Abstract
The bacterial flagellum is a motile organelle composed of thousands of protein subunits. The filamentous part that extends from the cell membrane is called the axial structure and consists of three major parts, the filament, hook, and rod, and other minor components. Each of the three main parts shares a similar self-assembly mechanism and a common basic architecture of subunit arrangement while showing quite distinct mechanical properties to achieve its specific function. Structural and molecular mechanisms to produce these various mechanical properties of the axial structure, such as the filament, the hook, and the rod, have been revealed by the complementary use of X-ray crystallography and cryo-electron microscopy. In addition, the mechanism of growth of the axial structure is beginning to be revealed based on the molecular structure.
Keywords: Bacterial flagellum, Axial structure, X-ray crystallography, Cryo-electron microscopy
Introduction
Gram-negative bacteria, such as Escherichia coli and Salmonella, swim by rotating helical filamentous organelles called the flagellum. The flagellum rotates at a speed of 200–300 Hz driven by a reversible rotary motor embedded in the cell membrane at the base of the filament. To drive rotation the motor utilizes an electrochemical potential gradient generated by differential proton accumulation across the cytoplasmic membrane.
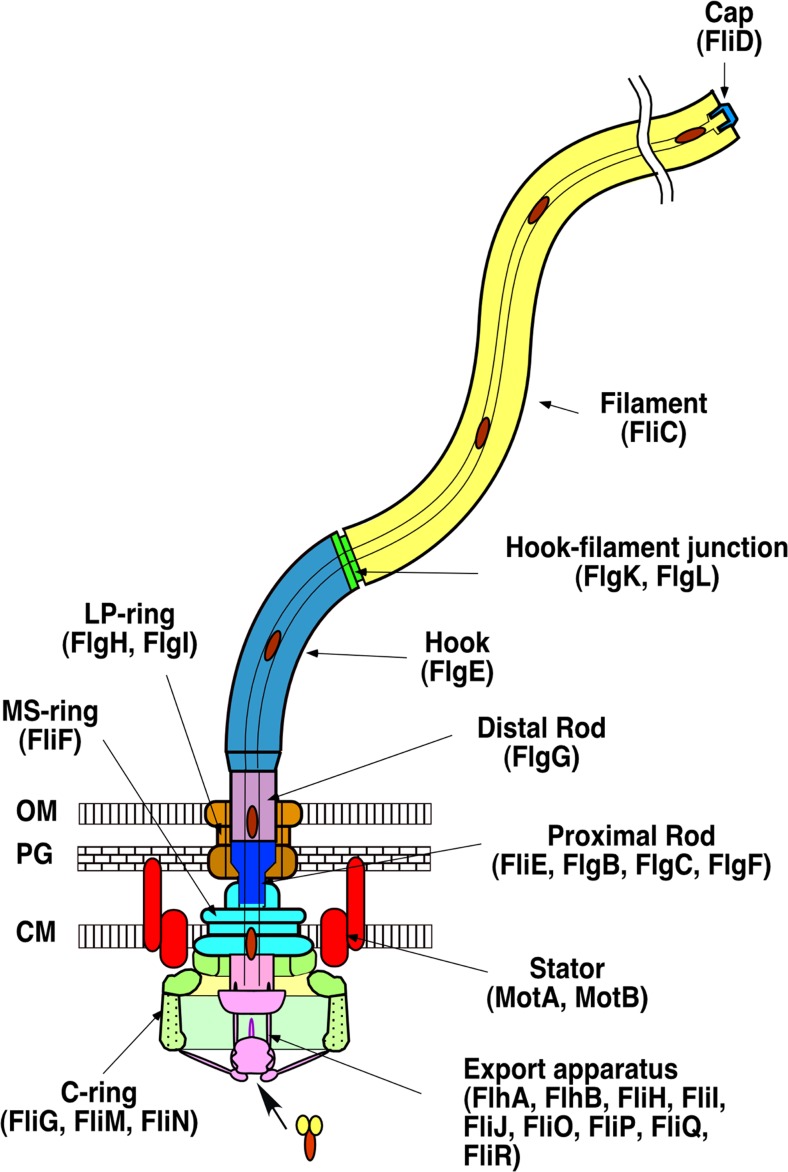
The flagellum is a huge molecular complex made of 20–30 thousand of protein subunits of about 30 different proteins. The structural components can be divided into two classes, the basal body rings and the tubular axial structure. The basal body rings form the rotary motor with the stator complex composed of cytoplasmic membrane proteins, MotA and MotB. The basal body rings consist of four major ring structures, the L-ring, the P-ring the MS-ring, and the C-ring. The MS- and C-ring form a rotor (Francis et al. 1994; Suzuki et al. 2004; Thomas et al. 2006) (Fig. 1), and torque is generated by the rotor–stator interaction that is coupled with proton flow through the proton pathway in the stator complex (Manson et al. 1977; Blair and Berg 1988).
Fig. 1.
Schematic drawing of the bacterial flagellum. Different colors represent different functional units. The names of the component proteins in each functional unit are shown in parentheses. CM Inner membrane, PG peptidoglycan layer, OM outer membrane
The axial structure consists of three major parts: the filament, the hook, and the rod (Fig. 1) (DePamphilis and Adler 1971a, b). The filament is a thin helical structure with a diameter of about 20 nm and typically grows to around 15 μm in length. It is composed of about 20,000 flagellar filament protein, flagellin (FliC), subunits and acts as a rigid propeller to produce thrust for the cell to swim in viscous environments. The hook is a short, curved segment whose length is approximately 55 nm. It is a helical assembly of flagellar hook protein (FlgE) subunits and acts as a universal joint that smoothly transmits the motor torque to the filament regardless of its orientation. For efficient transmission of torque, the hook is flexible with regards to bending but rigid against twisting. The rod is a drive shaft with an approximate length of 30 nm that connects the rotor rings and the hook. It rod is a rather complex helical cylinder composed of four flagellar rod proteins, FlgB, FlgC, FlgF, and FlgG (Homma et al. 1990; Kubori et al. 1992). The rod penetrates the peptidoglycan (PG) layer and the outer membrane through the lipoprotein (LP) ring complex, which works as a bushing. Between the filament and the hook, there is a short axial structural segment with thinner and smoother appearance, called the hook–filament junction. The junction is composed of FlgK and FlgL and thought to be an adaptor connecting the hook and the filament, which have mechanically distinct characteristics from each other.
All flagellar axial proteins synthesized in the cytoplasm are translocated into the central channel of the growing flagellum by the flagellar type III protein export apparatus. The axial proteins travel through the narrow channel to the distal end and are the incorporated into the growing structure (Macnab 2004; Minamino et al. 2008).
In this review, current understanding of the molecular mechanism of each part of the flagellar axial structure and its assembly mechanism are summarized based on the structure.
