Abstract
Viruses have developed intricate molecular machines to infect, replicate within and escape from their host cells. Perhaps one of the most intriguing of these mechanisms is the pyramidal egress structure that has evolved in archaeal viruses, such as SIRV2 or STIV1. The structure and mechanism of these virus-associated pyramids (VAPs) has been studied by cryo-electron tomography and complementary biochemical techniques, revealing that VAPs are formed by multiple copies of a virus-encoded 10-kDa protein (PVAP) that integrate into the cell membrane and assemble into hollow, sevenfold symmetric pyramids. In this process, growing VAPs puncture the protective surface layer and ultimately open to release newly replicated viral particles into the surrounding medium. PVAP has the striking capability to spontaneously integrate and self-assemble into VAPs in biological membranes of the archaea, bacteria and eukaryotes. This renders the VAP a universal membrane remodelling system. In this review, we provide an overview of the VAP structure and assembly mechanism and discuss the possible use of VAPs in nano-biotechnology.
Keywords: Archaeal virus, Nanostructure, Archaea, Sulfolobus, Cryo-electron tomography, Virion egress
Introduction
Members of all domains of life, archaea, bacteria and eukaryotes, are infected by viruses. A hallmark of archaeal viruses is the high morphological diversity of the capsids that enclose their genetic material (Prangishvili et al. 2006; Pina et al. 2011; Prangishvili 2013; Dellas et al. 2014). Even though viruses have only been isolated from a limited set of archaeal species, the high structural diversity of those isolated has led to the description of several new viral families (Pina et al. 2011; Krupovic et al. 2016; Adriaenssens et al. 2017). For this reason, it is anticipated that the isolation of new viruses from other species will result in the discovery of novel, as yet unknown virion (viral particle) structures. Archaeal viruses display unique structures that are not encountered among bacterial or eukaryotic viruses, including spindle, two tailed, egg, bacilliform, spiral or even bottle-like shapes (Schleper et al. 1992; Haring et al. 2005; Häring et al. 2005; Mochizuki et al. 2010, 2011, 2012). In recent years, cryo-electron microscopy (cryoEM) has been employed to study the detailed three-dimensional (3D) structure of a range of archaeal virions (Hong et al. 2015; DiMaio et al. 2015; Kasson et al. 2017). The many unusual virion structures of archaeal viruses are a rich illustration of the diversity of solutions for the same purpose that can evolve in nature. Archaeal virions usually consist of one or a limited set of capsid proteins. The major capsid protein can sometimes self-assemble into (a part of) the virion (Vestergaard et al. 2008). While many archaeal hosts have evolved a highly robust machinery to thrive under harsh conditions, such as extreme temperature, pH and salinity, their virions have co-evolved to adopt equally stable properties (Witte et al. 1997; Porter et al. 2005; Prangishvili 2006; Pina et al. 2011). These remarkable characteristics make archaeal viruses attractive subjects for structural biology research. Owing to their unique stability, proteins derived from archaeal viruses are generally easy to purify. For example, the heat-stable proteins of several archaeal viruses can be heterologously produced in Escherichia coli, and the removal of contaminant proteins is facilitated by a heat step to denature and remove native bacterial proteins (Larson et al. 2006; Guillière et al. 2009).
The unusual capsid structures of archaeal viruses underpin very surprising infection pathways. An especially striking example is the virion release (egress) mechanism shared by the STIV1 (Sulfolobus turreted icosahedral virus 1) and SIRV2 (Sulfolobus islandicus rod-shaped virus 2). Both viruses infect members of the hyperthermophilic order Sulfolobales that grow optimally in hot acidic environments (~ 75 °C, pH 3) (Bize et al. 2009; Brumfield et al. 2009), are lytic and are released from the cell via remarkable pyramidal egress structures that allow the formation of defined apertures in the cell membrane (Bize et al. 2009; Brumfield et al. 2009; Prangishvili and Quax 2011). These virus-associated pyramids (VAPs) display sevenfold symmetry and form stable assemblies that can be isolated from the cell (Quax et al. 2011). The expression of a single viral membrane protein is the only requirement for the formation of these pyramids, which can reach hundreds of nanometres in diameter (Quax et al. 2011; Snyder et al. 2011). Overexpression of this protein in archaeal, bacterial and eukaryotic model organisms has shown that self-assembly of the pyramidal structures is independent of the origin and type of the lipid membrane (Daum et al. 2014). As such, the VAPs represent a universal membrane remodelling system with potential nano-biotechnological applications. In this short review, we introduce the basic principles of the VAP-based egress mechanism and discuss its diversity in nature as well as its potential as an assembly mechanism. In addition, we highlight several applications in synthetic biology where VAPs could be employed.
