPS (EC 4.3.3.7)
The Schiff base formation is initiated by nucleophilic attack of the ε-amino group of a highly conserved lysine residue (Lys161 in E. coli DHDPS) to the keto carbon of pyruvate via a tetrahedral transition state (Laber et al. 1992). A catalytic triad (Thr44, Tyr133 and Tyr107, E. coli numbering) has been proposed to transfer protons to and from the active site through a water-filled channel leading to bulk solvent (Dobson et al. 2004). The Schiff base (imine) is converted to its enamine form to allow (S)-ASA to bind and undergo an aldol-like condensation reaction, leading to cyclisation and the release of the product, HTPA (Laber et al. 1992).
Allosteric regulation
As described in the section entitled DAP pathway, DHDPS represents a key point of regulation in the DAP pathway. This occurs via a classic feedback inhibition mechanism by the final product of the pathway, lysine (Fig. 1). This phenomenon has been investigated in several plants, Gram-negative and Gram-positive bacterial species. In general, plant DHDPS enzymes are highly sensitive to lysine inhibition, with IC 50 values between 10 and 50 μM (Atkinson et al. 2013; Dereppe et al. 1992; Frisch et al. 1991; Griffin et al. 2012; Kumpaisal et al. 1987; Matthews and Widholm 1979; Wallsgrove and Mazelis 1980). It is, therefore, not surprising that lysine is one of the most limiting amino acids in plants (Frizzi et al. 2008; Ufaz and Galili 2008). In contrast, bacterial DHDPS are moderately inhibited by lysine, with IC 50 values ranging from 53 μM to 1 mM (Bakhiet et al. 1984; Christensen et al. 2016; Devenish et al. 2009; Joerger et al. 2003; Laber et al. 1992; Skovpen and Palmer 2013; Soares da Costa et al. 2010; Tam et al. 2004; Yugari and Gilvarg 1965).
Interestingly, not all bacterial DHDPS enzymes are subject to allosteric inhibition (Dogovski et al. 2009, 2012; Soares da Costa et al. 2015). The original accepted dogma suggested that DHDPS from Gram-negative bacteria were inhibited by lysine (Bakhiet et al. 1984; Devenish et al. 2009; Dobson et al. 2005; Joerger et al. 2003; Kaur et al. 2011; Laber et al. 1992; Skovpen and Palmer 2013; Soares da Costa et al. 2010; Tam et al. 2004; Yugari and Gilvarg 1965), whereas those from Gram-positive bacteria were thought to be insensitive to allosteric inhibition (Burgess et al. 2008a; Cahyanto et al. 2006; Cremer et al. 1988; Domigan et al. 2009; Halling and Stahly 1976; Voss et al. 2010; Webster and Lechowich 1970; Yamakura et al. 1974). Recently, this dogma has been dispelled by the discovery that DHDPS from some Gram-negative species, including the enzyme from the pathogen Legionella pneumophila, lack allosteric inhibition by lysine, whilst DHDPS enzymes from some Gram-positive species, including S. pneumoniae, are inhibited by lysine (Soares da Costa et al. 2016). This prompted a re-evaluation of the original dogma and has led to the identification of a key residue at position 56 (E. coli numbering) that defines lysine-mediated allostery in DHDPS (Soares da Costa et al. 2016). The presence of a histidine or glutamate at position 56 imbues allosteric inhibition, whereas the presence of a basic residue results in no inhibition (Soares da Costa et al. 2016).
Structure
The first DHDPS structure was determined by Mirwaldt, Korndorfer and Huber in 1995 (Mirwaldt et al. 1995). Since then, there have been more than 75 structures determined from approximately 25 bacterial species and three plant species [refer to the Protein Data Bank (PDB)]. Most of the DHDPS enzymes exist as a homotetramer in both crystal structure and solution. This section will explore the architectural diversity of DHDPS enzymes, starting with a description of the subunit structure.
TIM-barrel
Each monomer in DHDPS is composed of two distinct domains (Dobson et al. 2005; Mirwaldt et al. 1995). The amino-terminal domain adopts a (β/α)8- or TIM-barrel, with the active site located at the centre of the barrel (Dobson et al. 2005; Mirwaldt et al. 1995). The carboxyl-terminal domain forms three α-helices that contain key residues mediating tetramerisation (Dobson et al. 2005; Mirwaldt et al. 1995).
Active site
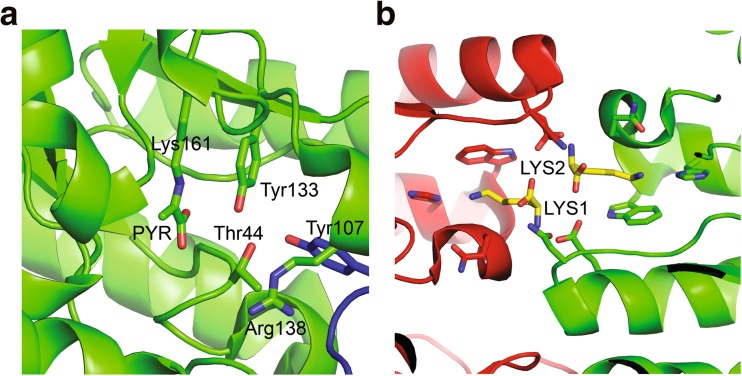
The active site is located in cavities formed by two monomers of the dimer (Mirwaldt et al. 1995) (Fig. 3a). The Schiff base forming Lys161 residue is found within the β-barrel adjacent to the catalytic triad, comprised of the residues Thr44, Tyr107 and Tyr133 (E. coli numbering), that acts as a proton shuttle (Blickling et al. 1997b). The carboxyl group of the bound pyruvate orientates towards Thr44 and Ty133. Thr44 forms a hydrogen bonding network with Tyr133 and Tyr107, which is critical for Schiff base formation and cyclisation (Dobson et al. 2005). Tyr133 acts as a proton donor during Schiff base formation and also accepts a proton while coordinating the amino group of (S)-ASA (Dobson et al. 2005). Tyr107 interdigitates across the dimerisation interface between adjacent subunits to complete the catalytic triad (Dobson et al. 2005). Arg138, located at the entrance to the active site cavity, plays a major role in binding the carboxyl group of (S)-ASA, as demonstrated by mutagenesis studies (Blickling et al. 1997b; Dobson et al. 2005), and also mediates entry of pyruvate into the active site as demonstrated by molecular dynamics simulations (Gordon et al. 2016).
