Abstract
In the last two decades, a wealth of structural and functional knowledge has been obtained for the three major cytoskeletal motor proteins, myosin, kinesin and dynein, which we review here. The cytoskeletal motor proteins myosin and kinesin are structurally similar in the core architecture of their motor domains and have similar force-producing mechanisms that are coupled with the chemical cycles of ATP binding, hydrolysis, Pi release and subsequent ADP release. The force is generated through conformational changes in the motor domain during Pi release and ATP binding in myosin and kinesin, respectively, and then converted into the rotation of the lever arm or neck linker (referred to as a power stroke) through the common structural pathways. On the other hand, the dynein cytoskeletal motor is an AAA+ protein and has a different structure and power stroke mechanism from those of myosins and kinesins. The linker protruding from the AAA+ ring of dynein swings according to the ATPase states, which, presumably, generates force to carry cargos within a cell. The communication mechanism between the track-binding and ATPase domains of dynein is unique because the two helices that presumably slide with respect to each other work as coordinators for these domains. Details of the mechanism underlying the power stroke and interdomain communication were revealed through recent progress in the structural studies of myosin, kinesin and dynein.
Keywords: Cytoskeletal motor, Myosin, Kinesin, Dynein, Structure, Force generation
Introduction
The first cytoskeletal motor, myosin, was discovered from muscle by Szent-Györgyi and colleagues in the early 1940s (Table 1) (Banga and Szent-Györgyi 1941–1942). Two decades later, the first microtubule motor, dynein, was isolated from cilia of Tetrahymena pyriformis by Gibbons and colleagues (Gibbons and Rowe 1965). In the middle of the 1980s, the third class of cytoskeletal motors, kinesin, was discovered by two independent groups (Brady 1985; Vale et al. 1985). These cytoskeletal motors move on the cytoskeleton by producing force that propel them and their cargo forward by using the free energy obtained from the hydrolysis of ATP. In other words, these motors possess mechanisms in which chemical energy is converted to mechanical work. Myosin moves on actin filaments, whereas kinesin and dynein move on microtubules toward the plus and minus ends, respectively (Kull and Endow 2013; Roberts et al. 2013), while a class of kinesins that moves toward the minus end was discovered later (McDonald et al. 1990; Walker et al. 1990; Thiede et al. 2012). Various subtypes of myosins have pivotal roles in various cellular events, including muscle contraction, cell motility and formation of a contractile ring (Weber et al. 2004; Schuh 2011). The sliding filament theory of muscle contraction was proposed, in which thick filaments composed of myosin and thin filaments composed of actin slide relative to each other to produce contractile force (Huxley and Niedergerke 1954; Huxley and Hanson 1954). Varieties of kinesins and dyneins also contribute to various events, including axonal transport and cell division (Lawrence et al. 2004; Roberts et al. 2013; Levy and Holzbaur 2006). In addition, various kinds of flagellar dyneins together produce the beating of cilia/flagella (Kamiya 2002). The crystal structures of myosins and kinesins revealed their structural similarity despite the difference in their tracks and function in cells (Kull et al. 1996). Structural studies of the motors greatly contributed to the understanding of the force-producing mechanisms, including the power stroke. The overall structure of dynein is different from myosin and kinesin (Burgess et al. 2003), which suggests that there is also a difference in the mechanism of the power stroke. The present review discusses proposed mechanisms for these motors based on structural studies.
Table 1.
Timeline of the studies of cytoskeletal motors
| 1941, 1942 | Discovery of myosin and actomyosin (Banga and Szent-Györgyi 1941–1942) |
| 1943 | Discovery of G- and F-actin (Straub 1943) |
| 1954 | Proposal of the sliding filament theory of muscle contraction (Huxley and Niedergerke 1954; Huxley and Hanson 1954) |
| 1965 | Discovery of the first microtubule motor, dynein (Gibbons and Rowe 1965) |
| 1967 | Discovery of tubulin (Borisy and Taylor 1967) |
| 1985 | Discovery of the kinesin microtubule motor (Brady 1985; Vale et al. 1985) |
| 1989 | Single-molecule assay for kinesin (Howard et al. 1989) |
| 1993 | Structures of myosin and actomyosin (Rayment et al. 1993a, b) |
| 1994 | Single-molecule assay for myosin (Finer et al. 1994) |
| 1995 | Structures of the kinesin microtubule complexes (Hirose et al. 1995; Hoenger et al. 1995; Kikkawa et al. 1995) |
| 1996 | Crystal structure of the kinesin motor (Kull et al. 1996) |
| 1998 | Visualisation of the power stroke of myosin (Dominguez et al. 1998) |
| 1999 | Single-molecule study of dynein (Sakakibara et al. 1999) |
| 1999 | Visualisation of the power stroke of kinesin (Rice et al. 1999) |
| 2003 | Visualisation of the power stroke of dynein (Burgess et al. 2003) |
| 2011, 2012 | Crystal structures of the dynein motors (Carter et al. 2011; Kon et al. 2012) |
Myosin and kinesin
Overview
Myosins and kinesins act as cytoskeletal motors via repeated mechanochemical cycles on actin filaments and microtubules, respectively (Kull and Endow 2013). These motor proteins hydrolyse ATP to drive the motor cycles. Myosin was originally identified in skeletal muscle as a motor protein that controls muscle contraction using ATP (Engelhardt and Ljubimowa 1939). Eukaryotic myosins are now classified into 35 different classes according to their gene sequence homologies (Odronitz and Kollmar 2007) and function in a wide range of cellular events, such as vesicle transport and spindle assembly (Weber et al. 2004; Schuh 2011). Myosin is composed of three common domains: the N-terminal motor domain, lever arm and C-terminal tail domain (Mermall et al. 1998). The motor domain can hydrolyse ATP and interact with actin; therefore, it plays a central role of myosin in the mechanochemical cycles. The lever arm amplifies the motion of the motor domain. In contrast, the tail domain has different functions associated with the cellular roles of different myosin classes (e.g. skeletal muscle myosin II assembles into myosin filaments using a long coiled-coil region of the tail domain, and the class V myosins have specific binding sites for different cargos in the tail domain) (Krendel and Mooseker 2005).
Kinesins are classified into 14 different classes and most of them are motile to transport cargos along microtubules (Lawrence et al. 2004). All kinesins have a conserved motor domain that offers an ATP-binding site and a microtubule-binding site, a neck linker that is an amplifier of the motor and a diverse tail domain for interactions with cargo (Hirokawa and Takemura 2005). The motor domain of kinesins is located on the N-terminus (N-type), middle region (M-type) or C-terminus (C-type), unlike myosin. The N-type kinesins, e.g. kinesin-1 (KIF5B) and kinesin-5 (Eg5), are the most common and move to the plus end along microtubule tracks to transport cargos; the C-type kinesin, e.g. Ncd, moves to the minus end; and the M-type kinesin travels to both ends and depolymerises microtubules (Hirokawa 1998). However, it has been recently discovered that kinesins are more divergent: Cin8-like kinesin-5s are minus-end-directed motors (Thiede et al. 2012), and the fungal kinesin-14 KlpA is a plus-end-directed motor on single microtubules (Popchock et al. 2017).
