Abstract
Sucrose phosphate synthase (SPS) is believed to be the key enzyme for controlling the biosynthesis of sucrose. SPSs consist of a functional glycosyltransferase domain that shares conserved residues with the glycosyltransferase domain of sucrose biosynthesis-related protein. The formation of sucrose-6-phosphate is catalyzed by SPS with the transfer of a glycosyl group of uridine diphosphate glucose (UDP-G) as an activated donor sugar to a fructose-6-phosphate as a sugar acceptor. However, understanding of the mechanism of catalytic and substrate binding in SPS is very limited. Based on amino acid sequence alignments with several enzymes that belong to the glycosyltransferase family, the UDP-G binding sites that might be critical for catalytic mechanism were identified. Here, we report that single point mutation of R496, D498, and V570 located in the proposed UDP-G binding site led to less active or complete loss of enzyme activity. Through structure-based site-directed mutagenesis and biochemical studies, the results indicated that these residues contribute to the catalytic activity of plant SPS. Moreover, understanding of the UDP-G binding site provides an insight into new strategies for enzyme engineering and redesigning a catalytic mechanism for UDP.
Keywords: Sucrose phosphate synthase, Glycosyltransferase, Uridine diphosphate glucose, Sugarcane, Site-directed mutagenesis
Introduction
Sucrose is synthesized in cytosol and is the main form of transporting photosynthesis products throughout plant cells (Gifford et al. 1984). Sucrose occupies many significant roles for plant cell metabolism, including cyanobacteria and unicellular algae. The biochemical and molecular studies reported on sucrose biosynthesis in prokaryotic offer new insights into the origin of sucrose metabolism (Lunn and MacRae 2003; Salerno and Curatti 2003).
One of the key enzymes to catalyze the first step of the sucrose synthesis pathway is sucrose phosphate synthase (SPS; EC 2.4.1.14). Plant SPS has a molecular size of approximately 120 kDa which consists of three domains, including a glycosyltransferase domain that regulates the catalytic function of SPS (Castleden et al. 2004). In contrast, recombinant sugarcane SPS (SoSPS1) expression in Escherichia coli mostly appears to be a shorter form (100 kDa) with retained enzyme activity, and the shorter form was predicted to have a truncated N-terminal 20-kDa region. It has recently been revealed that the expression of N-terminal truncated form (∆N-SPS) tends to increase the specific activity 10-fold compared to full-length SPS. In our previous results, we reported that the regulation of the N-terminal domain might function as a suppressor domain for plant SPS activity (Sawitri et al. 2016). The crystal structure of non-photosynthetic bacterial SPS has been solved with different characteristics compared to plant SPS. The lack of N-terminal and C-terminal domains of plant SPS are shown in Halothermothrix orenii SPS. Therefore, the structure of bacterial SPS contains only a glycosyltransferase domain for substrate binding and catalytic reaction (Salerno and Curatti 2003; Chua et al. 2008). However, the precise regulatory function of SPS in plants still remains unclear since there is no crystal structure of plant SPS.
In this study, we report the identification of substrate-binding sites for uridine diphosphate glucose (UDP-G) as a sugar donor at the glycosyltransferase domain of recombinant SoSPS1. As noted above, SoSPS1 has been used in a previous study for elucidating SPS regulation in sugarcane. The expression of SoSPS1 is actively regulated in the photosynthetic tissue and representative for the sucrose biosynthesis enzyme (Sugiharto et al. 1997). The knowledge of substrate binding sites would provide an important insight into the catalytic mechanism of SPS in plants. Moreover, it is interesting to determine the regulation of plant SPS involved in carbon partitioning which is a critical process in distributing chemical energy converted through photosynthesis.
Diphosphate moiety binding of UDP to SoSPS1
The enzymes responsible for regulation in sucrose metabolism are grouped into sucrose-biosynthesis-related proteins (SBRPs), and the conserved domain of glycosyltransferase has been determined in SPS and sucrose synthase (SuSy) (Pontis 1978; Cumino et al. 2002; Salerno and Curatti 2003). Other enzymes that are involved in the synthesis of polysaccharides are likewise members of glycosyltransferase, such as glycogen synthase (GS) and starch synthase (Coutinho et al. 2003; Sheng et al. 2009). The glycosyltransferases are generally utilizing sugar moiety from activated nucleotide sugar as a donor to promote enzymatic reaction (Breton and Imberty 1999; Tarbouriech et al. 2001).