Biological function of VAPs
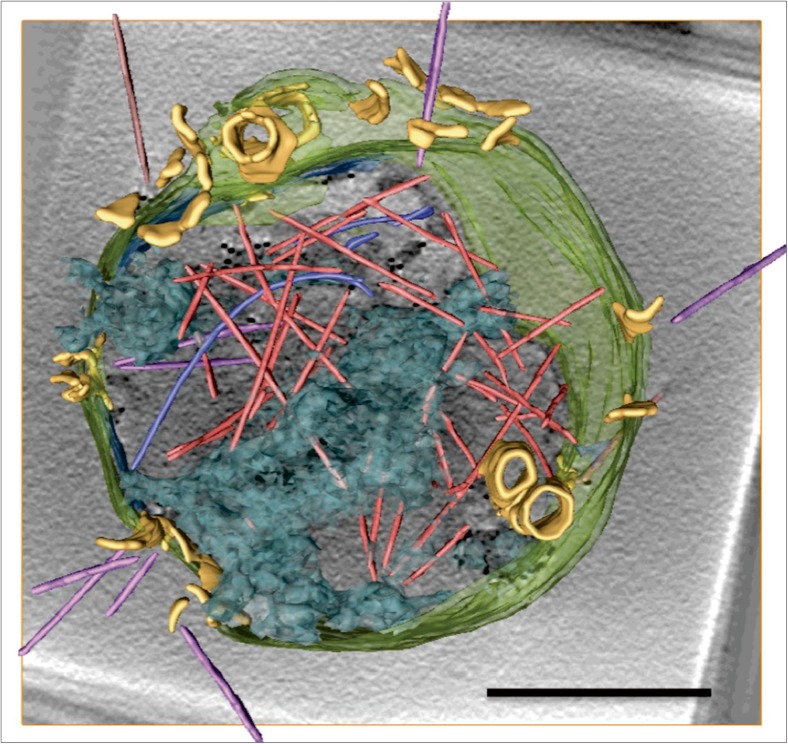
Virus-associated pyramids are the central feature of the egress mechanism of at least two archaeal viruses: SIRV2 and STIV1 (Bize et al. 2009; Brumfield et al. 2009). SIRV2 is member of the family Rudiviridae and its linear double-stranded (ds)DNA genome is wrapped in a rod-shaped capsid of ~ 900 nm in length and 23 nm in diameter (Zillig et al. 1993; Prangishvili et al. 1999; Peng et al. 2001; DiMaio et al. 2015). A high-resolution cryoEM study of the SIRV2 virion showed that the genomic DNA is uniquely packaged in A-form (DiMaio et al. 2015). The virions are not enveloped in a lipid membrane and possesses three tail fibres on each end of the capsid, which bind filamentous appendages on the cell surface of its host S. islandicus to aid the viral entry process (Quemin et al. 2013; Quemin and Quax 2015). The turreted icosahedral particles of STIV1 enwrap a circular dsDNA genome (Rice et al. 2004; Maaty et al. 2006). This member of the family Turriviridae contains an inner lipid layer and infects Sulfolobales solfataricus (Maaty et al. 2006). The structure, genomic organisation and content are distinct between the two viruses and, therefore, the replication processes are quite different. However, infection of both viruses results in the production of about a dozen VAPs on the surface of the host cell towards the second half of the infection cycle [from 3 to 9 h (SIRV2) and from 24 to 36 h (STIV1) post infection] (Bize et al. 2009; Brumfield et al. . 2009). This process is concomitant with the replication and production of approximately 50–150 new virions in the cytoplasm of the cell (Bize et al. 2009; Brumfield et al. 2009; Fu et al. 2010). As these VAPs are hollow and not closed at their base, their interior is continuous with the cytoplasm of the infected cell. Growing outward, their tips eventually penetrate the S-layer covering the cell (Bize et al. 2009; Brumfield et al. 2009). Towards the end of the infection cycle, when the virions inside the cell have matured, all VAPs open approximately at the same stage of infection, much like the petals of a flower (Fig. 1) (Fu et al. 2010; Quax et al. 2011; Daum et al. 2014). This results in apertures of ~ 150 nm in diameter, through which the virions can leave the cell (Fig. 1). After releasing the virions, the VAPs remain open, thus causing complete lysis of host cell (Bize et al. 2009; Brumfield et al. 2009; Fu et al. 2010; Daum et al. 2014).
Fig. 1.
CryoET of a Sulfolobus islandicus cell infected by SIRV2. Three-dimensional surface representation showing multiple virions in the cytoplasm (light red), in the process of egress (magenta) and outside the cell (brown). VAPs are yellow, DNA is shown in transparent blue and the cell envelope is transparent green. Adapted with permission from Daum et al. (2014). Scale bar 500 nm
Comparison of the membrane protein content of infected and non-infected cells led to the discovery of a ~ 10-kDa virus-encoded protein forming VAPs (PVAP), which has subsequently been identified as the sole constituent of the VAPs (Quax et al. 2010). Indeed, overexpression of SIRV2 or STIV1-derived PVAP in the archaea Sulfolobus acidocaldarius, S. solfataricus, the bacterium E. coli and the eukaryote Saccharomyces cerevisiae resulted in VAPs in all membranes (Snyder et al. 2011; Daum et al. 2014). In S. solfataricus, opening of the VAPs occurred after heterologous expression of both STIV1- and SIRV2-derived PVAP. However, VAPs that had been expressed heterologously in E. coli never opened, even after incubating of the cells for up to 2 weeks at room temperature (Daum et al. 2014). This indicates that VAP opening is not a spontaneous, independent process and suggests that a specific factor may have to be present to trigger the unfolding mechanism.