Several of the structures for myosins and kinesins were determined by X-ray crystallography and cryo-electron microscopy (cryo-EM) over the past two decades, which revealed a structural similarity and a common mechanism of force generation in the motor domains between myosin and kinesin, despite an almost complete lack of sequence identity.
Overall structure
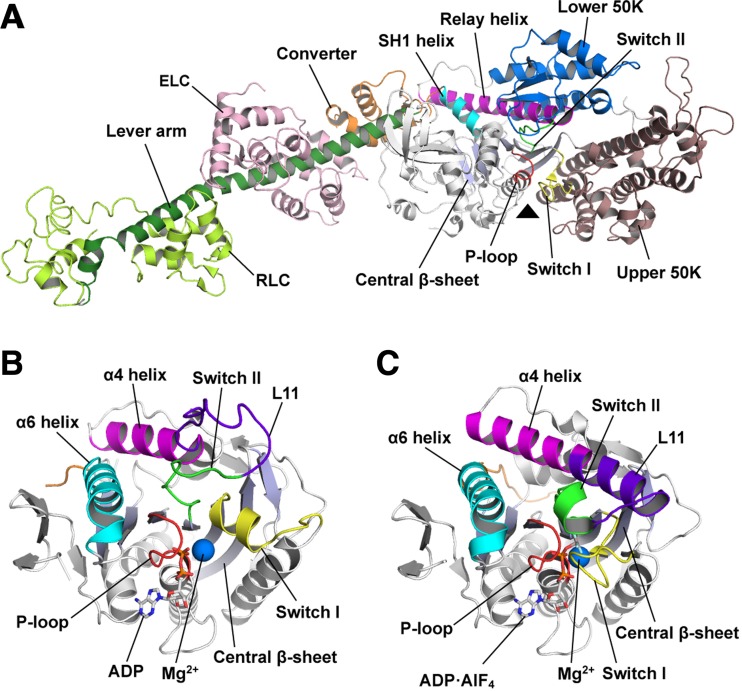
The first crystal structure of myosin to be determined was the nucleotide-free state of the motor domain and the lever arm (Rayment et al. 1993a). The motor domain has a globular structure with ~ 850 amino acids, in which a core architecture is composed of seven-stranded central β-sheet and six α-helices (with three on each side of the β-sheet) (Fig. 1a). The nucleotide-binding site is located on the central β-sheet and is surrounded by three nucleotide-recognition elements, specifically the P-loop, switch I and switch II. There are two subdomains (upper 50K domain and lower 50K domain) on the opposite face of the central β-sheet from the nucleotide-binding site. Actin can bind to the cleft formed between these two subdomains. An additional subdomain, which is called the converter subdomain, is found in the C-terminal region of the motor domain and is directly connected to the lever arm, which adopts a long α-helix and provides interaction surfaces for two calmodulin-like light chains in the cytoskeletal muscle myosin II.
Fig. 1.
Structures of myosin and kinesin. a Ribbon diagram of the motor domain, lever arm, essential light chain (ELC) and the regulatory light chain (RLC) of myosin II (scallop myosin S1; PDB 1SR6). The triangle indicates the nucleotide-binding site. b The motor domain of ADP-bound kinesin (KIF5B; PDB 1BG2). ADP and Mg2+ are represented by stick and sphere models, respectively. c The motor domain of ATP-bound KIF5B (PDB 4HNA). ADP·AlF4 (an ATP analogue) and Mg2+ are represented by stick and sphere models, respectively. The individual architectures related to motor function are shown with different colours in each panel
The motor domain of kinesin is composed of ~ 340 amino acids, which is much smaller than that of myosin. However, the crystal structure of kinesin revealed a structural homology to the motor domain of myosin, especially the core architecture with the nucleotide-binding site (Kull et al. 1996). The structures of ADP- and ATP-bound states show that the P-loop interacts with the α-, β- and γ-phosphates of nucleotides and Mg2+ ions, and switch I and switch II act as sensors that recognise the existence of γ-phosphate (Fig. 1b, c) (Kull et al. 1996; Parke et al. 2010; Gigant et al. 2013). These roles of nucleotide-recognition elements are conserved in the myosin motor domain (Houdusse et al. 2000). The microtubule-binding site of kinesin is also located on the opposite side of the central β-sheet from the nucleotide-binding site, which is similar to myosin. The α4 and α6 helices of kinesin are spatially conserved as the relay helix and the SH1 helix in myosin, respectively. These helices give rise to conformational changes in response to each nucleotide-binding state (Kull and Endow 2013). These structural similarities imply that there is a common force-producing mechanism in the myosin and kinesin motors.
Mechanisms for motor mechanochemical cycles
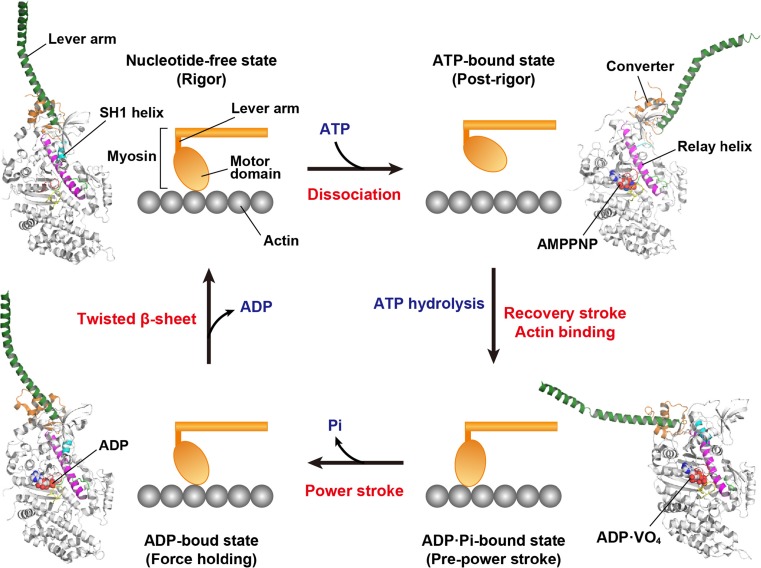
Myosin and kinesin interlock in a chemical cycle of ATP binding, hydrolysis and subsequent phosphate (Pi) and ADP releases through the mechanical cycle of motor motion and their interactions with filaments (Bustamante et al. 2004). The structures of myosin are well defined in four states of the cycle: the actin-detached ATP-bound state (M·ATP), the post-recovery ADP·Pi-bound state (M·ADP·Pi), the ADP-bound state (M·ADP) and the nucleotide-free myosin in complex with actin (Rayment et al. 1993a; Houdusse et al. 2000; Himmel et al. 2002; Yang et al. 2007). These structures and the molecular dynamic simulations have proposed a detailed mechanism of force generation in the mechanochemical cycle of myosin (Fig. 2) (Fischer et al. 2005; Cecchini et al. 2010; Kühner and Fischer 2011). The P-loop, switch I and switch II are closed by ATP binding to the motor domain. The switch I closure leads to the opening of the actin-binding cleft between the upper 50K and lower 50K subdomains to dissociate myosin from actin. Switch II tightly connects to the relay helix and its closure induces rotation of the lever arm through the SH1 helix and the converter subdomain. After closing both the switch I and switch II, ATP hydrolysis proceeds to complete the ‘recovery stroke’, which repositions the lever arm into the pre-power conformation by 5–7.5 nm of movement (Sugi et al. 2008). In the M·ADP·Pi state, the lower 50K subdomain can interact with actin to close the actin-binding cleft, which leads to a Pi release accompanying the ‘power stroke’. The stroke motion involves the rotation of the lever arm in an actin-bound state. In skeletal muscle, myosin filaments slide 5–10 nm on actin filaments through the power stroke. Therefore, the Pi release is a force generation step in the motor mechanochemical cycle of myosin. Subsequent ADP release is relatively slow, so the myosins adopt a long-lived force-holding state (Veigel et al. 2005; Greenberg et al. 2014).