Fig. 3.
Structure of (a) active site and (b) allosteric site of DHDPS. a The active site of pyruvate-bound Vitis vinifera DHDPS [Protein Data Bank (PDB) ID: 3TUU] with labelled residues annotated according to E. coli DHDPS numbering. b The allosteric binding cleft of V. vinifera DHDPS co-crystallised with lysine (PDB ID: 4HNN). Labelled are the two bound lysine ligands that mediate allosteric inhibition of the enzyme
Allosteric site
In the allosteric binding site, two lysine molecules bind in a bis-conformation, with the side chain ε-amino groups projecting away from each other (Dobson et al. 2005) (Fig. 3b). Lysine has been shown to be a partial inhibitor of DHDPS at saturating concentrations (Yugari and Gilvarg 1965). Isothermal titration calorimetry experiments indicate that lysine binding is cooperative, as the second molecule interacts more tightly than the first one (Atkinson et al. 2013; Muscroft-Taylor et al. 2010; Phenix and Palmer 2008).
Structural analyses allow the mapping of key residues forming interactions with the allosteric ligand, namely Ser48, Ala49, His53, His56, Asn80, Glu84 and Tyr106 (E. coli numbering) in bacterial enzymes (Dobson et al. 2005; Soares da Costa et al. 2016), with plant DHDPS commonly harbouring a Trp instead of His at the equivalent of position 53 (Fig. 3b). In most DHDPS crystal structures, there is very little conformational change associated with lysine binding. Therefore, the mechanism of lysine-mediated allosteric inhibition has not been fully understood (Dobson et al. 2005). Recently, studies performed by Atkinson et al. (2013) on Vitis vinifera DHDPS in the presence of lysine has shed some light onto this mechanism (Atkinson et al. 2013). Upon ligand binding, a shift on the hydroxyl group of Tyr131 (Tyr106 in E. coli numbering) is observed towards the carboxyl group of lysine. This re-orientation has been proposed to disrupt the hydrophobic stack this residue makes with the interdigitating catalytic triad Tyr132 (Tyr107 in E. coli numbering) from the adjacent monomer, leading to a disruption of the proton relay (Atkinson et al. 2013).
Dimeric versus tetrameric DHDPS
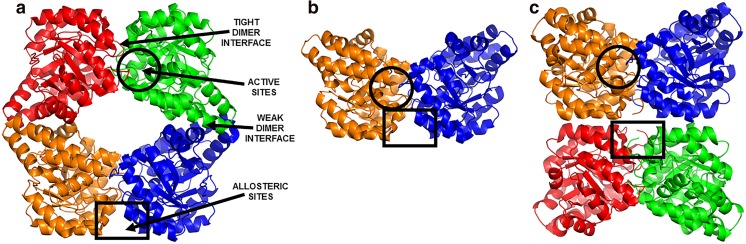
Most DHDPS enzymes from bacteria adopt a tetrameric structure or a dimer-of-dimers, consisting of a tight-dimer interface between two monomers and a weak-dimer interface with hydrogen bonds and non-covalent interactions (Dobson et al. 2005; Griffin et al. 2008, 2010; Perugini et al. 2005) (Fig. 4a). Each of the four monomers has contacts with only two neighbouring monomers, resulting in a large solvent-filled cavity in the centre of the tetramer (Dobson et al. 2005). The active site is located within the (β/α)8-barrel and the allosteric site at the tight-dimer interface, at the top and bottom of the tetramer (Dobson et al. 2005) (Fig. 4a). The buttressing of the two dimers together to form the homotetrameric structure has been proposed to stabilise the tight-dimer interface, including the key active site residues (Griffin et al. 2008, 2010; Voss et al. 2010).
Fig. 4.
Diverse quaternary structures of bacterial and plant DHDPS enzymes. a Bacterial ‘head-to-head’ DHDPS tetramer (PDB ID: 3HIJ), b Staphylococcus aureus DHDPS dimer (PDB ID: 3DAQ) and c plant ‘back-to-back’ DHDPS tetramer (PDB ID: 3TUU)
In E. coli DHDPS, 1400 Å2 of surface area from one monomer in each dimer is buried at the tight-dimer interface (Dobson et al. 2005). This interface is made up of 25 residues from each monomer, with hydrogen bonds formed between Ser111 and Cys141, and hydrophobic interactions between Leu51 and Ala81, among others (Dobson et al. 2005). In addition, Tyr107 of one monomer is coordinated with Tyr106 from the other monomer, interdigitating across the tight-dimer interface and, thus, forming a hydrophobic, sandwich-like stacking of aromatic rings. The importance of Tyr107 at this interface has been demonstrated, with mutations resulting in a monomeric species in equilibrium with the tetramer (Pearce et al. 2008).
