Abstract
In his Nobel Lecture, Anfinsen stated “the native conformation is determined by the totality of interatomic interactions and hence by the amino acid sequence, in a given environment.” As aqueous solutions and membrane systems co-exist in cells, proteins are classified into membrane and non-membrane proteins, but whether one can transform one into the other remains unknown. Intriguingly, many well-folded non-membrane proteins are converted into “insoluble” and toxic forms by aging- or disease-associated factors, but the underlying mechanisms remain elusive. In 2005, we discovered a previously unknown regime of proteins seemingly inconsistent with the classic “Salting-in” dogma: “insoluble” proteins including the integral membrane fragments could be solubilized in the ion-minimized water. We have thus successfully studied “insoluble” forms of ALS-causing P56S-MSP, L126Z-SOD1, nascent SOD1 and C71G-Profilin1, as well as E. coli S1 fragments. The results revealed that these “insoluble” forms are either unfolded or co-exist with their unfolded states. Most unexpectedly, these unfolded states acquire a novel capacity of interacting with membranes energetically driven by the formation of helices/loops over amphiphilic/hydrophobic regions which universally exit in proteins but are normally locked away in their folded native states. Our studies suggest that most, if not all, proteins contain segments which have the dual ability to fold into distinctive structures in aqueous and membrane environments. The abnormal membrane interaction might initiate disease and/or aging processes; and its further coupling with protein aggregation could result in radical proteotoxicity by forming inclusions composed of damaged membranous organelles and protein aggregates. Therefore, environment-transformable sequence–structure relationship may represent a general mechanism for proteotoxicity.
Keywords: Neurodegenerative diseases, Aging, Proteotoxicity, Membrane interaction, Liquid–liquid phase separation (LLPS), Prion-like domains
Sequence-structure relationships of protein folding and environments
Proteins, the most important functional players for all forms of life, are linear heteropolymers composed of 20 α-amino acids. Proteins represent one of the best examples to illustrate the self-assembly of biomolecules with diverse mechanisms. Remarkably, proteins can spontaneously fold into unique three-dimensional structures via protein folding processes (Anfinsen 1973). In his Nobel Lecture in 1972, Anfinsen stated “the native conformation is determined by the totality of interatomic interactions and hence by the amino acid sequence, in a given environment.” This implies that protein folding is not only specified by amino acid sequence but might also be influenced by the environment.
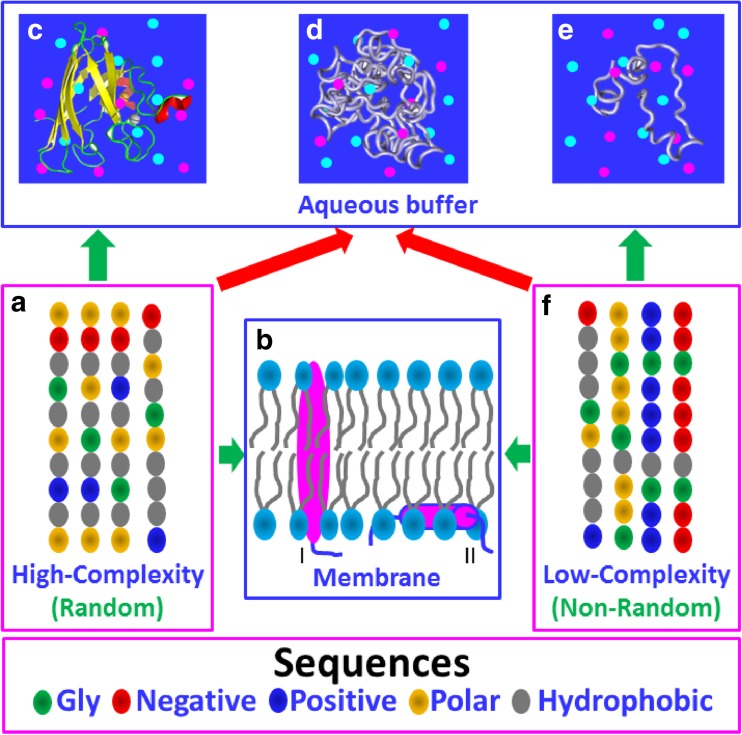
Amazingly, it has been demonstrated that a minimal set of five amino acids is sufficient to encode foldable proteins (Riddle et al. 1997; Plaxco et al. 1998). Based on the amino acid sequences, it is now clear that proteins encoded by higher eukaryotic genomes are in fact constituted of two categories: those with random and high-complexity (Fig. 1a), and those with non-random and low-complexity (Fig. 1f) sequences (Wootton 1994). Intriguingly, in cell-like buffers with high concentrations of salts, only a portion of sequences within the first category is able to fold into well-defined and soluble structures (Fig. 1c) (Li et al. 2013; Rocklin et al. 2017), while many of the second category are fully functional, but lack well-defined structures (Fig. 1e), and are thus called intrinsically disordered proteins (IDPs) (van der Lee et al. 2014). On the other hand, ~30% proteins are associated to lipid domains (Fig. 1b), and are thus called membrane proteins (MPs).
Fig. 1.
Sequence–structure relationship of proteins. Based on the sequences as represented by five types of amino acids, proteins can be classified into high-complexity or random (a), and low-complexity or non-random (f) sequences. A portion of proteins of the high-complexity sequence can fold into uniquely folded structures soluble in vivo with high concentrations of salts (c), while many of proteins of the low-complexity sequence remain intrinsically disordered (e). Interestingly, a large amount of proteins of both high- and low-complexity sequences appear to be aggregation-prone or even insoluble in vivo with high concentrations of salts (d). Furthermore, ~30% proteins of both high- and low-complexity sequences can fold in the membrane environments (b)
Previously, our studies revealed that a large amount of proteins from both categories of eukaryotic genomes appear to be highly aggregated or even “insoluble” in vivo with high salts (Fig. 1d), thus I designated them as “intrinsically insoluble proteins” (IIPs) (Song 2013). Unlike ‘misfolded proteins’, which still have the capacity to fold into uniquely-defined structures once under favorable conditions, IIPs lack the intrinsic ability to fold due to either low-complexity sequences, as well as splicing variation, and/or insertion/mutation, and/or premature termination in the well-folded domains. I also speculated that the majority of the “wastefully synthesized” proteins or defective ribosomal products (DRiPs) have to be degraded immediately after synthesis (Schubert et al. 2000; Duttler et al. 2013), are IIPs (Song 2013). Indeed, recent results in vivo revealed that DRiPs is the predominant source of aggregated proteins in living cells (Ganassi et al. 2016; Mateju et al. 2017). Recently, it was also found that 44–54% of the proteome in eukaryotes and viruses are dark, which has never been observed by experimental structure determination and is inaccessible to homology modeling (Perdigão et al. 2015). So, in the future, it is of fundamental interest to explore whether these dark proteins lacking similarity to any known structure in fact belong to IIPs (Song 2013).
Aggregation and self-assembly into liquid droplets and fibrils of the prion-like domains
There exists a subgroup of proteins within the low-complexity sequences (Fig. 1f), which are enriched in polar and uncharged amino acids such as Gln, Asn, Ser, Gly and Tyr. As they share compositional similarity to the yeast prions such as Sup35, they are thus called “prion-like” domains (Shorter and Lindquist 2005; Michelitsch and Weissman 2000; Chien and Weissman 2001; Han et al. 2012; Harrison and Shorter 2017). Recently, ~240 out of ~20,000 human protein-coding genes (~1.2%) were shown to contain at least one prion-like domain, and, interestingly, human proteins containing the prion-like domains are over-represented by those critically interacting with RNA/DNA including TDP-43 and FUS proteins. These proteins have now been identified as causing various human diseases, particularly age-related neurodegenerative diseases such as Alzheimer’s disease, Parkinson’s disease, amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), and multisystem proteinopathy (MSP). Most unexpectedly, almost all disease-causing mutations are located within their low-complexity domains (Han et al. 2012; Harrison and Shorter 2017; Ling et al. 2013; Lim et al. 2016a; Conicella et al. 2016).