Fig. 2.
Mechanochemical cycle of myosin. The structural models with ribbon diagrams are created by the crystal structures of myosin II (scallop myosin S1; upper left, PDB 1SR6; upper right, PDB 1KQM; lower right, 1DFL; lower left, 3I5F). The lever arm (dark green), converter subdomain (orange) and SH1 helix (cyan) adopt different orientations and conformations in each state. The nucleotide-binding regions, including the P-loop, switch I and switch II, are coloured in red, yellow and green, respectively. Each nucleotide in the motor domain is represented by a sphere model. A power stroke corresponds to a step of force generation
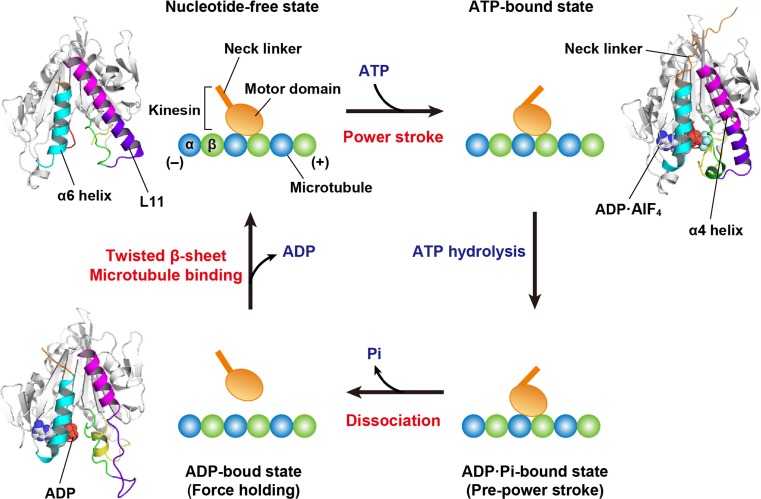
The motor mechanochemical cycle of kinesin has been well elucidated by crystallographic studies on the nucleotide-bound structures of the motor domain (Kull et al. 1996; Sablin et al. 1996; Kozielski et al. 1997; Kikkawa et al. 2001; Endres et al. 2006). More recently, the motor mechanism has been clarified using structures of motor domains complexed with microtubules (or tubulins) in their nucleotide-free state and ATP-bound state (Gigant et al. 2013; Cao et al. 2014; Atherton et al. 2014; Shang et al. 2014). Kinesin is thought to use similar structural architectures for the force generation and transduction of mechanical movements to myosins. However, the force generation is induced by ATP binding to kinesin, unlike the force generation step of myosin, which corresponds to a Pi release (Fig. 3) (Rice et al. 1999; Endres et al. 2006). The motor domain of kinesin can attach to a microtubule in the nucleotide-free state, where ATP initiates the mechanochemical cycle through binding to the motor domain. ATP binding induces conformational changes of the P-loop, switch I and switch II into closed states. Switch II connects to the α4 helix (referred to as the relay helix in myosin) through the L11 loop, which is stabilised into a helical structure as an extension of the α4 helix through an interaction with microtubules (Hirose et al. 2006; Kikkawa and Hirokawa 2006). The α4 helix provides the binding interface for microtubules and shows different orientations according to individual nucleotide-binding modes. Additionally, this helix is spatially close to the α6 helix (referred to as the SH1 helix in myosin) that is followed by the neck linker in kinesin. In the ATP-bound state, the rotated α4 helix leads to the conformational changes of the α6 helix, which allows the neck linker to dock to the small pocket of the motor domain (Kozielski et al. 1997). This movement of the neck linker is a crucial event that serves as the ‘power stroke’ and leads to the plus-end direction of the motor domain. ADP can bind to the motor domain at a relatively high affinity, even when switch I and switch II adopt the open state. Therefore, ADP release is thought to be coupled with microtubule binding accompanying the formation of twisted conformations in the central β-sheet of the motor domain, which was also observed in the structures of nucleotide-free myosin (Coureux et al. 2003; Reubold et al. 2003). Therefore, microtubules would act as a nucleotide exchanger from ADP to ATP, unlike actin filament, which triggers a Pi release from myosin to generate force.
Fig. 3.
The mechanochemical cycle of N-type kinesin. The structural models with ribbon diagrams are created by the crystal structures of kinesin-1 (KIF5B; upper left, PDB 4LNU; upper right, PDB 4HNA; lower left, 1BG2). The structures in individual steps adopt the different conformations of the neck linker (orange), α6 helix (cyan), α4 helix (magenta), L11 loop (violet), switch I (yellow), switch II (green) and P-loop (red). Each nucleotide in the motor domain is represented by a sphere model. Microtubules are composed of heterooligomers composed of α-tubulin (α) and β-tubulin (β). The neck linker moves from the minus-end direction (left) along a microtubule to the plus-end direction (right) via the power stroke, which corresponds to the step of force generation
In the C-type kinesin Ncd, the ATP-induced rotation of the neck mimic (referred to as the neck linker in the N-type) is converted into the inverted swing of the neck helix to trigger the movement of Ncd toward the minus-end direction (Yamagishi et al. 2016). The neck helix is specifically contained in the N-terminus of the Ncd motor domain. After ATP hydrolysis and a subsequent Pi release, the switch II would be open and the motor domain detaches from microtubules and tilts toward the minus direction. The folding–unfolding transition of the coiled-coil stalk of Ncd might be important for retrograde movement, and Arisaka contributed research into this transition along with our group (Makino et al. 2007). Ncd belongs to the kinesin-14 motor family that contains divergent members with a variety of N-terminal tails, providing some of the most crucial functional cues, such as binding sites of other interaction partners (She and Yang 2017). Most of the kinesin-14s are homodimeric, whereas yeast Kar3 forms heterodimer with Vik1 or Cik1 that contains the ATP-independent motor domain at the C-terminus. A recent study has proposed a common power stroke mechanism for homodimeric and heterodimeric kinesin-14s (Zhang et al. 2015). According to the conventional mechanism, the power stroke is generated by microtubule binding and one ATP turnover in only one motor domain. However, in the homodimeric and heterodimeric kinesin-14s, one process of force generation is required for cooperative interaction between both motor domains and two ATP turnovers (Zhang et al. 2015; She and Yang 2017).
Mechanochemical cycles of myosin and kinesin are similar, whereas the timing of actin or microtubule binding differs from each other. Power stroke means the movement of the lever arm (myosin) or neck linker (kinesin) in the state bound to actin filament or microtubule (Figs. 2 and 3). Myosin and kinesin use distinct structural elements of the motor domains as binding interfaces, the actin-binding cleft between the upper 50K and lower 50K subdomains, and the α4 helix containing the L11 loop, respectively. Therefore, the binding sites suitable for interaction partners and their conformational changes in mechanochemical cycles are indispensable factors to adjust the optimal timing of the power stroke.