The weak-dimer interface associates via two isologous interfaces formed between corresponding monomers (Dobson et al. 2005; Griffin et al. 2008, 2010; Voss et al. 2010). The interface is stabilised by hydrophobic contacts between Leu167, Thr168 and Leu197 (E. coli numbering) (Mirwaldt et al. 1995). The importance of Leu197 at the interface has been demonstrated, with mutations to an aspartate or tyrosine resulting in a dimeric species with reduced enzyme activity and attenuated binding to pyruvate (Griffin et al. 2008). Interestingly, the crystal structures for both mutants were determined with the substrate analogue α-ketoglutarate covalently trapped in the active site, which was not added during the enzyme preparation (Griffin et al. 2008). Small-angle X-ray scattering (SAXS) analysis further confirmed that these mutants were dimers in solutions, but also demonstrated differences between the solution and crystal structures as assessed by an interatomic distance profile comparison (Griffin et al. 2008). It has been proposed that these deviations and a reduction in substrate specificity can be attributed to an increase in ‘breathing motion’ associated with the movement between the subunits in the dimeric enzymes (Griffin et al. 2008). Additional mutations to Asn196, Asp193 and Asn234 in the weak-dimer interface have also been shown to destabilise the tetrameric structure (Griffin et al. 2010).
Interestingly, the DHDPS enzymes from S. aureus (Burgess et al. 2008a) (Fig. 4b) and P. aeruginosa (Kaur et al. 2011) have been crystallised as dimers with enzymatic activity that is equivalent to tetrameric orthologues. Absorbance-detected sedimentation velocity experiments conducted on S. aureus DHDPS indicates that enzyme exists as a 4.2 S dimeric species (Demeler and van Holde 2004; Van Holde and Weischet 1978), compared to the typical 7.0 S tetramer calculated for DHDPS enzymes from B. anthracis (Voss et al. 2010), E. coli (Burgess et al. 2008a) and S. pneumoniae (Dogovski et al. 2013). Additional sedimentation velocity experiments equipped with a fluorescence detection system (Nelson et al. 2016) indicates that Alexa Fluor 488 labelled S. aureus DHDPS in the presence of the substrate, pyruvate, sediments with a s 20,W value of 4.0 S, consistent with the DHDPS dimer (Perugini et al. 2005). Computational analysis of the S. aureus crystal structure using PISA software (Krissinel and Henrick 2007) demonstrates that the dimer interface has a greater proportion of buried surface area and incorporates more non-covalent contacts compared to the tight-dimer interface of tetrameric orthologues (Burgess et al. 2008b). For P. aeruginosa DHDPS, the dimer is yet to be confirmed in solution, but the crystal structure also contains an increased number of interactions at the dimer interface, which correlates to an increase in the buried surface area (Kaur et al. 2011). The data for these two dimeric enzymes indicate that they have evolved an alternative mechanism to overcome increased dynamics in the dimeric unit and, therefore, do not require tetramerisation to increase stability and maintain catalytic activity (Burgess et al. 2008a; Soares da Costa et al. 2015).
Architecture diversity between bacterial and plant DHDPS
The structures of all tetrameric bacterial DHDPS enzymes determined to date have been formed by the buttressing of the two tight dimers in a ‘head-to-head’ configuration (Fig. 4a). As described in the previous section, the active site is located within the (β/α)8-barrel of each monomer and the allosteric site is located at the tight-dimer interface, such that lysine binding sites are found at the top and bottom of the bacterial tetramer (Dobson et al. 2005; Voss et al. 2010) (Fig. 4a). Similarly, plant DHDPS structures are also homotetramers. However, they have an alternative dimer-of-dimers arrangement that can be described as a ‘back-to-back’ conformation (Fig. 4c). In this conformation, the active site is still located within the (β/α)8-barrel, but the allosteric cleft in the tight-dimer interface is located in the interior of the structure (Atkinson et al. 2012b; Blickling et al. 1997a; Griffin et al. 2012) (Fig. 4c). The self-association to form a ‘head-to-head’ or ‘back-to-back’ dimer-of-dimers has been proposed by Griffin et al. (2008, 2010) to be a mechanism of reducing the ‘breathing motion’ at the tight-dimer interface and stabilise the enzyme.
Confirmation of quaternary architectures in solution
Biophysical experiments have been employed to validate the diverse quaternary architectures observed for the DHDPS crystal structures. Atkinson et al. (2012b) initially set out to confirm the ‘back-to-back’ tetrameric conformation observed for the plant orthologues by characterising V. vinifera DHDPS using sedimentation velocity experiments. Two-dimensional spectrum (Brookes et al. 2010) and van Holde–Weischet (Demeler and van Holde 2004; Van Holde and Weischet 1978) analyses yielded a s 20,W value of 7.3 S, indicative of a tetramer in solution (Atkinson et al. 2012b). SAXS analyses show that the scattering profile for plant DHDPS overlayed well with the theoretical profile computed from the crystal structure (χ2 v = 1.5), but a poor fit resulted when the theoretical scattering profile derived from B. anthracis DHDPS structure was overlayed with the experimental data (χ2 v = 7) (Atkinson et al. 2012b). This study provided the first evidence that the ‘back-to-back’ dimer-of-dimers exists in solution and, therefore, cannot be attributed to crystal packing artefacts. To further confirm the differences in quaternary architecture observed between the structures of the bacterial and plant orthologues, scattering data for B. anthracis DHDPS was fitted to both the theoretical profile derived from the crystal structure of the enzyme and also to V. vinifera DHDPS (Atkinson et al. 2012b). Indeed, the experimental data fit more closely to that derived from the theoretical scattering profile of the bacterial crystal structure (χ2 v = 1.2) compared to the plant orthologue (χ2 v = 6.3) (Atkinson et al. 2012b). These data demonstrate that the different DHDPS quaternary architectures are also observed in solution.
Pyruvate stabilisation
Pyruvate has been shown to stabilise the structure of DHDPS enzymes (Burgess et al. 2008a; Gordon et al. 2016; Voss et al. 2010). For B. anthracis DHDPS, sedimentation equilibrium analyses demonstrated that the addition of pyruvate results in stabilisation of the tetrameric form with a 3-fold tighter tetramerisation dissociation constant compared to the apo enzyme (Voss et al. 2010). Utilising PISA analysis (Krissinel and Henrick 2007), comparison of the tetramerisation interface of pyruvate-bound B. anthracis DHDPS structure to the apo form showed a significant increase in the buried surface area at the weak-dimer interface (~ 90 Å2) (Voss et al. 2010). Additionally, the side chains of the active site residues orientate closer to one another, suggesting that pyruvate binding primes the catalytic triad residues for proton relay (Voss et al. 2010).
