Abstract
In multiscale structural biology, synthetic approaches are important to demonstrate biophysical principles and mechanisms underlying the structure, function, and action of bio-nanomachines. A central goal of “synthetic structural biology” is the design and construction of artificial proteins and protein complexes as desired. In this paper, I review recent remarkable progress of an array of approaches for hierarchical design of artificial proteins and complexes that signpost the path forward toward synthetic structural biology as an emerging interdisciplinary field. Topics covered include combinatorial and protein-engineering approaches for directed evolution of artificial binding proteins and membrane proteins, binary code strategy for structural and functional de novo proteins, protein nanobuilding block strategy for constructing nano-architectures, protein–metal–organic frameworks for 3D protein complex crystals, and rational and computational approaches for design/creation of artificial proteins and complexes, novel protein folds, ideal/optimized protein structures, novel binding proteins for targeted therapeutics, and self-assembling nanomaterials. Protein designers and engineers look toward a bright future in synthetic structural biology for the next generation of biophysics and biotechnology.
Keywords: Artificial protein and complex, Combinatorial library, Computational design, Directed evolution, Hierarchical design, Protein engineering
Introduction
Living organisms are maintained by self-assembling biomolecules, such as proteins, nucleic acids, sugars, and lipids. Chemical reconstitution of living matter is an ultimate goal of synthetic and systems biology. Design of structural and functional artificial proteins and complexes is a key challenge in “synthetic structural biology.” Synthetic biology is a field of research concerned with the design and construction of new biological parts, devices, and systems, and the redesign of existing, natural biological systems for useful purposes. Synthetic structural biology is a new interdisciplinary field of synthetic biology and structural biology. In multiscale structural biology, synthetic approaches are important to demonstrate biophysical principles and mechanisms underlying the structure, function, and action of bio-nanomachines.
In recent years, DNA origami has been developed as a synthetic approach to the design and construction of various supramolecular nanostructures. DNA base complementarity can be exploited in the rational design of artificial nanostructures with versatile two-dimensional (2D) and three-dimensional (3D) shapes, such as polyhedra (Ke 2014). However, nucleic acids generally comprise the bases A, T, G, and C, and the ensuing limitations on numbers of combinations and chemical features may confine the potential to produce molecules with advanced functions.
In contrast with DNA, proteins comprise 20 types of amino acids, allowing greater diversity of chemical properties. Accordingly, the enormous numbers of possible sequence combinations expand the probabilities to create diverse and advanced functions. Natural proteins are the most versatile biomacromolecules, and perform complex and functional tasks in all organisms, because of the formation of intricate and refined tertiary and quaternary structures with versatile chemical properties and functionalities. Protein functions are essentially determined by their 3D structures. Protein structures are constructed on four hierarchical levels. Specifically, primary structure refers to amino acid sequences, and secondary structures are local regular forms of α-helices or β-strands with hydrogen bonds. In globular forms of proteins, elements of α-helices and/or β-sheets and loops are folded into tertiary structures, and self-assembly of folded chains from multiple polypeptides produces quaternary structures. These complex and refined 3D structures produce the versatile functionalities of proteins.
A central goal of protein engineering and synthetic structural biology is to design and create novel structural and functional proteins and protein complexes as desired. The design of de novo proteins, which are not derived from natural protein sequences, has been in essence an exploration of untracked areas of amino-acid sequence space. This exploration can be challenging, both because sequence space is vast, and because the contribution of many cooperative and long-range interactions causes a significant gap between the primary structures and their resulting tertiary and quaternary structures. Research into de novo protein designs has progressed toward the construction of novel proteins, and has been achieved largely from combinatorial approaches (Keefe and Szostak 2001; Urvoas et al. 2012), rational and computational design approaches (Dahiyat and Mayo 1997; Kuhlman et al. 2003; Koga et al. 2012; Huang et al. 2016), and semirational approaches that include elements of both (Kamtekar et al. 1993; Hecht et al. 2004; Urvoas et al. 2012). Recent advances in science and technology, such as significant increases in the number of 3D protein structures deposited in the protein data bank (PDB), rapid advances in computer hardware and software, and the reduced costs of DNA synthesis for artificial genes, lead to further developments of design and construction of artificial proteins and complexes.
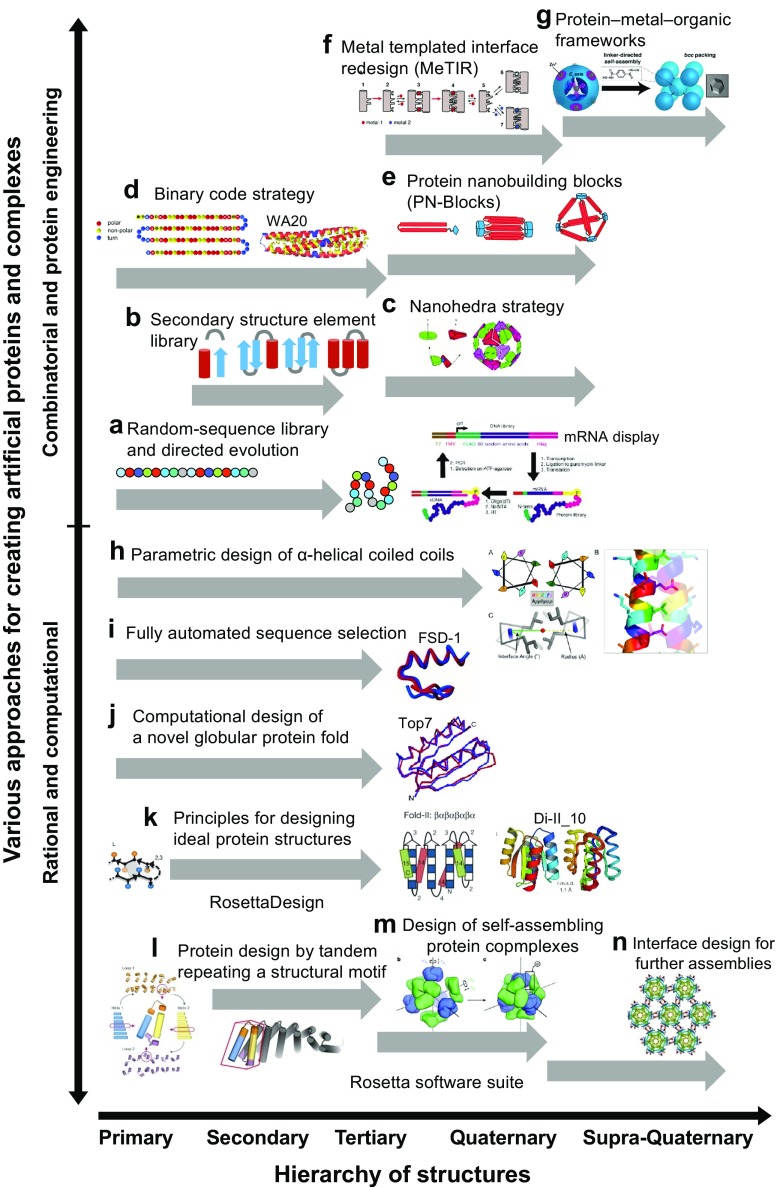
In this review, I describe recent progress in various approaches for designing and constructing artificial proteins and protein complexes. From the viewpoint of building blocks, I focus on the hierarchical design of artificial proteins and protein complexes from primary to quaternary structures using a range of combinatorial, protein engineering, rational and computational approaches for generating protein structures and functions. As shown in Fig. 1, there are two axes in the design of artificial proteins and complexes: the horizontal axis is the hierarchy of protein structures from primary to quaternary and supra-quaternary structures; and the vertical axis is how to design artificial proteins and complexes: a variety of combinatorial and protein-engineering (Fig. 1a–g) and rational and computational (Fig. 1h–n) approaches.
Fig. 1.
Hierarchical design approaches for creating artificial proteins and protein complexes toward synthetic structural biology. The horizontal axis is the hierarchy of protein structures from primary to quaternary and supra-quaternary structures. The vertical axis is how to design and create artificial proteins and complexes: a variety of combinatorial and protein-engineering (a–g) and rational and computational (h–n) approaches. a Random-sequence library and directed evolution: e.g., ATP-binding proteins were selected from a random-sequence library using mRNA display (Keefe and Szostak 2001). Reprinted with permission from Keefe and Szostak (2001); copyright © 2001, Nature Publishing Group (NPG). b Secondary structure element library: e.g., a library of secondary structural elements (α-helices, β-strands and loops) was designed and constructed for generating soluble and folded novel protein sequences containing secondary structures (Graziano et al. 2008). c Nanohedra strategy: a general strategy for designing fusion proteins that assemble into symmetric nanostructures (Padilla et al. 2001). Reprinted with permission from Padilla et al. (2001); copyright © 2001, the National Academy of Sciences, U.S.A. (NAS). d Binary code strategy: a semirational approach for creating structural and functional de novo proteins from focused libraries designed by the binary patterning of polar and nonpolar residues (Hecht et al. 2004; Smith and Hecht 2011). Reprinted with permission from Arai et al. (2012); copyright © 2012, American Chemical Society (ACS). e Protein nanobuilding blocks (PN-Blocks): an approach for constructing self-assembling polyhedral nanoarchitectures using an intermolecularly folded dimeric de novo protein (Kobayashi et al. 2015). Reprinted with permission from Kobayashi et al. (2015); copyright © 2015, ACS. f Metal templated interface redesign (MeTIR): an approach for engineering protein interfaces to form stable protein assemblies through an initial metal coordination event (Salgado et al. 2010a). Reprinted with permission from Salgado et al. (2010a). g Protein–metal–organic frameworks: a rational chemical design approach for constructing 3D protein complex crystals of ternary protein–metal–organic frameworks (Sontz et al. 2015; Bailey et al. 2017). Reprinted with permission from Sontz et al. (2015); copyright © 2015, ACS. h Parametric design of α-helical coiled coils: the parametric descriptions are implemented in an easy-to-use web application, e.g., CCBuilder, for modeling and optimizing α-helical coiled coils (Wood and Woolfson 2017). Reprinted from Wood and Woolfson (2017) under the terms of the Creative Commons Attribution License; copyright © 2017, the authors Protein Science. i Fully automated sequence selection: a computational design algorithm based on physical chemical potential functions and stereochemical constraints was used to screen a combinatorial library of possible amino acid sequences for compatibility with the design target (Dahiyat and Mayo 1997). Full sequence design 1 (FSD-1) is the first de novo protein created by fully automated design and experimental validation (PDB ID: 1FSD). Reprinted with permission from Dahiyat and Mayo (1997); copyright © 1997, American Association for the Advancement of Science (AAAS). j Computational design of a novel globular protein fold: a general computational strategy that iterates between sequence design and structure prediction to design an α/β protein called Top7 with a novel sequence and topology with atomic-level accuracy (PDB ID: 1QYS) (Kuhlman et al. 2003). Reprinted with permission from Kuhlman et al. (2003); copyright © 2003, AAAS. k Principles for designing ideal protein structures: an approach to designing ideal protein structures stabilized by completely consistent local and non-local interactions, based on a set of rules relating secondary structure patterns to protein tertiary motifs (Koga et al. 2012). Di-II_10 (PDB ID: 2LV8) is one of ideally structural de novo proteins. Reprinted with permission from Koga et al. (2012); copyright © 2012, NPG. l Protein design by tandem repeating structural motifs: Computational approaches for designing de novo repeat proteins generated by tandem repeating simple structural motifs (Brunette et al. 2015; Doyle et al. 2015). Reprinted with permission from Brunette et al. (2015); copyright © 2015, NPG. m Computational design of self-assembling protein copmplexes: a general approach for designing computationally self-assembling protein nanomaterials with atomic-level accuracy (King et al. 2012, 2014). Reprinted with permission from King et al. (2014); copyright © 2014, NPG. n Interface design for further assemblies: a computational approach for designing supra-quaternary structures of 2D protein arrays mediated by noncovalent protein–protein interfaces (Gonen et al. 2015). Reprinted with permission from Gonen et al. (2015); copyright © 2015, NPG. The approaches of (j–n) are implemented in the Rosetta software suite including RosettaDesign (Das and Baker 2008; Kaufmann et al. 2010; Leaver-Fay et al. 2011; Richter et al. 2011; Baker 2014; Bender et al. 2016; Huang et al. 2016)
Combinatorial and protein-engineering approaches for artificial protein and complex design
Hierarchical design of tertiary structures of artificial proteins
In general, protein structures are characterized by four hierarchical levels from primary to quaternary structures, and therefore adoption of hierarchical approaches is essential for protein design (Bryson et al. 1995). In this section, I briefly review several combinatorial and protein-engineering approaches for designing and constructing artificial proteins from the viewpoint of the hierarchical design of building blocks. Further details and specific topics are described in the following sections.
Using amino acid residues as building blocks in primary structures, huge combinatorial libraries of totally random polypeptide sequences are the primitive starting point of protein evolution. Due to the low frequency of soluble and folded proteins in random sequences, fully randomized sequence libraries have to be searched with powerful selection methods (Urvoas et al. 2012). One remarkable result was the isolation of a de novo ATP binding protein from fully randomized sequences by mRNA display (Fig. 1a) (Keefe and Szostak 2001). The frequency of folded and functional sequences in completely random sequences is very low when placed in comparison to our current experimental screening power. Thus, several strategies have been proposed to focus the exploration on limited sequence spaces expected to be rich in folded structures, using some designed patterns of amino acid sequences as building blocks (Urvoas et al. 2012). One of the most fruitful strategies is known as the binary code strategy which was developed to produce primary structure libraries for tertiary structures of de novo proteins using secondary structure units with binary patterns of polar and nonpolar residues (Fig. 1d), and various structural and functional de novo proteins with α-helices and/or β-sheets have been successfully created (Kamtekar et al. 1993; Hecht et al. 2004; Smith and Hecht 2011).