Dynein
Overview
Dyneins are cytoskeletal motor proteins that convert the chemical energy derived from ATP hydrolysis to mechanical force and move along microtubules toward the minus end (Kamiya 2002; Roberts et al. 2013). Dyneins are classified into two groups, including cytoplasmic and flagellar dyneins. Cytoplasmic dyneins contribute to cargo transport within a cell, whereas flagellar dyneins generate force to beat cilia/flagella. Dysfunction of dyneins is associated with widespread disorders and diseases, such as lissencephaly, hydrocephaly, motor neuron disease and ciliopathies (Fliegauf et al. 2007; Levy and Holzbaur 2006). Phylogenetic analysis revealed that dynein belongs to the AAA+ superfamily, many of which are hexameric enzymes with diverse functions, such as acting as a protease, chaperone, transcriptional activator proteins, as well as replication and recombination proteins (Neuwald et al. 1999). The force generation mechanism of dyneins has been anticipated to be fundamentally different from those of other cytoskeletal motors, such as myosins and kinesins, that evolved from a common ancestor protein (Vetter and Wittinghofer 2001). The molecular weight of the minimal unit of a dynein motor is ~ 380 kDa, which is roughly an order of magnitude greater than those of myosins and kinesins. Due to the large size of dynein, it has been more difficult to express, purify, manipulate and crystallise it compared to the other motors. Nevertheless, recent progress on dynein has revealed its elusive mechanism by overcoming such difficulties.
Overall structure
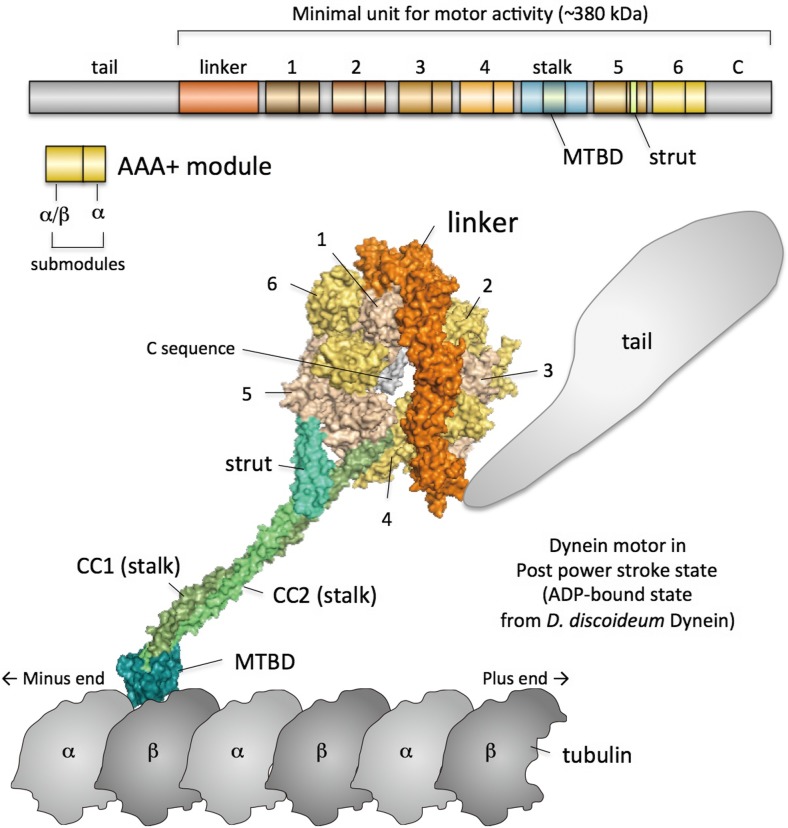
Unlike the other AAA+ proteins, dynein’s AAA+ ring is composed of a single polypeptide chain. The minimal unit for the motor activity of dynein contains the linker, six tandemly arranged AAA+ modules with various amino acid sequences, a stalk, a microtubule-binding domain (MTBD) and a C sequence (Fig. 4). The linker, which presumably produces the power stroke, is located at the N-terminal end of the AAA1 module (Roberts et al. 2009). The stalk and MTBD are located between AAA4 and AAA5. The C sequence follows AAA6. The overall structure of a dynein monomer is a ring-like structure with two protrusions of different sizes. The larger one contains the linker and tail. The N-terminus of the linker is connected to the tail that binds to cargos and other dynein monomer(s) in the case of multimer dyneins. The tail is not essential for the motor activity but is necessary for diverse biological events. It was suggested that the linker swings for the power stroke according to EM studies of a flagellar dynein (Burgess et al. 2003). The thinner and smaller protrusion is called the stalk. The stalk is composed of an anti-parallel coiled-coil. The MTBD is located at the distal globular end of the stalk. Thus, the overall configuration of dynein is characteristic, as the track-binding domain forms a globular domain and is separated by the thin stalk from an ATPase catalytic domain, the AAA+ ring. This is a remarkable difference compared with myosins and kinesins because their track-binding and ATPase components are within a single globular domain.
Fig. 4.
Overall structure of dynein. Upper panel Domain composition of dynein. The numbers of AAA+ modules (AAA1–AAA6) are indicated. ‘C’ indicates the C sequence. Lower panel The overall structure of dynein in the post-power stroke state. CC1 and CC2 indicate coiled-coil helices 1 and 2 of the stalk, respectively
Power stroke mechanism
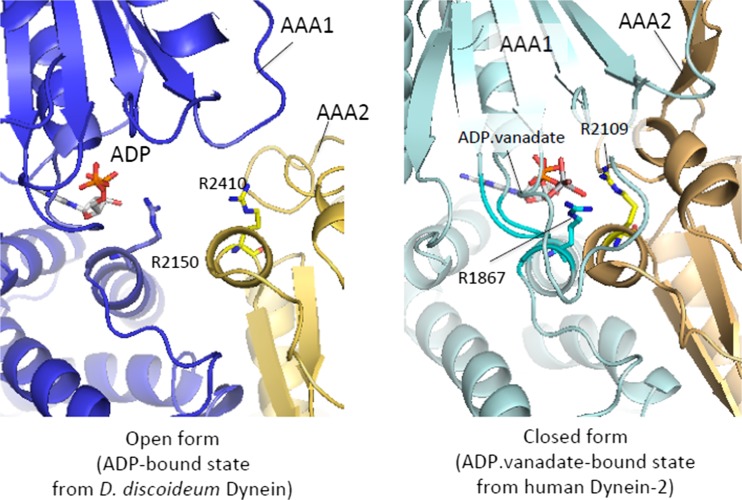
Dynein has a characteristic linker that presumably swings according to the ATPase cycle to produce the power stroke. Previous EM studies combined with protein engineering have inserted tags in various sites within the polypeptide chain of the motor unit of cytoplasmic dynein, which include studies that Arisaka contributed to, along with the Burgess group. Their results revealed that swing motion by the lever-like linker occurs between AAA2 and the stalk base (Roberts et al. 2009). However, the detailed mechanism of the linker swing was unknown until recent crystallographic studies of the motor unit. Pioneering crystallographic studies of the dynein AAA+ motor revealed that the motor was composed of the central AAA+ ring, liner, stalk, strut and C sequence (Carter et al. 2011; Kon et al. 2012). The AAA+ ring contained six AAA+ modules, AAA1–AAA6, which were arranged according to the order of their amino acid sequence. Each AAA+ module contained two submodules, α/β and α submodules, which were arranged on the opposite sides of the ring. AAA1–AAA4 contained characteristic insert sequences. The AAA5 module contained an additional globular domain. Active ATPase sites were found in AAA1–AAA4, while a previous biochemical analysis showed that the active site of AAA1 was critical for the power stroke. The ATPase sites of AAA5 and AAA6 are inherently inactive because the residues necessary for ATP hydrolysis were replaced. The ATPase site of AAA1 was on an interdomain interface between AAA1 and AAA2. While ADP was held within the AAA1 module, it lacked the binding residue for the γ-phosphate of ATP. Instead, the arginine finger sequence found in AAA2 was located close to the AAA1 catalytic site. It was, therefore, speculated that the cleft between AAA1 and AAA2 is narrowed after binding ATP to form a complete catalytic site. This speculation was supported by the structure of dynein-2 in complex with ADP.vanadate, in which the cleft between AAA1 and AAA2 narrowed to form the catalytic site (Schmidt et al. 2012). Thus, the AAA+ ring is presumed to convert to a closed conformation when it binds ATP (Fig. 5).