Similarly, in S. aureus DHDPS, the dimerisation dissociation constant is significantly tighter in the presence of pyruvate (Burgess et al. 2008a). Moreover, sedimentation equilibrium data generated in the AU-FDS were globally fitted to the monomer-dimer model, resulting in a dimerisation dissociation constant of 33 nM for the unliganded enzyme, compared to 1.6 nM in the presence of pyruvate (Burgess et al. 2008a). However, the dimerisation dissociation constant was unchanged in the presence of the other DHDPS substrate, (S)-ASA (Burgess et al. 2008a).
It remains to be seen whether substrate-mediated stabilisation is a more global phenomenon. It is speculated that, similarly to oligomerisation, pyruvate stabilisation has evolved to reduce protein dynamics, thus optimising the activity of DHDPS (Soares da Costa et al. 2015).
Conclusions
Here, we review the biophysical studies that have characterised the structure, function and regulation of the essential bacterial and plant enzyme, dihydrodipicolinate synthase. Studies to date show that, although DHDPS orthologues from bacteria and plants catalyse the same function, they are quite diverse in terms of allosteric regulation and quaternary structure architecture. This has provided insight into the molecular evolution of the enzyme and is informing the development of DHDPS-specific inhibitors as potential antibiotic and/or herbicide agents.
Acknowledgements
We would like to thank the present and past members of the Perugini laboratory for their contributions over the past 15 years and their helpful discussions during the preparation of this review. We also acknowledge the Australian Synchrotron and the La Trobe University—Comprehensive Proteomics Platform for providing infrastructure that has supported the primary research reviewed here, and the Australian Research Council, Defence Threat Reduction Agency, La Trobe University Research Focus Area—Securing Food, Water & the Environment, and National Health & Medical Research Council for contributing to funding over the past 15 years to the research described in this review.
Compliance with ethical standards
Conflict of interest
Tatiana P. Soares da Costa declares that she has no conflict of interest. Belinda M. Abbott declares that she has no conflict of interest. Anthony R. Gendall declares that he has no conflict of interest. Santosh Panjikar declares that he has no conflict of interest. Matthew A. Perugini declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is part of a Special Issue on ‘Biomolecules to Bio-nanomachines - Fumio Arisaka 70th Birthday’ edited by Damien Hall.
References
- Aghaie A, Lechaplais C, Sirven P, et al. New insights into the alternative D-glucarate degradation pathway. J Biol Chem. 2008;283:15638–15646. doi: 10.1074/jbc.M800487200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson SC, Dogovski C, Newman J, Dobson RC, Perugini MA. Cloning, expression, purification and crystallization of dihydrodipicolinate synthase from the grapevine Vitis vinifera. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:1537–1541. doi: 10.1107/S1744309111038395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson SC, Dogovski C, Dobson RC, Perugini MA. Cloning, expression, purification and crystallization of dihydrodipicolinate synthase from Agrobacterium tumefaciens. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68:1040–1047. doi: 10.1107/S1744309112033052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson SC, Dogovski C, Downton MT, et al. Crystal, solution and in silico structural studies of dihydrodipicolinate synthase from the common grapevine. PLoS One. 2012;7:e38318. doi: 10.1371/journal.pone.0038318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson SC, Dogovski C, Downton MT, et al. Structural, kinetic and computational investigation of Vitis vinifera DHDPS reveals new insight into the mechanism of lysine-mediated allosteric inhibition. Plant Mol Biol. 2013;81:431–446. doi: 10.1007/s11103-013-0014-7. [DOI] [PubMed] [Google Scholar]
- Atkinson SC, Hor L, Dogovski C, Dobson RC, Perugini MA. Identification of the bona fide DHDPS from a common plant pathogen. Proteins. 2014;82:1869–1883. doi: 10.1002/prot.24539. [DOI] [PubMed] [Google Scholar]
- Bakhiet N, Forney FW, Stahly DP, Daniels L. Lysine biosynthesis in Methanobacterium thermoautotrophicum is by the diaminopimelic acid pathway. Curr Microbiol. 1984;10:195–198. doi: 10.1007/BF01627254. [DOI] [Google Scholar]
- Barbosa JA, Smith BJ, DeGori R, et al. Active site modulation in the N-acetylneuraminate lyase sub-family as revealed by the structure of the inhibitor-complexed Haemophilus influenzae enzyme. J Mol Biol. 2000;303:405–421. doi: 10.1006/jmbi.2000.4138. [DOI] [PubMed] [Google Scholar]
- Becker D, Selbach M, Rollenhagen C, et al. Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature. 2006;440:303–307. doi: 10.1038/nature04616. [DOI] [PubMed] [Google Scholar]
- Blickling S, Beisel HG, Bozic D, Knäblein J, Laber B, Huber R. Structure of dihydrodipicolinate synthase of Nicotiana sylvestris reveals novel quaternary structure. J Mol Biol. 1997;274:608–621. doi: 10.1006/jmbi.1997.1393. [DOI] [PubMed] [Google Scholar]