VAP diversity
Interestingly, STIV1 and SIRV2 share only a handful of homologous proteins, of which PVAP is one (Peng et al. 2001; Rice et al. 2004). Expression of STIV1- and SIRV2-based PVAP chimeras in the STIV1 infection system showed that both proteins and all chimeras resulted in the formation of VAPs of the same size and geometry (Snyder et al. 2013a). However, neither the chimeras nor SIRV2_PVAP were sufficient to support STIV1 infection, suggesting that the two proteins are not completely interchangeable. One possibility is that the two proteins have different capabilities to bind interaction partners. For example, host-encoded ESCRT (Endosomal Sorting Complex Required for Transport) homologues are involved in the STIV1 infection cycle, but not in that of SIRV2 (Quax et al. 2013; Snyder et al. 2013b; Daum et al. 2014). The only other viruses to have homologues of the PVAP protein, besides STIV1 and SIRV2, are the archaeal viruses of the Rudiviridae family (Quax et al. 2010). As such, the VAP-based egress system seems to be restricted to a limited number of archaeal viruses.
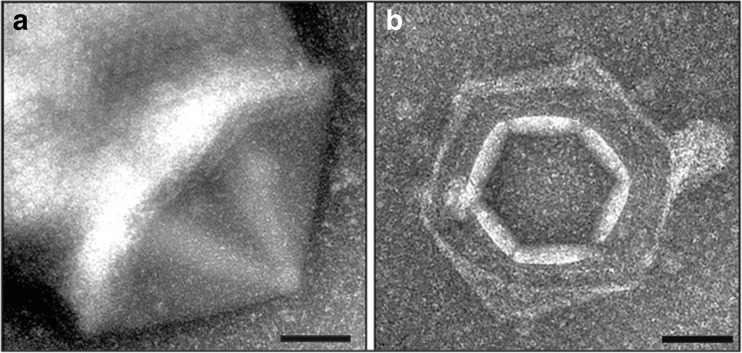
More recently, the formation of sixfold symmetric VAP-like structures was reported in an attempt to induce a putative virus of the hyperthermophilic archeon Pyrobaculum oguniense (Rensen et al. 2015). Irradiation with UV-light is a routine strategy to induce lysogenic viruses, which in archaea are commonly integrated into the host genome or exist as episomal genetic elements in the host cytoplasm (Prangishvili 2013). No viral particles were found after UV-irradiation of P. oguniense. However, dramatic morphological changes were observed on the surface of ~ 5% of the irradiated cells, which displayed one to nine pyramidal structures with sixfold symmetry (Fig. 2a) (Rensen et al. 2015). About 24 h after UV-irradiation, the pyramids eventually opened outwards in a fashion similar to that of the VAPs (Fig. 2b). Further biochemical characterisation of the VAP-like structures was not possible, as the virus could not be isolated (Rensen et al. 2015). Notably, despite the striking structual similatrity between VAPs and those found in SIRV2/STIV1 infected cells, the genome of the putative Pyrobaculum virus did not contain sequences homologous with the PVAP-encoding gene (Rensen et al. 2015). This highlights that VAP-like egress systems with different symmetries could be more widespread than previously thought.
Fig. 2.
Pyramidal structures on the surface of Pyrobaculum oguniense cells. Transmission electron micrographs of P. oguniense cells. a 16 h post UV-irradiation, with an external hexagonal pyramidal structure in closed conformation (side view). b 24 h post UV-irradiation, with a pyramidal structure in open conformation. Scale bars 100 nm. Adapted with permission from Rensen et al. (2015)