Surprisingly, despite being enriched in polar residues, the human prion-like domains appear to be intrinsically prone to aggregation to form amorphous structures at high concentrations of proteins and salts, as exemplified by the TDP-43 and FUS prion-like domains (Lim et al. 2016a; Lu et al. 2016, 2017a). On the other hand, under some conditions, like the yeast prion proteins (Shorter and Lindquist 2005; Michelitsch and Weissman 2000; Chien and Weissman 2001), the TDP-43 and FUS prion-like domains have been biophysically characterized to form fibril/hydrogel structures with cross-β structures (Han et al. 2012; Lim et al. 2016a; Lu et al. 2016; Kato and McKnight 2017; Murray et al. 2017). Very unexpectedly, we discovered that the self-assembly of both TDP-43 and FUS prion-like domains into the fibril structures is highly pH-dependent (Lim et al. 2016a; Lu et al. 2016): at low pH such as 4.0, they remained mostly monomeric for many weeks, while at neutral pH they showed a strong capacity to self-assemble into fibrils with cross-β structures as reflected by the development of intrinsic visible fluorescence, which is a novel protein fluorescence originating from the hydrogen-bonding network in β-rich secondary structures (Shukla et al. 2004; Chan et al. 2013; Lim et al. 2016a; Lu et al. 2016; Pinotsi et al. 2016). Furthermore, as indicated by low NMR temperature coefficients of most backbone amides, we found that, despite being intrinsically disordered, the TDP-43 prion-like domain contains a large number of intra-molecular hydrogen bonds between side chains of Ser, Thr, Asn, Gln and backbone atoms (I of Fig. 2a, b). Consequently, the backbone has many dynamic loop/turn conformations. At neutral pH, many hydrogen bonds between side chains and backbone atoms are disrupted because the dissociation of the amide protons become significantly enhanced (II of Fig. 2a, b). As such, these side chains are liberated and consequently available to form “hydrogen bonds/polar/steric zippers” (Lim et al. 2016a; Lu et al. 2016), as previously proposed for the amyloid fibrils formed by proteins enriched in Ser, Thr, Asn and Gln (Perutz et al. 1994; Michelitsch and Weissman 2000; Nelson et al. 2005; Han et al. 2012).
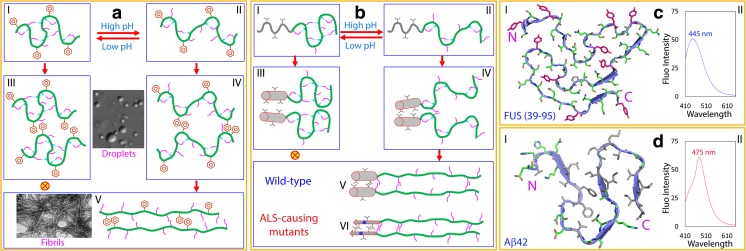
Fig. 2.
Mechanism for the prion-like domains to self-assemble into liquid droplets and fibril structures. a The self-assembly of the prion-like domain dominated by aromatic residues as exemplified by the FUS prion-like domain. The monomeric prion-like domain enriched in polar and uncharged residues (Ser, Thr, Asn, Gln and aromatic residues such as Tyr) exists in equilibrium between two conformational states: I under some conditions such as at low pH, the majority of Ser, Thr, Asn and Gln side chains are involved in forming hydrogen bonds with the backbone atoms; and consequently the backbone adopts a conformation with many dynamic loops/turns; II at neutral pH some hydrogen bonds become disrupted due to the rapid dissociation of the backbone amide protons, and consequently the side chains are liberated and the backbone adopts a more extended conformation. On the one hand, both states regardless at low pH (I) or neutral pH (II) can self-assemble into liquid droplets (III and IV) in which aromatic residues at strategical position provide relatively strong interactions to form dynamic oligomers with the backbone largely disordered which are sufficient to trigger liquid-liquid phase separation. On the other hand, only the state IV with a large portion of the Ser, Thr, Asn and Gln side-chains liberated will be further exaggerated into forming fibril structures with cross-β structures stabilized by “hydrogen-bond/polar/steric zippers” (V) under certain conditions such as long incubation. b The self-assembly of the prion-like domain dominated by hydrophobic residues as exemplified by the TDP-43 prion-like domain. The monomeric prion-like domain enriched in polar and uncharged residues (Ser, Thr, Asn and Gln) and further containing a hydrophobic region also exists in equilibrium between two conformational states I and II. Both states can also self-assemble into liquid droplets (III and IV) in which the formation of dynamic oligomers is mainly mediated by the self-association over the hydrophobic region. The state IV will be further exaggerated into forming fibril structures over the Ser, Thr, Asn and Gln rich region stabilized by “hydrogen-bond/polar/steric zippers” (V). Remarkably, the hydrophobic region remains to be helical even in the fibrils of the wild-type TDP-43 prion-like domain, while it transforms into the classic amyloid structure stabilized by hydrophobic interactions in the fibrils of the ALS-causing mutants. c The atomic structure of the fibrils formed by the FUS prion-like domain over residues 39–95 (I), which is characterized by the intrinsic visible fluorescence with the emission maximum at ~455 nm (II). d The atomic structure of the classic amyloid fibrils formed by Aβ42 (I), which is characterized by the intrinsic visible fluorescence with the emission maximum at ~475 nm (II). Green polar/charged residues, gray hydrophobic residues and pink aromatic residues
Recently, the formation of liquid droplets by liquid–liquid phase separation (LLPS) has been recognized as a general mechanism to form membraneless intracellular organelles that allow spatiotemporal control of molecular interactions in cells. In particular, the exaggeration or molecular aging of liquid droplets into aggregation or amyloid fibrils may lie at the heart of a variety of human diseases including ALS (Murakami et al. 2015; Patel et al. 2015; Shin and Brangwynne 2017). The TDP-43 and FUS prion-like domains appear to remain highly disordered in the liquid droplets (Burke et al. 2015; Lim et al. 2016a; Conicella et al. 2016; Lu et al. 2016). Consequently, it remains poorly understood what is the driving force for LLPS. Furthermore, the relationship between the formation of liquid droplets and fibrils/hydrogels is highly controversial (Han et al. 2012; Burke et al. 2015; Conicella et al. 2016; Kato and McKnight 2017; Murray et al. 2017).
We recently characterized the formation of both liquid droplets and fibrils/hydrogels of the TDP-43 and FUS prion-like domains under the same conditions. We found that both of them formed liquid droplets immediately after the protein powders were dissolved in buffers at both pH 4.0 and 6.8, revealing that this process is much less pH-dependent. By contrast, their formation of fibril structures with cross-β structures appeared to be much slower and highly pH-dependent, as monitored by the development of the intrinsic visible fluorescence and imaged by EM. Briefly, at pH 6.8, the formation of fibrils needed several days dependent on protein and salt concentrations, while at pH 4.0, no formation of fibrils has been observed for several months (Lim et al. 2016a; Lu et al. 2016). Furthermore, we have also characterized a truncated TDP-43 prion-like domain with only residues 342–414 which has a typical prion-like sequence enriched in polar and uncharged residues but lacking the middle hydrophobic region over region 311–341 (Lim et al. 2016a; Lu et al. 2016). Noticeably, although after long incubation it could still self-assemble into dynamic hydrogel with cross-β structures, it lost the ability to form liquid droplets under various conditions. This suggests that, for the TDP-43 prion-like domain, the hydrophobic region provides the key driving force for the formation of liquid droplets but is not essential for the formation of “hydrogen bonds/polar/steric zippers”.
Taking the results with the TDP-43 and FUS prion-like domains by us and other groups together, I would propose a mechanism here which integrates both liquid droplet and fibril structure formation for the prion-like domains. The formation of the liquid droplets and fibril structures appears to be driven by the different sets of forces/interactions, which may, however, have interplay/overlap to some degree. Consequently, the liquid droplets formed by the prion-like domains can be further exaggerated into the fibril structures, as they are enriched in Asn, Gln, Ser and Thr residues. Despite being weak and multivalent, the key driving forces for LLPS of the prion-like domains appear to come from relatively strong interactions involved in aromatic (as exemplified by the FUS prion-like domain in Fig. 2a), and/or hydrophobic (as exemplified by the TDP-43 prion-like domain in Fig. 2b) residues at strategical positions. For the FUS prion-like domain (Fig. 2a), Tyr residues have been shown to play a key role (Han et al. 2012; Patel et al. 2015), and thus various interactions with Tyr residues (Salonen et al. 2011) appear to be sufficient to trigger LLPS to form dynamic liquid droplets (III and IV of Fig. 2a) regardless of pH values because the liberation of Ser, Thr, Asn and Gln side chains is not essential for LLPS, and in fact the backbone conformations of most residues still remain highly similar to those in the monomeric states (I and II of Fig. 2a). The involvement of Ser, Thr, Asn and Gln side chains in hydrogen bonding with the backbone atoms might in fact act to inhibit the immediate formation of highly ordered fibril structures with cross-β secondary structures (V of Fig. 2a). Briefly, the presence of the hydrogen bonds between the side chains of Ser, Thr Asn and Gln and the backbone atoms might prevent those side chains from being immediately available as well as the backbones from becoming extended, both of which are required for further assembly into cross-β structures with “hydrogen bonds/polar/steric zippers” (V of Fig. 2a). Nevertheless, in vivo with neutral pH, if the liquid droplets formed by the prion-like domains remain unliquidized for a long time under pathological conditions, the dynamic liquid droplets may be exaggerated into dynamic fibril/hydrogel structures because the side chains of Ser, Thr, Asn and Gln residues will be slowly liberated from hydrogen bonding with the backbone atoms and subsequently to form “hydrogen bonds/polar/steric zippers” (V of Fig. 2a).