SPS catalyzes the synthesis of sucrose-6-phosphate (S6P) from UDP-G and fructose-6-phosphate (F6P) (Leloir and Cardini 1955; Amir and Preiss 1982; Huber and Huber 1996). The specific substrate for plant SPS is UDP-G, whereas the substrate for bacterial SPS is not specific for UDP-G. Unlike plant SPS, bacterial SPS is able to bind other nucleoside diphosphate glucose (NDP-G), such as adenosine diphosphate glucose (ADP-G) (Lunn et al. 1999; Chua et al. 2008). In spite of the fact that many glycosyltransferase enzymes recognize similar substrates, the amino acid sequences between different families have diverged (Ünligil and Rini 2000; Imberty et al. 2006). Thus, identifying the binding sites for UDP-G is necessary to elucidate the mechanism of enzyme regulation in plant SPS.
It has been reported that SPS, SuSy, and GS share a close similarity with the glycosyltransferase domain andthe stereochemistry of their substrates, such as UDP-G (Chua et al. 2008; Baskaran et al. 2010; Zheng et al. 2011; Wu et al. 2015). However, the amino acid sequence alignment with conserved residues among SPS, SuSy, and GS has not been well reported (Diricks et al. 2015).
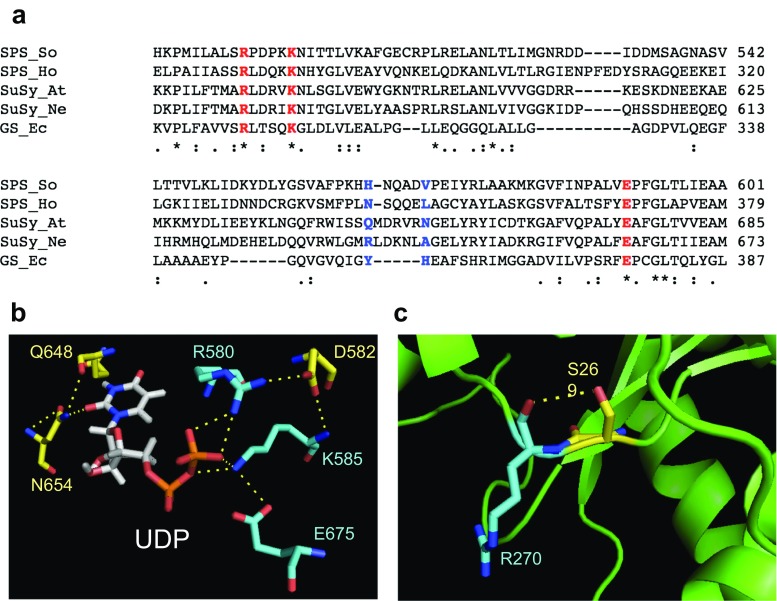
The deduced sequences of SoSPS1 and other representative enzymes of glycosyltransferases were aligned using ClustalW2 and compared to identify the predicted NDP binding pocket. The sequences were: sugarcane SPS (SoSPS1, GenBank ID: BAA19242.1), H. orenii SPS (HoSPS, GenBank ID: ACB11221.1), Arabidopsis thaliana SuSy (AtSuSy, GenBank ID: NP_197583.1), Nitrosomonas europaea SuSy (NeSuSy, GenBank ID: CAD85125.1), and Escherichia coli GS (EcGS, GenBank ID: EDU64330.1). The comparison of the SoSPS1 sequence alignment with its homologs is shown in Fig. 1a. There are three critical residues, R496, K501, and E591, in SoSPS1 (corresponding to R580, K585, and E675 in AtSuSy; R567, K572, and E663 in NeSuSy; R300, K305, and E377 in GS) (Fig. 1a). In previous studies, it has been reported that these critical residues may lead the conformational changes of enzyme architecture. The structure of N. europaea sucrose synthase (PDB ID: 4RBN), has proven that these three residues, Arg, Lys, and Glu, have a large effect on its catalytic and conformational change (Wu et al. 2015).
Fig. 1.
