Abstract
The low-density lipoprotein receptor (LDLR) and its homologs capture and internalize lipoproteins into the cell. Due to the fact that LDLR family members possess a modular ectodomain that undergoes dynamic conformational changes, multi-scale structural analysis has been performed so as to understand the ligand capture and release mechanism. For example, crystallographic analyses have provided models for both the entire ectodomain and high-resolution structures of individual modules. In addition, nuclear magnetic resonance spectroscopic analyses have shown the rigidity and flexibility of inter-module linkers to restrict the mobility of ectodomain. Accumulated structural data suggest that the ectodomains of LDLR family members are flexible at the cell surface and switch between two metastable conformations, that is, the extended and contracted conformations. Recent structural analysis of ApoER2, a close homolog of LDLR, raised the possibility that the receptor binds with the ligand in the contracted conformation. After transport to an endosome by endocytosis, the receptor undergoes a conformational change to the closed conformation for completion of ligand release. In contrast, LDLR has been reported to adopt the extended conformation when it binds with a inhibitory regulator that recruits LDLR toward the degradation pathway. These findings support a mechanism of different ectodomain conformations for binding the ligand versus binding the regulatory protein. In this review, I provide an overview of studies that analyze the structural and biophysical properties of the ectodomains of LDLR family members and discuss a hypothetical model for ligand uptake and receptor recycling that integrates the known ectodomain conformational variability.
Keywords: Low-density lipoprotein receptor, Endocytosis, Conformational change, X-ray crystallography, Molecular recognition
Introduction
Many single-pass transmembrane (1TM) receptors possess an ectodomain composed of multiple functional modules, and they display their physiological activities by adopting specific conformations. Hence, the elucidation of the entire ectodomain structure is important for understanding the mechanism by which the 1TM receptors receive extracellular signals from ligands and transduce them across the membrane. However, the presence of flexible inter-module linkers in many 1TM receptors has hampered crystallography or electron microscopy analyses at high resolutions and complicated structure analyses. Therefore, multi-scale approaches have been frequently employed to integrate the precise structural data collected on partial fragments with moderate-resolution models of the entire ectodomains. Nuclear magnetic resonance (NMR) spectroscopy has also been powerful for assessing such dynamic properties as the mobility and rigidity of inter-module linkers, which are influenced by solution conditions, such as pH and salt concentration.
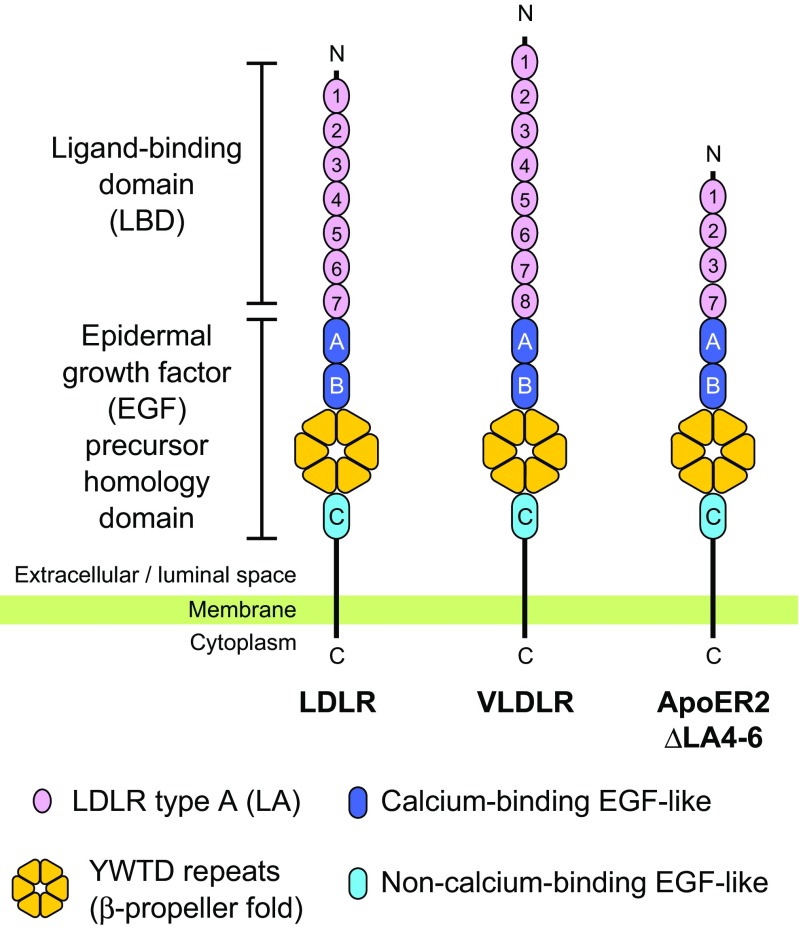
The low-density lipoprotein receptor (LDLR) is one of the 1TM receptors that have been the targets of such multi-scale structural analyses. At the cell surface, LDLR captures a lipoprotein particle, such as LDL as a ligand from the blood. Subsequently, the LDLR–LDL complex is internalized into the cell by endocytosis (Brown and Goldstein 1986). After being recruited to the endosome, LDLR releases LDL and is transported back to the cell surface for recycling. Since LDLR plays a pivotal role in clearing LDL from the blood, many mutations of the LDLR gene have been identified as the cause of familial hypercholesterolemia (FH) (Brown and Goldstein 1974; Usifo et al. 2012). As is often the case with 1TM receptors, LDLR possesses an ectodomain composed of multiple functional modules (Yamamoto et al. 1984) that are utilized for the regulation of the capture and release of LDL (Fig. 1). The ectodomain of LDLR is divided into the N-terminal ligand-binding domain and the C-terminal epidermal growth factor (EGF) precursor homology domain. The N-terminal domain contains seven repeats of the LDLR type A (LA) modules, which capture basic residues of the ligands in specific manners. The C-terminal domain is composed of two EGF-like modules, YWTD repeats, and an additional EGF-like module, in this order. LDLR is also involved in the uptake of apolipoprotein E (ApoE)-containing lipoprotein, such as β-very-low-density lipoprotein (VLDL) (Perrey et al. 2001). The EGF precursor homology domain is thought to contribute to recycling since deletion of this domain lowered the release efficiency of β-VLDL and surface expression level of the receptor (Davis et al. 1987).
Fig. 1.
