Abstract
Gliadins are well-known wheat grain proteins, particularly important in food science. They were studied as early as the 1700s. Despite their long history, it has been difficult to identify their higher-order structure as they aggregate in aqueous solution. Consequently, most studies have been performed by extracting the proteins in 70% ethanol or dilute acidic solutions. The carboxy-terminal half of α- and γ-gliadins have α-helix-rich secondary structures stabilized with intramolecular disulfide bonds, which are present in either aqueous ethanol or pure water. The amino-terminal-repeat region of α- and γ-gliadins has poly-L-proline II and β-reverse-turn structures. ω-Gliadins also have poly-L-proline II and β-reverse-turn structures, but no α-helix structure. The size and shape of gliadin molecules have been determined by assessing a variety of parameters: their sedimentation velocity in the analytical ultracentrifuge, intrinsic viscosity, small-angle X-ray scattering profile, and images of the proteins from scanning probe microscopes such as a tunneling electron microscope and atomic force microscope. Models for gliadins are either rods or prolate ellipsoids whether in aqueous ethanol, dilute acid, or pure water. Recently, gliadins have been shown to be soluble in pure water, and a novel extraction method into pure water has been established. This has made it possible to analyze gliadins in pure water at neutral pH, and permitted the characterization of hydrated gliadins. They formed hierarchical nanoscale structures with internal density fluctuations at high protein concentrations.
Keywords: Gliadin, Wheat protein, Protein aggregate, SAXS, Nanostructure
Introduction
Bread wheat (Triticum aestivum) is a globally important food crop, accounting for 20% of the calories consumed by humans. It is an important source of protein, vitamins, and minerals. Storage proteins in wheat are collectively referred to as gluten, but gluten is actually an aggregate formed from two major types of protein: gliadin and glutenin. The gluten in dough is created from these proteins by mixing wheat grain flour and water. The viscoelastic properties of bread dough, conferred by gluten, are important in the quality of common wheat foods, such as bread and noodles.
Gluten may be one of the first proteins to be studied by scientists. It was isolated in 1728 by Jacopo Beccari, a professor of chemistry at the University of Bologna. He described gluten as a sticky paste that resulted from washing dough made with wheat grain flour with dilute salt solution (Bailey 1941). About 90 years later, gluten was separated into two fractions that differed in their solubility in alcohol. The proteins in the soluble fraction were referred to as gliadins and the insoluble fraction was originally named zymon. Zymon was later renamed glutenin (Taddei 1819).
The biochemist Thomas Burr Osborne adopted the system of classification of proteins proposed by the Joint Physiological and Biochemical Committees (Chittenden et al. 1908) in his research on proteins. He categorized the plant tissue proteins as “simple proteins” and identified four major types: albumins, globulins, prolamins, and glutelins (Osborne 1924), referred to as the “Osborne fractions.” According to this system, the alcohol-soluble gliadin is a typical prolamin, soluble in 70–90% aqueous alcohol, and the alcohol-insoluble glutenins belong to the glutelin group.
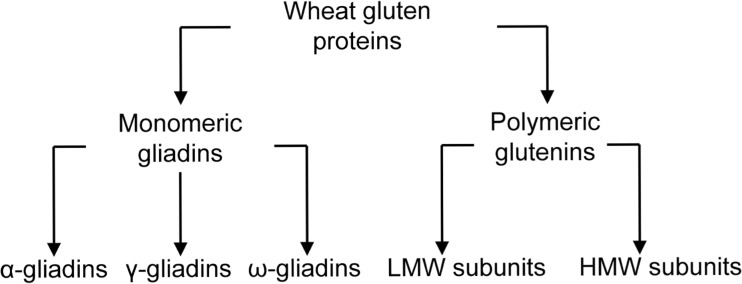
Gliadins and glutenins make up approximately 30 and 50%, respectively, of the total protein in wheat grain (Fig. 1). Considering bread dough’s rheological properties, it is believed that gliadin contributes to the flow properties, while glutenin contributes to its elasticity and strength.
Fig. 1.
Traditional (top) and molecular classification (bottom) of wheat grain proteins. (Adapted from Shewry et al. 1986)
Glutenin is composed of macropolymers, huge polymers of high- and low-molecular weight subunits crosslinked with disulfide bonds. These macropolymers intermingle randomly with individual particles of gliadins to form the aggregate, which is held together with non-covalent interactions. The arrangement of the gliadins in the aggregate has been demonstrated with immuno-localization transmission electron microscopy used with monoclonal and polyclonal antibodies selective for gliadins or the glutenin subunits (Lindsay and Skerritt 2000).
The elastic and strength properties of the dough has the greatest impact on bread making, generating considerable research into these properties. The variation in strength properties among wheat cultivars is caused by the “amount” and “quality” of glutenin. High-molecular weight glutenin appears to be particularly important to strength (Lawrence et al. 1988). Removal of the high-molecular weight glutenin had a negative effect on the quality of bread (Payne 1987). On the other hand, elevated gliadin levels decreased the dough strength and lowered the rupture viscosity (defined as the maximum elongational viscosity of the dough) but increased its rupture strain (Uthayakumaran et al. 2000).
Gluten proteins show extensive polymorphism within bread wheat cultivars. The number of genes encoding gluten proteins are believed to have been increased by duplication and translocation events. The amino acid sequences of these additional genes have altered due to substitution, deletion, and insertion events during their evolution, apparently in the absence of strong selection pressure (Shewry et al. 1984). These changes have resulted in complex mixtures of homologous proteins that vary widely in molecular mass and charge. This variation has made the isolation and study of these proteins difficult. To date, over-production studies of native gliadin with disulfide bonds in expression systems such as Escherichia coli have not been successful, preventing the identification of the structure of a single gliadin. Historically, most studies have prepared gliadins with conventional methods, which use aqueous alcohol or diluted hydrochloride solution. Alcohol and low pH cause denaturation of proteins in general, so it is unclear how accurately these studies reflect the true nature of the proteins in bread dough. Despite these difficulties, a considerable amount of effort has gone into clarifying the relationship between the properties of the dough and the structures within it, in the hope of understanding the mechanism of gluten formation and applying this knowledge to food processing technology. Here, we review these studies, beginning with the primary structure of the proteins and including the structures of the aggregates of gliadins on a nanoscale.