For the TDP-43 prion-like domain (Fig. 2b), on the one hand, unlike the FUS prion-like domain, the hydrophobic region appears to contribute the key driving force for LLPS to form liquid droplets (III and IV of Fig. 2b), as its deletion completely disrupted the ability to form liquid droplets but not cross-β structures stabilized by “hydrogen bonds/polar/steric zippers” (Lu et al. 2016). At neutral pH, the liquid droplets of the TDP-43 prion-like domain will also be exaggerated into fibril structures like the FUS prion-like domain. Noticeably, we previously found that the wild-type and ALS-causing mutants of the TDP-43 prion-like domain could form the fibrils with different secondary structures (Lim et al. 2016a; Song 2017): for the wild-type, the hydrophobic region appeared to remain helical even though the prion-like region formed “hydrogen bonds/polar/steric zippers” (V of Fig. 2b), while for the ALS-causing mutants, the hydrophobic region further transformed into the classic amyloid fibril (VI of Fig. 2b), as reported by different emission spectra of the intrinsic visible fluorescence (Lim et al. 2016a). Once this transformation occurs, the fibril structures formed by the TDP-43 prion-like domain become much less dynamic than those by the FUS prion-like domain. While the dynamic fibril/hydrogel structures formed by the FUS prion-like domain could be dissolved in buffers with low concentrations of detergents, the classic amyloid fibrils by Aβ peptides were even resistant to solubilization by buffers with 8 M urea (Han et al. 2012; Patel et al. 2015).
The mechanism suggests that the disease-causing mutations within the prion-like domains might affect the self-assembly of liquid droplets and fibril/hydrogel structures at least in two different ways dependent on the types of the mutations. If the mutations directly alter the residues providing the key driving forces for LLPS, as exemplified by deleting the hydrophobic region within the TDP-43 prion-like domain, the formation and dynamics of liquid droplets will be directly perturbed, such as disrupted or even eliminated, but the formation of dynamic cross-β structures still occurs despite becoming much slower (Lu et al. 2016). On the other hand, even if the mutations are not involved in the residues contributing the key driving force to LLPS, the formation and dynamics of liquid droplets, as well as its further exaggeration into fibril/hydrogel structures, can still be mediated because the mutations will shift the conformational distribution of the ensemble of the backbones and/or side chains through the global network of hydrogen bonds in the prion-like domains. More generally, the prion-like domains appear to have unique energy landscapes, which are very amenable to the remodeling by the genetic, pathological and environmental factors, as we previously proposed for the TDP-43 prion-like domain (Lim et al. 2016a; Lu et al. 2016).
Most amazingly, this mechanism offers the potential to generate conformational ensembles composed of enormous amounts of states for both liquid droplets and fibril structures, which are also extremely sensitive to various environmental factors such as pH and cellular processes, including phosphorylation of Ser, Thr and Tyr (Murray et al. 2017; Monahan et al. 2017; Shorter 2017). Therefore, it appears most likely that the biological functions of the prion-like domains request not just the formation of the liquid droplets but also the conformational dynamics, as well as further assembly into diverse fibril structures. Otherwise, residues, particularly Gln and Asn, are not needed in the prion-like domains because they trigger further exaggeration of the liquid droplets into fibril structures, which might trigger pathological consequences. This possibility is also supported by the intriguing observation that the mutations of the majority of residues of the intrinsically disordered prion-like domains unexpectedly have a disease-causing consequence. Indeed, although PR and GR dipeptide repeats encoded by C9orf72 hexanucleotide repeat expansions are sufficient to undergo LLPS and also to induce phase separation of a large set of other proteins, they are highly toxic and cause the pathogenesis of C9orf72 ALS/FTLD (Lin et al. 2016; Boeynaems et al. 2017). Therefore, PR and GR repeats appear to manifest toxicity by rephrasing the functional conformations and dynamics of liquid droplets and/or fibril structures formed by the proteins containing the prion-like domains.
So what could be the functions which require the ensembles of such enormous amounts of conformational states? One clue might come from the recent studies on the consolidation of long-term memory for both yeast and mammals (Shorter and Lindquist 2005; Rayman and Kandel 2017; Sudhakaran and Ramaswami 2017). It has been well established that, at a molecular level, long-term memory is characterized by its requirement for changing the profiles of gene expression which is mainly implemented by the RNA-binding proteins with the prion-like domains (Shorter and Lindquist 2005; Rayman and Kandel 2017; Sudhakaran and Ramaswami 2017). However, even in the human genome, it is likely that there exist <240 RNA-binding proteins with the prion-like domains (Harrison and Shorter 2017). So how can such a limited number of the RNA-binding proteins produce a giant amount of the gene expression profiles to consolidate long-term memory and/or even more generally cellular memory? The solution might be derived from the amazing ability of the prion-like domains to generate conformational ensembles containing a vast amount of states for both liquid droplets and fibril structures, which may subsequently produce a huge amount of gene expression profiles through a mechanism recently proposed for transcriptional control by LLPS-induced formation of super-enhancers sensitive to perturbation (Hnisz et al. 2017). Nevertheless, such systems appear to be maintained at a great cost: a slight exaggeration might be sufficient to trigger detrimental consequences such as neurodegenerative diseases.
Very recently, the structure of the dynamic fibril of the FUS prion-like domain over residues 39–95 has been successfully determined by solid-state NMR spectroscopy (Murray et al. 2017), which is almost completely constrained by interactions involved in polar (Ser, Thr, Asn and Gln) and aromatic (Tyr) residues (I of Fig. 2c). This is fundamentally different from the structure of the pathological and irreversible fibril of Aβ42 (Colvin et al. 2016), which is mostly stabilized by hydrophobic interactions (I of Fig. 2d). Recently, protein fibrils have been found to absorb light in the near-UV range and to emit a structure-specific intrinsic fluorescence in the visible range even in the absence of aromatic amino acids. Most strikingly, this intrinsic visible fluorescence has been characterized to originate from the hydrogen-bonding network of protein fibrils with cross-β structures (Chan et al. 2013; Lim et al. 2016a; Lu et al. 2016; Pinotsi et al. 2016). Based on previous studies by us and other groups, a diagnostic probe of this novel intrinsic visible fluorescence can be established which is able to distinguish two different types of the fibril structures. While the formation of the dynamic fibrils such as by the FUS and TDP-43 prion-like domains enriched with polar and uncharged residues (Lim et al. 2016a; Lu et al. 2016) is characterized by the development of the intrinsic visible fluorescence with the emission maximum at ~445 nm (II of Fig. 2c), the formation of the classic amyloid fibrils by Aβ42 (Chan et al. 2013) and by the hydrophobic region of the TDP-43 prion-like domain (Lim et al. 2016a) is indicated by the development of the intrinsic visible fluorescence with the emission maximum at ~475 nm (II of Fig. 2d). Noticeably, the emission maximum of the intrinsic visible fluorescence of the classic amyloid fibrils (~475 nm) is significantly red-shifted as compared to that of the dynamic fibrils formed by the prion-like domains (~445 nm). This is likely due to the significant difference in the amount and arrangement of the hydrogen bond networks involved in the backbone peptide bonds of two types of fibril structures: the higher the amount and the tighter of hydrogen bonds arranged in the cross-β structures, the larger the red-shift (Chan et al. 2013; Lim et al. 2016a; Lu et al. 2016; Song 2017; Pinotsi et al. 2016).
Transformation of well-folded and soluble cytosolic proteins into insoluble forms by disease- and aging-associated factors
Many human diseases including all neurodegenerative diseases are characterized by severe aggregation or even “insolubility” of specific proteins (Chiti and Dobson 2006; Song 2009; Auluck et al. 2010; Ling et al. 2013; Song 2013; Brender et al. 2012; Willis and Patterson 2013; McLendon and Robbins 2015; Song 2017). Recently, protein aggregation of non-specific proteins has been extensively identified to be the marker of aging down to unicellular organisms (Lindner et al. 2008; David et al. 2010). In particular, a list of ~20 genes has been linked to ALS (Ling et al. 2013), and the proteins encoded by these genes can be divided into two groups: (1) those with the high-complexity sequences whose wild-types can fold into highly soluble and well-defined structures including MSP, SOD1 and profilin1; and (2) those containing the low-complexity sequences such as the prion-like domains whose wild-types are intrinsically disordered and aggregation-prone including TDP-43 and FUS. While the wild-type itself of the second group is capable of triggering ALS, the ALS-causing mutations are needed to convert the first group into aggregation-prone or even “insoluble” forms which mysteriously gain toxicity to cause ALS (Ling et al. 2013). Previously, it has been challenging to define the mechanisms of their transformation because the ALS-causing forms of these proteins are highly aggregation-prone, or even completely insoluble.