Common features of the flagellar axial structure
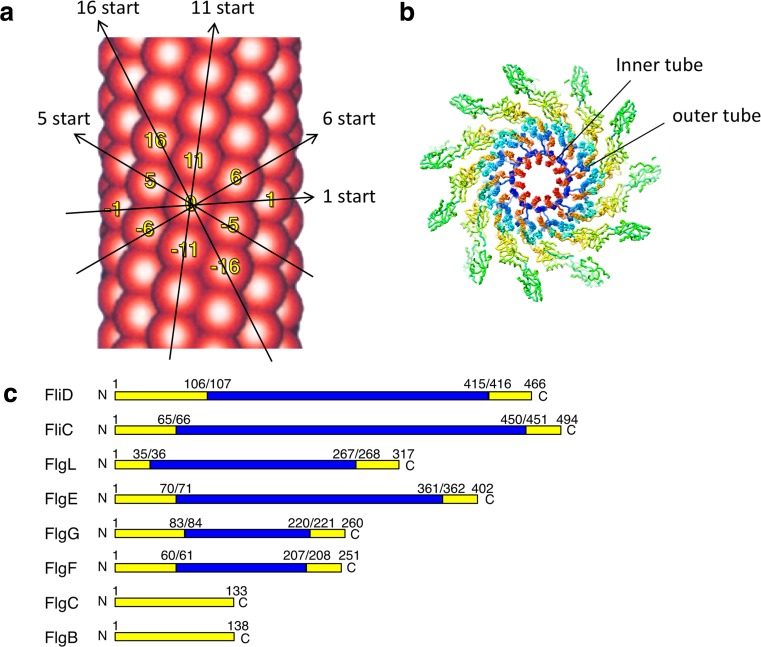
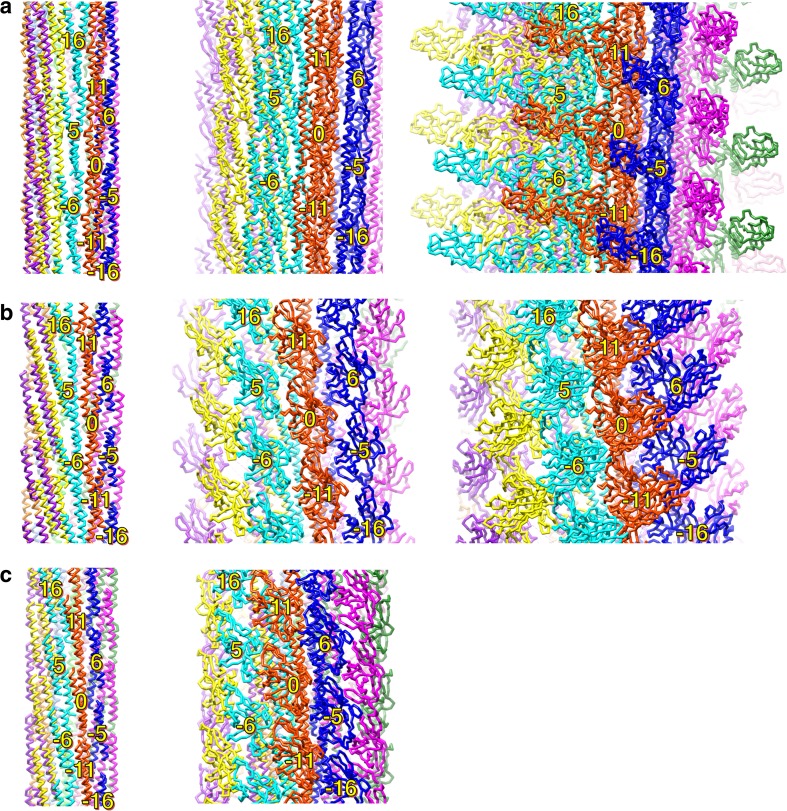
Early image analyses of electron micrographs revealed that the filament and the hook are tubular structures composed of 11 protofilaments, all of which are nearly axially aligned strands of subunit proteins (O’Brien and Bennett 1972; Wagenknecht et al. 1981, 1982). These tubular protofilaments can also be described as a helical array of 11 subunits in two turns of the 1-start helix (Fig. 2a). Subsequent structural studies indicated that they form a concentric multi-layer tube structure. The innermost tube is constructed of the N- and C- terminal regions of the component proteins (Mimori et al. 1995; Morgan et al. 1995; Mimori-Kiyosue et al. 1997; Shaikh et al. 2005) (Fig. 2b). Subunits of the rod and the hook–filament junction are also arranged in a similar helical manner in the flagellum (Shaikh et al. 2005).
Fig. 2.
Common structural feature of the flagellar axial structure. a Arrangement of the protein subunits in the flagellar filament. Major helical lines are indicated by arrows. The subunits along the 11-start helical line comprise the protofilament. The number labeled on the subunits represents the number of the subunit starting from the central subunit (subunit 0) along the 1-start helical line. The number also shows the direction of the helical line. The subunit monomers are sequentially assembled along the 1-start helix. b Cross section of the flagellar filament structure. Each flagellin subunit is colored in the rainbow sequence of colors from blue to red for the – to C-terminus. The N- (blue) and C-terminal (red) regions form the inner tube. c Disordered regions of flagellar axial proteins. Disordered region and folded domains are colored in yellow and blue, respectively. The disordered regions were determined by limited proteolysis (Saijo-Hamano et al. 2004)
The flagellar axial proteins share a conserved amino acid sequence motif in both terminal regions. In the N- and C-terminals about 65 and 45 residues, respectively, show heptad repeats of hydrophobic residues, suggesting that these regions form α-helical coiled-coil structures (Homma et al. 1990) The coiled-coil structures have been confirmed by high-resolution cryo-electron microscopy (cryo-EM) (Yonekura et al. 2003; Matsunami et al. 2016). However, these regions are unfolded in the monomeric state (Fig. 2c) and are folded into coiled-coils to form the inner tube structure only when the subunits assemble into the flagellum (Vonderviszt et al. 1992; Saijo-Hamano et al. 2004).