Primary structures of gliadins
Gliadins are large families of proteins with similar amino acid sequences. They have been classified as α-, β-, γ-, and ω-gliadins, based on their electrophoretic mobility in two-dimensional electrophoresis with isoelectric focusing in the first dimension and starch gel electrophoresis at acidic pH in the second dimension (Wrigley and Shepherd 1973). Cells of bread wheat are hexaploid and composed of genomes A, B, and D. Studies of genetic crosses reveal that the γ- and ω-gliadins are controlled by clusters of genes, Gli-A1, Gli-Bl, and Gli-Dl, located on the short arms of the group 1 chromosomes. The α-and β-gliadins are controlled by Gli-A2, Gli-B2, and Gli-D2 on the short arms of the group 2 chromosomes (Payne 1987). There are from 25 to 150 copies of the genes that encode the α-gliadins, depending on the variety (Anderson and Greene 1997). The γ-gliadins and ω-gliadins are encoded by clusters of gene families, composed of 15–40 and 15–18 copies, respectively (Sabelli and Shewry 1991; Anderson et al. 2001). Comparisons of amino acid sequences deduced from DNA sequences have demonstrated that α- and β-gliadins have similar sequences, leading to the proposal that they be combined, so that the gliadins would be subdivided into α, γ, and ω groups (Fig. 1; Shewry et al. 1986; Kasarda et al. 1987).
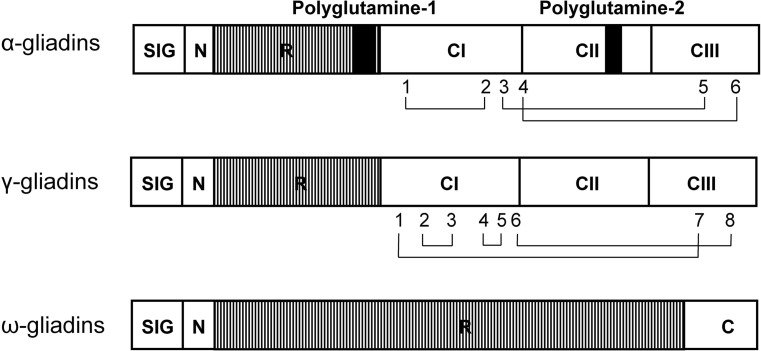
The α-gliadins usually contain a signal peptide of 20 amino acids, an N-terminal region of 5 residues, a repetitive domain of 110–130 residues, and a C-terminal region of 140–160 residues (Kasarda 1980). The C-terminal region is distinguished by a cysteine-rich region (CI) that contains four cysteine residues, a glutamine-rich region (CII) that contains stretches of glutamine residues, and a sequence of 35–39 residues (CIII) with the final two cysteine residues (Fig. 2; Shewry et al. 2009). Six of the cysteine residues form three intramolecular disulfide bonds (Müller and Wieser 1995). The N-terminal repetitive domains contain a repeat motif: P(F/Y)PQ3–5. Two stretches of polyglutamine are present in the C-terminal part of the repetitive domain and in CII of the C-terminal region. Alpha-gliadins vary in mass from 30 to 34 kD, and this variation is attributed to variation in the lengths of the repetitive domain and the two polyglutamine stretches.
Fig. 2.
Schematic representation of α-gliadins, γ-gliadins, and ω-gliadins. SIG signal peptide; N N-terminal region; R repetitive domain (small boxes indicate repeat motifs). C1, CII, CIII and C represent C-terminal subregions. Black boxes in α- gliadins represent polyglutamine peptides; numbers indicate cysteine residues; and lines connecting numeric characters indicate disulfide bonds
The γ-gliadins contain a signal peptide of 19 amino acids, an N-terminal region of 12 residues, a repetitive domain of 80–160 residues, and a C-terminal region of 140–150 residues. The C-terminal region is distinguished by a cysteine-rich region that contains six cysteine residues (CI), a glutamine-rich region (CII) containing stretches of glutamine residues, and a sequence of 41–43 residues (CIII) that contains the final two cysteine residues (Fig. 2). Disulfide bonds form between eight of the cysteine residues (Müller and Wieser 1997; Shewry and Tatham 1997). The N-terminal repetitive domains of γ-gliadins are composed of a repeat motif: PFPQQ0–1(PQQ)1–2 (Hsia and Anderson 2001). The mass of γ-gliadins ranges from 26 to 36 kD, due to variations in the length of the repetitive domain.
The ω-gliadins contain a signal peptide of 19 amino acids, an N-terminal region of 11 residues, a repetitive domain of approximately 238 residues, and a C-terminal region of 12 residues, and have no cysteine residues (Fig. 2). The repetitive domains of ω-gliadins are composed of a repeat motif: PFPQ1–2PQ1–2 that is similar to that of γ-gliadins.
The formation of the disulfide bonds of the γ-gliadins was catalyzed by proteins in the protein disulfide isomerase family in an in vitro wheat-germ translation system with dog pancreas microsomes (Bulleid and Freedman 1988). The evidence suggests that these disulfide bonds are necessary to correct the folding and stability of the protein structure. Kimura et al. (2015) showed that four groups of protein disulfide family proteins were highly expressed in the cotyledon cells of wheat grain during grain filling, and that the formation of these bonds in the endoplasmic reticulum is essential for their transport to the Golgi apparatus and deposition into protein bodies. Mutations that affect the cysteine residues or the reduction of the disulfide bonds resulted in precipitation into insoluble aggregates in the endoplasmic reticulum (Altschuler and Galili 1994; Shimoni and Galili 1996; Orsi et al. 2001).