Domain organization of human low-density lipoprotein receptor (LDLR) and its close homologs. The N-terminal domain of LDLR is termed as LBD while the C-terminal domain shows a homology to the EGF precursor. The number of LA modules differ between homologs, and the major splicing variant of human apolipoprotein E (ApoE)R2 contains four LA modules, lacking LA 4–6. The YWTD repeats assume a six-bladed β-propeller fold. The first and second EGF-like modules (EGF-A and -B) are classified as the calcium-binding type, and the third EGF-like module (EGF-C) is non-calcium-binding type. VLDLR Very-low-density lipoprotein receptor
Among the homologs of LDLR, ApoE receptor 2 (ApoER2) and VLDL receptor (VLDLR) are quite similar to LDLR in terms of domain composition, with differences being primarily in the number of the LA repeats (Schneider and Nimpf 2003). VLDLR contains eight LA repeats, and splicing variants of ApoER2 containing three to five LA repeats have been identified, although seven or eight LA repeats are coded in the gene (Kim et al. 1996, 1997; Novak et al. 1996; Takahashi et al. 1992). ApoER2 and VLDLR also show similarities to LDLR in function. For example, both receptors are known to bind with and internalize β-VLDL (Kim et al. 1996; Takahashi et al. 1992). On the other hand, ApoER2 and VLDLR also have a different aspect to their functions. Beyond lipoprotein uptake, the ApoER2 and VLDLR ectodomains bind with protein ligands. In particular, when these two receptors bind with the extracellular protein reelin on neurons, they perform signal transduction that regulates neuronal migration. This ultimately leads to the “inside–out” pattern of cortical layer formation in the brain (Herz and Beffert 2000; Tissir and Goffinet 2003; Trommsdorff et al. 1999).
It is also known that LDLR is degraded by proprotein convertase subtilisin/kexin type 9 (PCSK9) (Rashid et al. 2005), but the mechanism is not straightforward. Although PCSK9 is classified as an endoprotease, it does not display proteolytic activity toward LDLR that directly results in degradation (Li et al. 2007; McNutt et al. 2007). Rather, PCSK9 is thought to bind with the ectodomain of LDLR and recruit LDLR to the lysosome. The gain-of-function mutation of PCSK9 leads to excess degradation of LDLR and increases the level of LDL in the blood (Abifadel et al. 2003; Lagace et al. 2006), indicating that the balance between recycling and degradation of the receptor is important for the maintenance of LDL level. Recently, anti-PCSK9 monoclonal antibodies that inhibit interactions with LDLR have been developed for an anti-FH drug (Natarajan and Kathiresan 2016).
To gain an understanding of the mechanism by which the receptor captures the lipoprotein at the cell surface and releases it in the cell, multi-scale structural analyses have been extensively carried out on LDLR and its close homologs. In this review, I first describe the structural features of individual modules, such as the LA modules, EGF-like modules, and YWTD repeats. Subsequently, I review the structural analyses forcusing on the conformation of the entire ectodomain and outline a hypothetical mechanistic model for ligand uptake and receptor recycling.
Structure of LA modules and ligand recognition mechanism
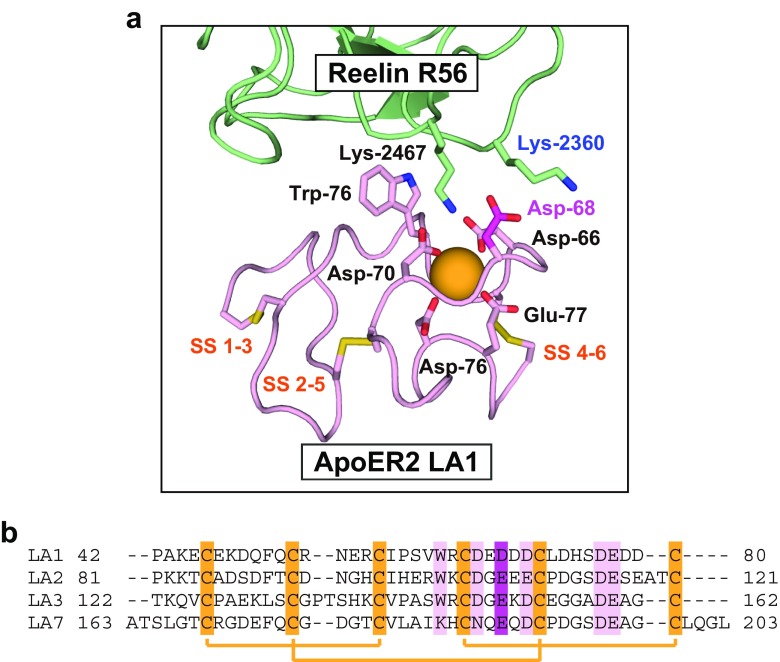
The LA modules generally consist of around 40 amino acid residues and include a calcium ion (Fig. 2). The first three-dimensional (3D) structure was determined by 2D NMR spectroscopy for the N-terminal first LA (LA1) from human LDLR (Daly et al. 1995). The NMR structure revealed that the LA module is composed of an N-terminal β-hairpin, followed by a series of β-turns where six conserved cysteines form three disulfide bonds with a 1–3, 2–5, 4–6 pairing pattern. Subsequently, the precise coordination structure of canonical calcium ion was revealed in the crystal structure of the fifth LA module (LA5) from human LDLR (Fass et al. 1997). The calcium ion was coordinated with octahedral geometry by side-chain oxygens on four conserved acidic residues and two main-chain oxygens. In this arrangement, these acidic residues come close together and form an electronegative surface. One of the calcium-coordinating main-chain oxygens belongs to an aromatic residue that is also conserved among the LA modules and present in the vicinity of the acidic cluster. The other residue coordinating the calcium ion with its main-chain oxygen is often an acidic residue, and its side-chain also contributes to the formation of the electronegative surface.
Fig. 2.