- Blickling S, Renner C, Laber B, Pohlenz HD, Holak TA, Huber R. Reaction mechanism of Escherichia coli dihydrodipicolinate synthase investigated by X-ray crystallography and NMR spectroscopy. Biochemistry. 1997;36:24–33. doi: 10.1021/bi962272d. [DOI] [PubMed] [Google Scholar]
- Brookes E, Cao W, Demeler B. A two-dimensional spectrum analysis for sedimentation velocity experiments of mixtures with heterogeneity in molecular weight and shape. Eur Biophys J. 2010;39:405–414. doi: 10.1007/s00249-009-0413-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari AI, Taylor AL. Genetic analysis of diaminopimelic acid- and lysine-requiring mutants of Escherichia coli. J Bacteriol. 1971;105:844–854. doi: 10.1128/jb.105.3.844-854.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess BR, Dobson RC, Bailey MF, et al. Structure and evolution of a novel dimeric enzyme from a clinically important bacterial pathogen. J Biol Chem. 2008;283:27598–27603. doi: 10.1074/jbc.M804231200. [DOI] [PubMed] [Google Scholar]
- Burgess BR, Dobson RC, Dogovski C, Jameson GB, Parker MW, Perugini MA. Purification, crystallization and preliminary X-ray diffraction studies to near-atomic resolution of dihydrodipicolinate synthase from methicillin-resistant Staphylococcus aureus. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64:659–661. doi: 10.1107/S1744309108016746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butour JL, Felenbok B, Patte JC. Synthesis of dihydrodipicolinate synthetase in Escherichia coli K12. Ann Microbiol. 1974;125:459–462. [PubMed] [Google Scholar]
- Cahyanto MN, Kawasaki H, Nagashio M, Fujiyama K, Seki T. Regulation of aspartokinase, aspartate semialdehyde dehydrogenase, dihydrodipicolinate synthase and dihydrodipicolinate reductase in Lactobacillus plantarum. Microbiology. 2006;152:105–112. doi: 10.1099/mic.0.28092-0. [DOI] [PubMed] [Google Scholar]
- Chen NY, Jiang SQ, Klein DA, Paulus H. Organization and nucleotide sequence of the Bacillus subtilis diaminopimelate operon, a cluster of genes encoding the first three enzymes of diaminopimelate synthesis and dipicolinate synthase. J Biol Chem. 1993;268:9448–9465. [PubMed] [Google Scholar]
- Christensen JB, Soares da Costa TP, Faou P, Pearce FG, Panjikar S, Perugini MA. Structure and function of cyanobacterial DHDPS and DHDPR. Sci Rep. 2016;6:37111. doi: 10.1038/srep37111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer J, Treptow C, Eggeling L, Sahm H. Regulation of enzymes of lysine biosynthesis in Corynebacterium glutamicum. Microbiol. 1988;134:3221–3229. doi: 10.1099/00221287-134-12-3221. [DOI] [PubMed] [Google Scholar]
- Demeler B, van Holde KE. Sedimentation velocity analysis of highly heterogeneous systems. Anal Biochem. 2004;335:279–288. doi: 10.1016/j.ab.2004.08.039. [DOI] [PubMed] [Google Scholar]
- Dereppe C, Bold G, Ghisalba O, Ebert E, Schär HP. Purification and characterization of dihydrodipicolinate synthase from pea. Plant Physiol. 1992;98:813–821. doi: 10.1104/pp.98.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devenish SR, Huisman FH, Parker EJ, Hadfield AT, Gerrard JA. Cloning and characterisation of dihydrodipicolinate synthase from the pathogen Neisseria meningitidis. Biochim Biophys Acta. 2009;1794:1168–1174. doi: 10.1016/j.bbapap.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Dobson RC, Griffin MD, Roberts SJ, Gerrard JA. Dihydrodipicolinate synthase (DHDPS) from Escherichia coli displays partial mixed inhibition with respect to its first substrate, pyruvate. Biochimie. 2004;86:311–315. doi: 10.1016/j.biochi.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Dobson RC, Griffin MD, Jameson GB, Gerrard JA. The crystal structures of native and (S)-lysine-bound dihydrodipicolinate synthase from Escherichia coli with improved resolution show new features of biological significance. Acta Crystallogr D Biol Crystallogr. 2005;61:1116–1124. doi: 10.1107/S0907444905016318. [DOI] [PubMed] [Google Scholar]
- Dogovski C, Atkinson SC, Dommaraju SR, et al. Lysine biosynthesis in bacteria: an unchartered pathway for novel antibiotic design. Encycl Life Supp Syst. 2009;11:116–136. [Google Scholar]
- Dogovski C, Atkinson SC, Dommaraju SR et al (2012) Enzymology of bacterial lysine biosynthesis. In: Ekinci D (ed) Biochemistry. InTech Open Access Publisher, pp 225–262
- Dogovski C, Gorman MA, Ketaren NE, et al. From knock-out phenotype to three-dimensional structure of a promising antibiotic target from Streptococcus pneumoniae. PLoS One. 2013;8:e83419. doi: 10.1371/journal.pone.0083419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domigan LJ, Scally SW, Fogg MJ, et al. Characterisation of dihydrodipicolinate synthase (DHDPS) from Bacillus anthracis. Biochim Biophys Acta. 2009;1794:1510–1516. doi: 10.1016/j.bbapap.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Dommaraju S, Gorman MA, Dogovski C, et al. Cloning, expression and crystallization of dihydrodipicolinate reductase from methicillin-resistant Staphylococcus aureus. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:57–60. doi: 10.1107/S1744309109047964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dommaraju SR, Dogovski C, Czabotar PE, Hor L, Smith BJ, Perugini MA. Catalytic mechanism and cofactor preference of dihydrodipicolinate reductase from methicillin-resistant Staphylococcus aureus. Arch Biochem Biophys. 2011;512:167–174. doi: 10.1016/j.abb.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Evans G, Schuldt L, Griffin MD, et al. A tetrameric structure is not essential for activity in dihydrodipicolinate synthase (DHDPS) from Mycobacterium tuberculosis. Arch Biochem Biophys. 2011;512:154–159. doi: 10.1016/j.abb.2011.05.014. [DOI] [PubMed] [Google Scholar]