Solvent solubility of gliadins
The consensus definition of gliadins is that they are proteins in wheat grain that are soluble in aqueous alcohols, but insoluble in water or neutral salt solutions (Kasarda et al. 1967). Most of the past studies about gliadins have used gliadin preparations extracted with aqueous alcohol. On the other hand, some researchers have reported that a considerable amount of gluten protein could be extracted with water from gluten to which NaCl had been added. For example, when gluten was homogenized in 1 M NaCl and repeatedly washed with pure water, both gliadin and glutenin could be extracted with distilled water. The amount depended on the concentration of NaCl with which the gluten had been pre-treated (Clements 1973; Fu et al. 1996). Ukai et al. (2008) and Sato et al. (2015) established a method to extract gliadins into pure water from bread dough containing 0.5 M NaCl; the method permitted the extraction of most of the gliadins with few contaminants.
The gliadins extracted by this procedure were soluble in neutral pure water that had been degassed. This method enables researchers to analyze gliadins in conditions close to those in real dough (made of wheat flour, water, and salt).
The question remains: why can gliadins only be extracted from salt-treated dough? It may be that interactions between ions, proteins and water are relevant to this question. Intramolecular disulfide bonds are essential for the solubility of gliadin in pure water. When the intramolecular disulfide bond of α- and γ-gliadins are cleaved by a reducing reagent, they become insoluble in pure water, but still soluble in 70% (v/v) aqueous ethanol (unpublished data). Apparently, the structures of α- and γ- gliadins that are stabilized by disulfide bonds are essential for the proteins to be soluble in water, while solubility in aqueous alcohol may depend only on the primary structure. The gliadins extracted with pure water are very sensitive to small amounts of a variety of ions, suggesting that the ions make gliadins aggregate by neutralizing the charged side chains of amino acid residues of gliadins (Ukai et al. 2008).
Secondary structure of gliadins
The circular dichroism (CD) spectra of α-gliadins was measured at pH 3 and 5 (in HCl; Kasarda et al. 1968). At pH 5, the spectra were consistent with approximately one-third of the protein being in a helical conformation, and there was a small conformational change at pH 3. The CD spectra were also assessed in 70% (v/v) ethanol (Tatham et al. 1985b, 1987), and indicate that the amount of the α-helix form in the α-gliadins and γ-gliadins are similar: 33–37 and 37%, respectively. In contrast, there was more β-sheet structure in the γ-gliadins (20–23%) than in the α-gliadins (11–12%). Based on a Raman optical activity spectrum of α-gliadin in 10 mM acetic acid (pH 3.5), most of the α-helical form lies within a C-terminal domain. This method revealed less of the β-sheet form than was reported by Tatham et al., and the results were consistent with the presence of a poly(l-proline) II (PPII) helix (Blanch et al. 2003). Based on these structural studies, it has been predicted that the repetitive domains of the α-gliadins consist of a mixture of poly-L-proline II and β-reverse-turn structures and that the non-repetitive domains are rich in α-helical structure (Matsushima et al. 1990; Tatham and Shewry 1995; Arêas and Cassiano 2001; Hsia and Anderson 2001; Matsuo et al. 2005; Altenbach and Kothari 2007). We have studied recombinant α-gliadin synthesized in an E. coli system. This protein has no disulfide bonds, and had a only very small amount of α-helical structure and more random coiling than reported by others (unpublished data), leading us to suggest that the disulfide bonds formed in the C-terminal region stabilize the α-helix. Addition of methanol to α-gliadin in aqueous HCl solution (pH 3.6) increased the amount of the α-helix form, apparently at the expense of some of the β-sheet and PPII structures (Blanch et al. 2003).
The CD spectra of ω-gliadins are quite different from those of the α- and γ-gliadins. The secondary structures of the ω-gliadins are rich in β-turns but they apparently have no α-helix or β-sheet (Tatham et al. 1985a). The Raman optical activity spectrum of ω-gliadins is consistent with a large amounts of well-defined PPII structure with some turns and no α-helical structure (Blanch et al. 2003). It is important to keep in mind that the structural studies of the gliadins are limited.
Size and shape of gliadin molecules
Details of the tertiary structure of gliadins are uncertain, as traditional techniques, such as X-ray crystallography and so on, have been unsuccessful with these proteins. Information acquired from other types of analyses has been conflicting. The α-gliadins in 0.01 M NaCl (pH 4) had low intrinsic viscosity (4.0 mL g- l), which is in the range observed for globular proteins, supporting the suggestion that α-gliadins have a compact structure (Cole et al. 1984). In another study, however, thermograms were acquired from freeze-dried gliadins extracted with 50% (v/v) 1-propanol by differential scanning calorimetry. The enthalpies associated with the protein transitions were very small (León et al. 2003). Ukai et al. have reported that, in gliadins dissolved in pure water, there was no signal of transition (Ukai et al. 2008). Both of these results are consistent with the conclusion that gliadins have unusual structures composed of multiple disordered regions, rather than a compact structure. In other studies, it appeared that α -, γ-, and ω-gliadins are extended molecules with axial ratios ranging from approximately 1 to 3. This conclusion is based on the proteins’ weight-average sedimentation coefficients and weight-average molecular weights from sedimentation equilibrium data obtained by analytical ultracentrifuge analysis (Ang et al. 2010). Alpha-gliadins appeared to be the most extended and γ-gliadins the least.