Structure of the LA module in complex with a ligand. The crystal structure of the ApoE receptor 2 (ApoER2) N-terminal first LA (LA1) in complex with the reelin R56 fragment is shown as a representative example. a Close-up view of the crystal structure of ApoER2 LA1 in complex with the reelin R56 fragment. In ApoER2 LA1, the conserved residues constituting the ligand-binding pocket and disulfide bonds are shown as a stick model. The orange sphere represents the calcium ion coordinated by the conserved acidic residues. The two basic residues recognized by LA1 in a double-Lys/Arg mode are also shown in the stick model. b Sequence alignment of LA modules in human ApoER2 ΔLA4–6. The disulfide pairs are connected with orange lines. The conserved acidic and aromatic residues are indicated in pink. The acidic residues that do not coordinate the calcium ion but contribute to the electrostatic interactions are highlighted in dark magenta
The structures of various LA modules have been analyzed in complex with the ligands, such as receptor-associated protein (RAP) (Fisher et al. 2006), the capsid protein of human rhinovirus HRV2 (Verdaguer et al. 2004), and the signaling-competent fragment of reelin (Yasui et al. 2010) (Fig. 2a). These structural analyses have confirmed a common recognition mechanism wherein a basic side-chain on the ligand interacts with the electronegative surface in the binding pocket formed by the conserved acidic and aromatic residues coordinating the calcium ion. In this canonical recognition mode, the side-chain protrudes out from the surface of ligand in an extended conformation and is accommodated into the binding pocket of the LA modules. Binding analyses have suggested that the length of the side-chain of the basic residue also affects the binding specificity with the LA module. For example, the substitution of the lysine residue at this position to arginine, which possesses a longer side-chain than lysine, abolishes the interactions between ApoER2 LA1 and reelin (Yasui et al. 2010). It has also been found that some of the ligands interact with the LA modules in the double-Lys/Arg recognition mode where an additional basic residue is present on the ligand surface and seems to stabilize the electrostatic interactions (Fisher et al. 2006; Yasui et al. 2010).
Structure of EGF-like modules and interactions with PCSK9
The EGF-like modules are frequently identified in secreted proteins and ectodomains of membrane proteins. These modules also contain six conserved cysteine residues forming three disulfide bonds, although their pairing pattern, 1–3, 2–4, 5–6, is different from that of the LA modules. There are two types of EGF-like modules, that is, calcium-binding EGF-like and non-calcium-binding EGF-like modules (Handford et al. 1991). LDLR and its close homologs commonly possess two calcium-binding EGF modules (EGF-A and -B) between the LA repeats and the YWTD repeats, as well as a non-calcium-binding EGF module (EGF-C) downstream of the YWTD repeats. The structure of human the LDLR–EGF-AB pair has been determined by NMR spectroscopy (Fig. 3a), which has shown that the EGF-B module coordinates a calcium ion with pentagonal bipyramidal geometry through a canonical calcium-binding consensus sequence consisting of Asp-333, Asp-335, Glu-336, Asn-349, and Tyr-354 (Malby et al. 2001). In contrast, the calcium-binding sequence of EGF-A is non-canonical since the conserved first Asp/Asn residue, which corresponds to Asp-333 of EGF-B, is absent.
Fig. 3.
Structure of the EGF-like modules and their interaction mode with proprotein convertase subtilisin/kexin type 9 (PCSK9). a The nuclear magnetic resonance structure of the LDLR–EGF-AB unit. EGF-A and -B, both calcium-binding EGF modules, are shown in light and dark colors, respectively. The residues contributing to the calcium-binding consensus and to the disulfide bonds are highlighted in stick model. Each EGF-like module contains a calcium ion, as shown in sphere model. EGF-A lacks the first consensus Asp/Asn residue corresponding to Asp-333 of EGF-B. b Close-up view of the PCSK9–EGF-A interface. PCSK9 docks to the surface close to the calcium-binding site of EGF-A. The side-chains of the residues involved in the inter-molecular interaction and the main-chains of the residues surrounding the calcium ion are shown in stick model. Since the structure was solved at acidic pH, the additional hydrogen bond was formed between His-306 of EGF-A and Asp-374 of PCSK9
The structure determination of the EGF-AB pair in complex with PCSK9 has shown that the calcium-binding site of EGF-A serves as the binding site for PCSK9 (Bottomley et al. 2009; Kwon et al. 2008; McNutt et al. 2009) (Fig. 3b). Asn-295 and Asp-310, which are the conserved residues in the calcium-binding sequence, make direct contact with PCSK9. In addition, the structure determination of the complex under different pH conditions has suggested that the His-306 of EGF-A forms an additional salt bridge with the Asp-374 of PCSK9 at acidic pH (McNutt et al. 2009), which should stabilize the PCSK9–LDLR complex under acidic conditions. In fact, the mutation of His-306 to Tyr is known to be associated with FH (Day et al. 1997), and crystallographic analysis has demonstrated that the hydroxyl group of Tyr forms a stable hydrogen bond with Asp-374 even at neutral pH (Bottomley et al. 2009; McNutt et al. 2009).
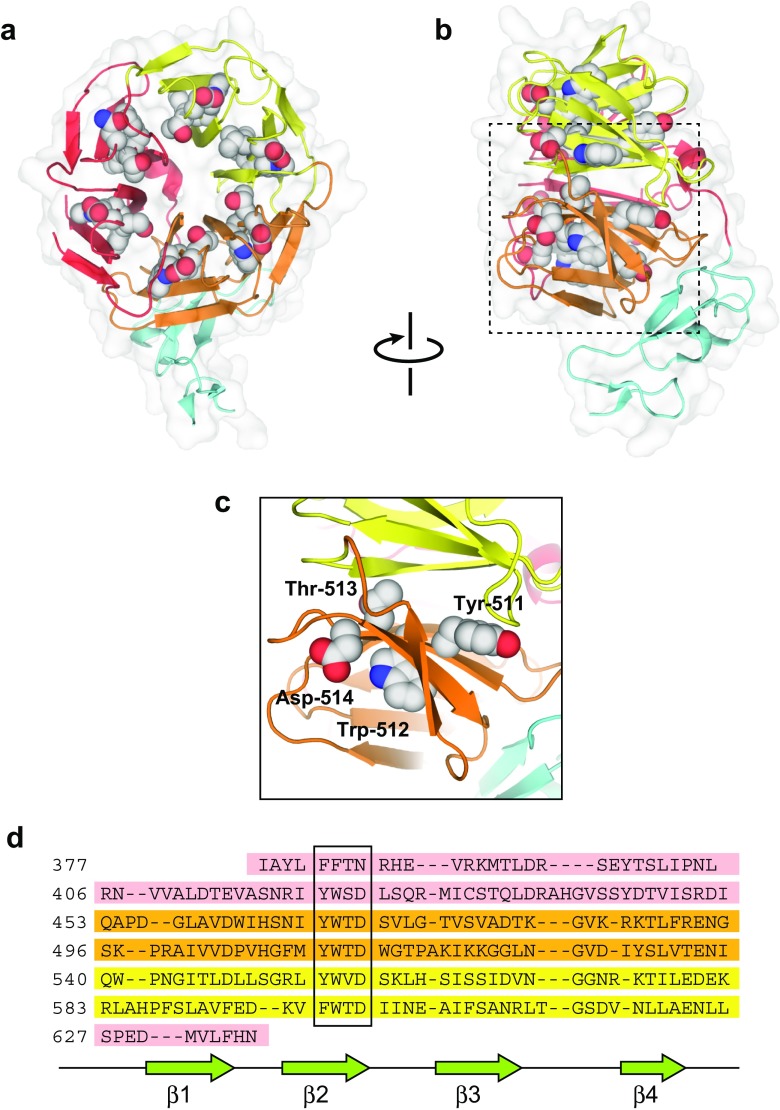
Structure of the YWTD repeats
The YWTD repeats, as the name suggests, contain repeats of a Tyr–Trp–Thr–Asp consensus sequence. As already predicted from the sequence (Springer 1998), structure determination has shown that the YWTD repeats assume a six-bladed β-propeller fold, in which each blade contains a YWTD sequence (Jeon et al. 2001) (Fig. 4a, b). Similar to other β-propellers, the blades are four-stranded anti-parallel β-sheets where the YWTD sequences serve as the second strand in the respective blades (Fig. 4c, d). The conserved residues at the Tyr and Trp positions are embedded inside the molecule and appear to contribute to the formation of the hydrophobic core, while the residues at the Asp position participate in several hydrogen bonds to maintain the structural integrity of the β-propeller fold. In the first structure determination of the LDLR YWTD repeats, the fragment, including the two flanking EGF modules (EGF-B and -C), were subjected to crystallization. While EGF-B was not visible in the electron density, EGF-C seemed to form intimate interactions with the bottom face of the propeller. This observation opens the possibility for intrinsic mobility in EGF-B, which might be related to regulation of the receptor functions, as discussed in following sections.