- Forsyth RA, Haselbeck RJ, Ohlsen KL, et al. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol. 2002;43:1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- Frisch DA, Gengenbach BG, Tommey AM, Sellner JM, Somers DA, Myers DE. Isolation and characterization of dihydrodipicolinate synthase from maize. Plant Physiol. 1991;96:444–452. doi: 10.1104/pp.96.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frizzi A, Huang S, Gilbertson LA, Armstrong TA, Luethy MH, Malvar TM. Modifying lysine biosynthesis and catabolism in corn with a single bifunctional expression/silencing transgene cassette. Plant Biotechnol J. 2008;6:13–21. doi: 10.1111/j.1467-7652.2007.00290.x. [DOI] [PubMed] [Google Scholar]
- Fullerton SW, Griffiths JS, Merkel AB, et al. Mechanism of the Class I KDPG aldolase. Bioorg Med Chem. 2006;14:3002–3010. doi: 10.1016/j.bmc.2005.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Rodríguez FM, Zekri S, Toro N. Characterization of the Sinorhizobium meliloti genes encoding a functional dihydrodipicolinate synthase (dapA) and dihydrodipicolinate reductase (dapB) Arch Microbiol. 2000;173:438–444. doi: 10.1007/s002030000169. [DOI] [PubMed] [Google Scholar]
- Gefflaut T, Blonski C, Perie J, Willson M. Class I aldolases: substrate specificity, mechanism, inhibitors and structural aspects. Prog Biophys Mol Biol. 1995;63:301–340. doi: 10.1016/0079-6107(95)00008-9. [DOI] [PubMed] [Google Scholar]
- Gerdes SY, Scholle MD, Campbell JW, et al. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M, Frankard V, Jacobs M. A dinucleotide mutation in dihydrodipicolinate synthase of Nicotiana sylvestris leads to lysine overproduction. Plant J. 1995;8:733–743. doi: 10.1046/j.1365-313X.1995.08050733.x. [DOI] [PubMed] [Google Scholar]
- Girish TS, Sharma E, Gopal B. Structural and functional characterization of Staphylococcus aureus dihydrodipicolinate synthase. FEBS Lett. 2008;582:2923–2930. doi: 10.1016/j.febslet.2008.07.035. [DOI] [PubMed] [Google Scholar]
- Gordon SE, Weber DK, Downton MT, Wagner J, Perugini MA. Dynamic modelling reveals ‘hotspots’ on the pathway to enzyme–substrate complex formation. PLoS Comput Biol. 2016;12:e1004811. doi: 10.1371/journal.pcbi.1004811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin MD, Dobson RC, Pearce FG, et al. Evolution of quaternary structure in a homotetrameric enzyme. J Mol Biol. 2008;380:691–703. doi: 10.1016/j.jmb.2008.05.038. [DOI] [PubMed] [Google Scholar]
- Griffin MD, Dobson RC, Gerrard JA, Perugini MA. Exploring the dihydrodipicolinate synthase tetramer: how resilient is the dimer–dimer interface? Arch Biochem Biophys. 2010;494:58–63. doi: 10.1016/j.abb.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Griffin MD, Billakanti JM, Wason A, et al. Characterisation of the first enzymes committed to lysine biosynthesis in Arabidopsis thaliana. PLoS One. 2012;7:e40318. doi: 10.1371/journal.pone.0040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunji Y, Tsujimoto N, Shimaoka M, Ogawa-Miyata Y, Sugimoto S, Yasueda H. Characterization of the L-lysine biosynthetic pathway in the obligate methylotroph Methylophilus methylotrophus. Biosci Biotechnol Biochem. 2004;68:1449–1460. doi: 10.1271/bbb.68.1449. [DOI] [PubMed] [Google Scholar]
- Halling SM, Stahly DP. Dihydrodipicolinic acid synthase of bacillus licheniformis. Quaternary structure, kinetics, and stability in the presence of sodium chloride and substrates. Biochim Biophys Acta. 1976;452:580–596. doi: 10.1016/0005-2744(76)90209-6. [DOI] [PubMed] [Google Scholar]
- Hudson AO, Bless C, Macedo P, Chatterjee SP, Singh BK, Gilvarg C, Leustek T. Biosynthesis of lysine in plants: evidence for a variant of the known bacterial pathways. Biochim Biophys Acta. 2005;1721:27–36. doi: 10.1016/j.bbagen.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Hutton CA, Perugini MA, Gerrard JA. Inhibition of lysine biosynthesis: an evolving antibiotic strategy. Mol BioSyst. 2007;3:458–465. doi: 10.1039/b705624a. [DOI] [PubMed] [Google Scholar]
- Izard T, Lawrence MC, Malby RL, Lilley GG, Colman PM. The three-dimensional structure of N-acetylneuraminate lyase from Escherichia coli. Structure. 1994;2:361–369. doi: 10.1016/S0969-2126(00)00038-1. [DOI] [PubMed] [Google Scholar]
- Joerger AC, Mayer S, Fersht AR. Mimicking natural evolution in vitro: an N-acetylneuraminate lyase mutant with an increased dihydrodipicolinate synthase activity. Proc Natl Acad Sci U S A. 2003;100:5694–5699. doi: 10.1073/pnas.0531477100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Held S, Ambrozevicius LP, Campbell M, Drumheller B, Harrington E, Leustek T. Two Arabidopsis thaliana dihydrodipicolinate synthases, DHDPS1 and DHDPS2, are unequally redundant. Funct Plant Biol. 2012;39:1058–1067. doi: 10.1071/FP12169. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Hashimoto T, Kumpaisal R, Yamada Y. Molecular cloning of wheat dihydrodipicolinate synthase. J Biol Chem. 1990;265:17451–17455. [PubMed] [Google Scholar]