Architectures of protein domains have evolved by the combinatorial assembly and exchange of pre-existing polypeptide modular segments, secondary structure elements and supersecondary structure motifs, derived from exon shuffling, nonhomologous recombination or alternative splicing (Fig. 1b) (Urvoas et al. 2012). This process can be experimentally simulated by selecting folded proteins from combinatorial libraries of shuffled secondary structure elements used as building blocks. In one example, a previously truncated protein sequence can be structurally rescued by fusion with random genomics sequences (Riechmann and Winter 2000). The recombined chimeric polypeptides were displayed on filamentous bacteriophage and folded polypeptides were selected for their proteolysis resistance. A truncated β-barrel domain from cold shock protein CspA was structurally rescued by a fragment of an Escherichia coli protein (de Bono et al. 2005). In another example, a library of secondary structural elements (α-helices, β-strands and loops) was designed and constructed by extraction from existing structures of bacterial proteins. A significant population of soluble and partially folded proteins with molten-globule-like behaviors suggested that the semirandom assembly of pre-existing secondary structural elements as building blocks could accelerate the emergence of novel proteins (Graziano et al. 2008).
Repeat proteins are defined by the repetition of a varying number of small structural units (20–50 amino acids). The modular and highly repetitive structures suggest that new artificial repeat protein repertoires can be constructed by concatenation of repetitive modules as idealized building blocks (Parmeggiani and Huang 2017). Various libraries of artificially designed repeat proteins have been generated to select protein binders, such as ankyrin repeat proteins (DARPins) (Jost and Pluckthun 2014). In addition, a “protein origami” strategy to design self-assembling polypeptide nanostructured polyhedra was proposed based on modularization using orthogonal dimerizing coiled-coil segments as building blocks. Polyhedra that self-assembles from a single polypeptide chain comprising concatenated coiled coil-forming segments connected with flexible peptide hinges were designed using graph theory and the formation of polyhedral nanostructures was demonstrated experimentally (Gradisar et al. 2013; Ljubetic et al. 2017).
Combinatorial and directed evolution approaches for creating functional artificial proteins
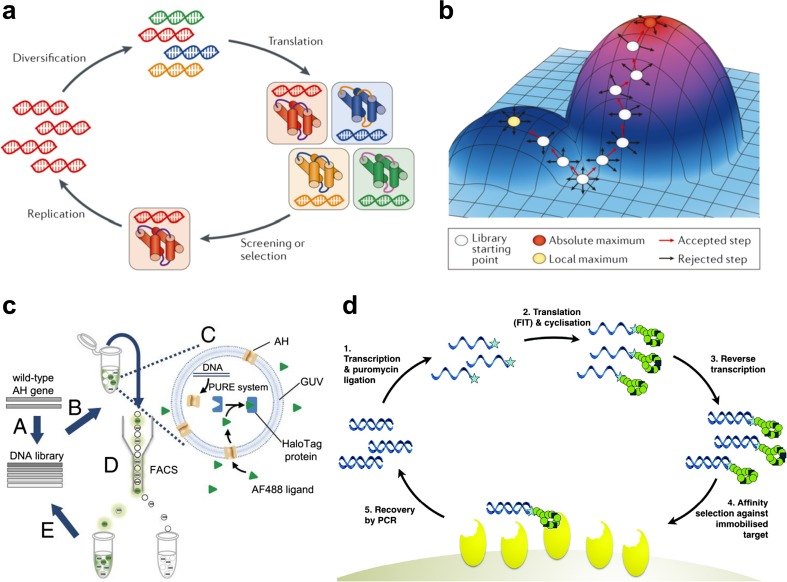
In general, combinatorial and directed evolution approaches do not require atomic-level structural information. Although a target protein structure is not necessary, a target function for proteins should be set for combinatorial and directed evolution approaches. However, in most combinatorial approaches, the experimental design should be based on the working hypothesis that functional proteins have some proper structures. It is important to select the screening method for good expression, solubility and assessment of molecular functions to properly take into account the role of folded 3D structures. Due to the low frequency of soluble and folded proteins in protein sequence space, protein libraries have to be searched with powerful selection methods for stably folded proteins (Matsuura et al. 2004; Urvoas et al. 2012) and binding proteins (Hoogenboom 2005). A variety of methods for the directed evolution of proteins are summarized in a recent review (Packer and Liu 2015). The cycle of directed evolution process in the laboratory mimics that of biological evolution. A diverse library of genes is translated into a corresponding library of gene products and screened or selected for functional variants in a manner that maintains the correspondence between genotype (genes) and phenotype (gene products and their functions). These functional genes are replicated and serve as starting points for subsequent rounds of diversification and high throughput screening (HTS) (Fig. 2a). Although the mutational space is multidimensional, it is conceptually helpful to visualize directed evolution as a series of steps within a 3D fitness landscape (Fig. 2b).
Fig. 2.
Schematics of key steps in cycles of combinatorial and directed evolution approaches. a The process of directed evolution in the laboratory mimics that of biological evolution (Packer and Liu 2015). A diverse library of genes is translated into a corresponding library of gene products and screened or selected for functional variants in a manner that maintains the correspondence between genotype (genes) and phenotype (gene products and their functions). These functional genes are replicated and serve as starting points for subsequent rounds of diversification and screening or selection. b Although the mutational space is multidimensional, it is conceptually helpful to visualize directed evolution as a series of steps within a three-dimensional fitness landscape (Packer and Liu 2015). Library generation samples the proximal surface of the landscape, and screening or selection identifies the genetic means to ‘climb’ toward fitness peaks. Directed evolution can arrive at absolute maximum activity levels but can also become trapped at local fitness maxima in which library diversification is insufficient to cross ‘fitness valleys’ and access neighboring fitness peaks. Reprinted with permission from Packer and Liu (2015); copyright © 2015, NPG. c Schematics of the in vitro evolution of α-hemolysin (AH) using liposome display. A Gene encoding wild-type AH was subjected to mutagenesis to generate a randomly mutagenized gene library. B Gene was encapsulated in giant unilamellar vesicles (GUVs) at the single-molecule level (i.e., one DNA molecule per GUV), together with the in vitro translation (IVT) system and HaloTag protein. C After GUV preparation, AH was synthesized and the fluorescence ligand (AF488) was added to the exterior of the GUVs. The ligand was cell-impermeable due to its negative charge and was expected to penetrate the membrane only through the AH pores. D GUVs with high-fluorescence signals were collected via FACS. E Finally, the recovered gene was amplified and included in the next round of selection. Reprinted with permission from Fujii et al. (2013). d RaPID selection cycle of macrocyclic peptides. The RaPID selection process starts with a semi-randomized DNA library (shown left in blue). Transcription of this library and conjugation to puromycin (represented by a star) leads to the formation of an mRNA library with linked 3′-puromycin (1). Translation of this library in a FIT reaction leads in turn to the synthesis of a library of macrocyclic peptides (green) incorporating nonstandard amino acids, each of which is covalently linked through the puromycin moiety to its cognate mRNA (2). Reverse transcription generates the cDNA of each mRNA (3), and the pooled mixture can then be panned against an immobilized protein target of interest (yellow) to identify peptides with high affinity (4). The cognate DNA molecules can then be recovered by PCR to generate an enriched library (5), and the process can be repeated iteratively until only peptides with high affinity to the target are represented in the library. Reprinted with permission from Passioura and Suga (2017); copyright © 2017, the Royal Society of Chemistry
In a typical target-binding selection, protein library members with desired binding activity and their encoding DNA sequences are captured using an immobilized target, whereas non-binding library members are washed away. In cell surface display or phage display methods, a cell or bacteriophage serves as a compartment to link genes and gene products and protein library members are expressed on the surface of the cell or the coat of the bacteriophage through fusion with endogenous cell surface proteins (Boder and Wittrup 1997; Bessette et al. 2004) or phage coat proteins (McCafferty et al. 1990). Ribosome display was also developed (Hanes and Pluckthun 1997). In the absence of a stop codon and under carefully controlled conditions, ribosomes remain stably bound to both the mRNA and the growing polypeptide, thereby coupling proteins with their encoding genes. Similarly, mRNA display covalently links a translated protein to its encoding mRNA through a puromycin analogue (Nemoto et al. 1997; Roberts and Szostak 1997; Wilson et al. 2001). Moreover, a cDNA display method was developed and improved as a robust in vitro display technology that converts an unstable mRNA display to a stable mRNA/cDNA–protein fusion whose cDNA is covalently linked to its encoded protein using a well-designed puromycin linker (Yamaguchi et al. 2009; Ueno et al. 2012; Nemoto et al. 2014). Bead display-based in vitro HTS methods were also reported (Stapleton and Swartz 2010; Zhu et al. 2015), and a liposome display method was recently developed for directed evolution of membrane proteins using an in vitro transcription-translation system (Fig. 2c) (Fujii et al. 2013, 2014; Uyeda et al. 2016).
In addition, a remarkable integrated method, the RaPID (random non-standard peptide integrated discovery) system has been developed to discover nonstandard macrocyclic peptide binders for drug targets (Yamagishi et al. 2011; Hipolito and Suga 2012; Passioura and Suga 2017) using the flexible in vitro translation technology (FIT) for genetic code reprogramming (Goto et al. 2011) with nonstandard amino acids and flexizymes (Murakami et al. 2006; Morimoto et al. 2011), greatly expanding functional artificial polypeptide sequence space (Fig. 2d).
Furthermore, the advent of next-generation sequencing (NGS) has revolutionized protein science, and the development of complementary methods enabling NGS-driven protein engineering have followed. In general, these experiments address the functional consequences of thousands of protein variants in a massively parallel manner using genotype–phenotype linked high-throughput functional screens followed by DNA counting via deep sequencing (Fowler et al. 2010; Hietpas et al. 2011; Fowler and Fields 2014; Boucher et al. 2016; Wrenbeck et al. 2017). Deep sequencing refers to sequencing of a specific DNA region multiple times. The NGS approach allows researchers to economically observe entire populations of molecules before, during, and after HTS. Deep mutational scanning approaches have been applied to affinity mature antibodies (Fujino et al. 2012; Forsyth et al. 2013; Miyazaki et al. 2015). Remarkably, a dual specificity antibody with high affinities for two unrelated proteins was also developed (Koenig et al. 2015). Furthermore, a germline-targeting immunogen was engineered by exhaustive deep mutational scanning against dozens of germline-reverted and mature broadly neutralizing antibodies to HIV-1 (Jardine et al. 2016). Evolutionary molecular engineering coupled with big data informatics from NGS analysis opens the door to developing the exciting field of NEO-Biomolecules (Newly Evolved and Optimized Biomolecules created from unexplored and expanded sequence spaces).
Artificial binding proteins generated by protein engineering and combinatorial approaches
Specific binding proteins are essential for diagnostic and therapeutic applications, and traditionally these have been antibodies. In recent years, development of new artificial binding proteins has been a major target in the field of protein engineering. An increasing number of alternative scaffolds for artificial binding proteins have been developed using protein engineering and combinatorial approaches: monobodies derived from fibronectin type III, anticalins derived from lipocalins, affibodies derived from the immunoglobulin binding protein A, and DARPins based on the ankyrin fold can be regarded as the established formats of alternative scaffolds (Gilbreth and Koide 2012; Jost and Pluckthun 2014). Especially, the modular and highly repetitive structures of repeat proteins suggest that new artificial repeat protein repertoires can be generated by concatenation of idealized structural modules as building blocks (Boersma and Pluckthun 2011; Urvoas et al. 2012; Parmeggiani and Huang 2017). Repeated structural motifs stack together to generate large protein surfaces usually involved in protein–protein interactions. Each module has a limited set of randomized positions that are juxtaposed in the structure. The variability at specific positions within each module associated with a combinatorial assembly of consecutive modules enables exploration of a large sequence space to generate hypervariable macrosurfaces. New repeat proteins binding specifically to any chosen target protein can be selected from a library, using ribosome display, phage display or functional complementation (Urvoas et al. 2012). Various types of artificial repeat families for specific binders have been reported, such as designed ankyrin repeat proteins (DARPins) (Jost and Pluckthun 2014), leucine-rich repeat (LRR)-based repebodies (Lee et al. 2012), Armadillo repeats (Varadamsetty et al. 2012; Reichen et al. 2014; Reichen et al. 2016), HEAT repeats (Urvoas et al. 2010), and pentatricopeptide repeats (PPR) (Gully et al. 2015). Furthermore, an artificial peptide library based on a cyclized helix-loop-helix as a molecular scaffold was developed for high-throughput de novo screening of receptor agonists with an automated single-cell analysis and isolation system (Yoshimoto et al. 2014) and inhibitors of intracellular protein-protein interactions by epitope and arginine grafting (Fujiwara et al. 2016).
Semirational approaches for creating structural and functional de novo proteins
The architectures of protein structural and functional domains have evolved by combinatorial assembly and exchange of pre-existing polypeptide modular segments, secondary structure elements and supersecondary structure motifs, derived from exon shuffling, nonhomologous recombination or alternative splicing (Urvoas et al. 2012). These processes can be experimentally simulated by semirational approaches for creating artificial proteins. For example, a library of shuffled secondary structural elements (α-helices, β-strands and loops) was designed, constructed, and screened, revealing a significant population of soluble and partially folded novel proteins with molten-globule-like behaviors (Graziano et al. 2008).
Honda et al. reported a thought-provoking study of “protein minimalism” on the polypeptide termed chignolin, consisting of only 10 amino acid residues (GYDPETGTWG), designed on the basis of statistics derived from more than ten thousands of protein segments (Honda et al. 2004). The polypeptide chignolin folds into a unique structure in water and shows a cooperative thermal transition, both of which may be hallmarks of a protein. They also described a designed variant of chignolin, CLN025, consisting of 10 amino acid residues (YYDPETGTWY). Despite its small size, its essential characteristics, revealed by its crystal structure, solution structure, thermal stability, free energy surface, and folding pathway network, are consistent with the properties of natural proteins (Honda et al. 2008). The performance of these short polypeptides yields insights into the raison d’etre of an autonomous element involved in a natural protein. This is of interest for the pursuit of folding mechanisms and evolutionary processes of proteins (Honda et al. 2004). These results deepen our understanding of proteins and also impel us to reach consensus of the definition of “ideal proteins” and “minimal size of proteins” without recourse to inquiring as to whether the molecule actually occurs in nature (Honda et al. 2008). Furthermore, Watababe et al. synthesized an artificial protein on the basis of an evolutionary hypothesis, segment-based elongation starting from the autonomously foldable short polypeptide, chignolin. The structural guidance facilitates structural organization and gain-of-function of a generated 25-residue artificial protein with nanomolar affinity against the Fc region of immunoglobulin G (Watanabe et al. 2014). They also proposed a combinatorial approach, termed adaptive assembly, which provides a tailor-made protein scaffold for a given functional polypeptide. A combinatorial library was designed to create a tailor-made scaffold, which was generated from β-hairpins derived from chignolin and randomized amino acid sequences. The adaptive assembly achieved significant functional enhancement of a peptide with low affinity for the Fc region of human immunoglobulin G, generating a 54-residue artificial protein with a greatly enhanced affinity without relying on known protein structures (Watanabe and Honda 2015).