Fig. 4.
Structure of the YWTD repeat and EGF-C, a non-calcium-binding EGF module. a Full view from the top-face of the β-propeller. b Full view from the side of the β-propeller. The structure is shown in ribbon model with a transparent surface. The residues in the YWTD consensus sequences are highlighted in sphere model. The six blades of the β-propeller are shown in the three different colors, and the EGF-C modules are shown in cyan. c Close-up view of the region around the fourth YWTD consensus. d Sequence alignment of the six blades of the β-propeller. The YWTD sequences form the second β-strand (β2), as highlighted with the box. The C-terminal region is incorporated into the blade containing the first YWTD sequence as the first β-strand (β1)
Structure of the entire ectodomain of LDLR and ApoER2
To understand the mechanism of ligand release as well as of ligand capture, it is of great importance to obtain information on the relative spatial arrangement of the modules constituting the ectodomain. The first detailed structural data for an entire ectodomain was elucidated by x-ray crystallography for human LDLR (Rudenko et al. 2002) (Fig. 5a). In this structure, the N-terminal LA repeats are extended and cover the top-face of the β-propeller of the YWTD repeats. The hairpin-like structure of the entire ectodomain is maintained through the intramolecular interactions between the LA repeats and the YWTD repeats. Two LA modules, LA4 and LA5, recognize two lysine residues on the YWTD repeats, Lys-560, and −582, respectively (Fig. 5b). Their interaction mechanisms are very similar to those of the canonical ligand recognition mode where the side-chains of the two lysine residues are accommodated into the conserved ligand-binding pockets of two LA modules. Of note, this structure was determined by using crystals generated in a crystallization buffer of pH 5.3, which would correspond to the environment of endosomal lumen. Therefore, it has been proposed that this structure reflects the binding-inactive, or “closed,” state after LDLR is transported to the endosome and releases LDL (Fig. 5c).
Fig. 5.
Structure of the entire ectodomain of LDLR. a Full view of the ligand-unbound LDLR ectodomain at acidic pH. b Close-up view of the intramolecular binding interface. The structure is shown in ribbon model with the calcium ions in sphere model. The residues involved in the intramolecular contacts are shown in stick model. Both LA4 and LA5 interact with the top-face of the β-propeller in the canonical ligand-binding mechanism. The protonated histidine residues, His-190, 560, and 582, are thought to stabilize the interactions. c Surface model of the ligand-unbound LDLR ectodomain at acidic pH. d Surface model of the PCSK9-bound LDLR ectodomain at neutral pH. PCSK9 is shown with transparent surface. e, f Diagram of the domain arrangement of ligand-unbound LDLR (e) and PCSK9-bound LDLR (f). The ligand-bound LDLR ectodomain adopts the contracted-closed conformation, in which the EGF-AB unit is located beside Blade 5–6, and draws the LA modules close to the top-face of β-propeller. In contrast, PCSK9-bound LDLR adopts the extended-open conformation, in which the EGF-AB unit is located beside Blade 1–2 and keeps the LA modules away from the β-propeller. The LA1–6 modules are outlined with a dotted line since they were not assigned in the crystal structure
Subsequently, another crystal structure of the human LDLR ectodomain was also reported for the PCSK9-bound state (Lo Surdo et al. 2011) (Fig. 5d). These authors reported that PCSK9 docked to the N-terminal region of EGF-A through the same interaction mechanism as observed in the PCSK9–EGF-AB complex. While an interaction was observed between PCSK9 and the YWTD β-propeller, that interface appeared to be auxiliary, with only a small contribution. In contrast to ligand-unbound LDLR at acidic pH, none of the LA modules made any direct contacts with the YWTD β-propeller, and disorder in the electron densities prevented assignment of atomic models for most of the LA repeats. These observations indicate that the LA repeats assume multiple conformations in the crystal. This structural analysis raised the possibility that the LA repeats of LDLR at the cell surface are liberated from the YWTD β-propeller and can explore a large conformational space to capture the ligand in the “extended” state since the crystals used for this structural analysis were obtained from crystallization buffer at neutral pH, which would correspond to the extracellular environment. However, it remains to be elucidated which conformation the receptor adopts when it captures the ligand. LDLR bound to LDL has been subjected to electron microscopy analysis, with the results indicating that the LDLR adopts the extended conformation (Ren et al. 2010). However, the precise arrangement of the respective modules could not be determined due to low resolution. In fact, determination of a high-resolution 3D structure of LDLR in the LDL-bound state is generally regarded as an intrinsically difficult task due to the heterogeneity in the protein:lipid stoichiometry for LDL particles.