Structure of flagellin and the flagellar filament
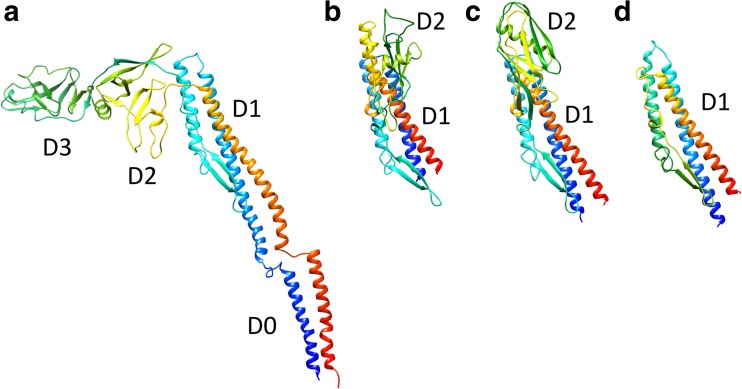
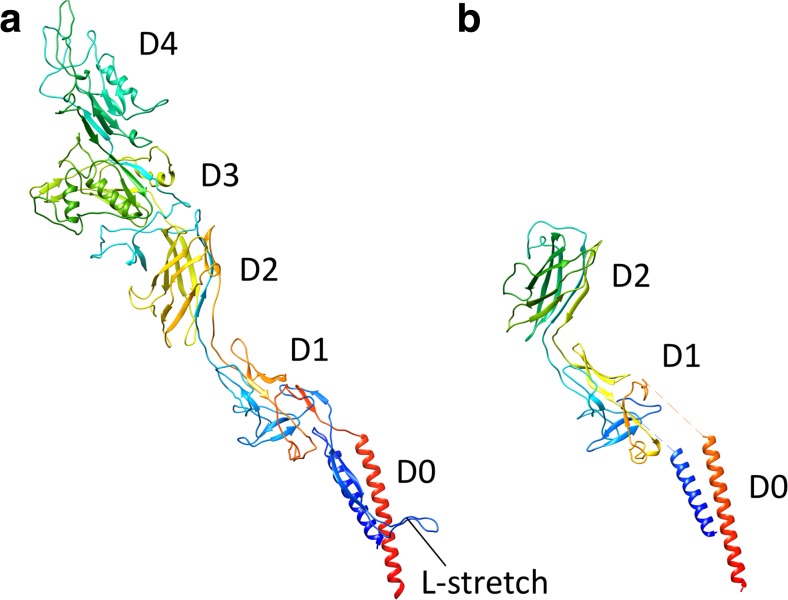
The FliC protein from Salmonella enterica serovar Typhimurium (St) consists of four domains, D0, D1, D2, and D3. The first atomic structure of FliC was determined by X-ray crystallography at 2.0-Å resolution from a 41-kDa fragment of StFliC that lacks the D0 domain (Samatey et al. 2001). Complete atomic structure models of straight filaments of mutant strains (SJW1655 and SJW1660) were subsequently obtained by cryo-EM at 4-Å resolution (Yonekura et al. 2003; Maki-Yonekura et al. 2010).
The amino acid chain of FliC begins from the D0 domain, goes through the D1, D2, and D3 domains, then back to D2 and D1, and terminates in D0. These four domains are radially arranged in the filament. The D0 domain is composed of the N-terminal and C-terminal helices, which form a coiled-coil structure (Fig. 2b). The D1 domain is made up of three helices and a β-hairpin, and forms the outer tube. The D0 and D1 domains are connected by the extended loop structures with a length of 20 Å, called the spoke. The D2 and D3 domains consist mainly of β-strands and protrude from the outer tube (Fig. 3). The helices in the D0 and D1 domains of neighboring subunits are closely packed, existing nearly parallel to the filament axis; they contribute to the stable filament formation. In contrast, the D2 and D3 domains are well separated from one another (Figs. 4, 5).
Fig. 3.
Structural comparison of flagellin from various bacteria. a Cryo-electron microscopy (Cryo-EM) structure of full-length flagellar filament protein, flagellin (FliC), from Salmonella (PDB ID 1ucu). b–d Crystal structures of Burkholderia pseudomallei FliC (BpFliC; PDB ID 4cfi) (b), Pseudomonas aeruginosa FliC (PaFliC; PDB ID 4nx9) (c), and Bacillus subtilis FliC (BsFliC; (PDB ID 5gy2) (d). The D0 domains of Bp-, Pa and Bs-FliC were removed for crystallization. The chains are colored in the rainbow sequence of colors from blue to red for the N- to C-terminus
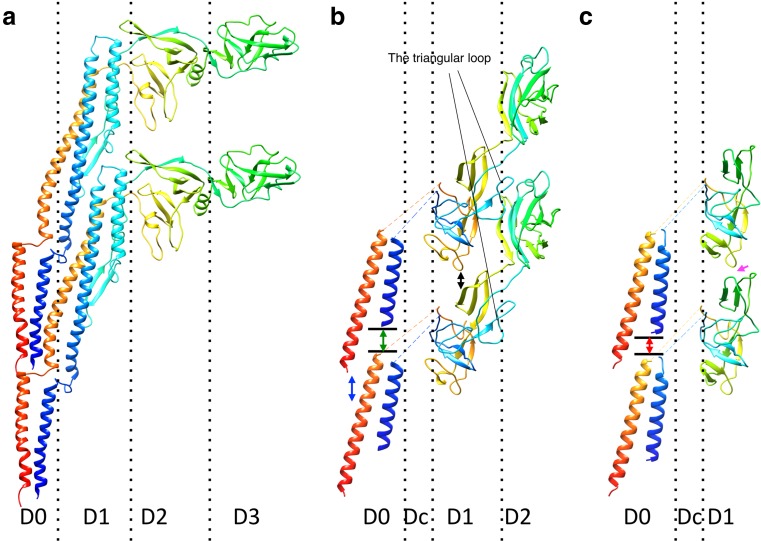
Fig. 4.
Comparison of the protofilament structures of the filament (a), the hook (b), and the rod (c). Two protein subunits in a protofilament are shown in each panel. The chains are colored in the rainbow sequence of colors from blue to red for the N- to C-terminus. The D0 and D1 helices of FliC are arranged nearly parallel to the filament axis and densely packed along the protofilament. The D0 helices of flagellar hook protein FlgE are tilted to form a gap between the axially neighboring subunits (blue two-direction arrow). The N-terminal helix of flagellar rod protein FlgG is one turn longer than that of FlgE; therefore the gap between the N-terminal helix and the axially neighboring subunit (red two-direction arrow) is shorter than that in the hook (green two-direction arrow). The D1 domains of FlgG are arranged upright to make an interaction with the neighboring subunit (magenta arrow), while those of FlgE in the hook are tilted, producing an axial gap along the protofilament (black two-direction arrow)
Fig. 5.
Intersubunit domain interactions in the axial structure. Side views of the R-type filament (a), the hook (b), and the rod (c) showing the subunit arrangement and packing of the different layers: left panel shows the array of the D0 domains; middle panel shows the D1 domains; right panel shows the D2 and D3 domains. Individual protofilaments are shown in different colors. The subunits surrounding subunit 0 are labeled with the number showing the direction of the helical line
From the helical symmetry of the filament, six nearest neighbor subunit interactions are expected for one subunit, two each along the 5-, 6-, and 11-start helical directions (Figs. 2a, 5). All these interactions were observed between the D0 domains in the inner tube. There is a slight inclination in the outer tube structure due to the nearly rhombohedral shape of the D1 domain; however, no contacts are observed along the 6-start between the D1 domains. Instead, there are 16-start interactions, which are thought to be important for polymorphic transition of the helical filament form (Samatey et al. 2001; Yonekura et al. 2003; Kitao et al. 2006). The subunit interactions between the D0 domains are hydrophobic, while the D1 domains mainly interact in a hydrophilic manner. These findings are consistent with the roles of each domain; the D0 domain stabilizes the filament structure, whereas the D1 domain is responsible for polymorphic transition by changing the interactions with the neighboring subunits.