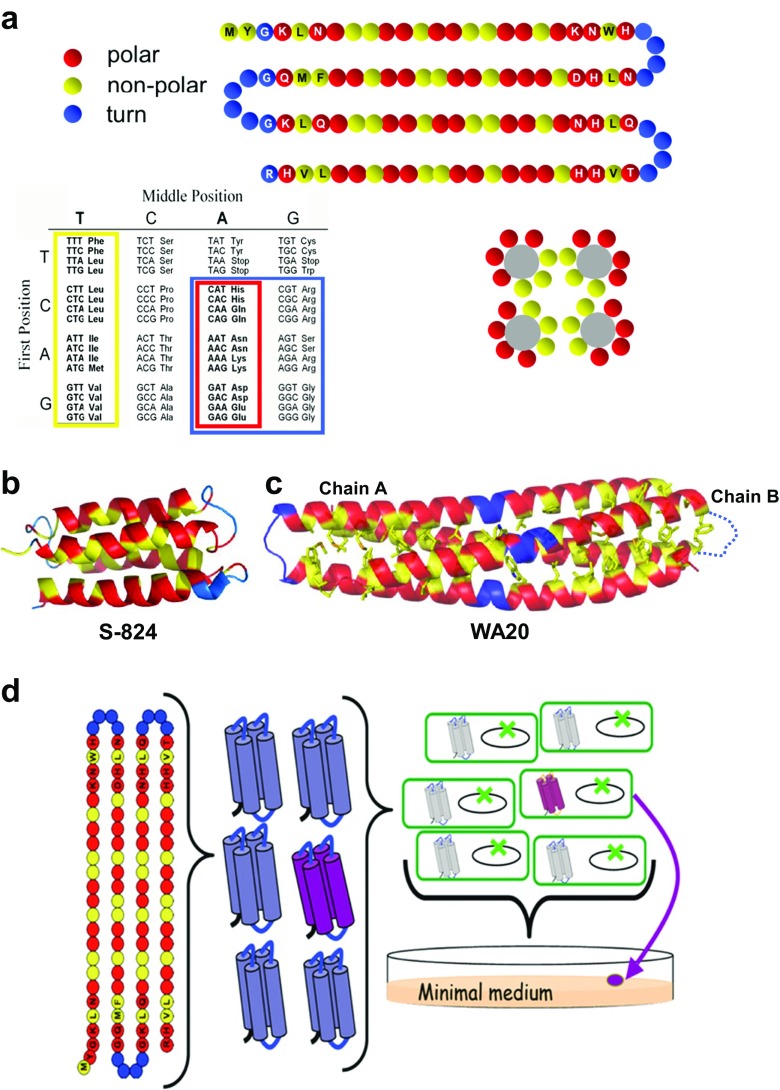
As a fruitful semirational approach, the binary code strategy has been developed to produce primary structure libraries for tertiary structures of de novo proteins using secondary structure units with binary patterns of polar and nonpolar residues (Kamtekar et al. 1993; Hecht et al. 2004) (Figs. 1d, 3a), and de novo proteins with α-helices and/or β-sheets have been successfully created (Kamtekar et al. 1993; West et al. 1999; Hecht et al. 2004; Jumawid et al. 2009). The monomeric structures of 4-helix bundle de novo proteins S-824 and S-836 from the second-generation binary-patterned combinatorial library were solved by NMR spectroscopy (Fig. 3b) (Wei et al. 2003; Go et al. 2008). Also, a stable and functional de novo protein WA20 was isolated from the third-generation library (Bradley et al. 2005; Patel et al. 2009) and the crystal structure of the de novo protein WA20 was solved (Arai et al. 2012). The WA20 structure was an unusual dimeric structure with an intermolecularly folded (domain-swapped) 4-helix bundle: each monomer (“nunchaku”-like structure) comprises two long α-helices that are intertwined with the helices of another monomer (Fig. 3c). From the third-generation combinatorial library of 4-helix bundle de novo proteins with the binary patterns, a variety of functional de novo proteins with cofactor binding, drug binding, and enzyme-like functions in vitro (Patel et al. 2009; Cherny et al. 2012; Patel and Hecht 2012) and life-sustaining functions in vivo (Fisher et al. 2011; Smith et al. 2015; Digianantonio and Hecht 2016; Hoegler and Hecht 2016; Digianantonio et al. 2017) have been successfully produced. Notably, the several binary-patterned de novo proteins which can rescue conditionally lethal gene deletions in E. coli auxotrophs (∆serB, ∆gltA, ∆ilvA, and ∆fes) were reported for the first time (Fisher et al. 2011) (Fig. 3d). In some cases, the de novo proteins can provide life-sustaining functions by altering gene regulation in cells: SynSerB3 rescued the serine auxotrophy in ∆serB cells by increasing expression of HisB, a promiscuous phosphatase (Digianantonio and Hecht 2016); and SynGltA rescued the deletion of citrate synthase in ∆gltA cells by increasing expression of PrpC, methylcitrate synthase with weak promiscuous activity (Digianantonio et al. 2017). Interestingly, further molecular evolution experiments demonstrated that a de novo protein Syn-IF, an original bifunctional generalist which can rescue two different auxotrophic strains, ∆ilvA and ∆fes, was evolved into two distinctive monofunctional specialist proteins with enhanced activity (Smith et al. 2015). In addition, a de novo protein rescued E. coli from toxic levels of copper (Hoegler and Hecht 2016). These advances in synthetic biology will open the possibility of creating “artificial proteomes” comprising sequences that did not arise in nature, but which nonetheless sustain the growth of living organisms.
Fig. 3.
Binary code strategy: a semirational approach for creating structural and functional de novo proteins. a Design template for binary-patterned 4-helix bundles. The binary code strategy was described in detail in a review paper (Hecht et al. 2004). The strategy was applied to the pattern design of four amphipathic α-helices, and these helices were connected by turns. Upon folding, the non-polar residues (yellow) are buried in the protein core and the polar residues (red) lie on the exterior. Library construction is facilitated by the organization of the genetic code: six polar residues (Lys, His, Glu, Gln, Asp, and Asn) are encoded by the degenerate codon VAN, and five nonpolar residues (Met, Leu, Ile, Val, and Phe) are encoded by the degenerate codon NTN. Combinatorial turn positions (blue) are encoded by the degenerate codon VRS, which encodes Gln, Glu, Asn, Asp, His, Lys, Arg, Ser, and Gly. (V = A, G, or C; R = A or G; N = A, G, C, or T). By defining each residue as either non-polar or polar—and varying the actual identity of each residue—a large and diverse collection of sequences encoding 4-helix bundles can be created. Reprinted with permission from Supporting Fig. 2 (Patel et al. 2009); copyright © 2009, the Protein Society. b Ribbon representation of the solution structure of a de novo protein, S-824, from the second-generation binary-patterned library of 4-helix bundle de novo proteins. The monomeric 4-helix bundle structure of S-824 was determined by NMR (PDB ID: 1P68) (Wei et al. 2003). c Ribbon representation of the crystal structure of a de novo protein, WA20, from the third-generation binary-patterned library of 4-helix bundle de novo proteins. Unexpectedly, the WA20 structure is not a monomeric 4-helix bundle, but a dimeric 4-helix bundle (PDB ID: 3VJF) (Arai et al. 2012). Each monomer comprises two long α-helices that intertwist with the helices of the other monomer. The two monomers together form a 3D domain-swapped 4-helix bundle dimer. Nonpolar residues in the hydrophobic core of WA20 are shown as stick models. The binary-patterned color coding is the same as (a). The disordered loop (turn) region in chain B is shown as a dashed curve line. Reprinted with permission from Arai et al. (2012); copyright © 2012, ACS. d Design of a collection of novel proteins and rescue of E. coli auxotrophs. Binary patterned library of polar (red) and nonpolar (yellow) residues designed to fold into 4-helix bundles as shown in (a). Circles with letters indicate fixed residues, and empty circles indicate combinatorially diverse positions. The experimental library (1.5 × 106 synthetic genes cloned on a high expression plasmid) was transformed into a strain of E. coli in which an endogenous gene had been deleted (green X). Colony formation on minimal media indicates a novel sequence (purple) enables cell growth under selective conditions. Reprinted from Fisher et al. (2011) under the terms of the Creative Commons Attribution License; copyright © 2011, Fisher et al.
Hierarchical design of quaternary and supra-quaternary structures of artificial protein complexes
In recent years, several approaches for hierarchical design of artificial protein complexes have been developed to construct protein quaternary structures using artificial and fusion proteins as nanoscale building blocks (Kobayashi and Arai 2017). These approaches include protein complexes and nanostructures constructed from self-assembling designed coiled-coil peptide modules, symmetrically self-assembling fusion proteins, 3D domain-swapped oligomers, metal-directed self-assembling proteins, and protein nanobuilding blocks (PN-Blocks).
Alpha-helical coiled coils are ubiquitous protein–protein interaction domains wherein folding and assembly of amphipathic α-helices direct a wide variety of protein assemblies (Woolfson et al. 2012; Lupas and Bassler 2017). Various self-assembling nanostructures have been constructed using designed coiled-coil peptide modules as building blocks: self-assembling cages from coiled-coil peptide modules (Fletcher et al. 2013); modular designed self-assembling peptide nanotubes using α-helical barrels with tunable internal cavities as building blocks (Burgess et al. 2015); and peptide nanotubes with control of the lateral association and the longitudinal assembly (Thomas et al. 2016); and self-assembling protein cages using de novo-designed short coiled-coil domains to mediate assembly by a flexible, symmetry-directed approach (Sciore et al. 2016). In addition, the design of protein–protein interactions through modification of inter-molecular helix–helix interface residues was reported (Yagi et al. 2016, 2017).
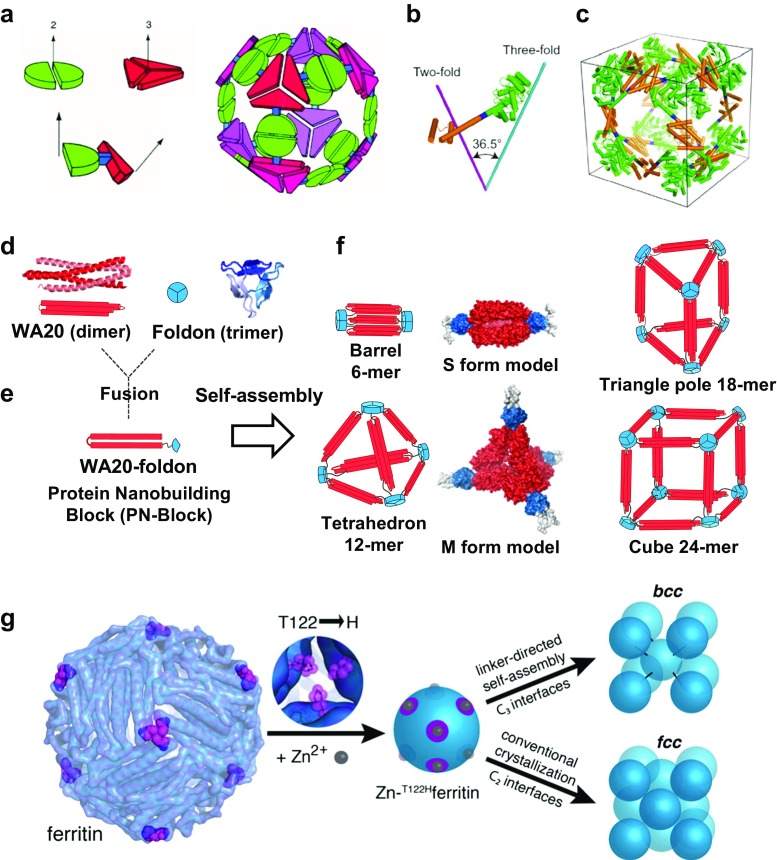
In natural protein complexes (Ahnert et al. 2015; Pieters et al. 2016), nanostructures self-assemble into symmetric polyhedral shapes, such as tetrahedrons, hexahedrons, octahedrons, dodecahedrons, and icosahedrons. Since symmetry provides a powerful tool for building large regular objects, a pioneering general strategy using symmetry to construct protein nanomaterials as “nanohedra” was described (Padilla et al. 2001; Yeates et al. 2016) (Figs. 1c; 4a–c). In this strategy, a protein that naturally forms a self-assembling oligomer is rigidly fused to another protein that forms another self-assembling oligomer, and the fusion protein self-assembles with other identical copies of itself into a designed nanohedral particle or material. The nanohedra strategy allows for the construction of a wide variety of potentially useful protein-based materials. Accordingly, the crystal structures of designed nanoscale protein cages were solved (Lai et al. 2012; Lai et al. 2014).
Fig. 4.