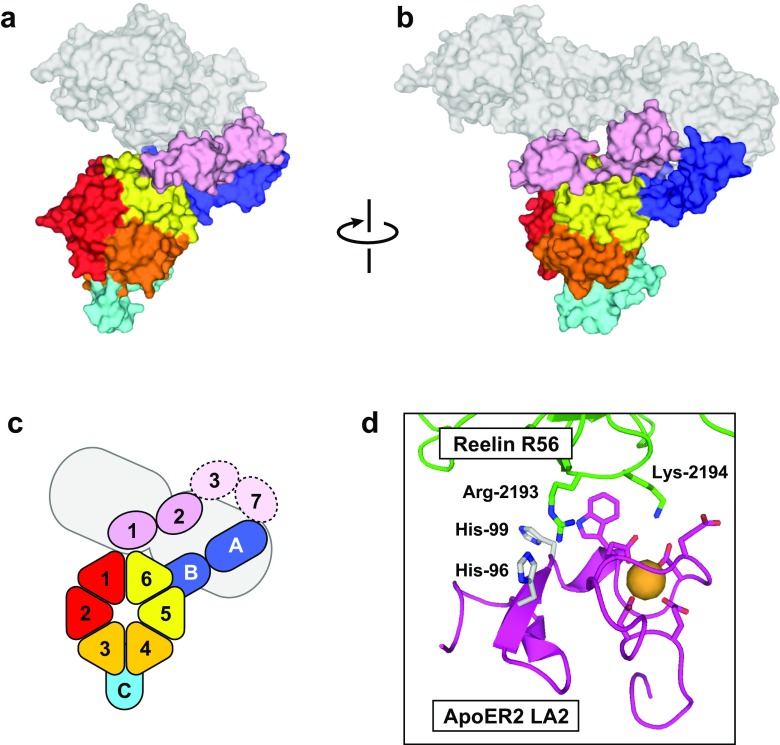
To circumvent this technical problem, structural analysis has also been performed on LDLR family receptors that can bind protein ligands. As mentioned above, ApoER2 and VLDLR are close homologs of LDLR and are known to bind the extracellular protein reelin during signal transduction. Reelin is composed of 3461 amino acid residues with characteristic repeats, termed reelin repeats (D’Arcangelo et al. 1995). Although reelin possesses eight reelin repeats in total, the fragment containing the fifth and sixth repeats, R56, is a minimal unit that can bind with ApoER2 and VLDLR and induce the signal transduction (Yasui et al. 2007). The R56 fragment can be produced as recombinant protein with high homogeneity, and crystallography analysis has successfully elucidated the structure of ApoER2 ectodomain in the ligand-bound state (Hirai et al. 2017) (Fig. 6a, b). Unexpectedly, the ApoER2–reelin complex structure resulting from a crystal obtained under neutral pH conditions bore many similarities to that of ligand-unbound LDLR under acidic pH conditions. In the ApoER2–reelin complex, the N-terminal LA repeats were located close to the top-face of the YWTD β-propeller. Without direct contacts with the β-propeller, however, the LA repeats were still open and therefore capable of binding with reelin. These observations suggested that the receptor adopts an intermediate conformation, which is neither extended nor closed, when it binds with the ligand.
Fig. 6.
Structure of the ApoER2 ectodomain in complex with the fragment containing the fifth and sixth repeats of reelin (R56). a Surface model viewed from the top-face of the β-propeller. b Side-view of the surface model. The reelin R56 fragment is shown as a transparent surface. c Diagram of the domain arrangement. Similar to ligand-unbound LDLR at acidic pH, the EGF-AB unit is located at the Blade 5–6 side (see text for description of “blade”). This conformation was defined as the contracted-open conformation as the LA modules are located close to the β-propeller, but they are still capable of binding with the ligand. d Close-up view of the binding interface between ApoER2 LA2 and reelin R56. The structure is shown in ribbon model, and the residues involved in the binding interfaces and calcium coordination are shown in stick model. The two histidine residues, His-96 and -99, on the binding surface of LA are presumed to contribute to the pH dependency of the binding interactions
Classification of ectodomain conformation based on the orientation of EGF-AB pair
Comparison of the three available structures of LDLR and ApoER2 ectodomains indicates that the position of the LA repeats relative to the YWTD β-propeller is determined by the orientation of the intervening EGF-AB modules. In the LDLR, the blade historically defined as W1 actually contains the second YWTD repeat in primary sequence because of strand exchange, whereby the C-terminal region of the YWTD repeat region is incorporated as the first β-strand (Jeon et al. 2001; Springer 1998). In this review, however, the first blade is defined as the one containing the first YWTD repeat and the six blades are grouped into three pairs from the N-terminus so as to be able to simply explain the positions of the EGF modules relative to the YWTD repeats (Fig. 4d). To avoid misunderstanding, the first blade is hereafter termed as “Blade 1”, and the subsequent five blades are similarly termed as “Blades 2 to 6”. Superposition of the three structures based on the β-propeller places EGF-B in two different orientations, whereas EGF-C is located between Blades 3 and 4 in all three structures. Specifically, EGF-B approaches the center of the bottom face of the β-propeller proximal to Blades 5–6 in ligand-unbound LDLR (Fig. 5e) and in reelin-bound ApoER2 (Fig. 6c), but it approaches proximal to the Blade 1–2 in PCSK9-bound LDLR (Fig. 5f). It cannot be ruled out that the EGF-B modules are inherently flexible and that the orientations are primarily affected by crystal packing; however, structural comparison, including other LDLR-related proteins (LRPs), suggests that the two orientations of EGF-B might be metastable states conserved in the LDLR family. The structural unit containing YWTD repeats with the incoming and exiting EGF modules (EGF–YWTD–EGF unit) are frequently identified in family members other than LDLR and ApoER2, and several crystal structures are also available for the EGF–YWTD–EGF units, such as EGF2–YWTD1–EGF3 of LRP4 (Zong et al. 2012), and EGF1–YWTD2–EGF2 and EGF3–YWTD4–EGF4 of LRP6 (Chen et al. 2011; Cheng et al. 2011). The orientations of the incoming EGF modules in these three structures are also classified into two groups similarly to those of LDLR and ApoER2 (Fig. 7). For example, EGF2 of LRP4 is located at the Blade 5–6 side, like ligand-unbound LDLR and reelin-bound ApoER2, while EGF1 and EGF3 of LRP6 approach the YWTD repeats from the Blade 1–2 side, like PCSK9-bound LDLR. In contrast, the orientations of the exiting EGF modules are fixed at the Blade 3–4 side in all structures. These observations raise the possibility that the EGF-B module in LDLR and ApoER2 serves as the switch to induce a conformational change throughout the entire ectodomain when it exchanges between the Blade 5–6 and Blade 1–2 positions.
Fig. 7.