Comparison of residues in glycosyltransferases group. a Multiple sequence alignment of SPS in sugarcane (SoSPS1), SPS in non-photosynthetic bacterial H. orenii (HoSPS), sucrose synthase in A. thaliana (AtSuSy), sucrose synthase in bacteria N. europaea (NeSuSy), and glycogen synthase from E. coli (EcGS). The three highly conserved and critical residues that consist of arginine, lysine, and glutamic acid are shown in red. The residues that are predicted to be involved in nucleotide sugar interaction is shown in blue. b The UDP binding site residues at positions corresponding to Arg-580, Asp-582, Lys-585, and Glu-675 for diphosphate moiety binding and Gln-648 and Asn-654 for uridine moiety binding of A. thaliana sucrose synthase (PDB ID: 3S28). c Predicted interaction hydrogen bonding between Ser-269 and Arg-270 in bacterial SPS (PDB ID: 2R60)
The above-mentioned highly conserved glycosyltransferases have been identified at R496 in SoSPS1. These conserved Arg residues are essential for interaction between enzyme and substrate UDP-G, notably the diphosphate moiety. The multiple sequence alignment in Fig. 1a shows that plant SuSy is significantly more similar to bacterial SuSy and SPS compared to glycogen synthase. To date, the crystal structure of sucrose synthase from A. thaliana (PDB ID: 3S28) in complex with product UDP-G has been solved and the diphosphate group of UDP-G showed to interact with residue R580 (Zheng et al. 2011). Interestingly, the residue D582 in AtSuSy exhibits a salt bridge interaction with R580 and K585 (Fig. 1b). Asp residue (D582 in AtSuSy) is not directly involved in the catalytic process, but D582 appears to stabilize R580 and K585 for binding its substrate. Therefore, R496, K501, and D498 in SoSPS1 might have a crucial contribution to interact with the pyrophosphate group of UDP-G.
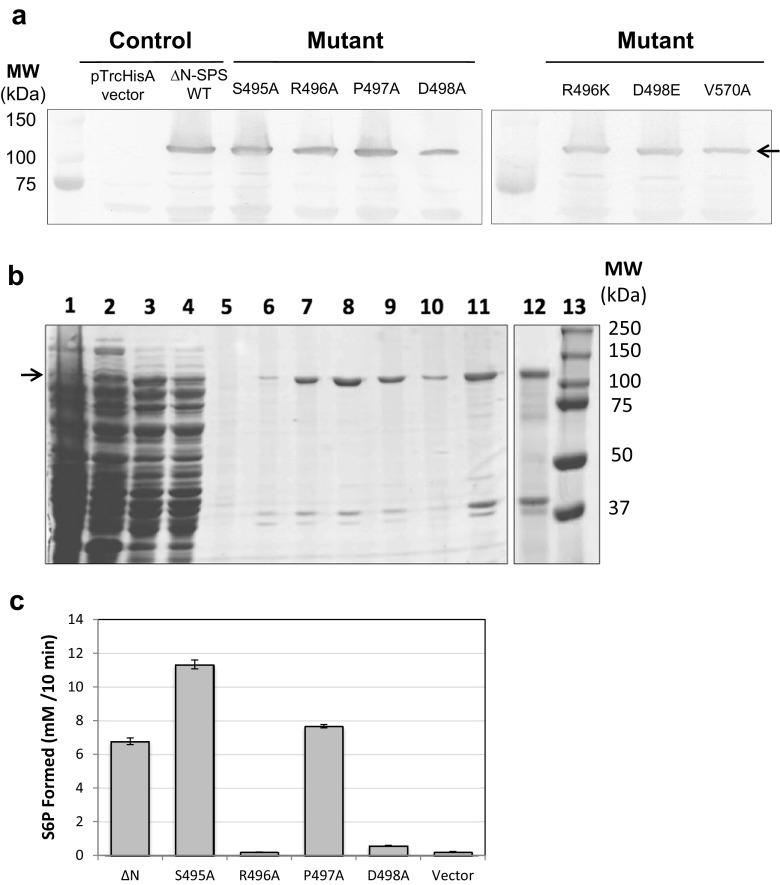
In this present work, structure-based site-direct mutagenesis is demonstrated to confirm experimentally the involvement of any residues in UDP-G binding sites of plant SPS. The point mutation was specified as S495A, R496A, P497A, D498A, R496K and D498E for the diphosphate moiety and V570A for the uridine moiety. The QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, USA) was utilized to introduce specific point mutations in the wild-type ∆N-SoSPS1 as previously described (Sawitri et al. 2016). In order to obtain biochemical characteristics, the wild-type and mutants were expressed into E. coli BL21 (DE3) using expression vector pTrcHisA (Life Technologies, Invitrogen, USA) and the expressions of wild-type and mutants were monitored by immunoblotting analysis using polyclonal antibody against the SPS protein. As shown in Fig. 2a, the bands of wild-type and mutants with apparent molecular size around 100 kDa were detected as expected. The yield of recombinant production was similar for all expressed proteins, indicating that wild-type and mutants were expressed in E. coli BL21 (DE3) cells. The sequential purification of wild-type and mutated proteins were carried out by DE52 anion exchange cellulose (Whatman, UK) and affinity system Complete His-Tag Purification Resin (Roche, Switzerland). The protein in the fraction obtained during the purification steps was monitored using sodium dodecyl sulfate polyacrylamide-gel electrophoresis (SDS-PAGE) as described by Laemmli (1970) and Coomassie Brilliant Blue staining as shown in Fig. 2b. Afterwards, the eluted samples were concentrated in 10 K Amicon Ultra Centrifugal Filter (Merck Millipore, Ireland) and the purified samples were further assayed for biochemical analysis to characterize the identified residues.