The D2 and D3 domains show variation in sequence and structure, whereas the D0 and D1 domains are conserved among flagellins of various bacteria. Pseudomonas aeruginosa FliC (PaFliC) and Burkholderia pseudomallei FliC (BpFliC) lack the D3 domain (Song and Yoon 2014; Nithichanon et al. 2015), and Bacillus subtilis FliC (BsFliC) is composed of only the D0 and D1 domains (Song et al. 2017, b). The D2 domains of PaFliC and BpFliC project in the opposite direction from that of StFliC. Moreover, the D2 structures of PaFliC, BpFliC, and StFliC are quite different from each other (Fig. 3).
Bacterial flagellin induces an innate immune response by binding to the Toll-like receptor 5 (TLR5), an innate immune receptor present on the host cell surface (Hayashi et al. 2001). The structures of TLR5 in complex with FliC from Salmonella enterica serovar Dublin (SdFliC) or BsFliC revealed that TLR5 recognizes the conserved three D1 helices, N1 N2, and C1, of FliC. On the other hand, the variable D2 and D3 domains are considered to be involved in adaptive immunity (Yoon et al. 2012; Song et al. 2017, b).
Molecular mechanism of the polymorphic transition
The most interesting and important property of the flagellar filament is that it shows various polymorphic supercoiled forms, although it is composed of a single protein, flagellin. In response to various environmental stimuli, bacteria alternate their swimming behavior between the swimming and the tumbling mode. In the swimming mode, each filament with a left-handed supercoil rotates counter-clockwise and forms a bundle to thrust the cell (Macnab and Koshland 1972). The tumbling mode is triggered by a quick reversal of rotation (Larsen et al. 1974), which produces a twisting force that transforms the left-handed supercoil of the filament into a right-handed one, causing the bundle to fall apart rapidly (Macnab and Ornston 1977; Turner et al. 2000). Polymorphic transition of the supercoiled forms is also induced by changing the pH and ionic strength (Kamiya and Asakura 1976, 1977) and by applying mechanical load (Macnab and Ornston 1977; Hotani 1982).
The mechanism of the polymorphic transition was first proposed independently by Asakura and Calladine (Asakura 1970; Calladine 1975, 1976, 1978). These authors demonstrated that the supercoiled structures can be constructed by mixing the two distinct protofilaments and be switched into other supercoiled forms by cooperative transition of each protofilament between the two modes, thereby changing the number ratio of the two types of protofilaments. Various FliC mutants that produce distinct supercoiled filaments have been isolated, including straight filament forms. The mutant straight filaments can be divided into only two distinct types, the L- and R-type, respectively (Kamiya et al. 1979), based on their subunit packing. The L-type is composed of protofilaments tilted to the left with a longer inter-subunit repeat distance, whereas the R-type exhibit a tilt to the right with a shorter inter-subunit repeat distance. Careful measurements of the helical packing parameters of the straight filaments by X-ray fiber diffraction revealed that the difference in the inter-subunit distance along the protofilament between L- and R-type filaments is only 0.8 Å (Yamashita et al. 1998). Applying the obtained packing parameters of the two straight filaments, supercoiled structures were modeled and found to show good agreement with experimentally observed forms (Hasegawa et al. 1998), supporting the Asakura and Calladine theory. To change the supercoiled form, some of the protofilaments switch their repeat distance and tilt in a cooperative manner (Asakura 1970; Calladine 1975, 1976, 1978). Comparison of the local subunit packing of L- and R-type filaments revealed only subtle differences in local packing at the inner tube radius, whereas these differences are significant at the outer tube radius (Mimori-Kiyosue et al. 1996; Maki-Yonekura et al. 2010). Moreover, even after removal of the terminal residues forming the D0 domain, fragments of FliC were still able to form a helical structure called the “coil” (Vonderviszt et al. 1991). These results indicate that inter-subunit interactions of the D1 domain in the outer tube are primarily responsible for supercoiling. Simulated extension of the atomic model of the protofilament revealed that the β-hairpin in the D1 domain shows a small but significant conformational change, indicating that this is a possible lengthwise switching unit responsible for curvature formation (Samatey et al. 2001). This switch is supposed to be tightly coupled with another switch that regulates the twist of supercoils, which presumably resides in the lateral interactions between neighboring protofilaments.
The hook
The hook allows the synchronous rotation of several filaments driven by their motors in a bundle formed behind the cell (swimming) as well as the uncoordinated rotation of individual, unbundled filaments in different orientations (tumbling) (Macnab and Ornston 1977; Turner et al. 2000). The bending flexibility of the hook enables it to smoothly transmit torque from the motor to the filament when the two are not coaxial (Berg and Anderson 1973). In every revolution of the motor, each protofilament changes its conformation from the longer repeat state to the shorter one and from the shorter to the longer. Therefore, well-organized conformational changes are essential for smooth and continuous rotation.
The length of the hook is regulated to be 55 ± 6 nm (Hirano et al. 1994; Moriya et al. 2006). The appropriate length of the hook seems to be important for its universal joint function. Mutant cells with a longer hook, known as polyhook-filament mutants, or with a shorter hook show poor motility (Suzuki and Iino 1981; Hirano et al. 1994; Williams et al. 1996; Moriya et al. 2006).
Structure of the hook
The hook is a helical assembly of about 120 copies of a single protein, FlgE (Kutsukake et al. 1979; Jones et al. 1990). StFlgE is a 402-amino acid protein composed of three domains, D0, Dc, D1, and D2. The atomic structure of the D1 and D2 domains were determined by X-ray crystallography (Samatey et al. 2004). The D1 and D2 domains consist mostly of β-structures and are completely different from those of FliC (Figs. 4, 6). The D2 domain is composed of eight-stranded β-barrel with extra loops. The D1 domain has an unusual fold composed of complex β-hairpins and a unique triangular loop. The D1 and D2 domains are connected by a short stretch of a two-stranded anti-parallel β-sheet (Samatey et al. 2004) (Fig. 6). The arrangement of the FlgE subunits in the hook was revealed by docking the crystal structure into the cryo-EM density (Shaikh et al. 2005; Fujii et al. 2009). Compared with the filament structure, the subunits are loosely packed in the hook (Figs. 4, 5). The D2 domains strongly contact each other along the 6-start direction on the outer surface of the hook, but no D2–D2 interactions are found along the 11- and the 5-start direction (Fig. 5). Inter-subunit interactions between D1 domains are very weak in all directions. The main axial intermolecular interactions that hold the protofilament structure are between the D1 domain of the distal subunit and the D2 domain of the proximal subunit through the triangular loop of domain D1 (Fig. 4). The N- and C-terminal helices of FlgE form the D0 domain in a similar manner to those of FliC in the filament, but they are significantly tilted (Figs. 4, 5). This arrangement produces a gap of about 5 Å between the D0 helices of the axially neighboring subunits in the hook, allowing axial compression and extension of each protofilament (Fujii et al. 2009) (Fig. 4). These loose axial packing properties of the FlgE subunits are primarily responsible for high flexibility in bending and rigidity against twisting (Samatey et al. 2004; Fujii et al. 2009).