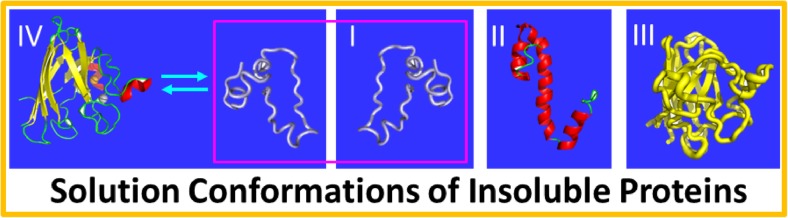
In 2005, we discovered a previously unknown regime associated with proteins (Li et al. 2006; Song 2009, 2013; Song 2017): unlike the well-folded proteins following the classic “Salting-in” rule that protein solubility first increases upon adding salts over the range of low salt concentrations (usually <300–500 mM), “insoluble” proteins including even the most hydrophobic integral membrane protein fragment in nature could only be solubilized in aqueous solution with minimized salts. This discovery has now been extensively confirmed, and become a powerful tool for us and other groups to biophysically characterize “insoluble” protein (Delak et al. 2009; Aguado-Llera et al. 2010; Futami et al. 2014). Initially, by CD and NMR characterization, we found that “insoluble” proteins could be divided into three groups according to their conformations in unsalted water (Li et al. 2006; Song 2009, 2013): group 1, which has no stable secondary and tertiary structures (I of Fig. 3); group 2, which has secondary but no tertiary structure (II of Fig. 3); and group 3, which has secondary structure as well as dynamic tertiary packing which is molten globule-like (III of Fig.3) (Song et al. 1999; Wei and Song 2005). Very recently, we also found that, in salt-minimized buffers, aggregation-prone TDP-43 N-domain (Qin et al. 2014) and C71G-PFN1 mutant (Lim et al. 2017) coexisted in equilibrium between the folded and unfolded states (VI of Fig. 3). Marvelously only in unsalted water, proteins can manifest their intrinsic conformations, regardless of their hydrophobicity and whether well-folded, partial folded or predominantly unstructured. One scenario that I proposed to rationalize this phenomenon is that the prebiotic aqueous medium where proteins originally emerged was largely unsalted (Song 2013). Indeed, there exists evidence indicating that, when primitive proteins were made, the ocean was highly unsalted and slightly acidic (Pinti 2005).
Fig. 3.
Solution conformations of insoluble proteins solubilized in unsalted water. Four groups of conformations have been observed so far on the insoluble proteins solubilized in unsalted water: I those without stable secondary and tertiary structures; II those with secondary but without tertiary structures; III those with secondary structures as well as dynamic tertiary packing which is molten globule-like; and IV those with coexistence between the folded and unfolded states in equilibrium
With this discovery, several years ago we started to explore the mechanisms for transforming the well-structured and soluble wild-types into insoluble and toxic forms of ALS-associated proteins by utilizing various biophysical methods in particular NMR spectroscopy. We first characterized the ALS-causing P56S mutant of the VAPB-MSP domain that forms very tight aggregates in vivo even resistant to solubilization by buffers containing nonionic detergents such as TritonX-100 (Teuling et al. 2007). Strikingly, the residue-specific NMR characterization revealed that the P56S point mutation was sufficient to convert the well-folded and highly soluble MSP domain adopting a seven-strand immunoglobulin-like β sandwich into a highly disordered form lacking of any stable secondary and tertiary structures, which was only soluble in the salt-minimized water (Shi et al. 2010). Furthermore, we showed that the splicing variant of VAPB was also insoluble in buffers and highly disordered in the salt-minimized water (Qin et al. 2013a). Remarkably, this splicing variant has been found only to become accumulated upon proteasomal inhibition, a condition commonly found in neurodegenerative diseases (Nachreiner et al. 2010).
Subsequently, we studied L126Z-SOD1, which is a truncation mutant with the deletion of the last 28 residues. It also forms tight aggregates resistant to detergent solubilization in vivo and, most importantly, it has much more elevated toxicity as compared to other SOD1 point mutants (Zu et al. 1997). We have discovered that this truncation completely abolished the intrinsic ability to fold into the native SOD1 fold even in the presence of zinc ions, and, consequently L126Z-SOD1 also became highly disordered without any stable secondary and tertiary structures and with the backbone atoms having unrestricted motions on a ps–ns time scale (Lim et al. 2015). Strikingly, we found that the nascent SOD1 depleted of zinc ions was also highly disordered but reached equilibrium of the unfolded and folded conformations in the presence of zinc. Residue-specific NMR parameters clearly indicate that the nascent WT-SOD1 has almost the same conformations and dynamics as L126Z-SOD1 over the identical 125 residues, thus implying that the wild-type and ALS-linked SOD1 mutants may trigger ALS by a common mechanism (Lim and Song 2016). Very recently, we revealed that the ALS-causing C71G mutation also rendered the well-folded profilin1 into the co-existence between the folded and unfolded states (Lim et al. 2017).
So the mechanism for ALS-causing mutations to convert the well-folded and soluble MSP domain, SOD1 and profilin1 into insoluble forms is clear: ALS-causing mutations represent a subgroup of protein mutations which carry the ability to radically destabilize the folded state, and/or stabilize the unfolded state of a protein. As a result, the unfolded state becomes populated to different degrees for the mutants, which unavoidably have the significant exposure of hydrophobic or amphiphilic patches universally existing in these proteins. These states are thus only soluble in unsalted water, but become severely aggregated in vivo with high concentrations of salts (Song 2009, 2013; Song 2017). Fundamentally, such highly-disordered mutants of MSP, SOD1 and profilin1 are indistinguishable from the intrinsically-disordered alpha-synuclein (Ulmer et al. 2005) and mellitin, a membrane-active toxin from honeybee venom, which is also highly disordered in unsalted water (Lauterwein et al. 1980) but prone to aggregation in buffers (Brown et al. 1980).
This appears to be a general mechanism for genetic, pathological/aging and environmental factors to transform well-folded and highly soluble wild-types into insoluble forms in all organisms, because we recently demonstrated that the fragmentation to mimic over-oxidation during aging also resulted in generating insoluble fragments of the well-folded and soluble E. coli S1 ribosomal protein (Lim et al. 2016b). Noticeably, the proteins which are converted into insoluble forms by these factors appear to be significantly over-represented by β-dominant structures (David et al. 2010), as exemplified by the MSP domain, SOD1 and profilin1, as well as E. coli S1 ribosomal protein (Song 2017). This phenomenon is most likely due to the fact that, unlike the formation of the helix which is mostly stabilized by local interactions, the formation of the β-sheet is stabilized by long-range interactions, or even covalent linkage such as disulfide bridges in SOD1, and consequently the β-dominant folds are highly vulnerable to the destabilizing factors such as mutation, deletion, breaking of the disulfide bridge and/or depletion of cofactor including depletion of zinc ions for SOD1 (Lim and Song 2016; Song 2017).
Transformation from cytosolic proteins into membrane-interacting proteins
Although we have successfully uncovered the mechanism by which the ALS-causing mutations/truncation/depletion of zinc ions transform the well-folded and highly soluble MSP domain, profilin1 and SOD1 into the insoluble forms in vivo with high salt concentrations, it still remained unknown how this transformation is associated with the gain of toxicity to initiate ALS pathogenesis. In particular, there were reports which challenged the notion that the protein aggregate itself is the cause of neurodegenerative diseases and showed instead that, surprisingly, the promotion of inclusion formation reduced the risk of neuronal death (Arrasate et al. 2004; Bodner et al. 2006).
As inspired by the diagram of the bilayer membrane structure, which is composed of both polar (~15 Å thick) and hydrophobic (~30 Å thick) phases (White and Wimley 1999), I realized that the insoluble proteins should interact with biological membranes as the “insolubility” of proteins is mostly resulting from the exposure of amphiphilic/hydrophobic patches to salted water (Song 2013, 2017). Indeed, the disordered P56S MSP and its splicing variant did interact with the DPC micelle (Qin et al. 2013a), as well as with the DMPC/DHPC bicelle and DMPC vesicle mimicking the membrane environment (Qin et al. 2013b). Subsequently, we determined the three-dimensional structure of P56S-MSP in the DPC micelle by a large set of NMR constraints, together with long-distance constraints derived from the paramagnetic relaxation enhancement. In the structure, several well-defined α-helices are formed, but the three-dimensional topology appears to be mostly constrained by the DPC micelle. Examination of the helical fragments formed in the membrane environment immediately revealed that they are highly amphiphilic or hydrophobic, fundamentally indistinguishable from those formed by α-synuclein (Ulmer et al. 2005) and mellitin in micelles (Lauterwein et al. 1980).
We further determined the conformations of L126Z-SOD1 (Lim et al. 2015), nascent SOD1 (Lim et al. 2016a) and C71G-profilin1 (Lim et al. 2017) in aqueous solution and membrane environments. While their unfolded states have no stable secondary and tertiary structures, as well as unrestricted ps–ns backbone motions, they could all transform into non-native helical structures in membrane environments. However, the properties of the helices are very diverse: most are either amphiphilic or hydrophobic, but a portion of them appeared to contain only short hydrophobic segments (Lim et al. 2017). This observation is completely consistent with the extreme complexity for protein folding in the membrane environments, which have both polar and hydrophobic phases dynamically interacting with each other (White and Wimley 1999; Ladokhin and White 1999). Another interesting feature is that none of these non-native helices have tight packing and consequently their orientations are not defined and thus highly dynamic, as evident from the lack of any long-range NOEs.