Fig. 5.
Cleft closure with an ADP.Pi analogue. Left panel The distance between ADP and R2410 is long, and AAA1 (blue) and AAA2 (gold) are detached from each other. Right panel The distance between ADP and R2109 corresponding to R2410 in the left panel is short, and the cleft between AAA1 (pale cyan) and AAA2 (light orange) is closed
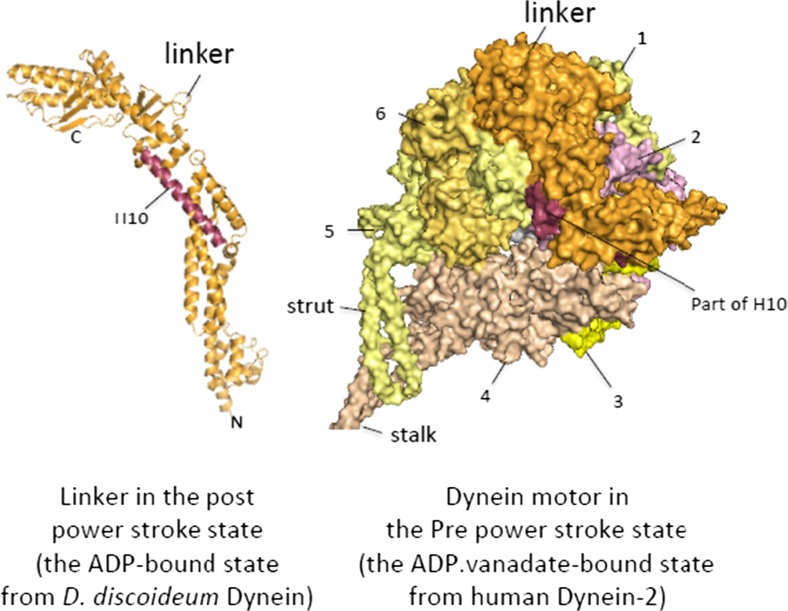
It has been suggested that the linker movement produces the power stroke based on the EM and fluorescence resonance energy transfer analysis (Burgess et al. 2003; Kon et al. 2005). Crystal structures of the ADP-bound dynein motor displayed the post-power stroke state. In this state, the contact area between the linker and AAA+ ring was remarkably small compared to the sizes of the linker. Schmidt et al. (2012) found the contact of the linker with AAA5 in yeast dynein, whereas Kon et al. (2012) found contact of the linker with AAA2 in Dictyostelium discoideum dynein but no contact with AAA5. Nevertheless, these structures together showed that most parts of the linker, except for its proximal part, are detached from the AAA+ ring. Proximal and distal parts of the linker were connected only by a long α-helix, H10, which prompted a proposal that this region might bend when dynein assumes the pre-power stroke state (Kon et al. 2012). This proposal was supported by a recent crystal structure of dynein-2 in complex with ADP.vanadate (Schmidt et al. 2015), in which the linker was bent at H10 to prime the power stroke (Fig. 6).
Fig. 6.
Structural change of the linker. Left panel Ribbon model of the linker. The hinge helix H10 is highlighted. Right panel The AAA+ ring in the pre-power stroke state has the linker bent at H10
Communication between the domains for ATP hydrolysis and track binding
For directed movement of dynein, it is required that various activities in a mechanochemical cycle of dynein, such as ATP hydrolysis, microtubule binding, the power stroke and microtubule dissociation, occur in the correct order. Therefore, communication between the MTBD for track binding and the AAA+ ring that contains the ATPase catalytic sites is essential for directed movement. The MTBD of dynein is separated from the AAA+ ring by the stalk with a ~ 140 Å length. For kinesins and myosins, changes of the structures of the ATPase components directly link structural changes of their track-binding components because the components for track binding and ATP hydrolysis are located within an inseparable structural domain. Thus, the communication mechanism between the domains for track binding and ATP hydrolysis of dyneins has been presumed to be fundamentally different from those of kinesins and myosins.
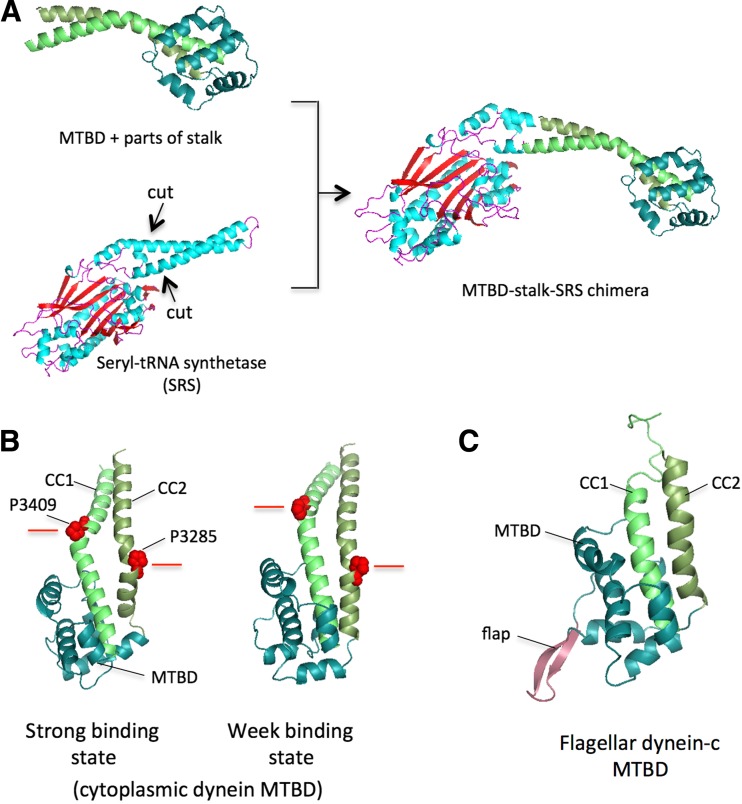
Analysis of an amino acid sequence of dynein predicted the existence of two long helical sequences that may form a long coiled-coil together (Gee et al. 1997). It was demonstrated that the recombinant 45-kDa protein that included the hypothetical coiled-coil region binds to microtubules without the AAA+ ring, which suggested that the MTBD is within this region. Later, cryo-EM studies demonstrated that the region sandwiched between the two long helical sequences directly binds to a microtubule (Mizuno et al. 2007). Gibbons et al. (2005) proposed that the two helices that compose the anti-parallel coiled-coil slide over each other. They produced constructs that contained the hypothetical coiled-coil and MTBD with different coiled-coil registries by connecting the coiled-coils of dynein and seryl-tRNA synthetase (SRS) and found that the binding affinity to microtubules changed depending on the registry (Fig. 7a, b). The crystal structure of one of those constructs with weak affinity revealed that the stalk was composed of an anti-parallel coiled-coil and that the MTBD was located at the distal end of the coiled-coil (Carter et al. 2008). We demonstrated that flagellar dynein had a similar overall structure of the distal stalk region containing the MTBD in aqueous solution (Kato et al. 2014). The flagellar MTBD contained a characteristic flap structure that might have an impact on microtubule binding (Fig. 7c) and showed trivial changes in the affinity, depending on the registry change.