Classification of domain arrangement in the EGF–YWTD–EGF unit from the LDLR family. a Structures with the incoming EGF at the Blade 5–6 side. b Structures with the incoming EGF beside Blades 1–2. The structures are shown in ribbon model with a transparent surface. The structures are viewed from the top-face of β-propeller, as shown in Figs. 5 and 6
Based on the comparison among the LDLR and ApoER2 structures, the classification of ectodomain conformations can be modified as follows. In PCSK9-bound LDLR, EGF-B at the Blade1–2 side separates the LA repeats from the YWTD repeats. This conformation can be defined as “extended-open” (Fig. 5f) because the entire ectodomain is extended and the LA repeats are open for the ligand. In reelin-bound ApoER2, the positioning of EGF-B beside Blades 5–6 draws the LA repeats close to the top-face of the β-propeller, resulting in a contracted structure for the entire ectodomain. However, the LA repeats remain open and accessible to the ligand as they make no direct contacts with the β-propeller. Therefore, this conformation can be defined as “contracted-open” (Fig. 6c). In contrast, the conformation of ligand-unbound LDLR at acidic pH can be defined as “contracted-closed” (Fig. 5) since the ectodomain assumes a contracted EGF-B conformation and the LA repeats are also inaccessible to the ligand due to direct intramolecular interactions with the β-propeller.
Modulation of binding affinities using histidine as the pH sensor
A series of structural analyses on LDLR family members have also revealed the importance of several histidine residues in modulating the binding affinities of both inter- and intra-molecular interactions. The closed state of ligand-unbound LDLR is maintained by intramolecular electrostatic interactions with protonated histidine side-chains (Rudenko et al. 2002) (Fig. 5b). For example, the YWTD β-propeller of LDLR possesses two histidine residues, His-562 and -586, on the top-face of YWTD, and the protonation of their side chains is thought to enhance interactions with LA4 at acidic pH. Furthermore, His-190 on LA5 is also presumed to stabilize the intramolecular interactions in the closed state as it can form a salt bridge with Glu-581 on the YWTD β-propeller. In fact, the H190Y and H562Y mutations, which should weaken the pH-dependent interaction between YWTD and LA4–5, are known to cause FH (Hopkins et al. 1999; Sun et al. 1994). Cell-based binding assays indeed have confirmed the decrease in LDL release activity in the mutant H562Y at acidic pH (Beglova et al. 2004). The histidine residues corresponding to His-562 and -586 are present in the ApoER2 YWTD, suggesting that it may also adopt a “contracted-closed” state.
In addition, histidine residues appear to contribute to the ligand dissociation process in ApoER2. Structural analysis of the ApoER2–reelin complex elucidated that LA1 contributes to the complex formation as the primary binding site while LA2 serves as an auxiliary binding site (Hirai et al. 2017). The LA2 module of ApoER2 possesses two histidine residues on the binding site for reelin, and the protonation of their side-chains at acidic pH should weaken the interaction with reelin via electrostatic repulsion to the positively charged basic residues on the binding site of reelin (Fig. 6d). In fact, alanine substitution of these two histidines on LA2, which eliminates the positive charges, increased the intermolecular affinity at acidic pH. Conversely, substitutions to lysine, which introduce positive charges even at neutral pH, decreased the affinity. These observations raise the possibility that the LA2 module serves as a pH-dependent switch to destabilize the ApoER2–reelin complex during endocytosis. Modulation of binding affinity is also observed in the LDLR–PCSK9 complex although the pH shift causes the opposite effect as that in the interactions between reelin and PCSK9 (Bottomley et al. 2009; Kwon et al. 2008; McNutt et al. 2009). For the interaction between LDLR EGF-A and PCSK9, the additional salt-bridge formed by the protonated side-chain of EGF-A His-306 enhances the interaction, as discussed above (Fig. 3b).
Hypothetical model for ligand uptake and receptor recycling
Assuming that LDLR and ApoER2 undergo similar conformational changes, a hypothetical model can now be proposed for the relationship between the ectodomain conformations and receptor behaviors in the cell (Fig. 8). Presumably, the receptor ectodomain is flexible in the extended conformation at the cell surface. Identification of the newly determined ApoER2 structure in complex with reelin has suggested that the receptor ectodomain is exchangeable between the extended and contracted conformations at the cell surface and that the receptor adopts the contracted-open conformation when it binds with the ligand. Although it remains unclear whether or not reelin preferably docks to the contracted conformation of the ApoER2 ectodomain, the formation of the complex in this conformation may prime the receptor for ligand release after endocytosis. Even after dissociation in the endosomal compartment, re-binding of the ligand to the receptor will still occur with some certain probability. Therefore, a competitive intramolecular interaction between the LA and YWTD repeats should be quite important to promote ligand release from the receptor. With the contracted-open conformation, the LA repeats come close to the top-face of the β-propeller and seem to be ready for intramolecular interaction with it. In other words, there is a possibility that the LA repeats can rapidly adopt the contracted-closed conformation after ligand dissociation without exploring a large conformational space in search of a binding site on the β-propeller. The importance of proper positioning of the LA modules with respect to the YWTD β-propeller has also been suggested by mutational analysis of LDLR. An NMR study revealed that the structure of the LA7–EGF-A unit is rigid at both neutral and acidic pH and that the disruption of the LA7–EGF-A interface lowered LDL release activity (Beglova et al. 2004). As the disruption would favor an open conformation for the LA modules, it is presumed that they need to be positioned close to the YWTD β-propeller to release LDL efficiently. In contrast, point mutations interfering with the formation of the extended-open conformation decrease the binding affinity of LDLR to PCSK9, indicating that PCSK9 has a preference for binding with LDLR in the extended-open conformation. Since the interaction between PCSK9 and LDLR is stabilized at acidic pH, PCSK9 separates the LA modules from contact with the β-propeller during endocytosis. This model implicitly emphasizes the importance of receptor recycling on the adoption of the contracted-closed conformation (i.e., the intramolecular interaction between the LA and YWTD repeats). One prediction would be that PCSK9 inhibits recycling by preventing the receptor from adopting to the contracted-closed conformation in the endosome.
Fig. 8.