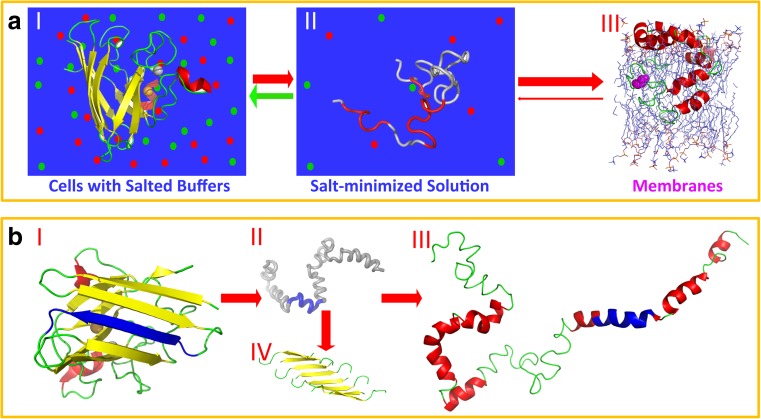
In my view, this is understandable, as SOD1 and VAPB-MSP domain are natively cytosolic proteins with completely different three-dimensional structures for implementing their physiological functions in cytosolic environments. Although the native profilin1 is reversibly localized to membranes, it only utilizes a small portion of residues on the N- and C-terminal helices to weakly interact with the membrane (Lim et al. 2017). It thus becomes clear for the mechanism of the ALS-causing mutations to transform well-folded and soluble proteins into insoluble and membrane-interacting forms (Fig. 4a): the ALS-causing factors act to reduce or even abolish the ability of the wild-type protein to fold into the well-structured native structures (I of Fig. 4a), and consequently the mutants becomes highly disordered (II of Fig. 4a), or co-existing with the unfolded state. The highly disordered state unavoidably has significant exposure of hydrophobic/amphiphilic regions universally existing in proteins including MSP, SOD1 and profilin1. Therefore, on the one hand, these highly disordered forms become aggregation-prone or even insoluble in vivo with high salt concentrations. On the other hand, they acquire novel or enhanced capacity to interact with membranes (III of Fig. 4a) energetically driven by forming stable helices/loops over hydrophobic/amphiphilic segments. The formation of helix/loop results in the establishment of a large number of hydrogen bonds, thus leading to gain of hydrogen bond energetics (White and Wimley 1999; Ladokhin and White 1999), which acts to drive the partitioning from aqueous solutions into membrane environments. Therefore, the driving forces for a protein to aggregate and to partition into membrane environments appear to be at least partly overlapped, or even two sides of the same coin as previously proposed (Qin et al. 2013b; Lim et al. 2015; Lim and Song 2016; Song 2017).
Fig. 4.
Transformation of a well-folded and soluble cytosolic protein into membrane-interacting form. a By disease- and aging-associated factors, a well-folded and highly soluble cytosolic protein (I) can be converted into a highly disordered form, which is only soluble in salt-minimized water (II). As this highly disordered form universally contains hydrophobic, and/or amphiphilic patches, it acquires the novel capacity to interact with membranes energetically driven by folding into non-native helix/loop structures in membranes (III). b Residues 28–38 (in blue) of SOD1 adopt a β-strand in the native structure (I), but become disordered in the ALS-causing L126Z-SOD1 (II). However, these residues transform into a helix in the membrane-embedded L126Z-SOD1 (III). Furthermore, eight isolated fragments of SOD1 (28–28) can assemble into an amyloid structure after incubation (IV)
On the other hand, the diverse physicochemical properties of the helices/loops and their lack of tight packing in the membrane environments may in fact offer the flexibility for these proteins to adjust the orientations, or lengths, or even secondary structure types, so as to effectively interact with various biological membrane systems of different compositions, structures and dynamics. In this context, unlike the native membrane proteins which require highly specific three-dimensional structures and dynamics to perform their biological functions, aggregation-prone proteins may simply use the acquired capacity in abnormally interacting with membranes to commonly trigger pathological events even forming distinctive structures and dynamics onto/into different membranes. This mechanism also appears to be a general mechanism in other organisms, because the insoluble fragments of the E. coli S1 ribosomal protein have also been characterized to strongly interact with various membrane mimetics (Lim et al. 2016b).
Very recently, a fragment of SOD1 (residues 28–38) has been characterized to be toxic to neurons, but its underlying mechanism still remains largely unknown (Sangwan et al. 2017). This fragment adopts a β-strand in the native structure of SOD1 (I of Fig. 4b), but becomes disordered (II of Fig. 4b) in the L126Z-SOD1 (Lim et al. 2015) and nascent SOD1 (Lim and Song 2016) in unsalted water. However, upon being embedded in DPC micelle, this region becomes highly helical (III of Fig. 4b). Most amazingly, eight isolated fragments have been recently found to self-assemble into an oligomer with an amyloid-like structure (VI of Fig. 4b), which has been proposed to be the toxic form (Sangwan et al. 2017). In this regard, it is of both fundamental and therapeutic interest to explore whether the isolated SOD1 fragment interacts with membranes andm if yes, what is its structure in the membrane environment? Remarkably, this observation strongly highlights that the sequence-structure relationship is highly environment-transformable for the proteins/peptides which manifest proteotoxicity. In other words, these proteins/peptides own a chameleon ability to fold (Minor and Kim 1996).
Coupled capacities in membrane interaction and aggregation/amyloid-formation: a general mechanism responsible for cell death
In fact, all intrinsically disordered and partly soluble proteins causing human diseases have been previously demonstrated to contain “intrinsic” regions which could insert into membranes by forming amphiphilic/hydrophobic helices, which include prions of spongiform transmissible encephalopathies (Elfrink et al. 2008), amyloid beta-peptides of Alzheimer’s disease (Kotler et al. 2014), tau tangles of Alzheimer’s disease (Künze et al. 2012), α-synuclein of Parkinson’s disease (Ulmer et al. 2005; Auluck et al. 2010), huntingtin of Huntington’s disease (Kegel et al. 2005), and the islet amyloid polypeptide of type II diabetes (Brender et al. 2012). Recently, we also identified a hydrophobic region in the middle of the TDP-43 prion-like domain which was capable of inserting into the membrane environment to transform from a partially folded helical conformation into a well-defined Ω–loop–helix motif (Lim et al. 2016a).
On the other hand, our studies on insoluble forms transformed from the well-folded and soluble wild-type proteins indicate that, due to significant impairment/disruption of the well-folded structures, these insoluble forms become highly disordered and their hydrophobic and/or amphiphilic regions universally existing in these proteins are unlocked from the well-folded native structures. As a consequence, these states have the “acquired” capacity in interacting with membranes, which is mechanistically indistinguishable from the “intrinsic” capacity in interacting with membranes owned by amyloid beta-peptides, α-synuclein, and even the membrane toxin mellitin. In fact, many years back, ALS-causing SOD1 mutants were extensively shown to be tightly associated with the mitochondrial membrane, even behaving as an integral membrane protein with the association resistant to high ionic strength and high pH (Liu et al. 2004; Vande Velde et al. 2008; Sun et al. 2015). Most remarkably, the abnormal insertion of SOD1 mutants into the ER membrane has been characterized to be sufficient to trigger ER stress, which represents an initial event of a cascade of cell-specific damage in ALS pathogenesis (Sun et al. 2015).
Taken together, all aggregation-prone proteins linked to human diseases and aging appear to share a common capacity in interacting with membranes regardless of being “partly” or “completely” insoluble. In fact, our studies indicate that “completely insoluble” proteins are not really insoluble but only progressively become aggregated in solution with high concentrations of salts. Strikingly, it was also demonstrated that slightly acidic pH was sufficient to disassemble the amyloid fibrils of microglobulin into soluble oligomers which were able to disrupt membranes (Tipping et al. 2015). Therefore, interactions with membranes appear to represent a common mechanism for these proteins, which have no consensus in physiological functions, to trigger a diverse spectrum of human diseases and aging. This also rationalizes the observation that, although the aggregation-prone proteins are membrane-toxic in general, the most popular human diseases caused by protein aggregation are neurodegenerative diseases and cardiac dysfunction. This may be due to the fact that neurons and cardiomyocytes are rarely replaced (Spalding et al. 2005).