The dynamic structure of the VAP
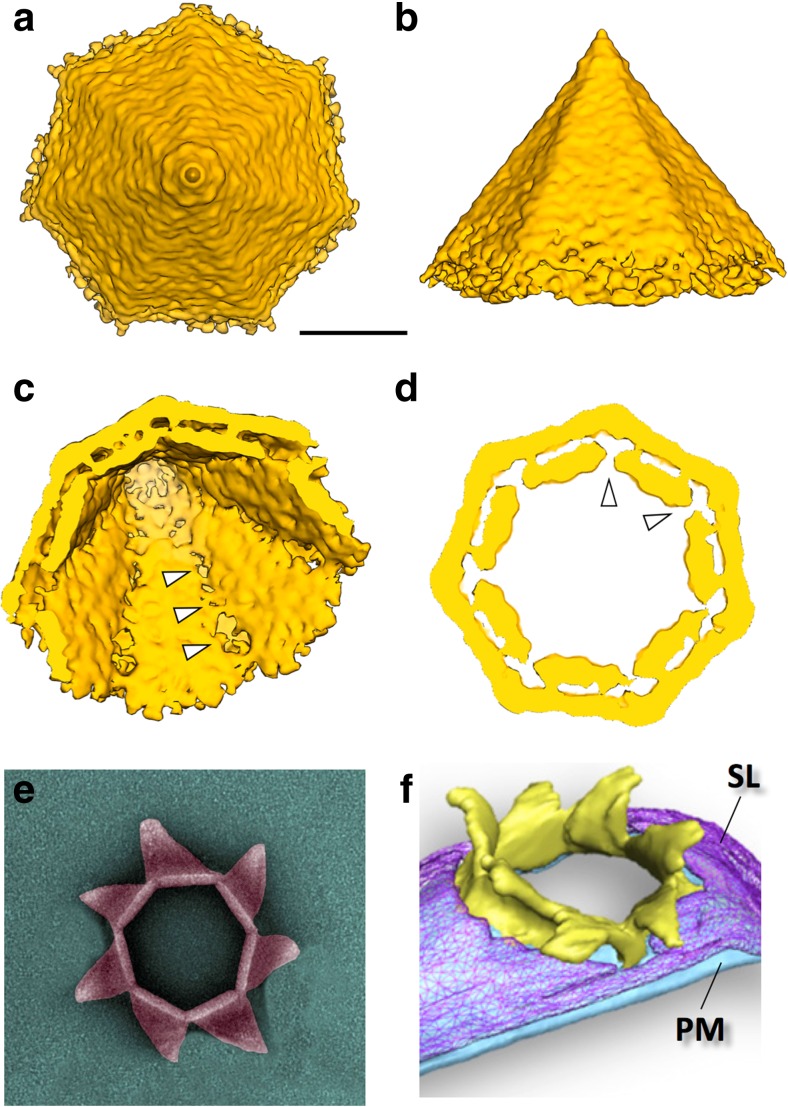
The structure of the open and closed form of the VAP was studied in situ by cryoET using whole SIRV2-induced S. islandicus cells (Quax et al. 2011; Daum et al. 2014). In addition, a 3D map of the closed VAP was obtained by sub-tomogram averaging (STA) (Daum et al. 2014). The STA map showed that the VAP is a baseless hollow pyramid, consisting of seven triangular facets with an angle of 35° at the tip (Fig. 3a-d; Quax et al. 2011; Daum et al. 2014). The inner opening angle of the pyramid was determined to be ~ 80°, and the triangular faces of the pyramids showed two parallel layers separated by a gap of ~ 5.8 nm (Fig. 3c, d) (Daum et al. 2014). The outer layer of the VAP was found to form a ~ 4.5-nm-thick sheet that was continuous with the cytoplasmic membrane. In contrast, the inner layer of ~ 4 nm thick was discontinuous with the membrane and extended ~ 15 nm beyond the outer layer into the cytoplasm (Daum et al. 2014). Upon infection, VAPs of different sizes (~ 20 – 150 nm) were observed in the cell membrane of S. islandicus and S. solfataricus (Bize et al. 2009; Brumfield et al. 2009; Quax et al. 2011). As their geometry is always the same, this observation suggests that VAPs develop by the formation of a small heptameric tip that grows into a pyramid by the gradual expansion of their seven triangular facets (Fu et al. 2010; Quax et al. 2011; Daum et al. 2014).
Fig. 3.
Structure of the VAP in closed and open state. a–d Sub-tomogram average of VAP (EMD-5844), shown in top (a), and side view (b) as seen from outside of the cell, in diagonal view showing the intracellular pyramidal cavity (c) and in cross-section (d). Low density at the inner seams of the VAPs suggest predetermined breaking points for VAP opening (white arrowheads). e, f, Open VAPs visualised by negative stain and transmission electron microscopy (e) and by cryoET of a VAP in the envelope of a SIRV2-infected S. islandicus cell (f). PM Plasma membrane, SL S-layer. Images adapted from Prangishvili and Quax (2011) and Daum et al. (2014). Scale bars (a–d) 25 nm
The sub-tomogram average of the mature, closed VAP displayed a slight anticlockwise handedness, with each facet being somewhat convex towards the inside (Daum et al. 2014). After VAP opening, this anticlockwise handedness is even more pronounced (Fig. 3e, f). These observations suggest that the VAP in closed state is under mechanical tension, which may provide the energy for the opening process. VAP opening usually commences when the VAPs have reached a diameter of ~ 150 nm. The process first manifests itself at the pyramidal tip and then progresses downward to the base by separation of triangular facets at their highly curved seams (Fig. 3e, f) (Fu et al. 2010; Daum et al. 2014).