Symmetrical self-assemblies of nanoscale building blocks. a Nanohedra strategy: a general strategy for designing fusion proteins that assemble into symmetric nanostructures (Padilla et al. 2001). The green semicircle represents a natural dimeric protein (i.e., a protein that associates with one other copy of itself), whereas the red shape represents a trimeric protein. The symmetry axes of the natural oligomers are shown. The two natural proteins are combined by genetic methods into a single fusion protein. Each of the original natural proteins serves as an “oligomerization domain” in the designed fusion protein. A fusion protein is shown to illustrate that the oligomerization domains can be joined rigidly at a geometric angle. The fusion proteins self-assemble into a cubic cage of a nanohedral structure. Reprinted with permission from Padilla et al. (2001); copyright © 2001, NAS. b, c Models of the engineered fusion protein and its assembled cage structure (Lai et al. 2014). b The designed fusion protein, with trimeric KDPGal aldolase (green), the four-residue helical linker (blue) and the dimeric domain of FkpA protein (orange) shown with lines for the three-fold (cyan) and two-fold (magenta) symmetry axes. c Model of the intended 24-subunit cage with octahedral symmetry in a bounding box. Reprinted with permission from (Lai et al. 2014). Copyright © 2014, NPG. d–f Schematics of construction and assemblies of the WA20-foldon fusion protein as a protein nanobuilding block (PN-Block) (Kobayashi et al. 2015). d Ribbon representation and schematics of the intermolecularly folded dimeric WA20 (PDB ID: 3VJF) (Arai et al. 2012) shown in red, and trimeric foldon domain of T4 phage fibritin (PDB ID: 1RFO) (Guthe et al. 2004) shown in blue. e Construction of the WA20-foldon fusion protein as a PN-Block. f Schematics of designed polyhedral nanoarchitectures by expected self-assemblies of the WA20-foldon. In the stable self-assembling complexes, the WA20-foldon is expected to form highly symmetric oligomers in multiples of 6-mer because of the possible combinations of the WA20 dimer and foldon trimer. The rigid-body model structures of the S form (6-mer) and M form (12-mer) of the WA20-foldon are also shown. Reprinted with permission from Kobayashi et al. (2015); copyright © 2015, ACS. g Schematics of protein–metal–organic frameworks: metal/linker-directed self-assembly of ferritin into 3D crystals (Sontz et al. 2015). Surface-exposed binding sites for Zn2+ (gray spheres) are engineered at the C 3 pores (magenta) through the T122H mutation. In the presence of ditopic organic linkers, the resulting T122Hferritin variant is expected to form a bcc lattice. Reprinted with permission from Sontz et al. (2015); copyright © 2015, ACS
Three-dimensional (3D) domain swapping involves exchanging one structural domain of a protein monomer with that of the identical domain from a second monomer, resulting in an intertwined oligomer. As a pioneering work, artificial domain-swapped proteins that formed dimers and fibrous oligomers were described and the design principle of 3D domain-swapped protein oligomers for biomaterials was proposed (Ogihara et al., 2001). Moreover, cytochrome c polymerization following successive domain swapping was described (Hirota et al. 2010). Recently, rational design of heterodimeric proteins using domain swapping for myoglobin (Lin et al. 2015b) and a nanocage encapsulating a Zn-SO4 cluster in the internal cavity of a domain-swapped cytochrome cb 562 dimer (Miyamoto et al. 2015) were also reported.
Recently, Kobayashi et al. designed and constructed a polyhedral protein nanobuilding block (PN-Block), WA20-foldon (Kobayashi et al. 2015), by fusing the intermolecularly folded dimeric de novo WA20 protein (Arai et al. 2012) as a rectilinear framework/edge and a trimeric foldon domain of the T4 phage fibritin as a corner vertex/node (Figs. 1e, 4d–f). The WA20-foldon formed several distinctive self-assembling nanoarchitectures in multiples of 6-mer (6-, 12-, 18-, and 24-mer) because of the possible combinations of dimer and trimer. The basic concept of the PN-Block strategy follows the construction of self-assembling nanostructures from a combination with a few types of simple and fundamental PN-Blocks. Further design and construction of new types of PN-Blocks and reconstruction of PN-Blocks are important steps to expand the PN-Block strategy. Accordingly, Kobayashi et al. designed and constructed de novo extender protein nanobuilding blocks (ePN-Blocks) by tandemly fusing two de novo WA20 proteins with various linkers. These comprise a new series of PN-Blocks for reconstruction of self-assembling cyclized or extended chain-like nanostructures, and the ePN-Block complexes further self-assembled into supra-quaternary nanostructures (Kobayashi et al., unpublished). A PN-Block strategy is proposed as a systematic design and construction tool for generating novel self-assembling supramolecular nanostructures with tertiary, quaternary, and supra-quaternary structures of artificial protein complexes. This general and systematic strategy for hierarchical design with highly compatible modularity will expand the possibilities of PN-Blocks as artificial building-block molecules for a wide range of fields including nanotechnology, supramolecular chemistry, and synthetic structural biology.
Metal ions are frequently found in natural protein–protein interfaces, where they stabilize quaternary and supramolecular protein structures, mediate transient protein–protein interactions, and serve as catalytic centers. Moreover, coordination chemistry of metal ions has been increasingly used to engineer and control the assembly of functional supramolecular peptide and protein architectures. In particular, design strategies of metal-directed protein self-assembly (MDPSA) and metal-templated interface redesign (MeTIR) were described (Fig. 1f) (Salgado et al. 2010a, b; Bailey et al. 2016). Using these strategies, a building block protein was designed and engineered to form homodimers bearing interfacial Zn-coordination sites, which enabled Zn-mediated self-assembly into 1D helical nanotubes and 2D and 3D crystalline arrays (Brodin et al. 2012). Moreover, the metal-coordination strategies have been expanded through the development of a designed functional assembly of a metalloprotein with in vivo β-lactamase activity (Song and Tezcan 2014), designed helical protein nanotubes with variable diameters from a single building block (Brodin et al. 2015), metal organic frameworks with spherical protein nodes (protein–metal–organic frameworks, i.e. rational chemical design of 3D protein crystals) (Figs. 1g, 4g) (Sontz et al. 2015; Bailey et al. 2017), and an allosteric metalloprotein assembly with strained disulfide bonds (Churchfield et al. 2016). Also, self-assembly of coherently dynamic, auxetic, two-dimensional protein crystals was reported (Suzuki et al. 2016).
Rational and computational approaches for hierarchical design of artificial proteins and complexes
Computational design of artificial proteins
Rational and computational approaches for protein design have been developed in recent years with advances in computer science. Fast calculation provides great advantages in optimizing parameters toward an ideal structure of proteins. In the computational design of proteins, a general approach is built on the thermodynamic hypothesis that proteins fold into the lowest energy states that are accessible to their amino acid sequences, as originally proposed by Anfinsen (1973). Given a suitably accurate method for computing free energy of a protein chain, as well as methods for sampling possible protein structures and sequences, it would be allowed to design protein sequences that fold into new structures. The rational and computational de novo design of coiled coil and helical bundle structures has been achieved by development of the parametric design of α-helical coiled coils in the early years (Fig. 1h) (Crick 1953a, b; Harbury et al. 1998; Grigoryan and Degrado 2011; Wood et al. 2014; Wood and Woolfson 2017).
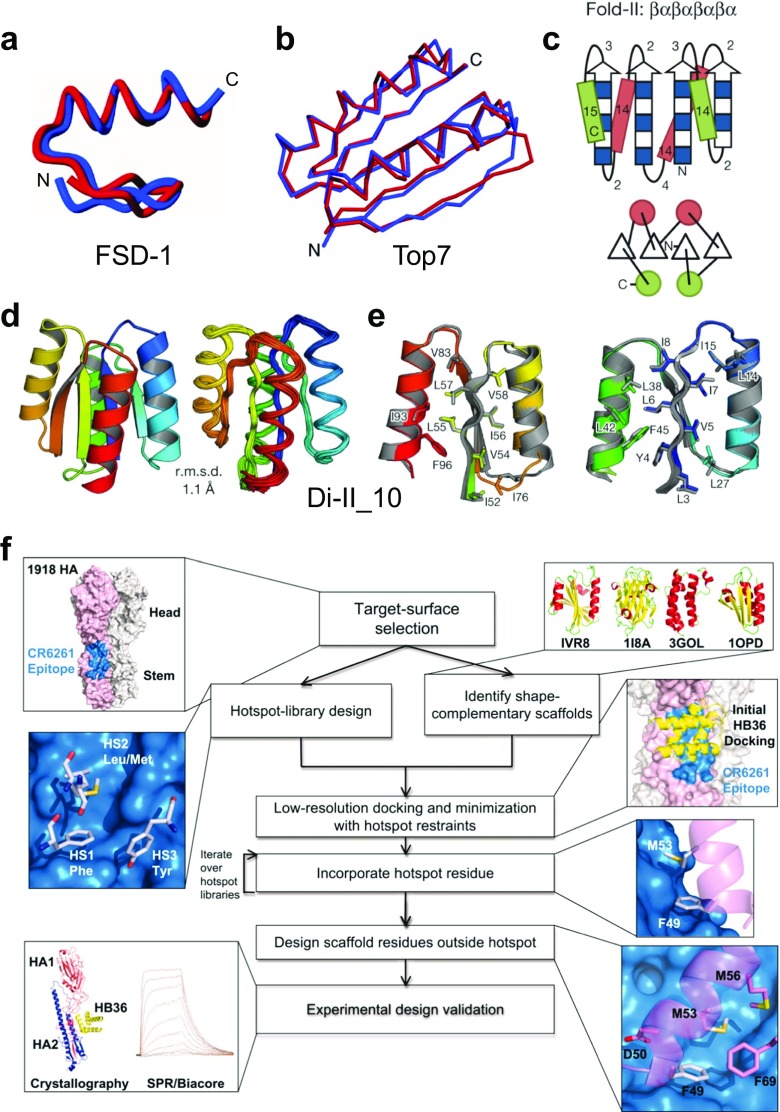
The first fully automated design and experimental validation of a novel sequence for an entire protein (FSD-1) was reported in 1997 (Fig. 1i) (Dahiyat and Mayo 1997). A computational design algorithm based on physical chemical potential functions and stereochemical constraints was used to screen possible amino acid sequences for compatibility with the design target, a zinc finger-like backbone structure (Fig. 5a). A successful computational design procedure of de novo proteins involves initial blueprint construction of the global backbone structure with optimized secondary structure topology followed by selection of side chain residues and rotamers within local ranges of stereochemical and physical constraints. Kuhlman et al. reported the design of a novel globular fold protein called Top7 with atomic-level accuracy, based on a general computational strategy that iterates between sequence design and structure prediction to design de novo proteins with a novel sequence and topology (Figs. 1j, 5b) (Kuhlman et al. 2003). The target 3D structural model of a novel protein fold satisfying the constraints were generated by assembling three- and nine-residue fragments from the PDB with secondary structures consistent with the Rosetta de novo structure prediction methodology (Bowers et al. 2000). These integrated approaches for computational design of artificial proteins can be implemented in Rosetta software suite (Das and Baker 2008; Kaufmann et al. 2010; DiMaio et al. 2011; Leaver-Fay et al. 2011; Richter et al. 2011; Baker 2014; Bender et al. 2016; Huang et al. 2016). The ability to design new protein folds makes possible the exploration of large regions of the protein universe not yet observed in nature.
Fig. 5.
Computational design of de novo proteins. a Comparison of the FSD-1 (full sequence design 1) structure (blue) (PDB ID: 1FSD) and the design target (red) (Dahiyat and Mayo 1997). The best-fit superposition of the restrained energy minimized average NMR structure of FSD-1 and the backbone of Zif268. Reprinted with permission from Dahiyat and Mayo (1997); copyright © 1997, AAAS. b Comparison of the computationally designed model (blue) to the solved X-ray structure (red) of Top7 (PDB ID: 1QYS) (Kuhlman et al. 2003). Cα overlay of the model and structure in stereo (backbone RMSD = 1.17 Å). Reprinted with permission from Kuhlman et al. (2003); copyright © 2003, AAAS. c Secondary structure lengths from the rules for Fold-II, Rossmann2 × 2 fold. In the upper illustrations, numbers represent the loop lengths following from the rules relating local structures to tertiary motifs (Koga et al. 2012). Strand lengths are represented by filled and open boxes. The filled boxes represent pleats coming out of the page, and the open boxes represent pleats going into the page. In the lower illustrations, the design topologies are represented with circles (helices) and triangles (strands) connected by solid lines (loops). d Comparison of overall topology of Di-II_10. Design models (left) and NMR structures (right) (PDB ID: 2LV8); the Cα root mean squared deviation (r.m.s.d.) between them is indicated. e Comparison of core side-chain packing in superpositions of design models (rainbow) and NMR structures (gray). The left and right panels show close-up views of the core packing and correspond to the left and right portions of the structures shown in (d). Reprinted with permission from Koga et al. (2012); copyright © 2012, NPG. f Flow chart illustrating the key steps in the computational design of novel binding proteins. The thumbnails illustrate each step in the creation of binders that target the stem of the 1918 hemagglutinin (HA). Reprinted with permission from Fleishman et al. (2011); copyright © 2011, AAAS
In the hierarchical design of protein tertiary structure, drawing blueprints for ideal topologies of secondary structures as building blocks is a key aim because the stereochemical and physical constraints of polypeptides formed by covalent bonds are fundamentally reflected in local backbone structures. Koga et al. described an approach to designing ideal protein structures stabilized by consistent local and non-local interactions (Fig. 1k) (Koga et al. 2012). The approach is based on a set of rules relating secondary structure patterns to protein tertiary motifs, which make possible the design of funnel-shaped protein folding energy landscapes leading into the target folded state. Simulations and analyses of protein structures have revealed sequence-independent design principles for ideal protein structures (Fig. 5c–e) (Koga et al. 2012). Guided by these rules, several de novo proteins were designed and folded into the ideal protein structures consisting of α-helices, β-strands and minimal loops (Lin et al. 2015a), and also the structures with cavities formed by curved β-sheets were reported (Marcos et al. 2017).
Moreover, computational design methods to explore the repeat protein universe of tandem repeat motif proteins with idealized units and internal symmetry were reported (Fig. 1l) (Brunette et al. 2015; Doyle et al. 2015; Park et al. 2015; Parmeggiani and Huang 2017). Mou et al. also reported a computationally designed symmetric protein homodimer (Mou et al. 2015a), and Voet et al. reported computationally designed symmetrical β-propeller proteins (Voet et al. 2014) and biomineralization of a cadmium chloride nanocrystal using a designed symmetrical protein (Voet et al. 2015). Furthermore, Woolfson and colleagues described computational design of water-soluble α-helical barrels (Thomson et al. 2014) and recently installed hydrolytic activity into a de novo α-helical barrel comprising seven helices with cysteine–histidine–glutamate catalytic triads (Burton et al. 2016). More studies of de novo protein design are also included in recent reviews (Woolfson et al. 2015; Huang et al. 2016).