Hypothetical model of ligand uptake and receptor recycling in the LDLR family members based on multi-scale structural biology. Presumably, the ectodomain of the receptor is flexible and can adopt both extended and contracted conformations at the cell surface. Structural analysis of ApoER2 raises the possibility that the receptor binds with the ligand in the contracted-open conformation, which seems to be primed for ligand release and adoption of the contracted-closed conformation when the receptor is recruited to the acidic endosomal compartment. In contrast, PCSK9 preferably binds with the receptor in the extended-open conformation. The interaction between the receptor and PCSK9 is stabilized in the endosome, and the receptor is subsequently transported to lysosome for degradation
Future prospect
Structural analyses on other receptor–ligand pairs need to be undertaken to expand our understanding of a ligand uptake mechanism that appears to be conserved among the LDLR family members. ApoER2 and VLDLR are known to bind with protein ligands other than reelin. Although ApoER2 binds with reelin in the contracted-open conformation, it should be examined whether the receptors bind with other ligands in similar conformations or in as-yet unidentified conformations. In addition, it is also unclear whether or not reelin preferably binds with ApoER2 in the contracted-open conformation. There is a possibility that binding to reelin induces a structural change in ApoER2 to the contracted-open conformation. Further biophysical binding analysis using mutants should be performed so as to reveal the relationship between the receptor conformation and the binding affinity to the ligand. The grand challenge will be to move beyond structures of LDLR family members in complex with protein ligands. To date, no high-resolution 3D structures of receptor bound to lipoprotein particle are available. Nevertheless, the fast-pace of recent innovative advancements in electron microscopic techniques lead us to expect structure determination of these complexes at near atomic resolutions in the future. Accumulation of structural data will help us not only obtain a deep understanding of the lipoprotein uptake mechanism, but also aid in developing new strategies for the treatment of FH.
Acknowledgements
The author would like to congratulate Prof. Fumio Arisaka on the occasion of his 70th birthday and thank him for his long-standing contributions in the development and promulgation of biophysical methods. The author thanks Samuel Thompson for editing the manuscript and Prof. Junichi Takagi for useful discussions.
Compliance with ethical standards
Conflict of interest
Terukazu Nogi declares that he has no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by the author.
Footnotes
This article is part of a Special Issue on ‘Biomolecules to Bio-nanomachines—Fumio Arisaka 70th Birthday’ edited by Damien Hall, Junichi Takagi and Haruki Nakamura.
References
- Abifadel M, Varret M, Rabès JP, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- Beglova N, Jeon H, Fisher C, Blacklow SC. Cooperation between fixed and low pH-inducible interfaces controls lipoprotein release by the LDL receptor. Mol Cell. 2004;16:281–292. doi: 10.1016/j.molcel.2004.09.038. [DOI] [PubMed] [Google Scholar]
- Bottomley MJ, Cirillo A, Orsatti L, et al. Structural and biochemical characterization of the wild type PCSK9-EGF(AB) complex and natural familial hypercholesterolemia mutants. J Biol Chem. 2009;284:1313–1323. doi: 10.1074/jbc.M808363200. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. Expression of the familial hypercholesterolemia gene in heterozygotes: mechanism for a dominant disorder in man. Science. 1974;185:61–63. doi: 10.1126/science.185.4145.61. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- Chen S, Bubeck D, MacDonald BT, et al. Structural and functional studies of LRP6 ectodomain reveal a platform for Wnt signaling. Dev Cell. 2011;21:848–861. doi: 10.1016/j.devcel.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Biechele T, Wei Z et al (2011) Crystal structures of the extracellular domain of LRP6 and its complex with DKK1. Nat Struct Mol Biol 18:1204–1210. 10.1038/nsmb.2139 [DOI] [PMC free article] [PubMed]
- Daly NL, Scanlon MJ, Djordjevic JT, Kroon PA, Smith R. Three-dimensional structure of a cysteine-rich repeat from the low-density lipoprotein receptor. Proc Natl Acad Sci USA. 1995;92:6334–6338. doi: 10.1073/pnas.92.14.6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T. A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature. 1995;374:719–723. doi: 10.1038/374719a0. [DOI] [PubMed] [Google Scholar]
- Davis CG, Goldstein JL, Sudhof TC, Anderson RG, Russell DW, Brown MS. Acid-dependent ligand dissociation and recycling of LDL receptor mediated by growth factor homology region. Nature. 1987;326:760–765. doi: 10.1038/326760a0. [DOI] [PubMed] [Google Scholar]
- Day IN, Whittall RA, O’Dell SD et al (1997) Spectrum of LDL receptor gene mutations in heterozygous familial hypercholesterolemia. Hum Mutat 10:116–127. 10.1002/(SICI)1098-1004(1997)10:2%3C116::AID-HUMU4%3E3.0.CO;2-I [DOI] [PubMed]
- Fass D, Blacklow S, Kim PS, Berger JM. Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module. Nature. 1997;388:691–693. doi: 10.1038/41798. [DOI] [PubMed] [Google Scholar]
- Fisher C, Beglova N, Blacklow SC. Structure of an LDLR-RAP complex reveals a general mode for ligand recognition by lipoprotein receptors. Mol Cell. 2006;22:277–283. doi: 10.1016/j.molcel.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Handford PA, Mayhew M, Baron M, Winship PR, Campbell ID, Brownlee GG. Key residues involved in calcium-binding motifs in EGF-like domains. Nature. 1991;351:164–167. doi: 10.1038/351164a0. [DOI] [PubMed] [Google Scholar]
- Herz J, Beffert U. Apolipoprotein E receptors: linking brain development and Alzheimer’s disease. Nat Rev Neurosci. 2000;1:51–58. doi: 10.1038/35036221. [DOI] [PubMed] [Google Scholar]
- Hirai H, Yasui N, Yamashita K et al (2017) Structural basis for ligand capture and release by the endocytic receptor ApoER2. EMBO Rep 18:982–999. 10.15252/embr.201643521 [DOI] [PMC free article] [PubMed]
- Hopkins PN, Wu LL, Stephenson SH, et al. A novel LDLR mutation, H190Y, in a Utah kindred with familial hypercholesterolemia. J Hum Genet. 1999;44:364–367. doi: 10.1007/s100380050179. [DOI] [PubMed] [Google Scholar]