Fig. 6.
Structural comparison of the Campylobacter jejuni flagellar hook protein FlgE (CjFlgE; PDB ID 5jxl) (a) and Salmonella enterica serovar Typhimurium FlgE (StFlgE; PDB ID 3A69) (b). The chains connecting the D0 and D1 domains are not determined in the StFlgE structure. The chains are drawn in rainbow sequence of colors from blue to red for the N- to C-terminus
The structure of the Dc domain of the Salmonella hook is still unclear because of the low resolution of the cryoEM density. However, the complete cryoEM structure of Campylobacter jejuni (Cj) hook has recently been reported (Matsunami et al. 2016). Although the Cj-hook has two extra domains, which are located radially outside of the D2 domain, the structures of D0, D1, and D2 in the Cj-hook are very similar to those in the St-hook (Fig. 6). The D0 and D1 domains are connected by an L-shaped stretch (L-stretch) composed of 50 residues following the N-terminal helix. The L-stretch is in extensive contact with the D1 domain and the L-stretch of other subunits in four different protofilaments, thereby stabilizing the structure of the Cj-hook (Matsunami et al. 2016). As the L-stretch is located at the corresponding position of the Dc domain of the St-hook, it may also be present in the St-hook.
The hook–filament junction
The hook–filament junction is a buffering structure connecting the hook with the filament. Since the hook and the filament have mechanically and structurally distinct characteristics, the junction proteins must form a special link between the flexible hook and the rigid filament. Two proteins, FlgK and FlgL, form the hook–filament junction—in this order toward the distal end (Homma and Iino 1985a; Ikeda et al. 1987). Single FlgK and FlgL molecules are expected to be present in each of the 11 protofilaments (Ikeda et al. 1987; Jones et al. 1990). Analysis of the structure of a core fragment of FlgK from Burkholderia pseudomallei has revealed that the D1 domain is similar to that of FliC (Gourlay et al. 2015). The structures of the core fragments of FlgK and FlgL from Salmonella have also been determined at a resolution of 1.9 and 2.1 Å, respectively (PDB ID 2d4y and 2d4x, respectively), and the hook–filament junction model will be published elsewhere.
The rod
The rod is a drive shaft to transmit the torque to the hook. The rod is a rigid and straight cylinder penetrating the PG layer and the outer membrane. It consists of two substructures, the proximal rod and the distal rod (Fig. 1). The proximal rod, with a diameter of about 7 nm, penetrates into the FliF ring, and the distal rod, with a diameter of 13 nm. is surrounded by the LP ring. The proximal rod is composed of FlgB, FlgC, and FlgF, each with about 6 subunits, and the distal rod comprises approximately 26 subunits of the single protein FlgG (Homma et al. 1990; Kubori et al. 1992; Jones et al. 1990). FlgB and FlgC consist of less than 140 residues (Homma et al. 1990) and contain heptad repeats of hydrophobic residues in their terminal regions (Homma et al. 1990). These proteins are completely unfolded in solution, and their molecular sizes correspond to the size of the disordered regions of FlgE and FliC (Fig. 2c). Therefore, they are probably folded into α-helical coiled-coils to form the tube structure, similar to the D0 inner tube of FliC (Saijo-Hamano et al. 2004). FlgF and FlgG are composed of 251 and 260 residues, respectively, and both have a compact proteolytic-resistant region of 16–17 kDa as well as the common disordered terminal regions (Fig. 2c). The amino acid sequences of the compact region show a high similarity with the D1 domain of FlgE, and the size of the compact region is almost the same as that of the D1 domain of FlgE (Homma et al. 1990; Saijo-Hamano et al. 2004), suggesting that FlgF and FlgG form a double tube structure in which the compact region forms the outer tube.
The cryo-EM structure of the distal rod has recently been reported (Fujii et al. 2017). The 7-Å resolution density map shows the radially arranged domains, D0, Dc, and D1. Although the crystal structure of FlgG has not yet been reported (Saijo-Hamano et al. 2013), a partial structure model has been built by docking a D1 structure of FlgG produced by homology modeling into the density map. The N- and C-terminal helices in the D0 domain have also modeled in the density map (Fujii et al. 2017). The helical symmetry and repeat distance of the distal rod are almost the same as those of the hook (Fig. 5). However, slight but significant differences are found in the orientation of each domain. The D1 domain of FlgG is arranged upright to make a tight interaction between the neighboring subunits in the protofilament of the distal rod, while the D1 domains of FlgE in the hook are tilted about 7° compared with those of FlgG in the distal rod, producing an axial gap along the protofilament (Fig. 4). The N- and C-terminal helices of FlgG in the D0 domain form a coiled-coil structure in a similar manner to those of FlgE in the hook. However, the N-terminal helix of FlgG is one turn longer than that of FlgE and fills the gap, allowing direct contact with the axially neighboring subunit (Fig. 4). These tight packing interactions in both the D1 and D0 domains of FlgG provide rigidity to the distal rod. The Dc domain, which connects the D0 and D1 domains, is not modeled due to the low resolution of the density map. The FlgG-specific insertion of 18 residues is located in the Dc domain. These 18 residues are important to making the rod rigid and straight because the FlgE mutant with the 18-residue insertion produces a straight and rigid hook (Hiraoka et al. 2017). Therefore, a high-resolution structure of the rod is needed to further our understanding of the molecular basis of its mechanical properties.
Construction of the axial structure
Construction of the bacterial flagellum is a highly organized process. Assembly of the flagellar substructures proceeds in a sequential manner, starting from the proximal basal body ring and continuing to the distal axial structures (Kubori et al. 1992). The distal substructure is built just after completion of the proximal structure. The initial event of the flagellar construction is the formation of the MS ring together with the flagellar protein export apparatus in the cytoplasmic membrane; this is followed in sequential order by the construction of the rod, the hook, the hook–filament junction, and the filament. The axial component proteins exported through the central channel of the growing structure assemble onto the distal tip (Macnab 2003).