Small-angle X-rays scattering (SAXS), which provides ensemble-averaged structural information even with non-crystallized molecules in a non-destructive manner, has also been used in the structural analysis of gliadins. Thomson et al. (1999) investigated the structure of the gliadins in 70% (v/v) aqueous ethanol in the concentration range of 1 to 10 mg of protein/mL, and analyzed their SAXS profiles with Guinier and cross-section Guinier approximation. They measured the radii of gyration (Rg) and radii of gyration of the cross-section (Rc) of α-, γ-, and ω-gliadins in aqueous ethanol as 3.55, 3.80, and 4.60 nm and 1.10, 1.15, and 1.15 nm, respectively (Table 1). Random-coil and chemically denatured crosslink-free polypeptides consisting of 300–400 amino acid residues typically have Rg values of approximately 5–6.5 nm (Kohn et al. 2004). Since ω-gliadins do not have crosslinks and have an Rg of less than 5 nm, it would seem that they do not entirely extend in aqueous ethanol. The Rg of ω-gliadin increased markedly with increasing protein concentration within the range of 1 to 10 mg/mL, but the Rc values did not vary with the concentration, suggesting that ω-gliadins aggregate end to end.
Table 1.
Gliadin molecular parameters derived from SAXS
| Protein | Solvent | Rg (nm) | Rc (nm) | d a (nm) | L b (nm) | 2a c (nm) | Lcd (nm) | Reference |
|---|---|---|---|---|---|---|---|---|
| Gliadin mixture | Pure water | 4.10–6.46 | 2.65 | 7.50 | 10.8 | – | – | Sato et al. 2015 |
| α-gliadins | 70% (v/v) ethanol | 3.55 | 1.10 | 3.20 | 11.7 | 15.1 | 15.8 | Thomson et al. 1999 |
| γ-gliadins | 70% (v/v) ethanol | 3.80 | 1.15 | 3.25 | 12.5 | 16.2 | 17.7 | Thomson et al. 1999 |
| 1% acetic acid | 2.66 | 1.22 | 3.40 | 9.2 | – | – | Thomson et al. 1992 | |
| ω-gliadins | 70% (v/v) ethanol | 4.60 | 1.15 | 3.25 | 15.4 | 19.9 | 26.3 | Thomson et al. 1999 |
aThe diameter calculated assuming a rod model
bThe length calculated assuming a rod model
cTwice the major semi-axis calculated assuming a prolate ellipsoid
dThe contour length
Prediction of the shapes of gliadins has been undertaken by calculating the molecular dimensions (d) and semi-major axes (a) of the gliadins in 70% ethanol, assuming both a rod model and a prolate ellipsoid model. When the semi-major axes of α- and γ-gliadins for a prolate ellipsoid were multiplied by 2 (2a), the values were close to the contour length (Lc) for the proteins, suggesting that the prolate ellipsoid is a more appropriate model than a rod. The large discrepancy between 2a and Lc of the ω-gliadin at 1 mg protein/mL, the lowest concentration, may be due to a considerable amount of aggregation or interparticle interference.
These results from the studies just discussed shed light on the nanostructure of gluten protein molecules at low concentrations in alcohol solution, but their structure may differ in an alcohol-free solution. Only a few studies have been done of gliadin structure in acidic aqueous solutions. Thomson et al. (1992) undertook a study of the γ-gliadins in 1% acetic acid (pH approximately 2.5) at protein concentrations of 3 mg/mL. The Rg and Rc values of the γ-gliadins under this condition were 2.66 and 1.22 nm, respectively, based on the SAXS profiles with Guinier and cross-section Guinier plots. The molecular diameters and maximum length of the γ-gliadins were calculated, assuming a rod model, as 3.4 and 9.2 nm, respectively. Thomson et al. (1992) also performed scanning tunneling microscopy (STM) of single molecules of γ-gliadins. In the images acquired from this study, two molecules aggregated head to tail were observed. The individual molecules were approximately 12 nm long, with a broad end of approximately 4.7 nm diameter and a narrow end of approximately 2.5 nm diameter. These values are in reasonable agreement with the dimensions determined by SAXS. The STM images of α-gliadin revealed a much more compact conformation than observed for γ-gliadin. α-Gliadin had diameters of approximately 6.4 × 5.1 nm (Shewry et al. 1997; Tatham et al. 1999), which yielded an axial ratio of 1.25: 1. This is close to the axial ratio of 1.6:1 that was acquired from the intrinsic viscosity (Cole et al. 1984). Single, isolated molecules of ω-gliadins deposited on highly oriented pyrolytic graphite by evaporation from a solution in trifluoroethanol were also observed (Shewry et al. 1997). These were extended rods with approximate dimensions of 11–18 nm length and 3.5–6.3 nm diameter. However, these values conflict with measurements of the Lc of the barley (Hordeum vulgare) seed homolog of the ω-gliadins, C hordeins, which have a repeat motif: PQQPFPQQ that is similar to that of ω-gliadins. This was calculated by SAXS analysis as 71.5 nm in 0.1 M acetic acid (I'Anson et al. 1992). The data for C hordein are consistent with the idea that this protein has a stiff, “worm-like,” chain conformation. In contrast, the Lc of ω-gliadins in 70% (v/v) ethanol was 26.3 nm, as calculated by SAXS analysis (Thomson et al. 1999). The different results could be due to the proteins adopting different conformations at different degrees of hydration and different pH. Sato et al. (2015) performed SAXS analysis of gliadin solution in pure water. The size of gliadin molecules could not be described well by a simple Guinier approximation, even at 0.025 wt%, indicating polydispersity of the scattering components. Consequently, they employed a three-component Guinier equation:
which fits the scattering profiles well. The smallest Rg (R g1) values in this study were from 4.10 to 6.46 nm greater than those of the Rg values (3.55 and 3.80 nm) for α- and γ-gliadins in 70% (v/v) aqueous ethanol. In addition, the Rc value of 2.65 nm were greater by a factor of 2 than those for α-, γ-, and ω-gliadins in 70% (v/v) aqueous ethanol. By assuming a rod structure for the gliadins at 0.025 wt% in pure water, the shape parameters were calculated to be 10.8 nm long and 7.5 nm in diameter. In analytical ultracentrifugation analysis of gliadins in pure water, the sedimentation coefficient of the major peak, corresponded to a molecular weight of 25.7 kDa, indicating that most of the extracted gliadins were in the monomeric form (Sato et al. 2015). Therefore, these differences of Rg and Rc of gliadins may due to the structural difference between gliadins in aqueous ethanol, acidic solutions, and pure water. In fact, an increase in alcohol concentration causes an increase in the α-helix content at the expense of some of the β-sheet and PPII structures in gliadins (Blanch et al. 2003). In general, alcohols, especially trifluoroethanol, induce α-helical structures in proteins in a manner that affects the solvation structure and the free-energy change associated with the helix-to-coil transition of a polypeptide (Hirota et al. 1999, Imai et al. 2009).