Fig. 7.
Registry change and structure of the microtubule-binding domain (MTBD). a The coiled-coil of seryl-tRNA synthetase (SRS) was ligated with the stalk of the MTBD to produce constructs with various coiled-coil registries. b Coiled-coil registries were different between the strong and weak binding states of the MTBD. In the figure, the height of P3409 relative to that of P3285 changes in two different states. c The MTBD of flagellar dynein-c contains the characteristic flap
The crystal and solution structures of the distal stalk region showed a weak-binding state of the MTBD at atomic resolution. Later, cryo-EM studies reported a stalk–microtubule complex structure at sub-nanometre resolution in a strong binding state (Redwine et al. 2012). These authors also proposed the atomic structure model of the stalk in a strong binding state based on molecular dynamics flexible fitting and suggested that electrostatic interactions between the MTBD and a microtubule are important for strong binding. Indeed, loss of one of the electrostatic interactions due to a substitution of the amino acid of microtubules impaired activation of ATP hydrolysis and directed dynein movement (Uchimura et al. 2015).
Later, the crystal structures of the whole stalk and dynein motor revealed the structure of the full-length stalk and existence of the strut (Carter et al. 2011; Kon et al. 2012). The strut was composed of an anti-parallel coiled-coil and interacted with the proximal region of the stalk to support the base of the stalk. The interaction between the stalk and the strut appeared tight. Therefore, it was proposed that tension is generated between the stalk and strut upon structural change of the AAA+ ring, which causes change in the registry and structure of the stalk, and causes the MTBD to change its affinity to microtubules.
Concluding remarks
Numerous structural studies have elucidated the conformational changes of myosin and kinesin in the multiple steps of their mechanochemical cycles and have proposed possible mechanisms of force generation in the motor domains. However, the structure of M·ADP·Pi in complex with actin could provide further structural information related to force generation by the myosin motor. Structural studies of dynein have also rapidly advanced in the past decade and revealed detailed structural differences between before and after the power stroke. However, the mechanism of how the structural transition occurs remains unknown. In addition, detailed molecular mechanisms of dyneins in a multimer situation are unsolved as well, although recent electron microscopy (EM) and single-molecule studies have approached this problem (Gennerich et al. 2007; Imai et al. 2015). We hope that the understanding of the cytoskeletal motor proteins continues to improve over the next decade.
Compliance with ethical standards
Conflict of interest
Yusuke Kato declares that he has no conflict of interest. Takuya Miyakawa declares that he has no conflict of interest. Masaru Tanokura declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by the authors.
Open access
This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) as well as a link to the Creative Commons license, and indicate if changes were made.
Footnotes
Yusuke Kato and Takuya Miyakawa contributed equally to this work.
This article is part of a Special Issue on ‘Biomolecules to Bio-nanomachines - Fumio Arisaka 70th Birthday’ edited by Damien Hall, Junichi Takagi and Haruki Nakamura.
References
- Atherton J, Farabella I, Yu IM, Rosenfeld SS, Houdusse A, Topf M, Moores CA. Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins. Elife. 2014;3:e03680. doi: 10.7554/eLife.03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banga I, Szent-Györgyi A (1941–1942) Preparation and properties of myosin A and B. In: Szent-Györgyi A (ed) Studies from the Institute of Medical Chemistry—University of Szeged, vol. 1. S. Karger AG, Basel, pp 5–15
- Borisy GG, Taylor EW. The mechanism of action of colchicine. Binding of colchincine-3H to cellular protein. J Cell Biol. 1967;34:525–533. doi: 10.1083/jcb.34.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady ST. A novel brain ATPase with properties expected for the fast axonal transport motor. Nature. 1985;317:73–75. doi: 10.1038/317073a0. [DOI] [PubMed] [Google Scholar]
- Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Dynein structure and power stroke. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Chemla YR, Forde NR, Izhaky D. Mechanical processes in biochemistry. Annu Rev Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- Cao L, Wang W, Jiang Q, Wang C, Knossow M, Gigant B. The structure of apo-kinesin bound to tubulin links the nucleotide cycle to movement. Nat Commun. 2014;5:5364. doi: 10.1038/ncomms6364. [DOI] [PubMed] [Google Scholar]
- Carter AP, Garbarino JE, Wilson-Kubalek EM, Shipley WE, Cho C, Milligan RA, Vale RD, Gibbons IR. Structure and functional role of dynein’s microtubule-binding domain. Science. 2008;322:1691–1695. doi: 10.1126/science.1164424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Cho C, Jin L, Vale RD. Crystal structure of the dynein motor domain. Science. 2011;331:1159–1165. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini M, Alexeev Y, Karplus M. Pi release from myosin: a simulation analysis of possible pathways. Structure. 2010;18:458–470. doi: 10.1016/j.str.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coureux PD, Wells AL, Ménétrey J, Yengo CM, Morris CA, Sweeney HL, Houdusse A. A structural state of the myosin V motor without bound nucleotide. Nature. 2003;425:419–423. doi: 10.1038/nature01927. [DOI] [PubMed] [Google Scholar]
- Dominguez R, Freyzon Y, Trybus KM, Cohen C. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state. Cell. 1998;94:559–571. doi: 10.1016/S0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- Endres NF, Yoshioka C, Milligan RA, Vale RD. A lever-arm rotation drives motility of the minus-end-directed kinesin Ncd. Nature. 2006;439:875–878. doi: 10.1038/nature04320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt WA, Ljubimowa MN. Myosin and adenosine triphosphatase. Nature. 1939;144:668–669. doi: 10.1038/144668b0. [DOI] [PubMed] [Google Scholar]
- Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- Fischer S, Windshügel B, Horak D, Holmes KC, Smith JC. Structural mechanism of the recovery stroke in the myosin molecular motor. Proc Natl Acad Sci U S A. 2005;102:6873–6878. doi: 10.1073/pnas.0408784102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- Gee MA, Heuser JE, Vallee RB. An extended microtubule-binding structure within the dynein motor domain. Nature. 1997;390:636–639. doi: 10.1038/37663. [DOI] [PubMed] [Google Scholar]