Protein properties of PVAP
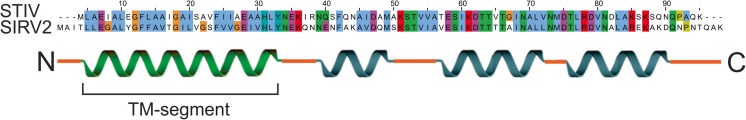
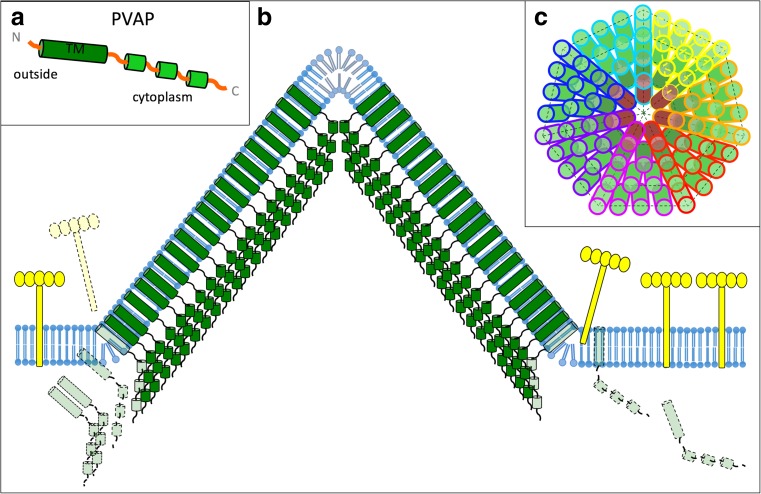
The unglycosylated membrane protein PVAP is the sole constituent of VAPs (Quax et al. 2010; Daum et al. 2014). Gel filtration analysis indicated that PVAP can form dimers, trimers and heptamers (Daum et al. 2014). The secondary structure of PVAP is predicted to contain three short α helixes on the C-terminus (Fig. 4). The N-terminus of PVAP contains a predicted transmembrane domain, but no apparent signal peptide. Edman degradation showed that the N-terminus was intact, indicating that the N-terminus of PVAP is not cleaved (Quax et al. 2010). VAP formation was observed upon PVAP overexpression in yeast, and in this case pyramids were observed in all intracellular membranes, including those of the Golgi apparatus, nucleus and mitochondria. In combination, these observations suggest that PVAP inserts into the membrane spontaneously and in a Sec-independent manner, similar to tail-anchored proteins and bacterial pore-forming toxins (Borgese and Fasana 2011; Bischofberger et al. 2012). Consequently, PVAP has the ability to self-assemble into VAPs in virtually any lipid bilayer and thus remodel membranes with fundamentally different lipid chemistry (ester or ether based for bacteria/eukarya or archaea, respectively) under a range of temperatures and acidities (37–80 °C, pH 3–7) (Albers and Meyer 2011; Quax et al. 2011; Daum et al. 2014). The expression of truncated PVAP variants showed that all, apart from the ten most C-terminal domains, amino acids are required for successful pyramid formation. Replacement of the PVAP transmembrane domain by another signal peptide free transmembrane domain did not result in pyramid formation, suggesting that the sequence of the transmembrane domain plays an important role in forming the VAP structure (Daum et al. 2014).
Fig. 4.
Secondary structure prediction of the virus-encoded protein forming VAPs (PVAP). Upper panel shows the sequence alignment of PVAP from STIV1 and SIRV2. Lower panel shows the in silico predicted secondary structure of PVAP [based on Tied Mixture Hidden Markov Model (TMHMM) and AmphipaSeek (amphipathic in-plane membrane anchors prediction); Kahsay et al. 2005; Sapay et al. 2006]. Transmembrane segment (TM) is shown in green, cytosolic α-helices are shown in blue.
VAP assembly and opening
The current model of VAP assembly suggests that the process is initiated by the cytoplasmic production of PVAP as new virions are replicated. Subsequently, the hydrophobic N-terminal transmembrane domain integrates into a lipid bilayer, similar to a tail-anchored protein (Daum et al. 2014). It is likely that this process is accompanied with a conformational change, as the transmembrane domain needs to be buried from the aqueous surrounding post expression and exposed towards the bilayer prior membrane insertion. In the membrane, PVAP monomers likely form a heptameric nucleation point in order to induce the assembly of a sevenfold symmetric pyramid (Fig. 5c). It is presently unknown if pyramid formation occurs by adding individual monomers or other types of PVAP oligomers to the central nucleation point. Comparison of the two-layer structure of the assembled VAP with the predicted domain organisation of the PVAP monomer (Fig. 5a) suggests that the outer, membrane-continuous VAP layer is formed by dense, parallel packing of PVAP transmembrane domains (Fig. 5b). Consequently, the inner VAP layer is formed by a tight array of cytosolic C-termin of PVAP (Fig. 5b) (Daum et al. 2014). It is conceivable that within each VAP facet, the close interactions between PVAP protomers lead to the formation of a two-dimensional crystalline array, which would exclude all other membrane proteins from the site of assembly, similar to holin rafts of bacterial viruses (Savva et al. 2014). This exclusion mechanism would consequently push aside or dislodge membrane-anchored S-layer subunits and thus allow the growing VAP to penetrate the protective cell wall (Albers and Meyer 2011).
Fig. 5.