- Kaur N, Gautam A, Kumar S, et al. Biochemical studies and crystal structure determination of dihydrodipicolinate synthase from Pseudomonas aeruginosa. Int J Biol Macromol. 2011;48:779–787. doi: 10.1016/j.ijbiomac.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Ehrlich SD, Albertini A, et al. Essential Bacillus subtilis genes. Proc Natl Acad Sci U S A. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Kumpaisal R, Hashimoto T, Yamada Y. Purification and characterization of dihydrodipicolinate synthase from wheat suspension cultures. Plant Physiol. 1987;85:145–151. doi: 10.1104/pp.85.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laber B, Gomis-Rüth FX, Romao MJ, Huber R. Escherichia coli dihydrodipicolinate synthase. Identification of the active site and crystallization. Biochem J. 1992;288(Pt 2):691–695. doi: 10.1042/bj2880691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MC, Barbosa JA, Smith BJ, et al. Structure and mechanism of a sub-family of enzymes related to N-acetylneuraminate lyase. J Mol Biol. 1997;266:381–399. doi: 10.1006/jmbi.1996.0769. [DOI] [PubMed] [Google Scholar]
- Matthews BF, Widholm JM. Expression of aspartokinase, dihydrodipicolinic acid synthase and homoserine dehydrogenase during growth of carrot cell suspension cultures on lysine- and threonine-supplemented media. Z Naturforsch C Biosci. 1979;34:1177–1185. doi: 10.1515/znc-1979-1216. [DOI] [PubMed] [Google Scholar]
- Mirwaldt C, Korndorfer I, Huber R. The crystal structure of dihydrodipicolinate synthase from Escherichia coli at 2.5 Å resolution. J Mol Biol. 1995;246:227–239. doi: 10.1006/jmbi.1994.0078. [DOI] [PubMed] [Google Scholar]
- Mitsakos V, Dobson RC, Pearce FG, et al. Inhibiting dihydrodipicolinate synthase across species: towards specificity for pathogens? Bioorg Med Chem Lett. 2008;18:842–844. doi: 10.1016/j.bmcl.2007.11.026. [DOI] [PubMed] [Google Scholar]
- Miyazaki T, Miyazaki J, Yamane H, Nishiyama M. α-Aminoadipate aminotransferase from an extremely thermophilic bacterium, Thermus thermophilus. Microbiology. 2004;150:2327–2334. doi: 10.1099/mic.0.27037-0. [DOI] [PubMed] [Google Scholar]
- Muscroft-Taylor AC, Soares da Costa TP, Gerrard JA. New insights into the mechanism of dihydrodipicolinate synthase using isothermal titration calorimetry. Biochimie. 2010;92:254–262. doi: 10.1016/j.biochi.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Nelson TG, Ramsay GD, Perugini MA (2016) Fluorescence-detection system. In: Uchiyama S, Arisaka F, Stafford WF, Laue TM (eds) Analytical ultracentrifugation: instrumentation, software, and application. Springer, Tokyo, pp 39-61
- Panchy N, Lehti-Shiu M, Shiu SH. Evolution of gene duplication in plants. Plant Physiol. 2016;171:2294–2316. doi: 10.1104/pp.16.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce FG, Dobson RC, Weber A, et al. Mutating the tight-dimer interface of dihydrodipicolinate synthase disrupts the enzyme quaternary structure: toward a monomeric enzyme. Biochemistry. 2008;47:12108–12117. doi: 10.1021/bi801094t. [DOI] [PubMed] [Google Scholar]
- Perugini MA, Griffin MD, Smith BJ, et al. Insight into the self-association of key enzymes from pathogenic species. Eur Biophys J. 2005;34:469–476. doi: 10.1007/s00249-005-0491-y. [DOI] [PubMed] [Google Scholar]
- Peverelli MG, Soares da Costa TP, Kirby N, Perugini MA. Dimerization of bacterial diaminopimelate decarboxylase is essential for catalysis. J Biol Chem. 2016;291:9785–9795. doi: 10.1074/jbc.M115.696591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phenix CP, Palmer DR. Isothermal titration microcalorimetry reveals the cooperative and noncompetitive nature of inhibition of Sinorhizobium meliloti L5-30 dihydrodipicolinate synthase by (S)-lysine. Biochemistry. 2008;47:7779–7781. doi: 10.1021/bi800629n. [DOI] [PubMed] [Google Scholar]
- Pisabarro A, Malumbres M, Mateos LM, Oguiza JA, Martin JF. A cluster of three genes (dapA, orf2, and dapB) of Brevibacterium lactofermentum encodes dihydrodipicolinate synthase, dihydrodipicolinate reductase, and a third polypeptide of unknown function. J Bacteriol. 1993;175:2743–2749. doi: 10.1128/jb.175.9.2743-2749.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray SS, Bonanno JB, Rajashankar KR, et al. Cocrystal structures of diaminopimelate decarboxylase: mechanism, evolution, and inhibition of an antibiotic resistance accessory factor. Structure. 2002;10:1499–1508. doi: 10.1016/S0969-2126(02)00880-8. [DOI] [PubMed] [Google Scholar]
- Richaud F, Richaud C, Ratet P, Patte JC. Chromosomal location and nucleotide sequence of the Escherichia coli dapA gene. J Bacteriol. 1986;166:297–300. doi: 10.1128/jb.166.1.297-300.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedlarski JG, Gilvarg C. The pyruvate-aspartic semialdehyde condensing enzyme of Escherichia coli. J Biol Chem. 1970;245:1362–1373. [PubMed] [Google Scholar]