The assembly processes of the filament and the hook are promoted by specific cap complexes composed of FliD and FlgD, respectively (Ikeda et al. 1987; Ohnishi et al. 1994). These caps are both homo-pentameric complexes stably attached to the distal end of the filament or the hook and they prevent leakage of their component proteins to the medium by capping the central channel (Ikeda et al. 1984, 1985, 1987, 1996; Homma and Iino 1985b). Thereby, the transported proteins are efficiently incorporated into the growing structure just beneath the cap complex. The rod is also expected to require its specific cap complex for assembly (Hirano et al. 2001).
The rod cap
Since the rod penetrates the PG layer, the rod cap has to possess PG-hydrolyzing activity to enable penetration of the PG layer. Among all of the flagella-related proteins identified to date, FlgJ is the only protein with a known PG-hydrolyzing activity , and it is required for rod formation. Thus, FlgJ is considered to be a capping protein for the rod (Hirano et al. 2001). StFlgJ is a 34-kDa protein composed of 316 residues. The N-terminal region has a heptad repeat of hydrophobic residues that is shared by other axial proteins, and the N-terminal half is essential for rod assembly, suggesting that the N-terminal half of FlgJ acts as the cap for rod assembly (Hirano et al. 2001). The C-terminal half has a PG-hydrolyzing activity (Nambu et al. 1999) and shows a sequence similarity to other muramidases, such as 1-4-b-N-acetylmuramidase and muramidase-2 (Joris et al. 1992; Buist et al. 1995). The structure of the muramidase domain of StFlgJ has recently been solved and resembles the structure of the lysozyme (Zaloba et al. 2016).
The hook cap and the hook formation
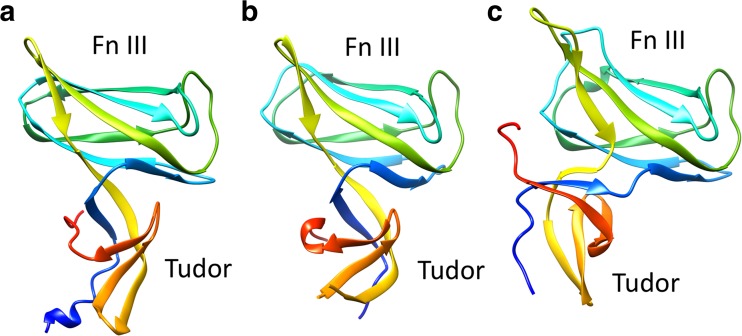
On the distal growing end of the hook, FlgD, a scaffolding protein, forms a capping complex for the flagellar hook assembly (Kubori et al. 1992; Ohnishi et al. 1994). The FlgD subunits may assemble onto the tip of the distal rod after removal of the rod cap, and then FlgE subunits are incorporated into the growing structure beneath the FlgD cap until the hook length reaches around 55 nm. The length of the hook is occasionally monitored by FliK through the interaction between FlgD and the extended N-terminal region of FliK (Moriya et al. 2006). After completion of the hook assembly, FlgD is replaced by a hook–filament junction protein FlgK (Ohnishi et al. 1994). StFlgD comprises 232 residues and its molecular weight is 24 kDa. Only the N-terminal 86 residues of StFlgD are sufficient for hook formation, suggesting that the N-terminal region interacts with the hook and is essential for promoting hook assembly (Kutsukake and Doi 1994). The remaining C-terminal region contributes to blocking the leakage of the transported FlgE into the medium for efficient assembly of the hook (Moriya et al. 2011). The crystal structures of the C-terminal fragments of FlgD from Xanthomonas campestris (XcFlgDc), Pseudomonas aeruginosa (PaFlgDc), and Helicobacter pylori (HpFlgDc) have been determined (Kuo et al. 2008; Zhou et al. 2011; Pulić et al. 2016) (Fig. 7). The structures of these fragments are basically identical and composed of two β-domains, the Tudor-like domain and the fibronectin type III-like domain. The HpFlgDc forms a tetramer both in crystal and in solution, suggesting species-specific variation of the oligomeric state.
Fig. 7.
Crystal structures of the C-terminal domain of FlgD (FlgDc), a scaffolding protein for flagellar hook assembly, from various bacteria. a Xanthomonas campestris (XcFlgDc; PDB ID 3c12), b Pseudomonas aeruginosa (PaFlgDc; PDB ID 3osv), c Helicobacter pylori (HpFlgDc; PDB ID 4zzf). The chains are shown in rainbow sequence of colors from blue to red for the N- to C-terminus
The filament cap and filament growth
At the distal tip of the filament, FliD forms a star-shaped homo-pentamer that caps the open end of the filament and assists inthe assembly of flagellin below the pentameric assembly (Ikeda et al. 1985, 1993, 1996; Maki et al. 1998) (Fig. 1). FliD was originally found to be an inhibitor of in vitro flagellin polymerization (Ikeda et al. 1984), but necessary for filament growth in vivo. (Homma et al. 1984). Before starting filament growth, FlgK and FlgL are assembled on the distal end of the hook in this order, then FliD is attached to FlgL (Homma and Iino 1985a; Ikeda et al. 1987). FliD can form a hetero-dimer with FlgL in solution (Furukawa et al. 2002), suggesting that FliD strongly binds to the distal end of the growing flagellum.
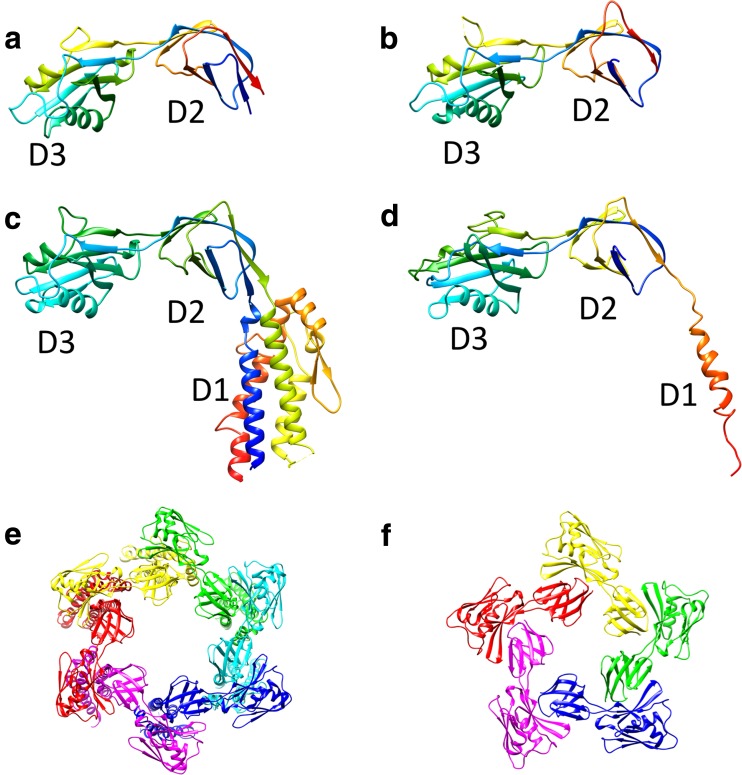
FliD is a 50-kDa protein with 466 residues and has flexible terminal regions that contain heptad repeats of hydrophobic amino acids (Homma et al. 1990). FliD exists in an equilibrium form between the monomer and the decamer, which is a bipolar pair of the pentamer, in solution (Imada et al. 1998). The terminal regions are responsible for interaction with the filament, as well as the association of the pentamer into the decamer (Vonderviszt et al. 1998). Cryo-EM studies have revealed that the FliD pentamer cap consists of a flat plate that is 145 Å in width and 30 Å in thickness, with five prolonged leg domains over a length of 125 Å (Maki-Yonekura et al. 2003). The crystal structure of E. coli FliD, which lacks both terminal regions, showed that FliD consists of three domains, D1, D2, and D3 (Song et al. 2017, b) (Fig. 8). The D1 domain consists of a four-helix bundle core decorated with one helix and two β-strands, and forms the leg of the cap. The D2 and D3 domains are mainly composed of β-strands, and form the flat plate. The crystal structure of the pentamer plate of StFliD revealed that the oligomerization interface is located between the D2 domain and the D3 domain of the neighboring subunit (Song et al. 2017, b) (Fig. 7).