Fig. 2.
Site-directed mutagenesis and expression of recombinant SoSPS1 in E. coli . a The resulting SPS mutation at glycosyltransferase domain was transformed into E. coli overnight and the total cell proteins were detected by western blotting using antibodies against polypeptide of SPS. b The affinity purification of His-tagged ∆N-SoSPS1 as monitored by SDS-PAGE and Coomassie blue staining. Lane 1 The eluted sample from DE52 anion exchange cellulose. Lanes 2–5 Washing steps of affinity His-tag purification with increasing concentration of imidazole 0 and 20 mM. Lanes 6–10 The fraction of samples eluted successively with the buffer containing 100 mM imidazole. Lanes 11–12 The purified SPSs were concentrated and exchanged to buffer composed of 50 mM Tris-HCl, pH 7.5 and 150 mM NaCl. Lane 13: Protein molecular marker (Biorad-Dual Color Standards). c Activity of SPS mutants (S495A, R496A, P497A, and D498A) compared to wild-type ∆N-SoSPS1 and vector pTrcHis as a control. Arrows represent the targeted proteins
In this report, alanine-scanning mutagenesis has been applied to investigate the predicted substrate-binding site in SoSPS1 by comparing the enzyme activity of wild-type and mutants (S495A, R496A, P497A, and D498A). SPS activity as previously described (Huber et al. 1985; Sawitri et al. 2016) was measured in the presence of substrate UDP-G and F6P at saturating concentrations (20 mM). As shown in Fig. 2c, enzyme activity of mutant P497A was comparable with wild-type. As expected, residue Pro at 497 is not involved in catalytic activity and does not induce conformational changes in SPS structure. Conversely, single mutation of S495 significantly altered activity of the enzyme. The residue Ser495–Arg496, which corresponds to Ser269–Arg270 in HoSPS, has been suggested to contribute a hydrogen-bonding interaction between Ser and Arg (Fig. 1c). If Ser at S495 is replaced with Ala, it adopts the sequence conserved in SuSy and the SPS activity was remarkably increased compared to wild-type ∆N-SoSPS1. The reason for this phenomenon was not clear, but we predicted that this effect might be attributed to the loss of the intramolecular hydrogen bond. Replacement of Ala at R496 results in complete loss of enzyme activity (Fig. 2c). These results were in good agreement with the previously mentioned mutagenic replacement at position R496 (corresponding to R580 in AtSuSy; R567 in NeSuSy; R300 in GS) emphasizing the critical role of catalytic regulation in plant SPS. Moreover, mutation at D498 displayed a tendency to be less active compared to the wild-type enzyme (Fig. 2c). The obtained result provides evidence that D498 stabilizes R496 to interact with UDP-G through hydrogen bond interaction. On the other hand, we conducted further mutational analysis on these two charged residues to examine another possible role of R496 and D498 by replacing each with Lys and Glu, respectively. We previously presumed that the salt bridge interaction between Arg as positively charged residue and Asp as negatively charged residue could be substituted to other potential salt bridge partners such as Lys and Glu. However, single mutation of R496K and D498E reveals an unexpected influence of these mutations without retaining the enzymatic activity (data not shown). The reason of these results is not yet clearly understood, but previous reports have suggested that these side chains play important roles to establish the structure of the enzymatic transition state and cannot be substituted to other residues by mutagenesis (Lairson et al. 2008; Wu et al. 2015).