- Jeon H, Meng W, Takagi J, Eck MJ, Springer TA, Blacklow SC. Implications for familial hypercholesterolemia from the structure of the LDL receptor YWTD-EGF domain pair. Nat Struct Biol. 2001;8:499–504. doi: 10.1038/88556. [DOI] [PubMed] [Google Scholar]
- Kim DH, Iijima H, Goto K, et al. Human apolipoprotein E receptor 2. A novel lipoprotein receptor of the low density lipoprotein receptor family predominantly expressed in brain. J Biol Chem. 1996;271:8373–8380. doi: 10.1074/jbc.271.14.8373. [DOI] [PubMed] [Google Scholar]
- Kim DH, Magoori K, Inoue TR, et al. Exon/intron organization, chromosome localization, alternative splicing, and transcription units of the human apolipoprotein E receptor 2 gene. J Biol Chem. 1997;272:8498–8504. doi: 10.1074/jbc.272.13.8498. [DOI] [PubMed] [Google Scholar]
- Kwon HJ, Lagace TA, MC MN, Horton JD, Deisenhofer J. Molecular basis for LDL receptor recognition by PCSK9. Proc Natl Acad Sci USA. 2008;105:1820–1825. doi: 10.1073/pnas.0712064105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagace TA, Curtis DE, Garuti R, et al. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J Clin Invest. 2006;116:2995–3005. doi: 10.1172/JCI29383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Tumanut C, Gavigan JA, et al. Secreted PCSK9 promotes LDL receptor degradation independently of proteolytic activity. Biochem J. 2007;406:203–207. doi: 10.1042/BJ20070664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Surdo P, Bottomley MJ, Calzetta A et al (2011) Mechanistic implications for LDL receptor degradation from the PCSK9/LDLR structure at neutral pH. EMBO Rep 12:1300–1305. 10.1038/embor.2011.205 [DOI] [PMC free article] [PubMed]
- Malby S, Pickering R, Saha S, Smallridge R, Linse S, Downing AK. The first epidermal growth factor-like domain of the low-density lipoprotein receptor contains a noncanonical calcium binding site. Biochemistry. 2001;40:2555–2563. doi: 10.1021/bi002322l. [DOI] [PubMed] [Google Scholar]
- McNutt MC, Kwon HJ, Chen C, Chen JR, Horton JD, Lagace TA. Antagonism of secreted PCSK9 increases low density lipoprotein receptor expression in HepG2 cells. J Biol Chem. 2009;284:10561–10570. doi: 10.1074/jbc.M808802200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNutt MC, Lagace TA, Horton JD. Catalytic activity is not required for secreted PCSK9 to reduce low density lipoprotein receptors in HepG2 cells. J Biol Chem. 2007;282:20799–20803. doi: 10.1074/jbc.C700095200. [DOI] [PubMed] [Google Scholar]
- Natarajan P, Kathiresan S. PCSK9 inhibitors. Cell. 2016;165:1037. doi: 10.1016/j.cell.2016.05.016. [DOI] [PubMed] [Google Scholar]
- Novak S, Hiesberger T, Schneider WJ, Nimpf J. A new low density lipoprotein receptor homologue with 8 ligand binding repeats in brain of chicken and mouse. J Biol Chem. 1996;271:11732–11736. doi: 10.1074/jbc.271.20.11732. [DOI] [PubMed] [Google Scholar]
- Perrey S, Ishibashi S, Kitamine T, et al. The LDL receptor is the major pathway for beta-VLDL uptake by mouse peritoneal macrophages. Atherosclerosis. 2001;154:51–60. doi: 10.1016/S0021-9150(00)00457-3. [DOI] [PubMed] [Google Scholar]
- Rashid S, Curtis DE, Garuti R, et al. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9. Proc Natl Acad Sci USA. 2005;102:5374–5379. doi: 10.1073/pnas.0501652102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Rudenko G, Ludtke SJ, Deisenhofer J, Chiu W, Pownall HJ. Model of human low-density lipoprotein and bound receptor based on cryoEM. Proc Natl Acad Sci USA. 2010;107:1059–1064. doi: 10.1073/pnas.0908004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko G, Henry L, Henderson K, et al. Structure of the LDL receptor extracellular domain at endosomal pH. Science. 2002;298:2353–2358. doi: 10.1126/science.1078124. [DOI] [PubMed] [Google Scholar]
- Schneider WJ, Nimpf J. LDL receptor relatives at the crossroad of endocytosis and signaling. Cell Mol Life Sci. 2003;60:892–903. doi: 10.1007/s00018-003-2183-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer TA. An extracellular beta-propeller module predicted in lipoprotein and scavenger receptors, tyrosine kinases, epidermal growth factor precursor, and extracellular matrix components. J Mol Biol. 1998;283:837–862. doi: 10.1006/jmbi.1998.2115. [DOI] [PubMed] [Google Scholar]
- Sun XM, Patel DD, Webb JC et al (1994) Familial hypercholesterolemia in China. Identification of mutations in the LDL-receptor gene that result in a receptor-negative phenotype. Arterioscler Thromb 14:85–94 [DOI] [PubMed]
- Takahashi S, Kawarabayasi Y, Nakai T, Sakai J, Yamamoto T. Rabbit very low density lipoprotein receptor: a low density lipoprotein receptor-like protein with distinct ligand specificity. Proc Natl Acad Sci USA. 1992;89:9252–9256. doi: 10.1073/pnas.89.19.9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissir F, Goffinet AM. Reelin and brain development. Nat Rev Neurosci. 2003;4:496–505. doi: 10.1038/nrn1113. [DOI] [PubMed] [Google Scholar]
- Trommsdorff M, Gotthardt M, Hiesberger T, et al. Reeler/disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999;97:689–701. doi: 10.1016/S0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- Usifo E, Leigh SE, Whittall RA, et al. Low-density lipoprotein receptor gene familial hypercholesterolemia variant database: update and pathological assessment. Ann Hum Genet. 2012;76:387–401. doi: 10.1111/j.1469-1809.2012.00724.x. [DOI] [PubMed] [Google Scholar]
- Verdaguer N, Fita I, Reithmayer M, Moser R, Blaas D. X-ray structure of a minor group human rhinovirus bound to a fragment of its cellular receptor protein. Nat Struct Mol Biol. 2004;11:429–434. doi: 10.1038/nsmb753. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Davis CG, Brown MS et al (1984) The human LDL receptor: a cysteine-rich protein with multiple Alu sequences in its mRNA. Cell 39:27–38 [DOI] [PubMed]
- Yasui N, Nogi T, Kitao T, Nakano Y, Hattori M, Takagi J. Structure of a receptor-binding fragment of reelin and mutational analysis reveal a recognition mechanism similar to endocytic receptors. Proc Natl Acad Sci USA. 2007;104:9988–9993. doi: 10.1073/pnas.0700438104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui N, Nogi T, Takagi J. Structural basis for specific recognition of reelin by its receptors. Structure. 2010;18:320–331. doi: 10.1016/j.str.2010.01.010. [DOI] [PubMed] [Google Scholar]
- Zong Y, Zhang B, Gu S et al (2012) Structural basis of agrin-LRP4-MuSK signaling. Genes Dev 26:247–258. 10.1101/gad.180885.111 [DOI] [PMC free article] [PubMed]