Fig. 8.
Crystal structures of the capping protein of the bacterial flagellar filament (FliD) from various bacteria. Ribbon drawings of StFliD (PDB ID 5h5t) (a), Serratia marcescens FliD (SmFliD; PDB ID 5xlk) (b), Escherichia coli FliD (EcFliD; PDB ID 5h5v) (b), and PaFliD (PDB ID 5fhy) (d) are shown in rainbow sequence of colors from blue to red for the N- to C-terminus. e, f Structure of the FliD cap assembly: e EcFliD hexamer (PDB ID 5h5v), f StFliD pentamer (PDB ID 5h5t). Subunits are represented in different colors
The structure of the cap attached onto the filament was observed by cryo-EM, and an assembly mechanism of the FliC with the help of the cap was proposed (Yonekura et al. 2000). In this mechanism, the leg domains are plugged into the filament and attached to the filament end. Since the filament is composed of 11 protofilamens, four pairs of up-and-down pattern and one double-down step are present at the distal end of the filament. The leg domains of FliD fill the indentations but not the double-down step. Therefore, a large opening is present on the filament surface. A new subunit is probably incorporated into the opening and pushes up the cap. Because of the symmetry mismatch between the fivefold cap and the helical filament with the array of 11 subunits per two turns of the 1-start helix, the five leg domains adopt different conformational interactions with the FliC subunits. As a result, the addition of each FliC subunit induces rotation of the cap to the opposite direction of filament addition (Yonekura et al. 2000 Maki-Yonekura et al. 2003). Recently, unexpected variation was found in the number of the subunits in the FliD cap complex. E. coli FliD (EcFliD) and PaFliD form a hexamer and SmFliD (FliD from Serratia marcescens) forms a tetramer in the crystal (Song et al. 2017, b; Postel et al. 2016; Cho et al. 2017), whereas StFliD assembles into a pentamer (Fig. 8). How EcFliD, PaFliD, and SmFliD fit on the tip of the filament is not yet known.
Perspective
The flagellum is composed of about 30 different proteins, each with copy number ranging from a few to a few tens of thousands. Consequently, structure analysis of this huge system is challenging. Since the mechanical property of each part of the flagellar axial structure appears only in the assembled form, the structure of whole complex is indispensable for an understanding of each function. The complementary approach using X-ray crystallography and cryo-EM has revealed the structures of the straight filament, the hook, and the rod, which have provided deep insights into the molecular mechanisms of their functions at the atomic level. However, for complete understanding of this system, further research efforts are required. The supercoil structures of the filament, the curved hook structure, and the structure of the junctions between the filament and the hook and between the hook and the rod will be the next targets. Recent developments in the cryo-EM techniques will greatly facilitate such research. The whole atomic model of the flagellar axial structure will be obtained in near future, and this structure will provide design principles of complex biomolecular assemblies. The principles will be applicable to bio-nanotechnology, such as the design of functional, self-assembling nanostructures and the construction of flagella-based nanotube systems.
Acknowledgements
The author thanks Keiichi Namba and Tohru Minamino for their encouragement and fruitful discussion. This work was partially supported by the Japanese Ministry of Education, Culture, Sports, Science and Technology.
Compliance with ethical standards
Conflict of interest
Katsumi Imada declares that he has no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by the author.
Footnotes
This article is part of a Special Issue on ‘Biomolecules to Bio-nanomachines—Fumio Arisaka 70th Birthday’ edited by Damien Hall, Junichi Takagi and Haruki Nakamura.