Assembly model of VAP. a Secondary structure prediction of PVAP suggests an N-terminal transmembrane helix (TM) and three α-helical cytoplasmic domains nearer the C-terminus. b Hypothetical model of VAP assembly in side view. Spontaneous integration of PVAP proteins (transparent green) leads to self-assembly of a VAP (solid green). The TM domains of PVAP proteins form a continuous, membrane-embedded sheet, while staggered arrangement of cytoplasmic domains pushes the VAP outwards. S-layer subunits (yellow) are displaced by the growing VAP. c Hypothetical model in top view, showing how closely associated PVAP subunits may form a sevenfold pyramid. VAP assembly may be nucleated by a central heptamer (brown cylinders), which then recruits additional PVAP monomers (green cylinders) at its periphery. PVAP monomers belonging to the seven facets of the VAP model are outlined in different colours
In order to assemble a 3D pyramid instead of a 2D sheet, at least two different interactions between PVAP protomers are required: one in-plane between protein subunits in the facets and one out-of-plane at the edges of the sheets (Fig. 5). The interaction at the edges has to be weaker, allowing opening along the seams while keeping the facets intact. A slight angle between the N-terminal transmembrane helix and the C-terminal domains of PVAP leading to a staggered arrangement between neighbouring monomers would provide a mechanism by which the VAP is pushed outwards from the membrane plane (Fig. 5b). Such an arrangement of the C-terminal PVAP domains would be consistent with the observed extension of the inner sheet beyond the outer layer of the VAP (Daum et al. 2014).
VAPs always grow to roughly the same size, regardless if they develop in their natural host or are expressed heterologously. This suggests that VAP assembly underlies certain size-limiting constraints (Quax et al. 2011; Snyder et al. 2011; Daum et al. 2014). It is conceivable that these limiting factors are connected to mechanical constrains of the membrane rather than the opening of the VAP, which never occurs in heterologous expression systems.
Although the trigger for VAP opening is unknown to date, the observation that it only efficiently occurs in the native infected host suggests that it has to involve a host or virus-specific factor. For example, VAP opening may be dependent on the specific growth conditions (temperature and pH). However, it is also possible that the opening mechanism is directly controlled by a factor encoded by the virions themselves, as they are dependent on the precise timing of VAP opening to avoid the release of immature virions (Quax et al. 2011; Snyder et al. 2013a; Daum et al. 2014).
Outlook
The unique geometry and high stability of the VAP will potentially be the impetus for future application in biotechnology. VAPs can be isolated as single particles and they can easily be produced in E. coli by heterologous overexpression of PVAP. To explore the technological potential of the VAP mechanimsm in detail, a high-resolution structure of PVAP as well as its assembly within the VAP is highly desirable. The recent identification of novel VAPs with different rotational symmetry expands the arsenal of applicable VAP systems (Rensen et al. 2015)—for example, in the nano-engineering of structured surfaces in materials research. EM analysis of E. coli cells expressing various putative Pyrobaculum viral proteins would be an effective strategy to identify the currently unknown protein constituent of the hexagonal VAPs. It would be highly interesting to compare its sequence and structure with that of PVAP, as this would provide new insight into the common structure and sequence requirements for VAP formation as well as the basis for the symmetry variations found in VAPs from different virions.
Identification of the factor triggering VAP opening is critically important for its applicability as a nanomaterial. Once this mechanism is identified, this system may be employed to create defined apertures of ~ 150 nm in any biological lipid layer. VAPs could be used in liposomes for targeted drug delivery or to time nano-sized pore generation in semi-permeable membranes. Alternatively, the PVAP transmembrane domain could be fused to proteins that otherwise cannot be reconstituted into membranes, in order to facilitate membrane insertion or promote their presentation on the cell surface.
Thus, VAPs have great application potential in research methods, biotechnology and therapy.
Acknowledgements
This work was supported by a Marie-Curie Intra-European fellowship, a Post-doctoral grant from the Carl-Zeiss-Stiftung to T.E.F. Quax and a Research Fellow’s Start-up Grant by the University of Exeter to B. Daum.
Compliance with ethical standards
Conflict of interest
Tessa E.F. Quax declares that she has no conflicts of interest. Bertram Daum declares that he has no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is part of a Special Issue on ‘Biomolecules to Bio-nanomachines—Fumio Arisaka 70th Birthday’ edited by Damien Hall, Junichi Takagi and Haruki Nakamura.