In various protein functions, the most important and basic function is probably molecular recognition and specific binding. The computational design strategies of molecular recognition and specific binding for various protein functions such as protein–protein interactions, ligand binding, and enzymatic reactions have been developed using Rosetta software suite. A remarkable computational method for designing proteins that bind a surface patch of interest on a target macromolecule was described (Fig. 5f) (Fleishman et al. 2011). Favorable interactions between disembodied amino acid residues and the target surface are identified and used to anchor de novo designed interfaces. The method was used to design the proteins that bind a conserved surface patch on the stem of the influenza hemagglutinin (Fleishman et al. 2011), and optimization of affinity, specificity and function of the designed influenza inhibitory proteins was achieved using deep sequencing (Whitehead et al. 2012). A general computational method was also developed for designing pre-organized and shape complementary small-molecule-binding sites, and used to generate protein binders to the steroid digoxigenin (DIG) (Tinberg et al. 2013). In addition, as an important goal of functional protein design, several pioneering and remarkable studies on rational and computational design of enzymes were reported (Jiang et al. 2008; Rothlisberger et al. 2008; Siegel et al. 2010; Giger et al. 2013) and summarized in reviews (Baker 2010; Kiss et al. 2013; Kries et al. 2013; Zanghellini 2014). These strategies expand possibilities to design novel structural and functional proteins, opening up a greater future for biomolecular engineering and synthetic structural biology.
Computational design of artificial protein complexes
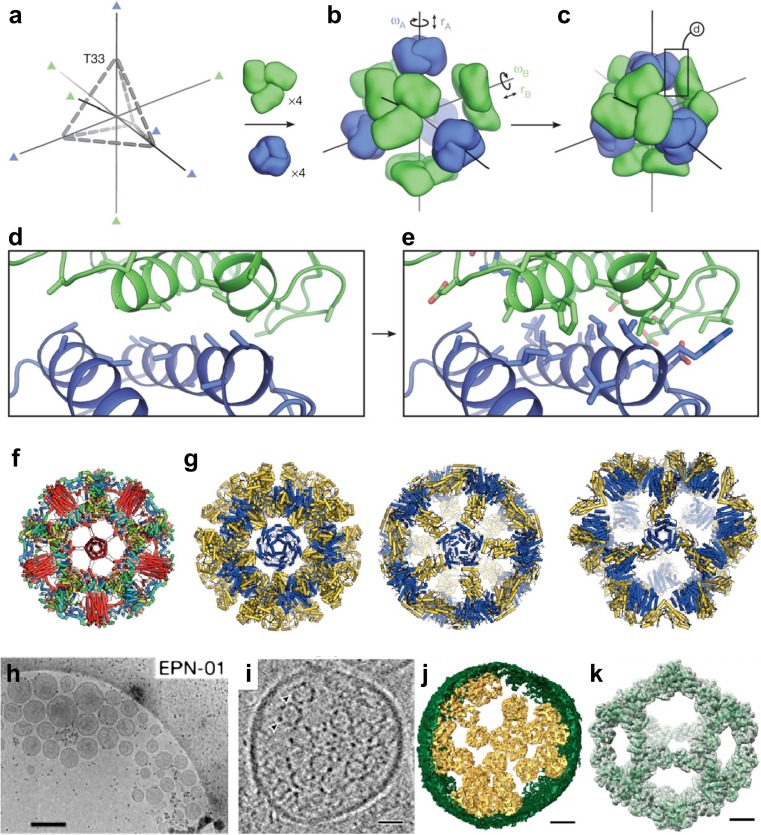
Since quaternary structures of proteins are constructed by a range of non-covalent bond interactions on protein–protein interfaces between tertiary structures of protein subunits, strategies for selection of contact surfaces with rotational symmetry and optimization of side chain packing, leading to the successful design of quaternary structures. King et al. developed a general approach for designing computationally self-assembling protein nanomaterials with atomic-level accuracy (Fig. 1m) (King et al. 2012, 2014). Their approach involves docking of protein building blocks in a target rotationally symmetric architecture followed by the design of a low-energy protein–protein interface that drives the symmetry of self-assembly using RosettaDesign calculations (Fig. 6a–e). The method can be applied to various symmetric architectures, including protein arrays and complexes that extend in 1, 2, or 3 dimensions. Significantly researchers have achieved computational design of a hyperstable self-assembling icosahedral nanocage from 60-subunit trimeric protein building blocks with atomic-level accuracy (Fig. 6f) (Hsia et al. 2016) and co-assembling two-component 120-subunit icosahedral protein complexes with molecular weights (1.8–2.8 MDa) and dimensions (24–40 nm in diameter) comparable to those of small viral capsids (Fig. 6g) (Bale et al. 2016). Moreover, there have been extensive reports on the parametric design of helical bundles with high thermodynamic stability (Huang et al. 2014), ordered two-dimensional arrays that are mediated by noncovalent protein–protein interfaces (Fig. 1n) (Gonen et al. 2015), de novo protein homo-oligomers with modular hydrogen-bond network-mediated specificity (Boyken et al. 2016), and self-assembling cyclic protein homo-oligomers (Fallas et al. 2017).
Fig. 6.
Computational design of various self-assembling protein complexes. a–e Overview of the computational design method of co-assembling multi-component protein nanomaterials (King et al. 2014). a The T33 architecture comprises four copies each of two distinct trimeric building blocks (green and blue) arranged with tetrahedral point group symmetry (24 total subunits; triangles indicate three-fold symmetry axes). b Each building block has two rigid-body degrees of freedom, one translational (r) and one rotational (ω), that are systematically explored during docking. c The docking procedure, which is independent of the amino acid sequence of the building blocks, identifies large interfaces with high densities of contacting residues formed by well-anchored regions of the protein structure. The details of such an interface, boxed, are shown in (d). e Amino acid sequences are designed at the new interface to stabilize the modeled configuration and to drive co-assembly of the two components. Reprinted with permission from King et al. (2014); copyright © 2014, NPG. f, g Designed self-assembling nanocages. f A one-component hyperstable icosahedron with a de novo helical bundle (red helices) fused in the center of the face (Hsia et al. 2016). g Two-component megadalton-scale icosahedra (Bale et al. 2016). The two components of each are colored in blue and yellow. Reprinted with permission from Huang et al. (2016); copyright © 2016, NPG. h–k Enveloped protein nanocages (EPNs) comprise cell-derived membrane envelopes containing multiple protein nanocages (Votteler et al. 2016). h Representative cryo-EM images showing extracellular vesicles/EPNs in culture supernatants from 293 T cells that expressed EPN-01. i Central slice from a cryo-EM tomographic reconstruction of a released EPN. Two internal protein nanocages are marked with arrowheads. j Isosurface model of the 3D cryo-EM reconstruction from (i). The EPN membrane is green and individual protein nanocages are gold. k Single-particle cryo-EM reconstruction of the nanocages released from EPNs following detergent treatment. Charge density from the 5.7 Å resolution electron microscopy reconstruction is shown in gray (contoured at 4.5σ). The I3–01 computational design model9 (green ribbon) was fitted into the density as a rigid body. Scale bars (h) 300 nm, (i, j) 25 nm, (k) 5 nm. Reprinted with permission from Votteler et al. (2016); copyright © 2016, NPG
Conclusions and perspectives
In this review, various approaches to the design and construction of artificial proteins and protein complexes have been described. Major issues on protein design and engineering in synthetic structural biology have been moving on to the next stages from structural design to functional design. In these years, rational, semirational, and combinatorial approaches have become successful methods for creating de novo proteins, functional in vitro and in vivo (Smith and Hecht 2011). Also, artificial proteins using computational design and directed evolution have led to various applications such as targeted therapeutics (Chevalier et al. 2017; Strauch et al. 2017) and plant environmental sensors for the opioid fentanyl (Bick et al. 2017). In addition, a key challenge of artificial proteins is functional design involving structural changes and dynamic motions, which essentially relate to active functions. Naturally occurring proteins provide numerous examples of the rich functionality, including allostery and signaling, that can emerge in protein systems with multiple low-energy states and moving parts that can be toggled by external stimuli. A notable pioneering work of functional protein design with dynamic structural properties was a zinc-transporting transmembrane protein (known as Rocker) that was designed to have two alternative states (Joh et al. 2014). Membrane-spanning α-helical barrels will become tantalizing and tractable protein-design targets (Niitsu et al. 2017).
Moreover, these approaches for designing supramolecular protein complexes will facilitate the development of functional nanobiomaterials. The high symmetry of self-assembling protein nanocages will likely enable multivalent presentation of antigens for vaccine applications, and the large volumes of their interior spaces are well suited to packaging of cargo and drugs for delivery to targets (Bale et al. 2016; Hsia et al. 2016; Huang et al. 2016). The design of protein complexes and crystals is a growing area of nanobiomaterial science and nanobiotechnology, and protein crystals have immense potential for use as new porous materials that allow for precise arrangement of exogenous compounds using metal coordination and chemical conjugation in solvent channels of crystals (Abe and Ueno 2015; Abe et al. 2016). Hybrid complexes of proteins with other materials, such as DNA (Mou et al. 2015b), carbon nanotubes (Grigoryan et al. 2011), and buckminsterfullerene (Kim et al. 2016), are also expected to provide novel attractive functional nanobiomaterials.
Furthermore, a key milestone in synthetic structural biology in the future will be the de novo design of artificial virus-like bio-nanomachines. One of target model systems should be natural bacteriophages such as T4 (Leiman et al. 2003; Arisaka et al. 2016). There is an exciting but challenging road ahead for the design of artificial virus-like bio-nanomachines. Recent developments in the accurate design of icosahedral protein cage complexes can be used for the design of core shell parts (Fig. 6f, g) (Bale et al. 2016; Hsia et al. 2016). The design of self-assembling protein nanocages that direct their own release from cells inside small vesicles in a manner that resembles some viruses was recently reported (Fig. 6h–k) (Votteler et al. 2016). The next challenges in the near future will be to incorporate nucleic acids (DNA or RNA) and to design an infection system to a host cell.
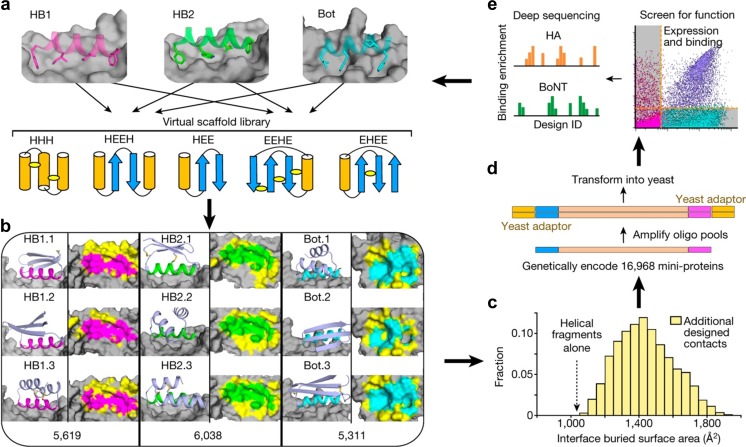
More recently, global analysis of protein folding (Rocklin et al. 2017) and massively parallel design of de novo binding proteins for targeted therapeutics (Chevalier et al. 2017) were reported by Baker and colleagues. They described massively parallel approaches integrating large-scale computational design, highly parallel DNA synthesis, high-throughput screening experiments, and NGS (Fig. 7) (Chevalier et al. 2017). The new integrated synthetic approach has achieved the long-standing goal of a tight feedback cycle between computation and experiment, and has the potential to transform computational protein design into a data-driven big science with deep learning.
Fig. 7.
Massively parallel approach for designing, manufacturing and screening de novo mini-protein binders (Chevalier et al. 2017). a Hundreds of 37- to 43-residue mini-protein backbones with different secondary structure elements, orientations and loop lengths were matched with hotspot binding motifs for hemagglutinin (HB1 and HB2) and botulinum neurotoxin (Bot) by identifying compatible mini-protein local backbone segments, superimposing them onto the hotspot motif–target complex, and discarding docks with mini-protein/target backbone clashes. Each topology included designs with many different disulfide configurations; several possibilities are illustrated. b For each non-clashing dock of each scaffold onto each target, the monomer and interaction energies were optimized with Rosetta sequence design. Representative models are shown at the left of each column. Right columns show a top view of the target with the hotspot interaction areas colored as above and new contact areas generated by Rosetta sequence design colored yellow; the total number of unique designs generated is indicated at the bottom. c Designed contacts substantially increase the interface buried surface area of the designs beyond the starting hotspot residues. d Genes encoding 16,968 mini-protein designs, including 6286 controls, were synthesized using DNA oligo pool synthesis. e The oligo pools were recombined into yeast display vectors and transformed into yeast, and binding of the designs hemagglutinin (HA) or botulinum neurotoxin (BoNT) at different concentrations was assessed by FACS. For each sorting condition, enriched designs were identified by comparing the frequencies in the original and sorted populations using deep sequencing. These data were used to guide improvement of the computational design model, and the entire design, synthesis and testing cycle was iterated. Reprinted with permission from Chevalier et al. (2017); copyright © 2017, NPG
In multiscale structural biology, synthetic structural biology becomes more important as an emerging interdisciplinary field to demonstrate biophysical principles and mechanisms underlying the structure, function, and action of proteins, complexes, and bio-nanomachines. As in the well-known dictum by famed physicist Richard Feynman, “What I cannot create, I do not understand,” successful design is a powerful way to show that a design principle has been understood. In conclusion, many protein designers and engineers look toward a bright future in synthetic structural biology for the next generation of biophysics and biotechnology.
Acknowledgements
I thank all colleagues and collaborators, especially, Dr. Naoya Kobayashi and Dr. Nobuyasu Koga at Institute for Molecular Science (IMS), Prof. Michael H. Hecht at Princeton University, Dr. Shinya Honda at Advanced Industrial Science and Technology, and Dr. Tomoaki Matsuura at Osaka University for their help and valuable discussion. I apologize to the protein designers and protein engineers whose works I was unable to acknowledge due to space and scope limitations. This work was supported by JSPS KAKENHI Grant Numbers JP16K05841 and JP16H00761 (an Innovative Area, “Dynamical Ordering and Integrated Functions”), and Joint Research by IMS.
Compliance with ethical standards
Conflict of interest
Ryoichi Arai declares that he has no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by the author.
Footnotes
This article is part of a Special Issue on ‘Biomolecules to Bio-nanomachines - Fumio Arisaka 70th Birthday’ edited by Damien Hall, Junichi Takagi and Haruki Nakamura.