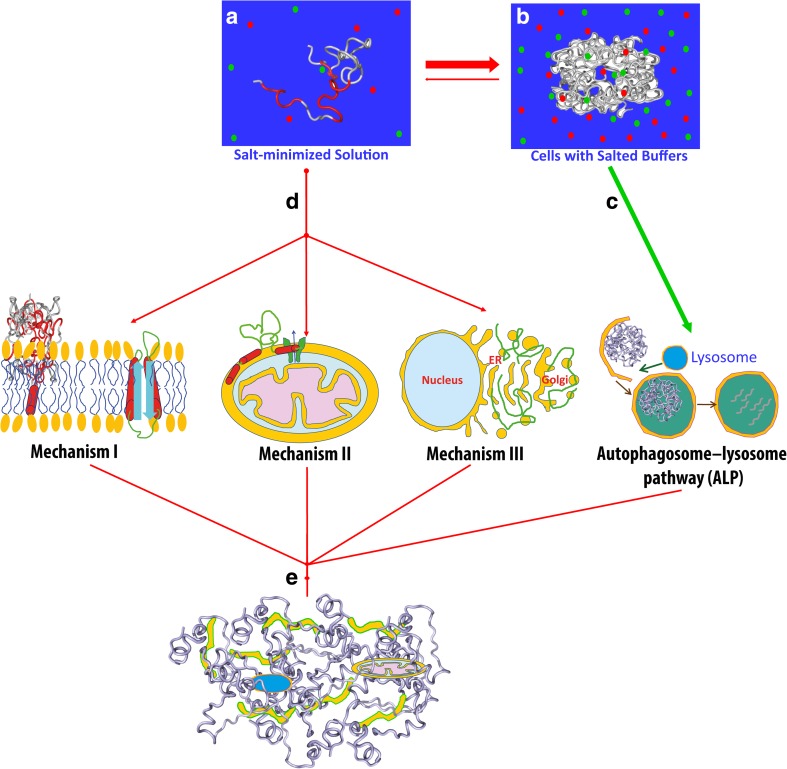
Previously, the mechanisms for pathogenesis by interaction with membranes have been extensively investigated, which are involved in all membrane-associated processes. Briefly, as summarized in Fig. 5, the disordered proteins with significant exposure of hydrophobic and/or amphiphilic patches are only soluble in salt-minimized aqueous solution (Fig. 5a), but become aggregated or even insoluble in vivo with ~150 mM salts (Fig. 5b). On the other hand, these aggregates may be degraded such as by the autophagosome–lysosome pathway (Fig. 5c). However, before they become completely insoluble, they may abnormally interact with membranes (Fig. 5d) by at least three mechanisms: (1) direct disruption of membrane structures and dynamics such as by forming ion channels/pores or large aggregates within membranes (Mechanism I of Fig. 5) (Brender et al. 2012; Kotler et al. 2014); and/or (2) perturbation/initiation of membrane-anchored signal pathways or machineries such as ion channels (Mechanism II of Fig. 5) (Liu et al. 2004; Vande Velde et al. 2008); and/or (3) interference in the membrane remodeling such as budding, shaping, trafficking and fusion (Mechanism III of Fig. 5) (Auluck et al. 2010; Sun et al. 2015). With consideration of the lack of membrane-interacting specificity for these protein forms, these three mechanisms are not necessarily mutually exclusive and may operate simultaneously in many cases. For E. coli cells which are largely absent from internal membrane networks, the dominant mechanism may be Mechanism I. For high eukaryotic organisms, Mechanisms II and III might be extremely critical as: (1) many membrane systems like ER and/or membrane-associated machineries in fact act as sensors for various stresses to initiate downstream signaling which may trigger pathogenesis; (2) membrane remodeling is an essential process constantly occurring in eukaryotic cells; and (3) perturbation of the membrane remodeling does not always request high affinity and specificity.
Fig. 5.
Mechanisms for proteins characterized by aggregation to manifest proteotoxicity by abnormally interacting with membranes. The disordered protein with significant exposure of hydrophobic and/or amphiphilic patches is only soluble in salt-minimized water (a), but become severely aggregated or even insoluble in vivo with ~150 mM salts (b), which might be degraded by complex machineries such as autophagosome-lysosome pathway (c). However, before it becomes completely insoluble, or under some conditions even the formed inclusions and amyloid fibrils might be disassembled into soluble oligomers, it can abnormally interact with membranes (d) by three mechanisms: I direct disruption of membrane structures and dynamics such as by forming ion channels/pores or large aggregates within membranes; and/or II perturbation/initiation of membrane-anchored signal pathways or machineries such as ion channels; and/or III interference in the membrane remodeling such as budding, shaping, trafficking and fusion. In particular, the coupled abilities of such a protein to aggregate and interact with membranes might radically damage various membranous organelles to manifest its proteotoxicity by forming inclusions composed of damaged membranous organelles and protein (e)
Previously, we have proposed that the coupled abilities in aggregation and interaction with membranes of SOD1 and profilin1 mutants as well as TDP-43 might radically enhance their toxicity in ALS pathogenesis. For FUS, although its N- and C-termini with the low complexity sequences have no detectable interaction with membrane mimetics, its RRM domain could interact with the large bicelle mimicking the bilayer membrane as well as formed fibril structures onto/into membrane mimetics (Lu et al. 2017a, b). Previously, Lewy bodies, the key hallmark for Parkinson’s disease, have been described as protein inclusions which are composed of α-synuclein aggregates/fibrils together with other “co-precipitated” proteins. Unexpectedly, however, a very recent study showed that Lewy bodies were in fact a crowded membranous medley together with α-synuclein aggregates, whose formation was proposed to result from the dual abilities of α-synuclein in aggregation and membrane interaction (Shahmoradian et al. 2017). Therefore, the coupling of non-specific capacities in membrane interaction and aggregation may be a general and key mechanism for aggregation-prone proteins to manifest high proteotoxicity to lead to further death of cells by disrupting the membranous organelles (Fig. 5e), as observed on Lewy bodies (Shahmoradian et al. 2017). In this context, the lower the membrane-interacting specificity these proteins have, the higher the membrane-toxicity they might manifest. This explains the emerging observation that aggregation-prone proteins are toxic in general, which not only cause all neurodegenerative diseases but also appear to drive aging of organisms down to E. coli cells.
Therefore, the membrane-interacting capacity appears to also be encoded in most well-folded non-membrane proteins, which is usually locked in the folded native structures. Nevertheless, upon disrupting their structures by disease, aging and/or even environmental factors, the membrane-interacting capacity will be released from the Pandora box to trigger diseases and aging. Additionally, the eukaryotic, particularly human genomes appear to contain a large amount of IIPs, most, if not all, of which are also capable of interacting with membranes. Consequently, they might also act to initiate diseases and aging when their expression is increased, and/or degradation is reduced under some pathological, aging and/or environmental conditions (Nachreiner et al. 2010; Qin et al. 2013b).
Concluding remarks
As all cellular functions are performed by proteins, the proper balance and integrity of the proteome, or proteostasis, is of central importance, and any uncontrolled perturbations to proteostasis might unavoidably have pathological consequences (Hartl 2016). When Anfinsen formulated the fundamental principle of protein folding that the amino acid sequence is sufficient to specify the three-dimensional structure of a protein, it was thought that all proteins will fold into the uniquely folded structures, and consequently the effect of the environments is relatively minor. Now it has become recognized that the foldable proteins only account for a portion of the high-complexity sequences, while many of the low-complexity sequences are intrinsically disordered. Furthermore, it has been discovered that to maintain proteostasis is a daunting and extremely challenging task even for the foldable proteins. In human cells, proteostasis has been characterized to be maintained by a complex network that involves at least ∼1400 proteins, which include molecular chaperones and their regulators, as well as proteins that modulate the oxidative stress defense and proteolytic degradation (Hartl 2016; Kim et al. 2013).
Particularly for the proteins that have no ability to fold into the unique structures, their conformations are highly environment transformable: even in aqueous conditions, their conformations can be significantly altered by different solution conditions. Furthermore, our studies decoded that disease and aging factors such as mutation, or deletion, or depletion of cofactors, or covalent modification including fragmentation are sufficient to convert many well-folded and soluble cytosolic proteins into the highly disordered and insoluble forms, which further transform into non-native structures upon being embedded in membrane environments. The capacity to abnormally perturb membranes regardless of being intrinsic or acquired might initiate disease/aging processes such as ALS even without the formation of detectable aggregates (Sun et al. 2015; Yang et al. 2016). Moreover, the coupled capacities of these proteins in membrane interaction and aggregation are anticipated to radically damage various membranous organelles to lead to death of cells by forming inclusions composed of damaged membranous organelles and protein aggregates. Therefore, an environment-transformable sequence–structure relationship, ranging from a slight exaggeration of dynamic liquid droplets into fibril structures of the prion-like domains to the dual capacity of most, if not all, proteins to fold in aqueous solution and membranes, appears to commonly provoke perturbations to proteostasis to different degrees, thus representing a general mechanism for proteotoxicity.
Acknowledgements
I would like to thank all laboratory members, which is a long list, for their critical contributions. The studies are supported by Ministry of Education of Singapore (MOE) Tier 2 Grants MOE2015-T2-1-111 to Jianxing Song. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Compliance with ethical standards
Conflict of interest
Jianxing Song declares that he has no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by the author.