References
- Adriaenssens EM, Krupovic M, Knezevic P, et al. Taxonomy of prokaryotic viruses: 2016 update from the ICTV bacterial and archaeal viruses subcommittee. Arch Virol. 2017;162:1153–1157. doi: 10.1007/s00705-016-3173-4. [DOI] [PubMed] [Google Scholar]
- Albers S-V, Meyer BH. The archaeal cell envelope. Nat Rev Microbiol. 2011;9:414–426. doi: 10.1038/nrmicro2576. [DOI] [PubMed] [Google Scholar]
- Bischofberger M, Iacovache I, Gisou van der Goot F. Pathogenic pore-forming proteins: function and host response. Cell Host Microbe. 2012;12:266–275. doi: 10.1016/j.chom.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Bize A, Karlsson EA, Ekefjärd K, et al. A unique virus release mechanism in the Archaea. Proc Natl Acad Sci USA. 2009;106:11306–11311. doi: 10.1073/pnas.0901238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese N, Fasana E. Targeting pathways of C-tail-anchored proteins. Biochim Biophys Acta Biomembr. 2011;1808:937–946. doi: 10.1016/j.bbamem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Brumfield SK, Ortmann AC, Ruigrok V, et al. Particle assembly and ultrastructural features associated with replication of the lytic Archaeal virus Sulfolobus turreted Icosahedral virus. J Virol. 2009;83:5964–5970. doi: 10.1128/JVI.02668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum B, Quax TEF, Sachse M, et al. Self-assembly of the general membrane-remodeling protein PVAP into sevenfold virus-associated pyramids. Proc Natl Acad Sci USA. 2014;111:3829–3834. doi: 10.1073/pnas.1319245111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellas N, Snyder JC, Bolduc B, Young MJ. Archaeal viruses: diversity, replication, and structure. Annu Rev Virol. 2014;1:399–426. doi: 10.1146/annurev-virology-031413-085357. [DOI] [PubMed] [Google Scholar]
- DiMaio F, Yu X, Rensen E, et al. A virus that infects a hyperthermophile encapsidates A-form DNA. Science. 2015;348(80):914–917. doi: 10.1126/science.aaa4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Wang K, Gan L, et al. In vivo assembly of an Archaeal virus studied with whole-cell electron cryotomography. Structure. 2010;18:1579–1586. doi: 10.1016/j.str.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillière F, Peixeiro N, Kessler A, et al. Structure, function, and targets of the transcriptional regulator SvtR from the hyperthermophilic archaeal virus SIRV1. J Biol Chem. 2009;284:22222–22237. doi: 10.1074/jbc.M109.029850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring M, Rachel R, Peng X, et al. Viral diversity in Hot Springs of Pozzuoli, Italy, and characterization of a unique Archaeal virus, Acidianus bottle-shaped virus, from a new family, the Ampullaviridae. J Virol. 2005;79:9904–9911. doi: 10.1128/JVI.79.15.9904-9911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häring M, Vestergaard G, Rachel R, et al. Virology: independent virus development outside a host. Nature. 2005;436:1101–1102. doi: 10.1038/4361101a. [DOI] [PubMed] [Google Scholar]
- Hong C, Pietilä MK, Fu CJ, et al. Lemon-shaped halo archaeal virus His1 with uniform tail but variable capsid structure. Proc Natl Acad Sci USA. 2015;112:2449–2454. doi: 10.1073/pnas.1425008112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahsay RY, Gao G, Liao L. An improved hidden Markov model for transmembrane protein detection and topology prediction and its applications to complete genomes. Bioinformatics. 2005;21:1853–1858. doi: 10.1093/bioinformatics/bti303. [DOI] [PubMed] [Google Scholar]
- Kasson P, DiMaio F, Yu X et al (2017) Model for a novel membrane envelope in a filamentous hyperthermophilic virus. eLIFE 6:e26268. 10.7554/eLife.26268 [DOI] [PMC free article] [PubMed]
- Krupovic M, Dutilh BE, Adriaenssens EM, et al. Taxonomy of prokaryotic viruses: update from the ICTV bacterial and archaeal viruses subcommittee. Arch Virol. 2016;161:1095–1099. doi: 10.1007/s00705-015-2728-0. [DOI] [PubMed] [Google Scholar]
- Larson ET, Reiter D, Young M, Lawrence CM. Structure of A197 from Sulfolobus turreted icosahedral virus: a crenarchaeal viral glycosyltransferase exhibiting the GT-a fold. J Virol. 2006;80:7636–7644. doi: 10.1128/JVI.00567-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaty WSA, Ortmann AC, Dlakic M, et al. Characterization of the Archaeal thermophile Sulfolobus turreted Icosahedral virus validates an evolutionary link among double-stranded DNA viruses from all domains of life. J Virol. 2006;80:7625–7635. doi: 10.1128/JVI.00522-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki T, Yoshida T, Tanaka R, et al. Diversity of viruses of the hyperthermophilic archaeal genus Aeropyrum, and isolation of the Aeropyrum pernix bacilliform virus 1, APBV1, the first representative of the family Clavaviridae. Virology. 2010;402:347–354. doi: 10.1016/j.virol.2010.03.046. [DOI] [PubMed] [Google Scholar]
- Mochizuki T, Sako Y, Prangishvili D. Provirus induction in hyperthermophilic archaea: characterization of Aeropyrum pernix spindle-shaped virus 1 and Aeropyrum pernix ovoid virus 1. J Bacteriol. 2011;193:5412–5419. doi: 10.1128/JB.05101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki T, Krupovic M, Pehau-Arnaudet G, et al. Archaeal virus with exceptional virion architecture and the largest single-stranded DNA genome. Proc Natl Acad Sci USA. 2012;109:13386–13391. doi: 10.1073/pnas.1203668109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Blum H, She Q, et al. Sequences and replication of genomes of the archaeal rudiviruses SIRV1 and SIRV2: relationships to the archaeal lipothrixvirus SIFV and some eukaryal viruses. Virology. 2001;291:226–234. doi: 10.1006/viro.2001.1190. [DOI] [PubMed] [Google Scholar]