The similarity of conserved motifs among SPS and SuSy suggest a possible regulation for the UDP binding mechanism. The interplay between R580 and D582 in AtSuSy is stronger than the interaction of Lys-Asp or Arg-Glu. The role of Asp is to maintain the mode of the UDP binding region through salt bridge interaction with Arg and to be considered as capable of binding substrate UDP-G. Thus, the predicted interaction between residues in SoSPS1 and UDP-G notably with the diphosphate moiety has been experimentally proven (Fig. 2c). However, our present work does not report the kinetic properties comparison between wild-type and each mutant due to the fact that some mutated SPSs become inactive.
Substrate preference for the uridine moiety of UDP in SoSPS1
The characterization of residues that are responsible for binding substrate preference is apparently necessary. Nucleotide binding at the glycosyltransferase domain has been identified in sucrose synthase (Zheng et al. 2011; Wu et al. 2015) and glycogen synthase (Sheng et al. 2009) from X-ray crystal structure analysis. The crystal structure of bacterial SPS (HoSPS) with the substrate F6P and the product S6P have been solved. However, the HoSPS structure and its complex with the substrate UDP-G could not be obtained. Thus, the predicted residues which bind to UDP-G was generated by computational docking, and residue Leu at position 342 in HoSPS was reported as a nucleotide-binding residue (Chua et al. 2008). The structural analysis of glycogen synthase from E. coli revealed that the residues Y355 and H356 interact with the ADP moiety. The role of residue Y355 is to stack the adenine ring, and adenine accepts a hydrogen bond from H356 (Sheng et al. 2009). Based on the crystal structure of AtSuSy with the UDP-G complex, the overlap structure comparison of AtSuSy and NeSuSy could be observed. The residues that interact with the phosphate and ribose moieties are well conserved, but those interacting with nucleotide are not (Zheng et al. 2011; Wu et al. 2015; Diricks et al. 2015).
Kinetic parameters of sucrose synthase from A. thaliana and N. europaea displays different substrate preferences between UDP and ADP, respectively. Likewise, plant SPS shows substrate specificity to UDP-G, whereas the HoSPS shows a tendency to accept NDP-G, such as ADP-G (Chua et al. 2008). As shown in Fig. 1a, the sequence alignment implies that residues H565 and V570 in SoSPS1 (corresponding to residues Q648 and N654 in AtSuSy; residues R636 and A642 in NeSuSy; and residues Y355 and H356 in EcGS) play a role in specific substrate preference. The previously reported SPS structure from H. orenii has shown us that residue L324, corresponding to V570 in SoSPS1, is predicted to interact with NDP-G, whereas the other residue corresponding to H565 in SoSPS1 is still not clear. Although these two residues involved with nucleotide sugar are not well conserved, the sequences are considered to have a functional value which is in good agreement with other reports (Sheng et al. 2009; Zheng et al. 2011; Wu et al. 2015; Diricks et al. 2015).
In addition, subsequent site-directed mutagenesis was directed particularly at V570 to substitute Val to Ala, and our result demonstrated that V570A mutation leads to loss of SPS activity (data not shown). This indicated that Val at 570 in SoSPS1 is an important residue to drive the catalytic mechanism of plant SPS. The sequence alignment in previous report showed that Val residue is highly conserved in other plant SPS sequences and predicted substrate specificity for UDP-G (Chua et al. 2008). In this report, we did not compare the kinetic properties of mutants for substrate specificity between UDP-G and ADP-G. On the contrary, engineering altered substrate specificity to switch from ADP to UDP has recently been reported in bacterial sucrose synthase for industrial applications. The single, double, and triple mutations of recombinant bacterial SuSy were made at corresponding positions to H565 and V570 in SoSPS1. However, the experiment did not succeed and the substrate preference showed no change, still preferring ADP (Diricks et al. 2015).