Aggregated structures of native gliadins
Gliadins are soluble in pure water when they are at concentrations of less than 10% by weight (wt%). At these concentrations, they yield a transparent solution. When they are in solutions of greater than approximately 15 wt%, gliadins form gel-like hydrated solids. These will not flow even if the container is inverted at 40 wt% or more. This behavior is consistent with the formation of a network structure at high gliadin concentrations. It is important to identify the structure of these hydrated gliadin aggregates to understand the molecular mechanism underlying the rheological properties that are so important in the food industry.
Atomic force microscopy (AFM), which can be applied to liquids, has been used to study the aggregative behavior of gliadins. The AFM images of fibril formation of α-gliadins in the presence of 10 mM ammonium acetate obtained in the tapping mode showed a network of fibrils with widths between 15 and 30 nm and lengths between 100 nm to 2 μm (McMaster et al. , b). As their concentration increased, the α-gliadins formed a dense network of fibrils. AFM can be used to map the “chemical” nature of a surface. When this ability was applied to α- and ω-gliadins, they were shown to form separate phases in a gliadin film. This may be relevant to their behavior in foods, particularly doughs.
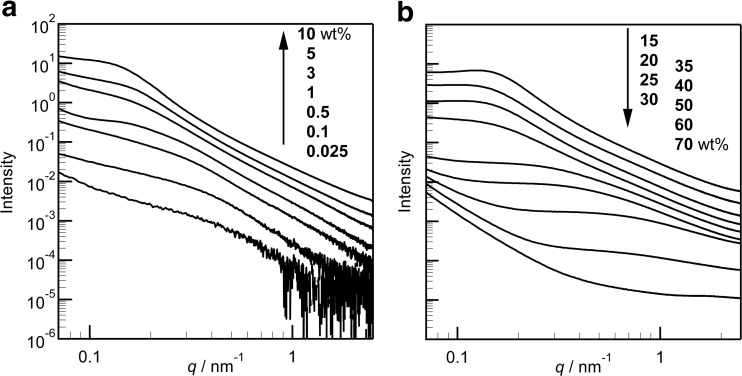
The SAXS analysis of the structures attained by gliadins over a wide range of their concentrations in aqueous solutions was assessed by Sato et al. 2015. A shoulder peak in the SAXS profiles appeared above 1 wt% of the solution (Fig. 3). The correlation length was calculated from the scattering vector value, q, of the shoulder peak, using the Bragg equation. The correlation length, L, increased with increasing gliadin concentration, suggesting that the size of the aggregates of gliadins increased with concentration and the distance between the aggregates also increased (Table 2). At concentrations above 15 wt%, a top-of-the-shoulder peak shifted to a higher q value, suggesting shortening of the distance between the nanoparticles of the gliadins compared to lower concentrations, and there was an upturn in the lower q region, indicative of the presence of aggregates larger than 100 nm. We consider that these data support the suggestion that the hydrated gliadins form hierarchical nanoscale structures at this concentration range (Fig. 4). When the concentration of gliadins was increased to 70 wt%, the broad shoulder peak was almost unrecognizable, and only the upturn in the lower q region remained.
Fig. 3.
SAXS profiles of gliadins in distilled water. a Solution components at low concentrations; b gel-like solid samples at high concentrations. The curves are vertically shifted for clarity. (From Sato et al. 2015)
Table 2.
Peak positions (qs) and correlation lengths (L) from Bragg’s law (from Sato et al. 2015)
| Gliadin concentration (wt%) | qs (nm−1) | L (nm) |
|---|---|---|
| 1 | 0.210 | 29.9 |
| 3 | 0.169 | 37.2 |
| 5 | 0.165 | 38.0 |
| 10 | 0.139 | 45.3 |
| 15 | 0.150 | 41.9 |
| 20 | 0.144 | 43.4 |
| 25 | 0.145 | 42.7 |
| 30 | 0.147 | 40.7 |
| 35 | 0.304 | 20.6 |
| 40 | 0.434 | 14.5 |
| 50 | 0.665 | 9.45 |
| 60 | 0.741 | 8.48 |
Fig. 4.