- Pina M, Bize A, Forterre P, Prangishvili D. The archeoviruses. FEMS Microbiol Rev. 2011;35:1035–1054. doi: 10.1111/j.1574-6976.2011.00280.x. [DOI] [PubMed] [Google Scholar]
- Porter K, Kukkaro P, Bamford JKH, et al. SH1: a novel, spherical halovirus isolated from an Australian hypersaline lake. Virology. 2005;335:22–33. doi: 10.1016/j.virol.2005.01.043. [DOI] [PubMed] [Google Scholar]
- Prangishvili D (2006) 14 hyperthermophilic virus–host systems: detection and isolation. In: Rainey FA, Oren A (eds) Methods in microbiology. Elsevier, Amsterdam, pp 331–347
- Prangishvili D. The wonderful world of Archaeal viruses. Annu Rev Microbiol. 2013;67:565–585. doi: 10.1146/annurev-micro-092412-155633. [DOI] [PubMed] [Google Scholar]
- Prangishvili D, Quax TEF. Exceptional virion release mechanism: one more surprise from archaeal viruses. Curr Opin Microbiol. 2011;14:315–320. doi: 10.1016/j.mib.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Prangishvili D, Arnold HP, Götz D, et al. A novel virus family, the Rudiviridae: structure, virus-host interactions and genome variability of the sulfolobus viruses SIRV1 and SIRV2. Genetics. 1999;152:1387–1396. doi: 10.1093/genetics/152.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prangishvili D, Forterre P, Garrett RA. Viruses of the Archaea: a unifying view. Nat Rev Microbiol. 2006;4:837–848. doi: 10.1038/nrmicro1527. [DOI] [PubMed] [Google Scholar]
- Quax TEF, Krupovic M, Lucas S, et al. The Sulfolobus rod-shaped virus 2 encodes a prominent structural component of the unique virion release system in Archaea. Virology. 2010;404:1–4. doi: 10.1016/j.virol.2010.04.020. [DOI] [PubMed] [Google Scholar]
- Quax TEF, Lucas S, Reimann J, et al. Simple and elegant design of a virion egress structure in Archaea. Proc Natl Acad Sci USA. 2011;108:3354–3359. doi: 10.1073/pnas.1018052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quax TEF, Voet M, Sismeiro O, et al. Massive activation of archaeal defense genes during viral infection. J Virol. 2013;87:8419–8428. doi: 10.1128/JVI.01020-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quemin ERJ, Quax TEF. Archaeal viruses at the cell envelope: entry and egress. Front Microbiol. 2015;6:552. doi: 10.3389/fmicb.2015.00552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quemin ERJ, Lucas S, Daum B, et al. First insights into the entry process of hyperthermophilic archaeal viruses. J Virol. 2013;87:13379–13385. doi: 10.1128/JVI.02742-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensen E, Krupovic M, Prangishvili D. Mysterious hexagonal pyramids on the surface of Pyrobaculum cells. Biochimie. 2015;118:365–367. doi: 10.1016/j.biochi.2015.06.007. [DOI] [PubMed] [Google Scholar]
- Rice G, Tang L, Stedman K, et al. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc Natl Acad Sci USA. 2004;101:7716–7720. doi: 10.1073/pnas.0401773101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapay N, Guermeur Y, Deléage G. Prediction of amphipathic in-plane membrane anchors in monotopic proteins using a SVM classifier. BMC Bioinf. 2006;7:255. doi: 10.1186/1471-2105-7-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savva CG, Dewey JS, Moussa SH, et al. Stable micron-scale holes are a general feature of canonical holins. Mol Microbiol. 2014;91:57–65. doi: 10.1111/mmi.12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleper C, Kubo K, Zillig W. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc Natl Acad Sci USA. 1992;89:7645–7649. doi: 10.1073/pnas.89.16.7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder JC, Brumfield SK, Peng N, et al. Sulfolobus turreted Icosahedral virus c92 protein responsible for the formation of pyramid-like cellular lysis structures. J Virol. 2011;85:6287–6292. doi: 10.1128/JVI.00379-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder JC, Brumfield SK, Kerchner KM, et al. Insights into a viral lytic pathway from an archaeal virus-host system. J Virol. 2013;87:2186–2192. doi: 10.1128/JVI.02956-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder JC, Samson RY, Brumfield SK, et al. Functional interplay between a virus and the ESCRT machinery in Archaea. Proc Natl Acad Sci USA. 2013;110:10783–10787. doi: 10.1073/pnas.1301605110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestergaard G, Shah SA, Bize A, et al. Stygiolobus rod-shaped virus and the interplay of Crenarchaeal Rudiviruses with the CRISPR antiviral system. J Bacteriol. 2008;190:6837–6845. doi: 10.1128/JB.00795-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte A, Baranyi U, Klein R, et al. Characterization of Natronobacterium magadii phage phi Ch1, a unique archaeal phage containing DNA and RNA. Mol Microbiol. 1997;23:603–616. doi: 10.1046/j.1365-2958.1997.d01-1879.x. [DOI] [PubMed] [Google Scholar]
- Zillig W, Kletzin A, Schleper C, et al. Screening for Sulfolobales, their plasmids and their viruses in Icelandic Solfataras. Syst Appl Microbiol. 1993;16:609–628. doi: 10.1016/S0723-2020(11)80333-4. [DOI] [Google Scholar]