References
- Abe S, Ueno T. Design of protein crystals in the development of solid biomaterials. RSC Adv. 2015;5:21366–21375. [Google Scholar]
- Abe S, Maity B, Ueno T. Design of a confined environment using protein cages and crystals for the development of biohybrid materials. Chem Commun. 2016;52:6496–6512. doi: 10.1039/c6cc01355d. [DOI] [PubMed] [Google Scholar]
- Ahnert SE, Marsh JA, Hernandez H, Robinson CV, Teichmann SA. Principles of assembly reveal a periodic table of protein complexes. Science. 2015;350:aaa2245. doi: 10.1126/science.aaa2245. [DOI] [PubMed] [Google Scholar]
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Arai R, Kobayashi N, Kimura A, Sato T, Matsuo K, Wang AF, Platt JM, Bradley LH, Hecht MH. Domain-swapped dimeric structure of a stable and functional de novo four-helix bundle protein, WA20. J Phys Chem B. 2012;116:6789–6797. doi: 10.1021/jp212438h. [DOI] [PubMed] [Google Scholar]
- Arisaka F, Yap ML, Kanamaru S, Rossmann MG. Molecular assembly and structure of the bacteriophage T4 tail. Biophys Rev. 2016;8:385–396. doi: 10.1007/s12551-016-0230-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JB, Subramanian RH, Churchfield LA, Tezcan FA. Metal-directed Design of Supramolecular Protein Assemblies. Methods Enzymol. 2016;580:223–250. doi: 10.1016/bs.mie.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JB, Zhang L, Chiong JA, Ahn S, Tezcan FA. Synthetic modularity of protein-metal-organic frameworks. J Am Chem Soc. 2017;139:8160–8166. doi: 10.1021/jacs.7b01202. [DOI] [PubMed] [Google Scholar]
- Baker D (2010) An exciting but challenging road ahead for computational enzyme design. Protein Sci 19:1817–1819 [DOI] [PMC free article] [PubMed]
- Baker D (2014) Protein folding, structure prediction and design. Biochem Soc Trans 42:225–229 [DOI] [PubMed]
- Bale JB, Gonen S, Liu Y, Sheffler W, Ellis D, Thomas C, Cascio D, Yeates TO, Gonen T, King NP, Baker D. Accurate design of megadalton-scale two-component icosahedral protein complexes. Science. 2016;353:389–394. doi: 10.1126/science.aaf8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender BJ, Cisneros A, 3rd, Duran AM, Finn JA, Fu D, Lokits AD, Mueller BK, Sangha AK, Sauer MF, Sevy AM, Sliwoski G, Sheehan JH, DiMaio F, Meiler J, Moretti R. Protocols for molecular Modeling with Rosetta3 and RosettaScripts. Biochemistry. 2016;55:4748–4763. doi: 10.1021/acs.biochem.6b00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessette PH, Rice JJ, Daugherty PS. Rapid isolation of high-affinity protein binding peptides using bacterial display. Protein Eng Des Sel. 2004;17:731–739. doi: 10.1093/protein/gzh084. [DOI] [PubMed] [Google Scholar]
- Bick MJ, Greisen PJ, Morey KJ, Antunes MS, La D, Sankaran B, Reymond L, Johnsson K, Medford JI, Baker D. Computational design of environmental sensors for the potent opioid fentanyl. elife. 2017;6:e28909. doi: 10.7554/eLife.28909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- Boersma YL, Pluckthun A. DARPins and other repeat protein scaffolds: advances in engineering and applications. Curr Opin Biotechnol. 2011;22:849–857. doi: 10.1016/j.copbio.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Boucher JI, Bolon DN, Tawfik DS. Quantifying and understanding the fitness effects of protein mutations: laboratory versus nature. Protein Sci. 2016;25:1219–1226. doi: 10.1002/pro.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers PM, Strauss CE, Baker D. De novo protein structure determination using sparse NMR data. J Biomol NMR. 2000;18:311–318. doi: 10.1023/a:1026744431105. [DOI] [PubMed] [Google Scholar]
- Boyken SE, Chen Z, Groves B, Langan RA, Oberdorfer G, Ford A, Gilmore JM, Xu C, DiMaio F, Pereira JH, Sankaran B, Seelig G, Zwart PH, Baker D. De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity. Science. 2016;352:680–687. doi: 10.1126/science.aad8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley LH, Kleiner RE, Wang AF, Hecht MH, Wood DW. An intein-based genetic selection allows the construction of a high-quality library of binary patterned de novo protein sequences. Protein Eng Des Sel. 2005;18:201–207. doi: 10.1093/protein/gzi020. [DOI] [PubMed] [Google Scholar]
- Brodin JD, Ambroggio XI, Tang C, Parent KN, Baker TS, Tezcan FA. Metal-directed, chemically tunable assembly of one-, two- and three-dimensional crystalline protein arrays. Nat Chem. 2012;4:375–382. doi: 10.1038/nchem.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin JD, Smith SJ, Carr JR, Tezcan FA. Designed, helical protein Nanotubes with variable diameters from a single building block. J Am Chem Soc. 2015;137:10468–10471. doi: 10.1021/jacs.5b05755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunette TJ, Parmeggiani F, Huang PS, Bhabha G, Ekiert DC, Tsutakawa SE, Hura GL, Tainer JA, Baker D. Exploring the repeat protein universe through computational protein design. Nature. 2015;528:580–584. doi: 10.1038/nature16162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson JW, Betz SF, Lu HS, Suich DJ, Zhou HX, O'Neil KT, DeGrado WF. Protein design: a hierarchic approach. Science. 1995;270:935–941. doi: 10.1126/science.270.5238.935. [DOI] [PubMed] [Google Scholar]
- Burgess NC, Sharp TH, Thomas F, Wood CW, Thomson AR, Zaccai NR, Brady RL, Serpell LC, Woolfson DN. Modular Design of Self-Assembling Peptide-Based Nanotubes. J Am Chem Soc. 2015;137:10554–10562. doi: 10.1021/jacs.5b03973. [DOI] [PubMed] [Google Scholar]
- Burton AJ, Thomson AR, Dawson WM, Brady RL, Woolfson DN. Installing hydrolytic activity into a completely de novo protein framework. Nat Chem. 2016;8:837–844. doi: 10.1038/nchem.2555. [DOI] [PubMed] [Google Scholar]
- Cherny I, Korolev M, Koehler AN, Hecht MH. Proteins from an unevolved library of de novo designed sequences bind a range of small molecules. ACS Synth Biol. 2012;1:130–138. doi: 10.1021/sb200018e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier A, Silva DA, Rocklin GJ, Hicks DR, Vergara R, Murapa P, Bernard SM, Zhang L, Lam KH, Yao G, Bahl CD, Miyashita SI, Goreshnik I, Fuller JT, Koday MT, Jenkins CM, Colvin T, Carter L, Bohn A, Bryan CM, Fernandez-Velasco DA, Stewart L, Dong M, Huang X, Jin R, Wilson IA, Fuller DH, Baker D. Massively parallel de novo protein design for targeted therapeutics. Nature. 2017;550:74–79. doi: 10.1038/nature23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchfield LA, Medina-Morales A, Brodin JD, Perez A, Tezcan FA (2016) De novo design of an allosteric metalloprotein assembly with strained disulfide bonds. J Am Chem Soc 138:13163–13166 [DOI] [PMC free article] [PubMed]
- Crick FHC. The Fourier transform of a coiled-coil. Acta Crystallogr. 1953;6:685–689. [Google Scholar]
- Crick FHC. The packing of α-helices: simple coiled-coils. Acta Crystallogr. 1953;6:689–697. [Google Scholar]
- Dahiyat BI, Mayo SL. De novo protein design: fully automated sequence selection. Science. 1997;278:82–87. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- Das R, Baker D. Macromolecular modeling with rosetta. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- de Bono S, Riechmann L, Girard E, Williams RL, Winter G. A segment of cold shock protein directs the folding of a combinatorial protein. Proc Natl Acad Sci U S A. 2005;102:1396–1401. doi: 10.1073/pnas.0407298102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digianantonio KM, Hecht MH. A protein constructed de novo enables cell growth by altering gene regulation. Proc Natl Acad Sci U S A. 2016;113:2400–2405. doi: 10.1073/pnas.1600566113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digianantonio KM, Korolev M, Hecht MH. A non-natural protein rescues cells deleted for a key enzyme in central metabolism. ACS Synth Biol. 2017;6:694–700. doi: 10.1021/acssynbio.6b00336. [DOI] [PubMed] [Google Scholar]
- DiMaio F, Leaver-Fay A, Bradley P, Baker D, Andre I. Modeling symmetric macromolecular structures in Rosetta3. PLoS ONE. 2011;6:e20450. doi: 10.1371/journal.pone.0020450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle L, Hallinan J, Bolduc J, Parmeggiani F, Baker D, Stoddard BL, Bradley P. Rational design of alpha-helical tandem repeat proteins with closed architectures. Nature. 2015;528:585–588. doi: 10.1038/nature16191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallas JA, Ueda G, Sheffler W, Nguyen V, McNamara DE, Sankaran B, Pereira JH, Parmeggiani F, Brunette TJ, Cascio D, Yeates TR, Zwart P, Baker D. Computational design of self-assembling cyclic protein homo-oligomers. Nat Chem. 2017;9:353–360. doi: 10.1038/nchem.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MA, McKinley KL, Bradley LH, Viola SR, Hecht MH. De novo designed proteins from a library of artificial sequences function in Escherichia coli and enable cell growth. PLoS ONE. 2011;6:e15364. doi: 10.1371/journal.pone.0015364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleishman SJ, Whitehead TA, Ekiert DC, Dreyfus C, Corn JE, Strauch EM, Wilson IA, Baker D. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JM, Harniman RL, Barnes FR, Boyle AL, Collins A, Mantell J, Sharp TH, Antognozzi M, Booth PJ, Linden N, Miles MJ, Sessions RB, Verkade P, Woolfson DN. Self-assembling cages from coiled-coil peptide modules. Science. 2013;340:595–599. doi: 10.1126/science.1233936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth CM, Juan V, Akamatsu Y, DuBridge RB, Doan M, Ivanov AV, Ma Z, Polakoff D, Razo J, Wilson K, Powers DB. Deep mutational scanning of an antibody against epidermal growth factor receptor using mammalian cell display and massively parallel pyrosequencing. MAbs. 2013;5:523–532. doi: 10.4161/mabs.24979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Fields S. Deep mutational scanning: a new style of protein science. Nat Methods. 2014;11:801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Araya CL, Fleishman SJ, Kellogg EH, Stephany JJ, Baker D, Fields S. High-resolution mapping of protein sequence-function relationships. Nat Methods. 2010;7:741–746. doi: 10.1038/nmeth.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Matsuura T, Sunami T, Kazuta Y, Yomo T. In vitro evolution of alpha-hemolysin using a liposome display. Proc Natl Acad Sci U S A. 2013;110:16796–16801. doi: 10.1073/pnas.1314585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Matsuura T, Sunami T, Nishikawa T, Kazuta Y, Yomo T. Liposome display for in vitro selection and evolution of membrane proteins. Nat Protoc. 2014;9:1578–1591. doi: 10.1038/nprot.2014.107. [DOI] [PubMed] [Google Scholar]
- Fujino Y, Fujita R, Wada K, Fujishige K, Kanamori T, Hunt L, Shimizu Y, Ueda T. Robust in vitro affinity maturation strategy based on interface-focused high-throughput mutational scanning. Biochem Biophys Res Commun. 2012;428:395–400. doi: 10.1016/j.bbrc.2012.10.066. [DOI] [PubMed] [Google Scholar]
- Fujiwara D, Kitada H, Oguri M, Nishihara T, Michigami M, Shiraishi K, Yuba E, Nakase I, Im H, Cho S, Joung JY, Kodama S, Kono K, Ham S, Fujii I (2016) A cyclized helix-loop-helix peptide as a molecular scaffold for the design of inhibitors of intracellular protein-protein interactions by epitope and arginine grafting. Angew Chem Int Ed 55:10612–10615 [DOI] [PubMed]
- Giger L, Caner S, Obexer R, Kast P, Baker D, Ban N, Hilvert D. Evolution of a designed retro-aldolase leads to complete active site remodeling. Nat Chem Biol. 2013;9:494–498. doi: 10.1038/nchembio.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbreth RN, Koide S. Structural insights for engineering binding proteins based on non-antibody scaffolds. Curr Opin Struct Biol. 2012;22:413–420. doi: 10.1016/j.sbi.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go A, Kim S, Baum J, Hecht MH. Structure and dynamics of de novo proteins from a designed superfamily of 4-helix bundles. Protein Sci. 2008;17:821–832. doi: 10.1110/ps.073377908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen S, DiMaio F, Gonen T, Baker D. Design of ordered two-dimensional arrays mediated by noncovalent protein-protein interfaces. Science. 2015;348:1365–1368. doi: 10.1126/science.aaa9897. [DOI] [PubMed] [Google Scholar]
- Goto Y, Katoh T, Suga H. Flexizymes for genetic code reprogramming. Nat Protoc. 2011;6:779–790. doi: 10.1038/nprot.2011.331. [DOI] [PubMed] [Google Scholar]
- Gradisar H, Bozic S, Doles T, Vengust D, Hafner-Bratkovic I, Mertelj A, Webb B, Sali A, Klavzar S, Jerala R. Design of a single-chain polypeptide tetrahedron assembled from coiled-coil segments. Nat Chem Biol. 2013;9:362–366. doi: 10.1038/nchembio.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano JJ, Liu W, Perera R, Geierstanger BH, Lesley SA, Schultz PG. Selecting folded proteins from a library of secondary structural elements. J Am Chem Soc. 2008;130:176–185. doi: 10.1021/ja074405w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan G, Degrado WF. Probing designability via a generalized model of helical bundle geometry. J Mol Biol. 2011;405:1079–1100. doi: 10.1016/j.jmb.2010.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan G, Kim YH, Acharya R, Axelrod K, Jain RM, Willis L, Drndic M, Kikkawa JM, DeGrado WF. Computational design of virus-like protein assemblies on carbon nanotube surfaces. Science. 2011;332:1071–1076. doi: 10.1126/science.1198841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gully BS, Shah KR, Lee M, Shearston K, Smith NM, Sadowska A, Blythe AJ, Bernath-Levin K, Stanley WA, Small ID, Bond CS. The design and structural characterization of a synthetic pentatricopeptide repeat protein. Acta Crystallogr D. 2015;71:196–208. doi: 10.1107/S1399004714024869. [DOI] [PubMed] [Google Scholar]