Perspective
Similar to sucrose synthase, plant SPS showed a strong preference for UDP-G as a substrate, as opposed to bacterial SPS, which are predicted to bind ADP-G. In plants, UDP-G and ADP-G are directed to the biosynthesis of sucrose, cellulose, and starch (Salerno and Curatti 2003; Baroja-Fernández et al. 2003). It is noteworthy that the enzymes involved in nucleotide sugar metabolism are essential to control the regulation of carbon partitioning in plants. This led to the hypothesis that one of the strategies to redesign the regulation of plant sugar metabolism is through substrate binding site engineering of plant SPS. This work is challenging since a crystal structure of plant SPS has not yet been obtained. Identifying the specific residues related to substrate nucleotide sugar by mutagenesis is proposed to overcome the lack of information about functional residues in SoSPS1. Following the analogy of glycosyltransferase group structures, our results have clarified that residues R496, D498, and V570 in SoSPS1 are important for catalysis by plant SPS. Further mutational analysis is required to alter substrate preference and offer new prospects for structural arrangement accommodating UDP-G or ADP-G.
Acknowledgements
This work was supported by Ministry of Research, Technology and Higher Education of the Republic of Indonesia (Penelitian Unggulan Strategis Nasional and Penelitian Dasar Unggulan Perguruan Tinggi) and by the International Collaborative Research Program of Institute for Protein Research, Osaka University, ICR-14-03 and 05, Japan.
Abbreviations
- F6P
Fructose-6-phophate
- G6P
Glucose-6-phosphate
- SPS
Sucrose phosphate synthase
- UDP-G
Uridine diphosphate glucose;
Compliance with ethical standards
Conflict of interest
Widhi Dyah Sawitri declares that she has no conflicts of interest. Siti Nurul Afidah declares that she has no conflicts of interest. Atsushi Nakagawa declares that he has no conflicts of interest. Toshiharu Hase declares that he has no conflicts of interest. Bambang Sugiharto declares that he has no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is part of a Special Issue on ‘Biomolecules to Bio-nanomachines - Fumio Arisaka 70th Birthday’ edited by Damien Hall, Junichi Takagi and Haruki Nakamura.
References
- Amir J, Preiss J. Kinetic characterization of spinach leaf sucrose-phosphate synthase. Plant Physiol. 1982;69:1027–1030. doi: 10.1104/pp.69.5.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroja-Fernández E, Muñoz FJ, Saikusa T, Rodríguez-López M, Akazawa T, Pozueta-Romero J. Sucrose synthase catalyzes the de novo production of ADPglucose linked to starch biosynthesis in heterotrophic tissues of plants. Plant Cell Physiol. 2003;44(5):500–509. doi: 10.1093/pcp/pcg062. [DOI] [PubMed] [Google Scholar]
- Baskaran S, Roach PJ, Depaoli-Roach AA, Hurley TD. Structural basis for glucose-6-phosphate activation of glycogen synthase. Proc Natl Acad Sci U S A. 2010;107(41):17563–17568. doi: 10.1073/pnas.1006340107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton C, Imberty A. Structure/function studies of glycosyltransferases. Curr Opin Struct Biol. 1999;9:563–571. doi: 10.1016/S0959-440X(99)00006-8. [DOI] [PubMed] [Google Scholar]
- Castleden CK, Aoki N, Gillespie VJ, MacRae EA, Quick WP, Buchner P, Foyer CH, Furbank RT, Lunn JE. Evolution and function of the sucrose phosphate synthase gene families in wheat and other grasses. Plant Physiol. 2004;135:1753–1764. doi: 10.1104/pp.104.042457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua TK, Bujnicki JM, Tan TC, Hyunh F, Patel BK, Sivaraman J. The structure of sucrose phosphate synthase from Halothermothrix orenii reveals its mechanism of action and binding mode. Plant Cell. 2008;20:1059–1072. doi: 10.1105/tpc.107.051193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–317. doi: 10.1016/S0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- Cumino A, Curatti L, Giarrocco L, Salerno GL. Sucrose metabolism: anabaena sucrose-phosphate synthase and sucrose-phosphate phosphatase define minimal functional domains shuffled during evolution. FEBS Lett. 2002;517:19–23. doi: 10.1016/S0014-5793(02)02516-4. [DOI] [PubMed] [Google Scholar]