Footnotes
This article is part of a Special Issue on ‘Biomolecules to Bio-nanomachines - Fumio Arisaka 70th Birthday’ edited by Damien Hall, Junichi Takagi and Haruki Nakamura
References
- Aguado-Llera D, et al. The basic helix-loop-helix region of human neurogenin 1 is a monomeric natively unfolded protein which forms a “fuzzy” complex. Biochemistry. 2010;49:1577–1589. doi: 10.1021/bi901616z. [DOI] [PubMed] [Google Scholar]
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Auluck PK, Caraveo G, Lindquist S. α-Synuclein: membrane interactions and toxicity in Parkinson’s disease. Annu Rev Cell Dev Biol. 2010;26:211–233. doi: 10.1146/annurev.cellbio.042308.113313. [DOI] [PubMed] [Google Scholar]
- Bodner RA, Outeiro TF, Altmann S, Maxwell MM, Cho SH, Hyman BT, McLean PJ, Young AB, Housman DE, Kazantsev AG. Pharmacological promotion of inclusion formation: a therapeutic approach for Huntington’s and Parkinson’s diseases. Proc Natl Acad Sci U S A. 2006;103:4246–4251. doi: 10.1073/pnas.0511256103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, et al. Phase separation of C9orf72 Dipeptide repeats perturbs stress granule dynamics. Mol Cell. 2017;65:1044–1055. doi: 10.1016/j.molcel.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brender JR, Salamekh S, Ramamoorthy A. Membrane disruption and early events in the aggregation of the diabetes related peptide IAPP from a molecular perspective. Acc Chem Res. 2012;45:454–462. doi: 10.1021/ar200189b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown LR, Lauterwein J, Wüthrich K. High-resolution 1H-NMR studies of self aggregation of melittin in aqueous solution. Biochim Biophys Acta. 1980;622:231–244. doi: 10.1016/0005-2795(80)90034-3. [DOI] [PubMed] [Google Scholar]
- Burke KA, Janke AM, Rhine CL, Fawzi NL. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol Cell. 2015;60:231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan FT, Kaminski Schierle GS, Kumita JR, Bertoncini CW, Dobson CM, Kaminski CF. Protein amyloids develop an intrinsic fluorescence signature during aggregation. Analyst. 2013;138:2156–2162. doi: 10.1039/c3an36798c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien P, Weissman JS. Conformational diversity in a yeast prion dictates its seeding specificity. Nature. 2001;410:223–227. doi: 10.1038/35065632. [DOI] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Colvin MT, et al. Atomic resolution structure of Monomorphic Aβ42 Amyloid fibrils. J Am Chem Soc. 2016;138:9663–9674. doi: 10.1021/jacs.6b05129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conicella AE, Zerze GH, Mittal J, Fawzi NL. ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure. 2016;24:1537–1549. doi: 10.1016/j.str.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, Kenyon C. Widespread protein aggregation as an inherent part of aging in C. Elegans. PLoS Biol. 2010;8:e1000450. doi: 10.1371/journal.pbio.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delak K, et al. The tooth enamel protein, porcine amelogenin, is an intrinsically disordered protein with an extended molecular configuration in the monomeric form. Biochemistry. 2009;48:2272. doi: 10.1021/bi802175a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttler S, Pechmann S, Frydman J. Principles of cotranslational ubiquitination and quality control at the ribosome. Mol Cell. 2013;50:379–393. doi: 10.1016/j.molcel.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfrink K, Ollesch J, Stöhr J, Willbold D, Riesner D, Gerwert K. Structural changes of membrane-anchored native PrP(C) Proc Natl Acad Sci U S A. 2008;105:10815–10819. doi: 10.1073/pnas.0804721105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futami J, et al. Denatured mammalian protein mixtures exhibit unusually high solubility in nucleic acid-free pure water. PLoS ONE. 2014;9:e113295. doi: 10.1371/journal.pone.0113295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganassi M, et al. A surveillance function of the HSPB8-BAG3-HSP70 chaperone complex ensures stress granule integrity and dynamism. Mol Cell. 2016;63:796–810. doi: 10.1016/j.molcel.2016.07.021. [DOI] [PubMed] [Google Scholar]
- Han TW, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Harrison AF, Shorter J. RNA-binding proteins with prion-like domains in health and disease. Biochem J. 2017;474:1417–1438. doi: 10.1042/BCJ20160499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU. Cellular homeostasis and aging. Annu Rev Biochem. 2016;85:1–4. doi: 10.1146/annurev-biochem-011116-110806. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A phase separation model for transcriptional control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, McKnight SL (2017) Cross-β polymerization of low complexity sequence domains. Cold Spring Harb Perspect Biol 9. 10.1101/cshperspect.a023598 [DOI] [PMC free article] [PubMed]
- Kegel KB, et al. Huntingtin associates with acidic phospholipids at the plasma membrane. J Biol Chem. 2005;280:36464–36473. doi: 10.1074/jbc.M503672200. [DOI] [PubMed] [Google Scholar]
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- Kotler SA, Walsh P, Brender JR, Ramamoorthy A. Differences between amyloid-β aggregation in solution and on the membrane: insights into elucidation of the mechanistic details of Alzheimer’s disease. Chem Soc Rev. 2014;43:6692–6700. doi: 10.1039/C3CS60431D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künze G, Barré P, Scheidt HA, Thomas L, Eliezer D, Huster D. Binding of the three-repeat domain of tau to phospholipid membranes induces an aggregated-like state of the protein. Biochim Biophys Acta. 2012;1818:2302–2313. doi: 10.1016/j.bbamem.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladokhin AS, White SH. Folding of amphipathic alpha-helices on membranes: energetics of helix formation by melittin. J Mol Biol. 1999;285:1363–1369. doi: 10.1006/jmbi.1998.2346. [DOI] [PubMed] [Google Scholar]
- Lauterwein J, Brown LR, Wüthrich K. High-resolution. 1H-NMR studies of monomeric melittin in aqueous solution. Biochim Biophys Acta. 1980;622:219–230. doi: 10.1016/0005-2795(80)90033-1. [DOI] [PubMed] [Google Scholar]
- Li M, Liu J, Ran X, Fang M, Shi J, Qin H, Goh JM, Song J. Resurrecting abandoned proteins with pure water: CD and NMR studies of protein fragments solubilized in salt-free water. Biophys J. 2006;91:4201–4209. doi: 10.1529/biophysj.106.093187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Yang Y, Zhan J, Dai L, Zhou Y. Energy functions in de novo protein design: current challenges and future prospects. Annu Rev Biophys. 2013;42:315–335. doi: 10.1146/annurev-biophys-083012-130315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L, Song J. SALS-linked WT-SOD1 adopts a highly similar helical conformation as FALS-causing L126Z-SOD1 in a membrane environment. Biochim Biophys Acta. 2016;1858:2223–2230. doi: 10.1016/j.bbamem.2016.06.027. [DOI] [PubMed] [Google Scholar]
- Lim L, Lee X, Song J. Mechanism for transforming cytosolic SOD1 into integral membrane proteins of organelles by ALS causing mutations. Biochim Biophys Acta. 2015;1848:1–7. doi: 10.1016/j.bbamem.2014.10.002. [DOI] [PubMed] [Google Scholar]
- Lim L, Wei Y, Lu Y, Song J. ALS-causing mutations significantly perturb the self-assembly and interaction with nucleic acid of the intrinsically disordered Prion-like domain of TDP-43. PLoS Biol. 2016;14:e1002338. doi: 10.1371/journal.pbio.1002338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L, Lu Y, Song J (2016b) Unlocked capacity of proteins to attack membranes characteristic of aggregation: the evil for diseases and aging from Pandora’s box. bioRxiv 071274. 10.1101/071274
- Lim L, Kang J, Song J. ALS-causing rofiling-1-mutant forms a non-native helical structure in membrane environments. Biochim Biophys Acta Biomembr. 2017;1859:2161. doi: 10.1016/j.bbamem.2017.08.013. [DOI] [PubMed] [Google Scholar]
- Lin Y, et al. Toxic PR poly-Dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell. 2016;167:789–802. doi: 10.1016/j.cell.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc Natl Acad Sci U S A. 2008;105:3076–3081. doi: 10.1073/pnas.0708931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, et al. Toxicity of familial ALS-linked SOD1 mutants from selective recruitment to spinal mitochondria. Neuron. 2004;43:5–17. doi: 10.1016/j.neuron.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Lu Y, Lim L, Tan Y, Wang L, Song J (2016) Mechanisms of self-assembly and fibrillization of the prion-like domains. bioRxiv 065631. 10.1101/065631
- Lu Y, Lim L, Song J. RRM domain of ALS/FTD-causing FUS characteristic of irreversible unfolding spontaneously self-assembles into amyloid fibrils. Sci Rep. 2017;7:1043. doi: 10.1038/s41598-017-01281-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Lim L, Song J. (2017b) RRM domain of ALS/FTD-causing FUS interacts with membrane: an anchor of membraneless organelles to membranes? bioRxiv 122671. 10.1101/122671
- Mateju D, et al. An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 2017;36:1669–1687. doi: 10.15252/embj.201695957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLendon PM, Robbins J. Proteotoxicity and cardiac dysfunction. Circ Res. 2015;116:1863–1882. doi: 10.1161/CIRCRESAHA.116.305372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci U S A. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor DL, Jr, Kim PS. Context-dependent secondary structure formation of a designed protein sequence. Nature. 1996;380:730–734. doi: 10.1038/380730a0. [DOI] [PubMed] [Google Scholar]