- Gennerich A, Carter AP, Reck-Peterson SL, Vale RD. Force-induced bidirectional stepping of cytoplasmic dynein. Cell. 2007;131:952–965. doi: 10.1016/j.cell.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons IR, Rowe AJ. Dynein: a protein with adenosine triphosphatase activity from cilia. Science. 1965;149:424–426. doi: 10.1126/science.149.3682.424. [DOI] [PubMed] [Google Scholar]
- Gibbons IR, Garbarino JE, Tan CE, Reck-Peterson SL, Vale RD, Carter AP. The affinity of the dynein microtubule-binding domain is modulated by the conformation of its coiled-coil stalk. J Biol Chem. 2005;280:23960–23965. doi: 10.1074/jbc.M501636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigant B, Wang W, Dreier B, Jiang Q, Pecqueur L, Plückthun A, Wang C, Knossow M. Structure of a kinesin–tubulin complex and implications for kinesin motility. Nat Struct Mol Biol. 2013;20:1001–1007. doi: 10.1038/nsmb.2624. [DOI] [PubMed] [Google Scholar]
- Greenberg MJ, Shuman H, Ostap EM. Inherent force-dependent properties of β-cardiac myosin contribute to the force–velocity relationship of cardiac muscle. Biophys J. 2014;107:L41–L44. doi: 10.1016/j.bpj.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmel DM, Gourinath S, Reshetnikova L, Shen Y, Szent-Györgyi AG, Cohen C. Crystallographic findings on the internally uncoupled and near-rigor states of myosin: further insights into the mechanics of the motor. Proc Natl Acad Sci U S A. 2002;99:12645–12650. doi: 10.1073/pnas.202476799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Takemura R. Molecular motors and mechanisms of directional transport in neurons. Nat Rev Neurosci. 2005;6:201–214. doi: 10.1038/nrn1624. [DOI] [PubMed] [Google Scholar]
- Hirose K, Lockhart A, Cross RA, Amos LA. Nucleotide-dependent angular change in kinesin motor domain bound to tubulin. Nature. 1995;376:277–279. doi: 10.1038/376277a0. [DOI] [PubMed] [Google Scholar]
- Hirose K, Akimaru E, Akiba T, Endow SA, Amos LA. Large conformational changes in a kinesin motor catalyzed by interaction with microtubules. Mol Cell. 2006;23:913–923. doi: 10.1016/j.molcel.2006.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenger A, Sablin EP, Vale RD, Fletterick RJ, Milligan RA. Three-dimensional structure of a tubulin–motor–protein complex. Nature. 1995;376:271–274. doi: 10.1038/376271a0. [DOI] [PubMed] [Google Scholar]
- Houdusse A, Szent-Györgyi AG, Cohen C. Three conformational states of scallop myosin S1. Proc Natl Acad Sci U S A. 2000;97:11238–11243. doi: 10.1073/pnas.200376897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J, Hudspeth AJ, Vale RD. Movement of microtubules by single kinesin molecules. Nature. 1989;342:154–158. doi: 10.1038/342154a0. [DOI] [PubMed] [Google Scholar]
- Huxley H, Hanson J. Changes in the cross-striations of muscle during contraction and stretch and their structural interpretation. Nature. 1954;173:973–976. doi: 10.1038/173973a0. [DOI] [PubMed] [Google Scholar]
- Huxley AF, Niedergerke R. Structural changes in muscle during contraction; interference microscopy of living muscle fibres. Nature. 1954;173:971–973. doi: 10.1038/173971a0. [DOI] [PubMed] [Google Scholar]
- Imai H, Shima T, Sutoh K, Walker ML, Knight PJ, Kon T, Burgess SA. Direct observation shows superposition and large scale flexibility within cytoplasmic dynein motors moving along microtubules. Nat Commun. 2015;6:8179. doi: 10.1038/ncomms9179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya R. Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int Rev Cytol. 2002;219:115–155. doi: 10.1016/S0074-7696(02)19012-7. [DOI] [PubMed] [Google Scholar]
- Kato YS, Yagi T, Harris SA, Ohki SY, Yura K, Shimizu Y, Honda S, Kamiya R, Burgess SA, Tanokura M. Structure of the microtubule-binding domain of flagellar dynein. Structure. 2014;22:1628–1638. doi: 10.1016/j.str.2014.08.021. [DOI] [PubMed] [Google Scholar]
- Kikkawa M, Hirokawa N. High-resolution cryo-EM maps show the nucleotide binding pocket of KIF1A in open and closed conformations. EMBO J. 2006;25:4187–4194. doi: 10.1038/sj.emboj.7601299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa M, Ishikawa T, Wakabayashi T, Hirokawa N. Three-dimensional structure of the kinesin head–microtubule complex. Nature. 1995;376:274–277. doi: 10.1038/376274a0. [DOI] [PubMed] [Google Scholar]
- Kikkawa M, Sablin EP, Okada Y, Yajima H, Fletterick RJ, Hirokawa N. Switch-based mechanism of kinesin motors. Nature. 2001;411:439–445. doi: 10.1038/35078000. [DOI] [PubMed] [Google Scholar]
- Kon T, Mogami T, Ohkura R, Nishiura M, Sutoh K. ATP hydrolysis cycle-dependent tail motions in cytoplasmic dynein. Nat Struct Mol Biol. 2005;12:513–519. doi: 10.1038/nsmb930. [DOI] [PubMed] [Google Scholar]
- Kon T, Oyama T, Shimo-Kon R, Imamula K, Shima T, Sutoh K, Kurisu G. The 2.8 Å crystal structure of the dynein motor domain. Nature. 2012;484:345–350. doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- Kozielski F, Sack S, Marx A, Thormählen M, Schönbrunn E, Biou V, Thompson A, Mandelkow EM, Mandelkow E. The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell. 1997;91:985–994. doi: 10.1016/S0092-8674(00)80489-4. [DOI] [PubMed] [Google Scholar]
- Krendel M, Mooseker MS. Myosins: tails (and heads) of functional diversity. Physiol. 2005;20:239–251. doi: 10.1152/physiol.00014.2005. [DOI] [PubMed] [Google Scholar]
- Kühner S, Fischer S. Structural mechanism of the ATP-induced dissociation of rigor myosin from actin. Proc Natl Acad Sci U S A. 2011;108:7793–7798. doi: 10.1073/pnas.1018420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kull FJ, Endow SA. Force generation by kinesin and myosin cytoskeletal motor proteins. J Cell Sci. 2013;126:9–19. doi: 10.1242/jcs.103911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature. 1996;380:550–555. doi: 10.1038/380550a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, Malmberg RL, McIntosh JR, Miki H, Mitchison TJ, Okada Y, Reddy AS, Saxton WM, Schliwa M, Scholey JM, Vale RD, Walczak CE, Wordeman L. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy JR, Holzbaur EL. Cytoplasmic dynein/dynactin function and dysfunction in motor neurons. Int J Dev Neurosci. 2006;24:103–111. doi: 10.1016/j.ijdevneu.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Makino T, Morii H, Shimizu T, Arisaka F, Kato Y, Nagata K, Tanokura M. Reversible and irreversible coiled coils in the stalk domain of ncd motor protein. Biochemistry. 2007;46:9523–9532. doi: 10.1021/bi700291a. [DOI] [PubMed] [Google Scholar]