- Diricks M, Bruyn FD, Daele PV, Walmagh M, Desmet T. Identification of sucrose synthase in nonphotosynthetic bacteria and characterization of the recombinant enzymes. Appl Microbiol Biotechnol. 2015;99:8465–8474. doi: 10.1007/s00253-015-6548-7. [DOI] [PubMed] [Google Scholar]
- Gifford RM, Thorne JH, Hitz WD, Giaquinta RT (1984) Crop productivity and photoassimilate partitioning. Science 225:801–808 [DOI] [PubMed]
- Huber SC, Huber JL. Role and regulation of sucrose-phosphate synthase in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:431–444. doi: 10.1146/annurev.arplant.47.1.431. [DOI] [PubMed] [Google Scholar]
- Huber SC, Kerr PS, Rufty TW. Diurnal changes in sucrose phosphate synthase activity in leaves. Plant Physiol. 1985;64:81–87. doi: 10.1111/j.1399-3054.1985.tb01216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imberty A, Wimmerova M, Koca J, Breton C. Molecular modeling of glycosyltransferases. In: Brockhausen I, editor. Methods in molecular biology, 347: Glycobiology protocols. Totowa: Humana; 2006. pp. 145–156. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- Leloir LF, Cardini CE. The biosynthesis of sucrose phosphate. J Biol Chem. 1955;214:157–165. [PubMed] [Google Scholar]
- Lunn JE, MacRae E. New complexities in the synthesis of sucrose. Curr Opin Plant Biol. 2003;6:208–214. doi: 10.1016/S1369-5266(03)00033-5. [DOI] [PubMed] [Google Scholar]
- Lunn JE, Price GD, Furbank RT. Cloning and expression of a prokaryotic sucrose-phosphate synthase gene from the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol. 1999;40:297–305. doi: 10.1023/A:1006130802706. [DOI] [PubMed] [Google Scholar]
- Pontis HG. On the scent of the riddle of sucrose. Trends Biochem Sci. 1978;3:137–139. doi: 10.1016/S0968-0004(78)80034-6. [DOI] [Google Scholar]
- Salerno GL, Curatti L. Origin of sucrose metabolism in higher plants: when, how and why? Trends Plant Sci. 2003;8:63–69. doi: 10.1016/S1360-1385(02)00029-8. [DOI] [PubMed] [Google Scholar]
- Sawitri WD, Narita H, Ishizaka-Ikeda E, Sugiharto B, Hase T, Nakagawa A. Purification and characterization of recombinant sugarcane sucrose phosphate synthase expressed in E. coli and insect Sf9 cells: an importance of the N-terminal domain for an allosteric regulatory property. J Biochem. 2016;159(6):599–607. doi: 10.1093/jb/mvw004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng F, Jia X, Yep A, Preiss J, Geiger JH. The crystal structures of the open and catalytically competent closed conformation of Escherichia coli glycogen synthase. J Biol Chem. 2009;284(26):17796–17807. doi: 10.1074/jbc.M809804200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiharto B, Sakakibara H, Sumadi, Sugiyama T. Differential expression of two genes for sucrose-phosphate synthase in sugarcane: molecular cloning of the cDNAs and comparative analysis of gene expression. Plant Cell Physiol. 1997;38(8):961–965. doi: 10.1093/oxfordjournals.pcp.a029258. [DOI] [PubMed] [Google Scholar]
- Tarbouriech N, Chamock SJ, Davies GJ. Three-dimensional structures of the Mn and mg dTDP complexes of the family GT-2 glycosyltransferase spsA: a comparison with related NDP-sugar glycosyltransferases. J Mol Biol. 2001;314:655–661. doi: 10.1006/jmbi.2001.5159. [DOI] [PubMed] [Google Scholar]
- Ünligil UM, Rini JM. Glycosyltransferase structure and mechanism. Curr Opin Struct Biol. 2000;10:510–517. doi: 10.1016/S0959-440X(00)00124-X. [DOI] [PubMed] [Google Scholar]
- Wu R, Diez MDA, Figueroa CM, Matchey M, Iglesias AA, Ballicora MA, Liu D. The crystal structure of Nitrosomonas europaea sucrose synthase reveals critical conformational changes and insights into sucrose metabolism in prokaryotes. J Bacteriol. 2015;197(17):2734–2746. doi: 10.1128/JB.00110-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Anderson S, Zhang Y, Garavito RM. The structure of sucrose synthase-1 from Arabidopsis thaliana and its functional implications. J Biol Chem. 2011;286(41):36108–36118. doi: 10.1074/jbc.M111.275974. [DOI] [PMC free article] [PubMed] [Google Scholar]