- Guthe S, Kapinos L, Moglich A, Meier S, Grzesiek S, Kiefhaber T. Very fast folding and association of a trimerization domain from bacteriophage T4 fibritin. J Mol Biol. 2004;337:905–915. doi: 10.1016/j.jmb.2004.02.020. [DOI] [PubMed] [Google Scholar]
- Hanes J, Pluckthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci U S A. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbury PB, Plecs JJ, Tidor B, Alber T, Kim PS. High-resolution protein design with backbone freedom. Science. 1998;282:1462–1467. doi: 10.1126/science.282.5393.1462. [DOI] [PubMed] [Google Scholar]
- Hecht MH, Das A, Go A, Bradley LH, Wei Y. De novo proteins from designed combinatorial libraries. Protein Sci. 2004;13:1711–1723. doi: 10.1110/ps.04690804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietpas RT, Jensen JD, Bolon DNA. Experimental illumination of a fitness landscape. Proc Natl Acad Sci U S A. 2011;108:7896–7901. doi: 10.1073/pnas.1016024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipolito CJ, Suga H. Ribosomal production and in vitro selection of natural product-like peptidomimetics: the FIT and RaPID systems. Curr Opin Chem Biol. 2012;16:196–203. doi: 10.1016/j.cbpa.2012.02.014. [DOI] [PubMed] [Google Scholar]
- Hirota S, Hattori Y, Nagao S, Taketa M, Komori H, Kamikubo H, Wang Z, Takahashi I, Negi S, Sugiura Y, Kataoka M, Higuchi Y (2010) Cytochrome c polymerization by successive domain swapping at the C-terminal helix. Proc Natl Acad Sci U S A 107:12854–12859 [DOI] [PMC free article] [PubMed]
- Hoegler KJ, Hecht MH. A de novo protein confers copper resistance in Escherichia Coli. Protein Sci. 2016;25:1249–1259. doi: 10.1002/pro.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Yamasaki K, Sawada Y, Morii H. 10 residue folded peptide designed by segment statistics. Structure. 2004;12:1507–1518. doi: 10.1016/j.str.2004.05.022. [DOI] [PubMed] [Google Scholar]
- Honda S, Akiba T, Kato YS, Sawada Y, Sekijima M, Ishimura M, Ooishi A, Watanabe H, Odahara T, Harata K. Crystal structure of a ten-amino acid protein. J Am Chem Soc. 2008;130:15327–15331. doi: 10.1021/ja8030533. [DOI] [PubMed] [Google Scholar]
- Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat Biotechnol. 2005;23:1105–1116. doi: 10.1038/nbt1126. [DOI] [PubMed] [Google Scholar]
- Hsia Y, Bale JB, Gonen S, Shi D, Sheffler W, Fong KK, Nattermann U, Xu C, Huang PS, Ravichandran R, Yi S, Davis TN, Gonen T, King NP, Baker D. Design of a hyperstable 60-subunit protein icosahedron. Nature. 2016;535:136–139. doi: 10.1038/nature18010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang PS, Oberdorfer G, Xu C, Pei XY, Nannenga BL, Rogers JM, DiMaio F, Gonen T, Luisi B, Baker D. High thermodynamic stability of parametrically designed helical bundles. Science. 2014;346:481–485. doi: 10.1126/science.1257481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang PS, Boyken SE, Baker D. The coming of age of de novo protein design. Nature. 2016;537:320–327. doi: 10.1038/nature19946. [DOI] [PubMed] [Google Scholar]
- Jardine JG, Kulp DW, Havenar-Daughton C, Sarkar A, Briney B, Sok D, Sesterhenn F, Ereno-Orbea J, Kalyuzhniy O, Deresa I, Hu X, Spencer S, Jones M, Georgeson E, Adachi Y, Kubitz M, deCamp AC, Julien JP, Wilson IA, Burton DR, Crotty S, Schief WR. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science. 2016;351:1458–1463. doi: 10.1126/science.aad9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Althoff EA, Clemente FR, Doyle L, Rothlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, 3rd, Hilvert D, Houk KN, Stoddard BL, Baker D. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joh NH, Wang T, Bhate MP, Acharya R, Wu Y, Grabe M, Hong M, Grigoryan G, DeGrado WF. De novo design of a transmembrane Zn2+-transporting four-helix bundle. Science. 2014;346:1520–1524. doi: 10.1126/science.1261172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost C, Pluckthun A. Engineered proteins with desired specificity: DARPins, other alternative scaffolds and bispecific IgGs. Curr Opin Struct Biol. 2014;27:102–112. doi: 10.1016/j.sbi.2014.05.011. [DOI] [PubMed] [Google Scholar]
- Jumawid MT, Takahashi T, Yamazaki T, Ashigai H, Mihara H (2009) Selection and structural analysis of de novo proteins from an α3β3 genetic library. Protein Sci 18:384–398 [DOI] [PMC free article] [PubMed]
- Kamtekar S, Schiffer JM, Xiong H, Babik JM, Hecht MH. Protein design by binary patterning of polar and nonpolar amino acids. Science. 1993;262:1680–1685. doi: 10.1126/science.8259512. [DOI] [PubMed] [Google Scholar]
- Kaufmann KW, Lemmon GH, Deluca SL, Sheehan JH, Meiler J. Practically useful: what the Rosetta protein modeling suite can do for you. Biochemistry. 2010;49:2987–2998. doi: 10.1021/bi902153g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y. Designer three-dimensional DNA architectures. Curr Opin Struct Biol. 2014;27:122–128. doi: 10.1016/j.sbi.2014.07.010. [DOI] [PubMed] [Google Scholar]
- Keefe AD, Szostak JW. Functional proteins from a random-sequence library. Nature. 2001;410:715–718. doi: 10.1038/35070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Ko DK, Kim YT, Kim NH, Paul J, Zhang SQ, Murray CB, Acharya R, DeGrado WF, Kim YH, Grigoryan G. Protein-directed self-assembly of a fullerene crystal. Nat Commun. 2016;7:11429. doi: 10.1038/ncomms11429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King NP, Sheffler W, Sawaya MR, Vollmar BS, Sumida JP, Andre I, Gonen T, Yeates TO, Baker D. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science. 2012;336:1171–1174. doi: 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King NP, Bale JB, Sheffler W, McNamara DE, Gonen S, Gonen T, Yeates TO, Baker D. Accurate design of co-assembling multi-component protein nanomaterials. Nature. 2014;510:103–108. doi: 10.1038/nature13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss G, Celebi-Olcum N, Moretti R, Baker D, Houk KN. Computational enzyme design. Angew Chem Int Ed. 2013;52:5700–5725. doi: 10.1002/anie.201204077. [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Arai R. Design and construction of self-assembling supramolecular protein complexes using artificial and fusion proteins as nanoscale building blocks. Curr Opin Biotech. 2017;46:57–65. doi: 10.1016/j.copbio.2017.01.001. [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Yanase K, Sato T, Unzai S, Hecht MH, Arai R. Self-assembling Nano-architectures created from a protein Nano-building block using an Intermolecularly folded Dimeric de novo protein. J Am Chem Soc. 2015;137:11285–11293. doi: 10.1021/jacs.5b03593. [DOI] [PubMed] [Google Scholar]
- Koenig P, Lee CV, Sanowar S, Wu P, Stinson J, Harris SF, Fuh G. Deep sequencing-guided Design of a High Affinity Dual Specificity Antibody to target two Angiogenic factors in Neovascular age-related macular degeneration. J Biol Chem. 2015;290:21773–21786. doi: 10.1074/jbc.M115.662783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga N, Tatsumi-Koga R, Liu G, Xiao R, Acton TB, Montelione GT, Baker D. Principles for designing ideal protein structures. Nature. 2012;491:222–227. doi: 10.1038/nature11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kries H, Blomberg R, Hilvert D. De novo enzymes by computational design. Curr Opin Chem Biol. 2013;17:221–228. doi: 10.1016/j.cbpa.2013.02.012. [DOI] [PubMed] [Google Scholar]
- Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- Lai YT, Cascio D, Yeates TO. Structure of a 16-nm cage designed by using protein oligomers. Science. 2012;336:1129. doi: 10.1126/science.1219351. [DOI] [PubMed] [Google Scholar]
- Lai YT, Reading E, Hura GL, Tsai KL, Laganowsky A, Asturias FJ, Tainer JA, Robinson CV, Yeates TO. Structure of a designed protein cage that self-assembles into a highly porous cube. Nat Chem. 2014;6:1065–1071. doi: 10.1038/nchem.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban YE, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popovic Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Park K, Han J, Lee JJ, Kim HJ, Hong S, Heu W, Kim YJ, Ha JS, Lee SG, Cheong HK, Jeon YH, Kim D, Kim HS. Design of a binding scaffold based on variable lymphocyte receptors of jawless vertebrates by module engineering. Proc Natl Acad Sci U S A. 2012;109:3299–3304. doi: 10.1073/pnas.1113193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman PG, Kanamaru S, Mesyanzhinov VV, Arisaka F, Rossmann MG. Structure and morphogenesis of bacteriophage T4. Cell Mol Life Sci. 2003;60:2356–2370. doi: 10.1007/s00018-003-3072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YR, Koga N, Tatsumi-Koga R, Liu G, Clouser AF, Montelione GT, Baker D. Control over overall shape and size in de novo designed proteins. Proc Natl Acad Sci U S A. 2015;112:E5478–E5485. doi: 10.1073/pnas.1509508112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YW, Nagao S, Zhang M, Shomura Y, Higuchi Y, Hirota S. Rational design of heterodimeric protein using domain swapping for myoglobin. Angew Chem Int Ed. 2015;54:511–515. doi: 10.1002/anie.201409267. [DOI] [PubMed] [Google Scholar]
- Ljubetic A, Lapenta F, Gradisar H, Drobnak I, Aupic J, Strmsek Z, Lainscek D, Hafner-Bratkovic I, Majerle A, Krivec N, Bencina M, Pisanski T, Velickovic TC, Round A, Carazo JM, Melero R, Jerala R. Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo. Nat Biotechnol. 2017;35:1094–1101. doi: 10.1038/nbt.3994. [DOI] [PubMed] [Google Scholar]
- Lupas AN, Bassler J. Coiled coils - a model system for the 21st century. Trends Biochem Sci. 2017;42:130–140. doi: 10.1016/j.tibs.2016.10.007. [DOI] [PubMed] [Google Scholar]
- Marcos E, Basanta B, Chidyausiku TM, Tang Y, Oberdorfer G, Liu G, Swapna GV, Guan R, Silva DA, Dou J, Pereira JH, Xiao R, Sankaran B, Zwart PH, Montelione GT, Baker D. Principles for designing proteins with cavities formed by curved beta sheets. Science. 2017;355:201–206. doi: 10.1126/science.aah7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura T, Ernst A, Zechel DL, Pluckthun A. Combinatorial approaches to novel proteins. Chembiochem. 2004;5:177–182. doi: 10.1002/cbic.200300755. [DOI] [PubMed] [Google Scholar]
- McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348:552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Kuribayashi M, Nagao S, Shomura Y, Higuchi Y, Hirota S. Domain-swapped cytochrome cb(562) dimer and its nanocage encapsulating a Zn-SO4 cluster in the internal cavity. Chem Sci. 2015;6:7336–7342. doi: 10.1039/c5sc02428e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki N, Kiyose N, Akazawa Y, Takashima M, Hagihara Y, Inoue N, Matsuda T, Ogawa R, Inoue S, Ito Y. Isolation and characterization of antigen-specific alpaca (Lama Pacos) VHH antibodies by biopanning followed by high-throughput sequencing. J Biochem. 2015;158:205–215. doi: 10.1093/jb/mvv038. [DOI] [PubMed] [Google Scholar]
- Morimoto J, Hayashi Y, Iwasaki K, Suga H. Flexizymes: their evolutionary history and the origin of catalytic function. Acc Chem Res. 2011;44:1359–1368. doi: 10.1021/ar2000953. [DOI] [PubMed] [Google Scholar]
- Mou Y, Huang PS, Hsu FC, Huang SJ, Mayo SL. Computational design and experimental verification of a symmetric protein homodimer. Proc Natl Acad Sci U S A. 2015;112:10714–10719. doi: 10.1073/pnas.1505072112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Y, Yu JY, Wannier TM, Guo CL, Mayo SL. Computational design of co-assembling protein-DNA nanowires. Nature. 2015;525:230–233. doi: 10.1038/nature14874. [DOI] [PubMed] [Google Scholar]
- Murakami H, Ohta A, Ashigai H, Suga H. A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nat Methods. 2006;3:357–359. doi: 10.1038/nmeth877. [DOI] [PubMed] [Google Scholar]
- Nemoto N, Miyamoto-Sato E, Husimi Y, Yanagawa H. In vitro virus: bonding of mRNA bearing puromycin at the 3′-terminal end to the C-terminal end of its encoded protein on the ribosome in vitro. FEBS Lett. 1997;414:405–408. doi: 10.1016/s0014-5793(97)01026-0. [DOI] [PubMed] [Google Scholar]
- Nemoto N, Fukushima T, Kumachi S, Suzuki M, Nishigaki K, Kubo T. Versatile C-terminal specific biotinylation of proteins using both a puromycin-linker and a cell-free translation system for studying high-throughput protein-molecule interactions. Anal Chem. 2014;86:8535–8540. doi: 10.1021/ac501601g. [DOI] [PubMed] [Google Scholar]
- Niitsu A, Heal JW, Fauland K, Thomson AR, Woolfson DN (2017) Membrane-spanning α-helical barrels as tractable protein-design targets. Philos Trans R Soc B 372:20160213 [DOI] [PMC free article] [PubMed]
- Ogihara NL, Ghirlanda G, Bryson JW, Gingery M, DeGrado WF, Eisenberg D (2001) Design of three-dimensional domain-swapped dimers and fibrous oligomers. Proc Natl Acad Sci U S A 98:1404–1409 [DOI] [PMC free article] [PubMed]
- Packer MS, Liu DR. Methods for the directed evolution of proteins. Nat Rev Genet. 2015;16:379–394. doi: 10.1038/nrg3927. [DOI] [PubMed] [Google Scholar]