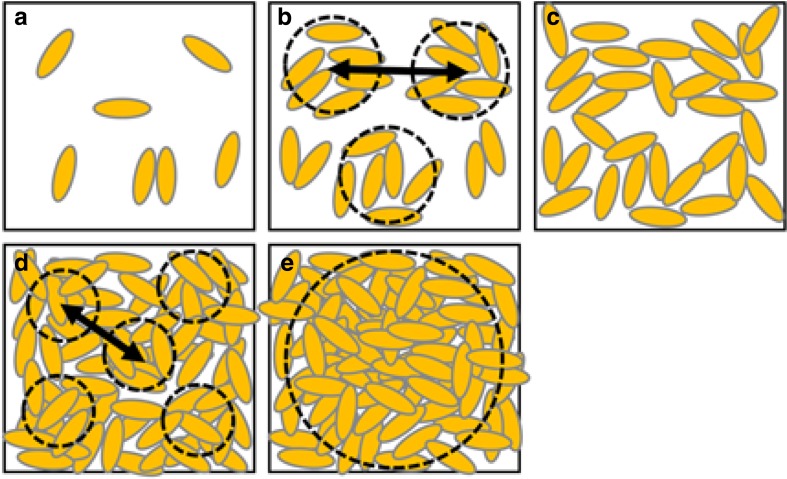
Schematic illustrations of the nanostructures of gliadin assemblies in distilled water over a wide range of concentrations. a Gliadins are present as water-soluble isolated monomers and a few dimers at 0.025–0.5 wt%. b Gliadin molecules self-assemble to form small aggregates (dashed line circles) at 0.5–10 wt%. The distance between the aggregates is estimated to be ∼40 nm. c Gliadins begin to form continuous networks at 15–20 wt%. d Gliadin molecules fill the space, but condensed regions due to density fluctuation (dashed line circles) appear above 30 wt%. The distance between dense domains is estimated to be 14 nm at 40 wt%. e Above 50 wt%, the density fluctuation almost vanishes, but large aggregates over 100 nm in size are formed
The existence of a nanoparticle ensemble of hydrated gliadins at high concentrations should provide a useful model for representing gliadins in the gluten of dough used in the making of bread and noodles. Salt is frequently added to breads and noodles in order to tighten the dough. Recently, we showed that NaCl shortens the distance between gliadin nanoparticles, resulting in a dramatic increase in the tensile strength, and changes the elongation property of the gliadin hydrate (submitted). We also noted that the effects of NaCl were not as pronounced on glutenins than they were on gliadins. It may be that the well-known tightening effect of salt on dough can be explained mainly by its effects of gliadin nanoparticles.
Future perspectives
The difficulty inherent in the preparation of a single type of gliadin has impeded studies on these proteins’ higher-order and agglomerated structures. Additionally, many studies of gliadins have been performed with the proteins suspended in aqueous ethanol or acidic solutions, since they are insoluble in neutral aqueous solution. These studies have revealed some intriguing and unusual properties of these proteins. However, the agglomerated structure of gliadins depends on their interactions with water and other low molecular weight compounds in the system. These can only be assessed when the gliadins are solubilized or hydrated in near-neutral aqueous solutions. Gliadins prepared in pure water or wheat grain dough containing NaCl is useful for this purpose, although only a few studies have used these conditions to date. We look forward to studies that combine these conditions with powerful methods such as quantum beam scattering and high-speed AFM, as such studies may reveal the true nature and behavior of gliadins in gluten.
Acknowledgements
This work was supported by JSPS KAKENHI Grant Number 26660111 to R.U, JP15H02042 to M.S. and a grant from the Tojuro Iijima Foundation for Food Science and Technology.
Compliance with ethical standards
Conflict of interest
Reiko Urade declares that she has no conflicts of interest. Nobuhiro Sato declares that he has no conflicts of interest. Masaaki Sugiyama declares that he has no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is part of a Special Issue on ‘Biomolecules to Bio-nanomachines - Fumio Arisaka 70th Birthday’ edited by Damien Hall, Junichi Takagi and Haruki Nakamura.
References
- Altenbach SB, Kothari KM. Omega gliadin genes expressed in Triticum aestivum cv. Butte 86: effects of post-anthesis fertilizer on transcript accumulation during grain development. J Cereal Sci. 2007;46:169–177. doi: 10.1016/j.jcs.2007.02.001. [DOI] [Google Scholar]
- Altschuler Y, Galili G. Role of conserved cysteines of a wheat gliadin in its transport and assembly into protein bodies in xenopus oocytes. J Biol Chem. 1994;269:6677–6682. [PubMed] [Google Scholar]
- Anderson OD, Greene FC. The α-gliadin gene family. II. DNA and protein sequence variation, subfamily structure, and origins of pseudogenes. Theor Appl Genet. 1997;95:59–65. doi: 10.1007/s001220050532. [DOI] [Google Scholar]
- Anderson OD, Hsia CC, Torres V. The wheat γ-gliadin genes: characterization of ten new sequences and further understanding of γ-gliadin gene family structure. Theor Appl Genet. 2001;103:323–330. doi: 10.1007/s00122-001-0551-3. [DOI] [Google Scholar]
- Ang S, Kogulanathan J, Morris AM, Kök MS, Shewry PR, Tatham AS, Adams GG, Rowe AJ, Harding SE. Structure and heterogeneity of gliadin: a hydrodynamic evaluation. Eur Biophys J. 2010;39:255–261. doi: 10.1007/s00249-009-0529-7. [DOI] [PubMed] [Google Scholar]
- Arêas EPG, Cassiano MM. Folding interpenetration in a gliadin model: the role of the characteristic octapeptide motif. Biophys Chem. 2001;90:135–146. doi: 10.1016/S0301-4622(01)00138-7. [DOI] [PubMed] [Google Scholar]
- Bailey CH. A translation of Beccari's lecture “concerning grain” (1728) J Cereal Chem. 1941;18:555–561. [Google Scholar]
- Blanch EW, Kasarda DD, Hecht L, Nielsen K, Barron LD. New insight into the solution structures of wheat gluten proteins from Raman optical activity. Biochemistry. 2003;42:5665–5673. doi: 10.1021/bi027059y. [DOI] [PubMed] [Google Scholar]