- McDonald HB, Stewart RJ, Goldstein LS. The kinesin-like ncd protein of drosophila is a minus end-directed microtubule motor. Cell. 1990;63:1159–1165. doi: 10.1016/0092-8674(90)90412-8. [DOI] [PubMed] [Google Scholar]
- Mermall V, Post PL, Mooseker MS. Unconventional myosins in cell movement, membrane traffic, and signal transduction. Science. 1998;279:527–533. doi: 10.1126/science.279.5350.527. [DOI] [PubMed] [Google Scholar]
- Mizuno N, Narita A, Kon T, Sutoh K, Kikkawa M. Three-dimensional structure of cytoplasmic dynein bound to microtubules. Proc Natl Acad Sci U S A. 2007;104:20832–20837. doi: 10.1073/pnas.0710406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007;8:R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parke CL, Wojcik EJ, Kim S, Worthylake DK. ATP hydrolysis in Eg5 kinesin involves a catalytic two-water mechanism. J Biol Chem. 2010;285:5859–5867. doi: 10.1074/jbc.M109.071233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popchock AR, Tseng KF, Wang P, Karplus PA, Xiang X, Qiu W. The mitotic kinesin-14 KlpA contains a context-dependent directionality switch. Nat Commun. 2017;8:13999. doi: 10.1038/ncomms13999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayment I, Rypniewski WR, Schmidt-Bäse K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Rayment I, Holden HM, Whittaker M, Yohn CB, Lorenz M, Holmes KC, Milligan RA. Structure of the actin–myosin complex and its implications for muscle contraction. Science. 1993;261:58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- Redwine WB, Hernández-López R, Zou S, Huang J, Reck-Peterson SL, Leschziner AE. Structural basis for microtubule binding and release by dynein. Science. 2012;337:1532–1536. doi: 10.1126/science.1224151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubold TF, Eschenburg S, Becker A, Kull FJ, Manstein DJ. A structural model for actin-induced nucleotide release in myosin. Nat Struct Mol Biol. 2003;10:826–830. doi: 10.1038/nsb987. [DOI] [PubMed] [Google Scholar]
- Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, Pate E, Cooke R, Taylor EW, Milligan RA, Vale RD. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- Roberts AJ, Numata N, Walker ML, Kato YS, Malkova B, Kon T, Ohkura R, Arisaka F, Knight PJ, Sutoh K, Burgess SA. AAA+ ring and linker swing mechanism in the dynein motor. Cell. 2009;136:485–495. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AJ, Kon T, Knight PJ, Sutoh K, Burgess SA. Functions and mechanics of dynein motor proteins. Nat Rev Mol Cell Biol. 2013;14:713–726. doi: 10.1038/nrm3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablin EP, Kull FJ, Cooke R, Vale RD, Fletterick RJ. Crystal structure of the motor domain of the kinesin-related motor ncd. Nature. 1996;380:555–559. doi: 10.1038/380555a0. [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Kojima H, Sakai Y, Katayama E, Oiwa K. Inner-arm dynein c of Chlamydomonas flagella is a single-headed processive motor. Nature. 1999;400:586–590. doi: 10.1038/23066. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Gleave ES, Carter AP. Insights into dynein motor domain function from a 3.3-Å crystal structure. Nat Struct Mol Biol. 2012;19:492–497. doi: 10.1038/nsmb.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Zalyte R, Urnavicius L, Carter AP. Structure of human cytoplasmic dynein-2 primed for its power stroke. Nature. 2015;518:435–438. doi: 10.1038/nature14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M. An actin-dependent mechanism for long-range vesicle transport. Nat Cell Biol. 2011;13:1431–1436. doi: 10.1038/ncb2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Z, Zhou K, Xu C, Csencsits R, Cochran JC, Sindelar CV. High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation. Elife. 2014;3:e04686. doi: 10.7554/eLife.04686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She ZY, Yang WX. Molecular mechanisms of kinesin-14 motors in spindle assembly and chromosome segregation. J Cell Sci. 2017;130:2097–2110. doi: 10.1242/jcs.200261. [DOI] [PubMed] [Google Scholar]
- Straub FB. Actin, II. Stud Inst Med Chem Univ Szeged. 1943;III:23–37. [Google Scholar]
- Sugi H, Minoda H, Inayoshi Y, Yumoto F, Miyakawa T, Miyauchi Y, Tanokura M, Akimoto T, Kobayashi T, Chaen S, Sugiura S. Direct demonstration of the cross-bridge recovery stroke in muscle thick filaments in aqueous solution by using the hydration chamber. Proc Natl Acad Sci U S A. 2008;108:17396–17401. doi: 10.1073/pnas.0809581105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiede C, Fridman V, Gerson-Gurwitz A, Gheber L, Schmidt CF. Regulation of bi-directional movement of single kinesin-5 Cin8 molecules. BioArchitecture. 2012;2:70–74. doi: 10.4161/bioa.20395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura S, Fujii T, Takazaki H, Ayukawa R, Nishikawa Y, Minoura I, Hachikubo Y, Kurisu G, Sutoh K, Kon T, Namba K, Muto E. A flipped ion pair at the dynein–microtubule interface is critical for dynein motility and ATPase activation. J Cell Biol. 2015;208:211–222. doi: 10.1083/jcb.201407039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/S0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veigel C, Schmitz S, Wang F, Sellers JR. Load-dependent kinetics of myosin-V can explain its high processivity. Nat Cell Biol. 2005;7:861–869. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- Walker RA, Salmon ED, Endow SA. The Drosophila claret segregation protein is a minus-end directed motor molecule. Nature. 1990;347:780–782. doi: 10.1038/347780a0. [DOI] [PubMed] [Google Scholar]
- Weber KL, Sokac AM, Berg JS, Cheney RE, Bement WM. A microtubule-binding myosin required for nuclear anchoring and spindle assembly. Nature. 2004;431:325–329. doi: 10.1038/nature02834. [DOI] [PubMed] [Google Scholar]
- Yamagishi M, Shigematsu H, Yokoyama T, Kikkawa M, Sugawa M, Aoki M, Shirouzu M, Yajima J, Nitta R. Structural basis of backwards motion in kinesin-1-kinesin-14 chimera: implication for kinesin-14 motility. Structure. 2016;24:1322–1334. doi: 10.1016/j.str.2016.05.021. [DOI] [PubMed] [Google Scholar]
- Yang Y, Gourinath S, Kovács M, Nyitray L, Reutzel R, Himmel DM, O’Neall-Hennessey E, Reshetnikova L, Szent-Györgyi AG, Brown JH, Cohen C. Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor. Structure. 2007;15:553–564. doi: 10.1016/j.str.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Zhang P, Dai W, Hahn J, Gilbert SP. Drosophila Ncd reveals an evolutionarily conserved powerstroke mechanism for homodimeric and heterodimeric kinesin-14s. Proc Natl Acad Sci U S A. 2015;112:6359–6364. doi: 10.1073/pnas.1505531112. [DOI] [PMC free article] [PubMed] [Google Scholar]