- Padilla JE, Colovos C, Yeates TO. Nanohedra: using symmetry to design self assembling protein cages, layers, crystals, and filaments. Proc Natl Acad Sci U S A. 2001;98:2217–2221. doi: 10.1073/pnas.041614998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K, Shen BW, Parmeggiani F, Huang PS, Stoddard BL, Baker D. Control of repeat-protein curvature by computational protein design. Nat Struct Mol Biol. 2015;22:167–174. doi: 10.1038/nsmb.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmeggiani F, Huang PS. Designing repeat proteins: a modular approach to protein design. Curr Opin Struct Biol. 2017;45:116–123. doi: 10.1016/j.sbi.2017.02.001. [DOI] [PubMed] [Google Scholar]
- Passioura T, Suga H. A RaPID way to discover nonstandard macrocyclic peptide modulators of drug targets. Chem Commun. 2017;53:1931–1940. doi: 10.1039/c6cc06951g. [DOI] [PubMed] [Google Scholar]
- Patel SC, Hecht MH. Directed evolution of the peroxidase activity of a de novo-designed protein. Protein Eng Des Sel. 2012;25:445–452. doi: 10.1093/protein/gzs025. [DOI] [PubMed] [Google Scholar]
- Patel SC, Bradley LH, Jinadasa SP, Hecht MH. Cofactor binding and enzymatic activity in an unevolved superfamily of de novo designed 4-helix bundle proteins. Protein Sci. 2009;18:1388–1400. doi: 10.1002/pro.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieters BJ, van Eldijk MB, Nolte RJ, Mecinovic J. Natural supramolecular protein assemblies. Chem Soc Rev. 2016;45:24–39. doi: 10.1039/c5cs00157a. [DOI] [PubMed] [Google Scholar]
- Reichen C, Hansen S, Pluckthun A. Modular peptide binding: from a comparison of natural binders to designed armadillo repeat proteins. J Struct Biol. 2014;185:147–162. doi: 10.1016/j.jsb.2013.07.012. [DOI] [PubMed] [Google Scholar]
- Reichen C, Hansen S, Forzani C, Honegger A, Fleishman SJ, Zhou T, Parmeggiani F, Ernst P, Madhurantakam C, Ewald C, Mittl PR, Zerbe O, Baker D, Caflisch A, Pluckthun A. Computationally designed armadillo repeat proteins for modular peptide recognition. J Mol Biol. 2016;428:4467–4489. doi: 10.1016/j.jmb.2016.09.012. [DOI] [PubMed] [Google Scholar]
- Richter F, Leaver-Fay A, Khare SD, Bjelic S, Baker D. De novo enzyme design using Rosetta3. PLoS ONE. 2011;6:e19230. doi: 10.1371/journal.pone.0019230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann L, Winter G. Novel folded protein domains generated by combinatorial shuffling of polypeptide segments. Proc Natl Acad Sci U S A. 2000;97:10068–10073. doi: 10.1073/pnas.170145497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RW, Szostak JW. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc Natl Acad Sci U S A. 1997;94:12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocklin GJ, Chidyausiku TM, Goreshnik I, Ford A, Houliston S, Lemak A, Carter L, Ravichandran R, Mulligan VK, Chevalier A, Arrowsmith CH, Baker D. Global analysis of protein folding using massively parallel design, synthesis, and testing. Science. 2017;357:168–175. doi: 10.1126/science.aan0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DS, Baker D. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- Salgado EN, Ambroggio XI, Brodin JD, Lewis RA, Kuhlman B, Tezcan FA. Metal templated design of protein interfaces. Proc Natl Acad Sci U S A. 2010;107:1827–1832. doi: 10.1073/pnas.0906852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado EN, Radford RJ, Tezcan FA. Metal-directed protein self-assembly. Acc Chem Res. 2010;43:661–672. doi: 10.1021/ar900273t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciore A, Su M, Koldewey P, Eschweiler JD, Diffley KA, Linhares BM, Ruotolo BT, Bardwell JC, Skiniotis G, Marsh EN. Flexible, symmetry-directed approach to assembling protein cages. Proc Natl Acad Sci U S A. 2016;113:8681–8686. doi: 10.1073/pnas.1606013113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel JB, Zanghellini A, Lovick HM, Kiss G, Lambert AR, St Clair JL, Gallaher JL, Hilvert D, Gelb MH, Stoddard BL, Houk KN, Michael FE, Baker D. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science. 2010;329:309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BA, Hecht MH. Novel proteins: from fold to function. Curr Opin Chem Biol. 2011;15:421–426. doi: 10.1016/j.cbpa.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Smith BA, Mularz AE, Hecht MH. Divergent evolution of a bifunctional de novo protein. Protein Sci. 2015;24:246–252. doi: 10.1002/pro.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WJ, Tezcan FA. A designed supramolecular protein assembly with in vivo enzymatic activity. Science. 2014;346:1525–1528. doi: 10.1126/science.1259680. [DOI] [PubMed] [Google Scholar]
- Sontz PA, Bailey JB, Ahn S, Tezcan FA. A metal organic framework with spherical protein nodes: rational chemical design of 3D protein crystals. J Am Chem Soc. 2015;137:11598–11601. doi: 10.1021/jacs.5b07463. [DOI] [PubMed] [Google Scholar]
- Stapleton JA, Swartz JR. Development of an in vitro compartmentalization screen for high-throughput directed evolution of [FeFe] hydrogenases. PLoS ONE. 2010;5:e15275. doi: 10.1371/journal.pone.0015275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch EM, Bernard SM, La D, Bohn AJ, Lee PS, Anderson CE, Nieusma T, Holstein CA, Garcia NK, Hooper KA, Ravichandran R, Nelson JW, Sheffler W, Bloom JD, Lee KK, Ward AB, Yager P, Fuller DH, Wilson IA, Baker D. Computational design of trimeric influenza-neutralizing proteins targeting the hemagglutinin receptor binding site. Nat Biotechnol. 2017;35:667–671. doi: 10.1038/nbt.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Cardone G, Restrepo D, Zavattieri PD, Baker TS, Tezcan FA. Self-assembly of coherently dynamic, auxetic, two-dimensional protein crystals. Nature. 2016;533:369–373. doi: 10.1038/nature17633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas F, Burgess NC, Thomson AR, Woolfson DN. Controlling the assembly of coiled-coil peptide Nanotubes. Angew Chem Int Ed. 2016;55:987–991. doi: 10.1002/anie.201509304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson AR, Wood CW, Burton AJ, Bartlett GJ, Sessions RB, Brady RL, Woolfson DN. Computational design of water-soluble alpha-helical barrels. Science. 2014;346:485–488. doi: 10.1126/science.1257452. [DOI] [PubMed] [Google Scholar]
- Tinberg CE, Khare SD, Dou J, Doyle L, Nelson JW, Schena A, Jankowski W, Kalodimos CG, Johnsson K, Stoddard BL, Baker D. Computational design of ligand-binding proteins with high affinity and selectivity. Nature. 2013;501:212–216. doi: 10.1038/nature12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno S, Kimura S, Ichiki T, Nemoto N. Improvement of a puromycin-linker to extend the selection target varieties in cDNA display method. J Biotechnol. 2012;162:299–302. doi: 10.1016/j.jbiotec.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Urvoas A, Guellouz A, Valerio-Lepiniec M, Graille M, Durand D, Desravines DC, van Tilbeurgh H, Desmadril M, Minard P (2010) Design, production and molecular structure of a new family of artificial alpha-helicoidal repeat proteins (αRep) based on thermostable HEAT-like repeats. J Mol Biol 404:307–327 [DOI] [PubMed]
- Urvoas A, Valerio-Lepiniec M, Minard P. Artificial proteins from combinatorial approaches. Trends Biotechnol. 2012;30:512–520. doi: 10.1016/j.tibtech.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Uyeda A, Nakayama S, Kato Y, Watanabe H, Matsuura T (2016) Construction of an in vitro gene screening system of the E. Coli EmrE transporter using liposome display. Anal Chem 88:12028–12035 [DOI] [PubMed]
- Varadamsetty G, Tremmel D, Hansen S, Parmeggiani F, Pluckthun A. Designed armadillo repeat proteins: library generation, characterization and selection of peptide binders with high specificity. J Mol Biol. 2012;424:68–87. doi: 10.1016/j.jmb.2012.08.029. [DOI] [PubMed] [Google Scholar]
- Voet AR, Noguchi H, Addy C, Simoncini D, Terada D, Unzai S, Park SY, Zhang KY, Tame JR. Computational design of a self-assembling symmetrical beta-propeller protein. Proc Natl Acad Sci U S A. 2014;111:15102–15107. doi: 10.1073/pnas.1412768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voet AR, Noguchi H, Addy C, Zhang KY, Tame JR. Biomineralization of a cadmium chloride Nanocrystal by a designed symmetrical protein. Angew Chem Int Ed. 2015;54:9857–9860. doi: 10.1002/anie.201503575. [DOI] [PubMed] [Google Scholar]
- Votteler J, Ogohara C, Yi S, Hsia Y, Nattermann U, Belnap DM, King NP, Sundquist WI. Designed proteins induce the formation of nanocage-containing extracellular vesicles. Nature. 2016;540:292–295. doi: 10.1038/nature20607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Honda S. Adaptive assembly: maximizing the potential of a given functional peptide with a tailor-made protein scaffold. Chem Biol. 2015;22:1165–1173. doi: 10.1016/j.chembiol.2015.07.015. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Yamasaki K, Honda S. Tracing primordial protein evolution through structurally guided stepwise segment elongation. J Biol Chem. 2014;289:3394–3404. doi: 10.1074/jbc.M113.530592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Kim S, Fela D, Baum J, Hecht MH. Solution structure of a de novo protein from a designed combinatorial library. Proc Natl Acad Sci U S A. 2003;100:13270–13273. doi: 10.1073/pnas.1835644100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MW, Wang W, Patterson J, Mancias JD, Beasley JR, Hecht MH. De novo amyloid proteins from designed combinatorial libraries. Proc Natl Acad Sci U S A. 1999;96:11211–11216. doi: 10.1073/pnas.96.20.11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead TA, Chevalier A, Song Y, Dreyfus C, Fleishman SJ, De Mattos C, Myers CA, Kamisetty H, Blair P, Wilson IA, Baker D. Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat Biotechnol. 2012;30:543–548. doi: 10.1038/nbt.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DS, Keefe AD, Szostak JW. The use of mRNA display to select high-affinity protein-binding peptides. Proc Natl Acad Sci U S A. 2001;98:3750–3755. doi: 10.1073/pnas.061028198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CW, Woolfson DN (2017) CCBuilder 2.0: powerful and accessible coiled-coil modeling. Protein Sci (in press) doi:10.1002/pro.3279 [DOI] [PMC free article] [PubMed]
- Wood CW, Bruning M, Ibarra AA, Bartlett GJ, Thomson AR, Sessions RB, Brady RL, Woolfson DN. CCBuilder: an interactive web-based tool for building, designing and assessing coiled-coil protein assemblies. Bioinformatics. 2014;30:3029–3035. doi: 10.1093/bioinformatics/btu502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfson DN, Bartlett GJ, Bruning M, Thomson AR (2012) New currency for old rope: from coiled-coil assemblies to α-helical barrels. Curr Opin Struct Biol 22:432–441 [DOI] [PubMed]
- Woolfson DN, Bartlett GJ, Burton AJ, Heal JW, Niitsu A, Thomson AR, Wood CW (2015) De novo protein design: how do we expand into the universe of possible protein structures? Curr Opin Struct Biol 33:16–26 [DOI] [PubMed]
- Wrenbeck EE, Faber MS, Whitehead TA. Deep sequencing methods for protein engineering and design. Curr Opin Struct Biol. 2017;45:36–44. doi: 10.1016/j.sbi.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi S, Akanuma S, Yamagishi M, Uchida T, Yamagishi A (2016) De novo design of protein–protein interactions through modification of inter-molecular helix–helix interface residues. Biochim Biophys Acta 1864:479–487 [DOI] [PubMed]
- Yagi S, Akanuma S, Yamagishi A (2017) Creation of artificial protein–protein interactions using α-helices as interface. Biophys Rev. 10.1007/s12551-017-0352-9 [DOI] [PMC free article] [PubMed]
- Yamagishi Y, Shoji I, Miyagawa S, Kawakami T, Katoh T, Goto Y, Suga H. Natural product-like macrocyclic N-methyl-peptide inhibitors against a ubiquitin ligase uncovered from a ribosome-expressed de novo library. Chem Biol. 2011;18:1562–1570. doi: 10.1016/j.chembiol.2011.09.013. [DOI] [PubMed] [Google Scholar]
- Yamaguchi J, Naimuddin M, Biyani M, Sasaki T, Machida M, Kubo T, Funatsu T, Husimi Y, Nemoto N. cDNA display: a novel screening method for functional disulfide-rich peptides by solid-phase synthesis and stabilization of mRNA-protein fusions. Nucleic Acids Res. 2009;37:e108. doi: 10.1093/nar/gkp514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates TO. Liu Y, Laniado J. The design of symmetric protein nanomaterials comes of age in theory and practice. Curr Opin Struct Biol. 2016;39:134–143. doi: 10.1016/j.sbi.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Yoshimoto N, Tatematsu K, Iijima M, Niimi T, Maturana AD, Fujii I, Kondo A, Tanizawa K, Kuroda S. High-throughput de novo screening of receptor agonists with an automated single-cell analysis and isolation system. Sci Rep. 2014;4:4242. doi: 10.1038/srep04242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanghellini A. de novo computational enzyme design. Curr Opin Biotechnol. 2014;29:132–138. doi: 10.1016/j.copbio.2014.03.002. [DOI] [PubMed] [Google Scholar]
- Zhu B, Mizoguchi T, Kojima T, Nakano H. Ultra-high-throughput screening of an in vitro-synthesized horseradish peroxidase displayed on microbeads using cell sorter. PLoS ONE. 2015;10:e0127479. doi: 10.1371/journal.pone.0127479. [DOI] [PMC free article] [PubMed] [Google Scholar]