- Bulleid NJ, Freedman RB. Defective co-translational formation of disulphide bonds in protein disulphide-isomerase-deficient microsomes. Nature. 1988;335:649–651. doi: 10.1038/335649a0. [DOI] [PubMed] [Google Scholar]
- Chittenden RH, Folin O, Gies WJ, Koch W, Osborne TB, Osborne TB, Levene PA, Mandel JA, Mathews AP, Mendel LB. Joint recommendations of the physiological and biochemical committees on protein nomenclature. Science. 1908;27:554–556. doi: 10.1126/science.27.692.554. [DOI] [PubMed] [Google Scholar]
- Clements RL. Effects of prior salt treatment on gluten dispersibility. Cereal Chem. 1973;50:87–100. [Google Scholar]
- Cole EW, Kasarda DD, Lafiandra D. The conformational structure of a-gliadin intrinsic viscosities under conditions approaching the native state and under denaturing conditions. Biochim Biophys Acta. 1984;787:244–251. doi: 10.1016/0167-4838(84)90315-7. [DOI] [Google Scholar]
- Fu BX, Sapirstein HD, Bushuk W. Salt-induced disaggregation / solubilization of gliadin and glutenin proteins in water. J Cereal Sci. 1996;24:241–246. doi: 10.1006/jcrs.1996.0056. [DOI] [Google Scholar]
- Hirota N, Mizuno K, Goto U. Group additive contributions to the alcohol-induced α-helix formation of melittin: implication for the mechanism of the alcohol effects on proteins. J Mol Biol. 1999;275:365–378. doi: 10.1006/jmbi.1997.1468. [DOI] [PubMed] [Google Scholar]
- Hsia CC, Anderson OD. Isolation and characterization of wheat -gliadin genes. Theor Appl Genet. 2001;103:37–44. doi: 10.1007/s00122-001-0552-2. [DOI] [Google Scholar]
- I'Anson KJ, Morris VJ, Shewry PR, Tatham AS. Small-angle X-ray-scattering studies of the C hordeins of barley (Hordeum vulgare) Biochem J. 1992;287:183–185. doi: 10.1042/bj2870183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Kovalemko A, Hirata F, Kidera A. Molecular thermodynamics of trifluoroethanol-induced helix formation: analysis of the solvation structure and free energy by the 3D-RISM theory. Interdiscip Sci. 2009;1:156–160. doi: 10.1007/s12539-009-0037-6. [DOI] [PubMed] [Google Scholar]
- Kasarda DD. Structure and properties of α-gliadin. Annal Technol Agric. 1980;29:151–173. [Google Scholar]
- Kasarda DD, Bernardin JE, Thomas RS. Reversible aggregation of α-gliadin to fibrils. Science. 1967;155:203–205. doi: 10.1126/science.155.3759.203. [DOI] [PubMed] [Google Scholar]
- Kasarda DD, Bernardin JE, Gaffield W. Circular dichroism and optical rotatory dispersion of α-gliadin. Biochemistry. 1968;7:3950–3957. doi: 10.1021/bi00851a023. [DOI] [PubMed] [Google Scholar]
- Kasarda DD, Adalstein AE, Laird NF. γ-Gliadins with α-type structure coded on chromosome 6B of the wheat (Triticum aestivum L.) cultivar ‘Chinese spring’. In: Lásztity R, Békés F, editors. Proc. 3rd Int. workshop on gluten proteins. Singapore: World Scientific Publishing; 1987. pp. 20–29. [Google Scholar]
- Kimura S, Higashino Y, Kitao Y, Masuda T, Urade R. Expression and characterization of protein disulfide isomerase family proteins in bread wheat. BMC Plant Biol. 2015;15:73. doi: 10.1186/s12870-015-0460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn JE, Millett IS, Jacob J, Zagrovic B, Dillon TM, Cingel N, Dothager RS, Seifert S, Thiyagarajan P, Sosnick TR, Hasan MZ, Pande VS, Ruczinski I, Doniach S, Plaxco KW. Random-coil behavior and the dimensions of chemically unfolded proteins. Proc Natl Acad Sci U S A. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence G, MacRitchie F, Wrigley CW. Dough and baking quality of wheat lines deficient in glutenin subunits controlled by the Glu-Al, Glu-Bland Glu-Dl loci. J Cereal Sci. 1988;7:109–112. doi: 10.1016/S0733-5210(88)80012-2. [DOI] [Google Scholar]
- León A, Rosell CM, Barber CB. A differential scanning calorimetry study of wheat proteins. Eur Food Res Technol. 2003;217:13–16. doi: 10.1007/s00217-003-0699-y. [DOI] [Google Scholar]
- Lindsay MP, Skerritt JH. Immunocytochemical localization of gluten proteins uncovers structural organization of glutenin macropolymer. Cereal Chem. 2000;77:360–369. doi: 10.1094/CCHEM.2000.77.3.360. [DOI] [Google Scholar]
- Matsuo H, Kohno K, Morita E. Molecular cloning, recombinant expression and IgE binding ω-5 gliadin is a major allergen in wheat-dependent exercise-induced anaphylaxis. FEBS J. 2005;272:4431–4438. doi: 10.1111/j.1742-4658.2005.04858.x. [DOI] [PubMed] [Google Scholar]
- Matsushima N, Creutz CE, Kretsinger RH. Polyproline, β-turn helices. Novel secondary structures proposed for the tandem repeat within rhodopsin, synaptophysin, synexin, gliadin, RNA polymerase II, hordein, and glute. Proteins Struct Funct Genet. 1990;7:125–155. doi: 10.1002/prot.340070204. [DOI] [PubMed] [Google Scholar]
- McMaster TJ, Miles MJ, Kasarda DD, Shewry PR, Tatham AS. Atomic force microscopy of A-gliadin fibrils and in situ degradation. J Cereal Sci. 1999;31:281–286. doi: 10.1006/jcrs.2000.0307. [DOI] [Google Scholar]
- McMaster TJ, Miles MJ, Wannerberger L, Eliasson A-C, Shewry PR, Tatham AS. Identification of microphases in mixed α- and ω-gliadin protein films investigated by atomic force microscopy. J Agric Food Chem. 1999;47:5093–5099. doi: 10.1021/jf9904057. [DOI] [PubMed] [Google Scholar]