- Siddiqui T, Paxman JJ, Dogovski C, Panjikar S, Perugini MA. Cloning to crystallization of dihydrodipicolinate synthase from the intracellular pathogen Legionella pneumophila. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69:1177–1181. doi: 10.1107/S1744309113024639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk GW, Matthews BF, Somers DA, Gengenbach BG. Cloning and expression of the soybean DapA gene encoding dihydrodipicolinate synthase. Plant Mol Biol. 1994;26:989–993. doi: 10.1007/BF00028865. [DOI] [PubMed] [Google Scholar]
- Skovpen YV, Palmer DR. Dihydrodipicolinate synthase from Campylobacter jejuni: kinetic mechanism of cooperative allosteric inhibition and inhibitor-induced substrate cooperativity. Biochemistry. 2013;52:5454–5462. doi: 10.1021/bi400693w. [DOI] [PubMed] [Google Scholar]
- Soares da Costa TP, Muscroft-Taylor AC, Dobson RC, Devenish SR, Jameson GB, Gerrard JA. How essential is the ‘essential’ active-site lysine in dihydrodipicolinate synthase? Biochimie. 2010;92:837–845. doi: 10.1016/j.biochi.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Soares da Costa TP, Christensen JB, Desbois S, et al. Quaternary structure analyses of an essential oligomeric enzyme. Methods Enzymol. 2015;562:205–223. doi: 10.1016/bs.mie.2015.06.020. [DOI] [PubMed] [Google Scholar]
- Soares da Costa TP, Desbois S, Dogovski C, et al. Structural determinants defining the allosteric inhibition of an essential antibiotic target. Structure. 2016;24:1282–1291. doi: 10.1016/j.str.2016.05.019. [DOI] [PubMed] [Google Scholar]
- Soares da Costa TP, Patel M, Desbois S, Gupta R, Faou P, Perugini MA. Identification of a dimeric KDG aldolase from Agrobacterium tumefaciens. Proteins. 2017;85:2058–2065. doi: 10.1002/prot.25359. [DOI] [PubMed] [Google Scholar]
- Tam PH, Phenix CP, Palmer DR. MosA, a protein implicated in rhizopine biosynthesis in Sinorhizobium meliloti L5-30, is a dihydrodipicolinate synthase. J Mol Biol. 2004;335:393–397. doi: 10.1016/j.jmb.2003.10.063. [DOI] [PubMed] [Google Scholar]
- Theodossis A, Walden H, Westwick EJ, et al. The structural basis for substrate promiscuity in 2-keto-3-deoxygluconate aldolase from the Entner–Doudoroff pathway in Sulfolobus solfataricus. J Biol Chem. 2004;279:43886–43892. doi: 10.1074/jbc.M407702200. [DOI] [PubMed] [Google Scholar]
- Torruella G, Suga H, Riutort M, Peretó J, Ruiz-Trillo I. The evolutionary history of lysine biosynthesis pathways within eukaryotes. J Mol Evol. 2009;69:240–248. doi: 10.1007/s00239-009-9266-x. [DOI] [PubMed] [Google Scholar]
- Ufaz S, Galili G. Improving the content of essential amino acids in crop plants: goals and opportunities. Plant Physiol. 2008;147:954–961. doi: 10.1104/pp.108.118091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Holde KE, Weischet WO. Boundary analysis of sedimentation-velocity experiments with monodisperse and paucidisperse solutes. Biopolymers. 1978;17:1387–1403. doi: 10.1002/bip.1978.360170602. [DOI] [Google Scholar]
- Vauterin M, Jacobs M. Isolation of a poplar and an Arabidopsis thaliana dihydrodipicolinate synthase cDNA clone. Plant Mol Biol. 1994;25:545–550. doi: 10.1007/BF00043882. [DOI] [PubMed] [Google Scholar]
- Velasco AM, Leguina JI, Lazcano A. Molecular evolution of the lysine biosynthetic pathways. J Mol Evol. 2002;55:445–449. doi: 10.1007/s00239-002-2340-2. [DOI] [PubMed] [Google Scholar]
- Voss JE, Scally SW, Taylor NL, et al. Substrate-mediated stabilization of a tetrameric drug target reveals Achilles heel in anthrax. J Biol Chem. 2010;285:5188–5195. doi: 10.1074/jbc.M109.038166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallsgrove RM, Mazelis M. The enzymology of lysine biosynthesis in higher plants: complete localization of the regulatory enzyme dihydrodipicolinate synthase in the chloroplasts of spinach leaves. FEBS Lett. 1980;116:189–192. doi: 10.1016/0014-5793(80)80640-5. [DOI] [PubMed] [Google Scholar]
- Webster FH, Lechowich RV. Partial purification and characterization of dihydrodipicolinic acid synthetase from sporulating Bacillus megaterium. J Bacteriol. 1970;101:118–126. doi: 10.1128/jb.101.1.118-126.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolterink-van Loo S, Levisson M, Cabrieres MC, Franssen MC, van der Oost J. Characterization of a thermostable dihydrodipicolinate synthase from Thermoanaerobacter tengcongensis. Extremophiles. 2008;12:461–469. doi: 10.1007/s00792-008-0152-z. [DOI] [PubMed] [Google Scholar]
- Wubben JM, Dogovski C, Dobson RC, et al. Cloning, expression, purification and crystallization of dihydrodipicolinate synthase from the psychrophile Shewanella benthica. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:1511–1516. doi: 10.1107/S1744309110036791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Andi B, Qian J, West AH, Cook PF. The α-aminoadipate pathway for lysine biosynthesis in fungi. Cell Biochem Biophys. 2006;46:43–64. doi: 10.1385/CBB:46:1:43. [DOI] [PubMed] [Google Scholar]
- Yamakura F, Ikeda Y, Kimura K, Sasakawa T. Partial purification and some properties of pyruvate-aspartic semialdehyde condensing enzyme from sporulating Bacillus subtilis. J Biochem. 1974;76:611–621. doi: 10.1093/oxfordjournals.jbchem.a130605. [DOI] [PubMed] [Google Scholar]
- Yugari Y, Gilvarg C. The condensation step in diaminopimelate synthesis. J Biol Chem. 1965;240:4710–4716. [PubMed] [Google Scholar]