- Monahan Z, et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017;36:2951–2967. doi: 10.15252/embj.201696394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, et al. ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible Hydrogels into irreversible Hydrogels impairs RNP granule function. Neuron. 2015;88:678–690. doi: 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray DT, Kato M, Lin Y, Thurber KR, Hung I, McKnight SL, Tycko R. (2017) Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains. Cell 171(3):615–627.e16. 10.1016/j.cell.2017.08.048 [DOI] [PMC free article] [PubMed]
- Nachreiner T, Esser M, Tenten V, Troost D, Weis J, Krüttgen A. Novel splice variants of the amyotrophic lateral sclerosisassociated gene VAPB expressed in human tissues. Biochem Biophys Res Commun. 2010;394:703–708. doi: 10.1016/j.bbrc.2010.03.055. [DOI] [PubMed] [Google Scholar]
- Nelson R, et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Perdigão N, et al. Unexpected features of the dark proteome. Proc Natl Acad Sci U S A. 2015;112:15898–15903. doi: 10.1073/pnas.1508380112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutz MF, Johnson T, Suzuki M, Finch JT. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc Natl Acad Sci U S A. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinotsi D, et al. Proton transfer and structure-specific fluorescence in hydrogen bond-rich protein structures. J Am Chem Soc. 2016;138:3046–3057. doi: 10.1021/jacs.5b11012. [DOI] [PubMed] [Google Scholar]
- Pinti DL (2005) The origin and evolution of the oceans. In Gargaud M et al. (eds) Lectures in Astrobiology. Springer, Berlin, pp 83–111. http://www.springer.com/gb/book/9783540262299
- Plaxco KW, Riddle DS, Grantcharova V, Baker D. Simplified proteins: minimalist solutions to the ‘protein folding problem’. Curr Opin Struct Biol. 1998;8:80–85. doi: 10.1016/S0959-440X(98)80013-4. [DOI] [PubMed] [Google Scholar]
- Qin H, Wang W, Song J. ALS-causing P56S mutation and splicing variation on the hVAPB MSP domain transform its β-sandwich fold into lipid-interacting helical conformations. Biochem Biophys Res Commun. 2013;431:398–403. doi: 10.1016/j.bbrc.2013.01.039. [DOI] [PubMed] [Google Scholar]
- Qin H, Lim L, Wei Y, Gupta G, Song J. Resolving the paradox for protein aggregation diseases: NMR structure and dynamics of the membrane-embedded P56S-MSP causing ALS imply a common mechanism for aggregation-prone proteins to attack membranes. F1000Res. 2013;2:221. doi: 10.12688/f1000research.2-221.v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Lim LZ, Wei Y, Song J. TDP-43 N terminus encodes a novel ubiquitin-like fold and its unfolded form in equilibrium that can be shifted by binding to ssDNA. Proc Natl Acad Sci U S A. 2014;111:18619. doi: 10.1073/pnas.1413994112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayman JB, Kandel ER (2017) Functional Prions in the Brain. Cold Spring Harb Perspect Biol 9(1). 10.1101/cshperspect.a023671 [DOI] [PMC free article] [PubMed]
- Riddle DS, et al. Functional rapidly folding proteins from simplified amino acid sequences. Nat Struct Biol. 1997;4:805–809. doi: 10.1038/nsb1097-805. [DOI] [PubMed] [Google Scholar]
- Rocklin GJ, et al. Global analysis of protein folding using massively parallel design, synthesis, and testing. Science. 2017;357:168–175. doi: 10.1126/science.aan0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen LM, Ellermann M, Diederich F. Aromatic rings in chemical and biological recognition: energetics and structures. Angew Chem Int Ed Engl. 2011;50:4808–4842. doi: 10.1002/anie.201007560. [DOI] [PubMed] [Google Scholar]
- Sangwan S, et al. Atomic structure of a toxic, oligomeric segment of SOD1 linked to amyotrophic lateral sclerosis (ALS) Proc Natl Acad Sci U S A. 2017;114:8770. doi: 10.1073/pnas.1705091114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Antón LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- Shahmoradian SH et al (2017) Lewy pathology in Parkinson’s disease consists of a crowded organellar membranous medley. bioRxiv. 10.1101/137976
- Shi J, Lua S, Tong JS, Song J. Elimination of the native structure and solubility of the hVAPB MSP domain by the Pro56Ser mutation that causes amyotrophic lateral sclerosis. Biochemistry. 2010;49:3887–3397. doi: 10.1021/bi902057a. [DOI] [PubMed] [Google Scholar]
- Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017;357:6357. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- Shorter J. Liquidizing FUS via prion-like domain phosphorylation. EMBO J. 2017;36:2925–2927. doi: 10.15252/embj.201798078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- Shukla A, Mukherjee S, Sharma S, Agrawal V, Radha Kishan KV, Guptasarma P. A novel UV laser induced visible blue radiation from protein crystals and aggregates: scattering artifacts or fluorescence transitions of peptide electrons delocalized through hydrogen bonding? Arch Biochem Biophys. 2004;428:144–153. doi: 10.1016/j.abb.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Song J. Insight into “insoluble proteins” with pure water. FEBS Lett. 2009;583:953. doi: 10.1016/j.febslet.2009.02.022. [DOI] [PubMed] [Google Scholar]
- Song J. Why do proteins aggregate? “Intrinsically insoluble proteins” and “dark mediators” revealed by studies on “insoluble proteins” solubilized in pure water. F1000 Res. 2013;2:94. doi: 10.12688/f1000research.2-94.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. Transforming cytosolic proteins into “insoluble” and membrane-toxic forms triggering diseases/aging by genetic, pathological or environmental factors. Protein Pept Lett. 2017;24:294–306. doi: 10.2174/0929866524666170209154001. [DOI] [PubMed] [Google Scholar]
- Song J, Jamin N, Gilquin B, Vita C, Ménez A. A gradual disruption of tight side-chain packing: 2D 1H-NMR characterization of acidinduced unfolding of CHABII. Nat Struct Biol. 1999;6:129–134. doi: 10.1038/5815. [DOI] [PubMed] [Google Scholar]
- Spalding KL, Bhardwaj RD, Buchholz BA, Druid H, Frisén J. Retrospective birth dating of cells in humans. Cell. 2005;122:133–143. doi: 10.1016/j.cell.2005.04.028. [DOI] [PubMed] [Google Scholar]
- Sudhakaran IP, Ramaswami M. Long-term memory consolidation: the role of RNA-binding proteins with prion-like domains. RNA Biol. 2017;14:568–586. doi: 10.1080/15476286.2016.1244588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, et al. Translational profiling identifies a cascade of damage initiated in motor neurons and spreading to glia in mutant SOD1-mediated ALS. Proc Natl Acad Sci U S A. 2015;112:E6993–E7002. doi: 10.1073/pnas.1520639112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuling E, et al. Motor neuron disease-associated mutant vesicle-associated membrane protein- associated protein (VAP) B recruits wild-type VAPs into endoplasmic reticulum-derived tubular aggregates. J Neurosci. 2007;27:9801–9815. doi: 10.1523/JNEUROSCI.2661-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipping KW, et al. pH-induced molecular shedding drives the formation of amyloid fibril-derived oligomers. Proc Natl Acad Sci U S A. 2015;112:5691–5696. doi: 10.1073/pnas.1423174112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer TS, et al. Structure and dynamics of micelle-bound human alpha-synuclein. J Biol Chem. 2005;280:9595–9603. doi: 10.1074/jbc.M411805200. [DOI] [PubMed] [Google Scholar]
- van der Lee R, et al. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114:6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Velde C, Miller TM, Cashman NR, Cleveland DW. Selective association of misfolded ALS-linked mutant SOD1 with the cytoplasmic face of mitochondria. Proc Natl Acad Sci U S A. 2008;105:4022–4027. doi: 10.1073/pnas.0712209105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Song J. Molecular mechanism underlying the thermal stability and pH-induced unfolding of CHABII. J Mol Biol. 2005;348:205–218. doi: 10.1016/j.jmb.2005.02.028. [DOI] [PubMed] [Google Scholar]
- White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- Willis MS, Patterson C. Proteotoxicity and cardiac dysfunction--Alzheimer’s disease of the heart? N Engl J Med. 2013;368:455–464. doi: 10.1056/NEJMra1106180. [DOI] [PubMed] [Google Scholar]
- Wootton JC. Sequences with ‘unusual’ amino acid compositions. Curr Opin Struct Biol. 1994;4:413–421. doi: 10.1016/S0959-440X(94)90111-2. [DOI] [Google Scholar]
- Yang C, et al. Mutant PFN1 causes ALS phenotypes and progressive motor neuron degeneration in mice by a gain of toxicity. Proc Natl Acad Sci U S A. 2016;113:E6209–E6218. doi: 10.1073/pnas.1605964113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu JS, Deng HX, Lo TP, Mitsumoto H, Ahmed MS, Hung WY, Cai ZJ, Tainer JA, Siddique T. Exon 5 encoded domain is not required for the toxic function of mutant SOD1 but essential for the dismutase activity: identification and characterization of two new SOD1 mutations associated with familial amyotrophic lateral sclerosis. Neurogenetics. 1997;1:65–71. doi: 10.1007/s100480050010. [DOI] [PubMed] [Google Scholar]