- Müller S, Wieser H. The location of disulphide bonds in γ-type gliadins. J Cereal Sci. 1995;22:21–27. doi: 10.1016/S0733-5210(05)80004-9. [DOI] [Google Scholar]
- Müller S, Wieser H. The location of disulfide bonds in monomeric γ-type gliadins. J Cereal Sci. 1997;26:169–176. doi: 10.1006/S0733-5210(97)90100-4. [DOI] [Google Scholar]
- Orsi A, Sparvoli F, Ceriotti A. Role of individual disulfide bonds in the structural maturation of a low molecular weight glutenin subunit. J Biol Chem. 2001;276:32322–32329. doi: 10.1074/jbc.M103833200. [DOI] [PubMed] [Google Scholar]
- Osborne TB. The vegetable proteins. 2. London: Longmans Green; 1924. [Google Scholar]
- Payne P (1987) Genetics of wheat storage proteins and the effect of allelic variation on bread-making quality. Ann Rev Plant Physiol 38:141–153
- Sabelli P, Shewry PR. Characterization and organization of gene families at the Gli-1 loci of bread and durum wheats by restriction fragment analysis. Theor Appl Genet. 1991;83:209–216. doi: 10.1007/BF00226253. [DOI] [PubMed] [Google Scholar]
- Sato N, Matsumiya A, Higashino Y, Funaki S, Kitao Y, Oba Y, Inoue R, Arisaka F, Sugiyama M, Urade R. Molecular assembly of wheat gliadins into nanostructures: a small-angle x-ray scattering study of gliadins in distilled water over a wide concentration range. J Agric Food Chem. 2015;63:8715–8721. doi: 10.1021/acs.jafc.5b02902. [DOI] [PubMed] [Google Scholar]
- Shewry PR, Tatham AS. Disulphide bonds in wheat gluten proteins. J Cereal Sci. 1997;25:207–227. doi: 10.1006/jcrs.1996.0100. [DOI] [Google Scholar]
- Shewry PR, Miflin BJ, Kasarda DD. The structural and evolutionary relationships of the prolamin storage proteins of barley, rye and wheat. Philos Trans R Soc Lond B. 1984;304:297–308. doi: 10.1098/rstb.1984.0025. [DOI] [Google Scholar]
- Shewry PR, Tatham AS, Forde J, Kreis M, Miflin BJ. The classification and nomenclature of wheat gluten proteins: a reassessment. J Cereal Sci. 1986;4:97–106. doi: 10.1016/S0733-5210(86)80012-1. [DOI] [Google Scholar]
- Shewry PR, Miles MJ, Tompson NH, Tatham AS. Scanning probe microscopes - applications in cereal science. Cereal Chem. 1997;74:193–199. doi: 10.1094/CCHEM.1997.74.3.193. [DOI] [Google Scholar]
- Shewry PR, D’Ovidio R, Lafiandra D, Jenkins JA, Mills ENC, Békés F. Wheat grain proteins. In: Khan K, Shewry PR, editors. Wheat: chemistry and technology. 4. Minnesota: AACC International; 2009. pp. 223–298. [Google Scholar]
- Shimoni Y, Galili G. Intramolecular disulfide bonds between conserved cysteines in wheat gliadins control their deposition into protein bodies. J Biol Chem. 1996;271:18869–18874. doi: 10.1074/jbc.271.31.18869. [DOI] [PubMed] [Google Scholar]
- Taddei G. Ricerchesulglutine del frumento. Giornale di fisica, chimica, e storianaturale. Brugnatelli. 1819;2:360–361. [Google Scholar]
- Tatham AS, Shewry PR. The S-poor prolamins. J Cereal Sci. 1995;2:99–103. [Google Scholar]
- Tatham AS, Drake AF, Shewry PR. A conformational study of a glutamine-rich and proline-rich cereal seed protein, C-hordein. Biochem J. 1985;226:557–562. doi: 10.1042/bj2260557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham AS, Miflin B, Shewry PR. The β-turn conformation in wheat gluten proteins - relationship to gluten elasticity. Cereal Chem. 1985;62:405–411. [Google Scholar]
- Tatham AS, Field JM, Smith SJ, Shewry PR. The conformations of wheat gluten proteins, II*, aggregated gliadins and low molecular weight subunits of glutenin. J Cereal Sci. 1987;5:203–214. doi: 10.1016/S0733-5210(87)80023-1. [DOI] [Google Scholar]
- Tatham AS, Tomson NH, McMaster TJ, Humphris ADL, Miles MJ. Scanning probe microscopy studies of cereal seed storage protein structures. Scanning. 1999;21:293–298. doi: 10.1002/sca.4950210502. [DOI] [Google Scholar]
- Thomson NH, Miles MJ, Tatham AS, Shewry PR. Molecular images of cereal proteins by STM. Ultramicroscopy. 1992;42-44(Pt B):1204–1213. doi: 10.1016/0304-3991(92)90425-J. [DOI] [PubMed] [Google Scholar]
- Thomson NH, Miles MJ, Popineau Y, Harries J, Shewry PR, Tatham AS. Small angle X-ray scattering of wheat seed-storage proteins: α-, γ- and ω-gliadins and the high molecular weight (HMW) subunits of glutenin. Biochim Biophys Acta. 1999;1430:359–366. doi: 10.1016/S0167-4838(99)00019-9. [DOI] [PubMed] [Google Scholar]
- Ukai T, Matsumura Y, Urade R. Disaggregation and reaggregation of gluten proteins by sodium chloride. J Agric Food Chem. 2008;56:1122–1130. doi: 10.1021/jf0725676. [DOI] [PubMed] [Google Scholar]
- Uthayakumaran S, Newberry M, Keentok M, Stoddard FL, Bekes F. Basic rheology of bread dough with modified protein content and glutenin-to-gliadin ratios. Cereal Chem. 2000;77:744–749. doi: 10.1094/CCHEM.2000.77.6.744. [DOI] [Google Scholar]
- Wrigley CW, Shepherd KW. Electrofocusing of grain proteins from wheat genotypes. Ann N Y Acad Sci. 1973;209:154–162. doi: 10.1111/j.1749-6632.1973.tb47526.x. [DOI] [PubMed] [Google Scholar]