References
- Asakura S. Polymerization of flagellin and polymorphism of flagella. Advan Biophys (Japan) 1970;1:99–155. [PubMed] [Google Scholar]
- Berg HC, Anderson RA. Bacteria swim by rotating their flagellar filaments. Nature. 1973;245:380–382. doi: 10.1038/245380a0. [DOI] [PubMed] [Google Scholar]
- Blair DF, Berg HC. Restoration of torque in defective flagellar motors. Science. 1988;242:1678–1681. doi: 10.1126/science.2849208. [DOI] [PubMed] [Google Scholar]
- Buist G, Kok J, Leenhouts KJ, Dabrowska M, Venema G, Haandrikman AJ. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J Bacteriol. 1995;177:1554–1563. doi: 10.1128/jb.177.6.1554-1563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calladine CR. Construction of bacterial flagella. Nature. 1975;225:121–124. doi: 10.1038/255121a0. [DOI] [PubMed] [Google Scholar]
- Calladine CR. Design requirements for the construction of bacterial flagella. J Theor Biol. 1976;57:469–489. doi: 10.1016/0022-5193(76)90016-3. [DOI] [PubMed] [Google Scholar]
- Calladine CR. Change of waveform in bacterial flagella: the role of mechanics at the molecular level. J Mol Biol. 1978;118:457–479. doi: 10.1016/0022-2836(78)90285-1. [DOI] [Google Scholar]
- Cho SY, Song WS, Hong HJ, Lee GS, Kang SG, Ko HJ, Kim PH, Yoon SI. Tetrameric structure of the flagellar cap protein FliD from Serratia Marcescens. Biochem Biophys Res Commun. 2017;489:63–69. doi: 10.1016/j.bbrc.2017.05.093. [DOI] [PubMed] [Google Scholar]
- DePamphilis ML, Adler J. Purification of intact flagella from Escherichia coli and Bacillus subtilis. J Bacteriol. 1971;105:376–383. doi: 10.1128/jb.105.1.376-383.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis ML, Adler J. Fine structure and isolation of the hook–basal body complex of flagella from Escherichia coli and Bacillus subtilis. J Bacteriol. 1971;105:384–395. doi: 10.1128/jb.105.1.384-395.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NR, Sosinsky GE, Thomas D, DeRosier DJ. Isolation, characterization, and structure of bacterial flagellar motors containing the switch complex. J Mol Biol. 1994;235:1261–1270. doi: 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- Fujii T, Kato T, Hiraoka KD, Miyata T, Minamino T, Chevance FF, Hughes KT, Namba K. Identical folds used for distinct mechanical functions of the bacterial flagellar rod and hook. Nat Commun. 2017;8:14276. doi: 10.1038/ncomms14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T, Kato T, Namba K. Specific arrangement of alpha-helical coiled coils in the core domain of the bacterial flagellar hook for the universal joint function. Structure. 2009;17:1485–1493. doi: 10.1016/j.str.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Furukawa Y, Imada K, Vonderviszt F, Matsunami H, Sano K-I, Kutsukake K, Namba K. Interactions between bacterial flagellar axial proteins in their monomeric state in solution. J Mol Biol. 2002;318:889–900. doi: 10.1016/S0022-2836(02)00139-0. [DOI] [PubMed] [Google Scholar]
- Gourlay LJ, Thomas RJ, Peri C, Conchillo-Solé O, Ferrer-Navarro M, Nithichanon A, Vila J, Daura X, Lertmemongkolchai G, Titball R, Colombo G, Bolognesi M. From crystal structure to in silico epitope discovery in the Burkholderia pseudomallei flagellar hook-associated protein FlgK. FEBS J. 2015;282:1319–1333. doi: 10.1111/febs.13223. [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Yamashita I, Namba K. Quasi- and nonequivalence in the structure of bacterial flagellar filament. Biophys J. 1998;74:569–575. doi: 10.1016/S0006-3495(98)77815-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, Eng JK, Akira S, Underhill DM, Aderem A. The innate immune response to bacterial flagellin is mediated by toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- Hirano T, Minamino T, Macnab RM. The role in flagellar rod assembly of the N-terminal domain of Salmonella FlgJ, a flagellum-specific muramidase. J Mol Biol. 2001;312:359–369. doi: 10.1006/jmbi.2001.4963. [DOI] [PubMed] [Google Scholar]
- Hirano T, Yamaguchi S, Oosawa K, Aizawa S-I. Roles of FliK and FlhB in the determination of flagellar hook length in Salmonella typhimurium. J Bacteriol. 1994;176:5439–5449. doi: 10.1128/jb.176.17.5439-5449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka KD, Morimoto YV, Inoue Y, Fujii T, Miyata T, Makino F, Minamino T, Namba K. Straight and rigid flagellar hook made by insertion of the FlgG specific sequence into FlgE. Sci Rep. 2017;7:46723. doi: 10.1038/srep46723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M, DeRosier DJ, Macnab RM. Flagellar hook and hook-associated proteins of Salmonella typhimurium and their relationship to other axial components of the flagellum. J Mol Biol. 1990;213:819–832. doi: 10.1016/S0022-2836(05)80266-9. [DOI] [PubMed] [Google Scholar]
- Homma M, Fujita H, Yamaguchi S, Iino T. Excretion of unassembled flagellin by Salmonella typhimurium mutants deficient in hook-associated proteins. J Bacteriol. 1984;159:1056–1059. doi: 10.1128/jb.159.3.1056-1059.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M, Kutsukake K, Hasebe M, Iino T, Macnab RM. FlgB, FlgC, FlgF and FlgG. A family of structurally related proteins in the flagellar basal body of Salmonella typhimurium. J Mol Biol. 1990;211:465–477. doi: 10.1016/0022-2836(90)90365-S. [DOI] [PubMed] [Google Scholar]
- Homma M, Iino T. Locations of hook-associated proteins in flagellar structures of Salmonella typhimurium. J Bacteriol. 1985;162:183–189. doi: 10.1128/jb.162.1.183-189.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M, Iino T. Excretion of unassebmled hook-associated proteins by Salmonella typhimurium. J Bacteriol. 1985;164:1370–1372. doi: 10.1128/jb.164.3.1370-1372.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotani H. Micro-video study of moving bacterial flagellar filaments III. Cyclic transformation induced by mechanical force. J Mol Biol. 1982;156:791–806. doi: 10.1016/0022-2836(82)90142-5. [DOI] [PubMed] [Google Scholar]
- Ikeda T, Asakura S, Kamiya R. "Cap" on the tip of Salmonella flagella. J Mol Biol. 1985;184:735–737. doi: 10.1016/0022-2836(85)90317-1. [DOI] [PubMed] [Google Scholar]
- Ikeda T, Homma M, Iino T, Asakura S, Kamiya R. Localization and stoichiometry of hook-associated proteins within Salmonella typhimurium flagella. J Bacteriol. 1987;169:1168–1173. doi: 10.1128/jb.169.3.1168-1173.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T, Kamiya R, Yamaguchi S. In vitro polymerization of flagellin excreted by a short-flagellum Salmonella typhimurium mutant. J Bacteriol. 1984;159:787–789. doi: 10.1128/jb.159.2.787-789.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T, Oosawa K, Hotani H. Self-assembly of the filament capping protein, FliD, of bacterial flagella into an annular structure. J Mol Biol. 1996;259:679–686. doi: 10.1006/jmbi.1996.0349. [DOI] [PubMed] [Google Scholar]
- Ikeda T, Yamaguchi S, Hotani H. Flagellar growth in a filament-less Salmonella fliD mutant supplemented with purfied hook-associated protein 2. J Bacteriol. 1993;114:39–44. doi: 10.1093/oxfordjournals.jbchem.a124136. [DOI] [PubMed] [Google Scholar]
- Imada K, Vonderviszt F, Furukawa Y, Oosawa K, Namba K. Assembly charcteristics of flagellar cap protein HAP2 of Salmonella: decamer and pentamer in the pH-sensitive equilibrium. J Mol Biol. 1998;277:883–891. doi: 10.1006/jmbi.1998.1662. [DOI] [PubMed] [Google Scholar]
- Jones CJ, Macnab RM, Okino H, Aizawa S-I. Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella typhimurium. J Mol Biol. 1990;212:377–387. doi: 10.1016/0022-2836(90)90132-6. [DOI] [PubMed] [Google Scholar]
- Joris B, Englebert S, Chu C-P, Kariyama R, Daneo-Moore L, Shockman GD, Ghuysen J-M. Modular design of the enterococcus hirae muramidase-2 and Streptococcus faecalis autolysin. FEMS Microbiol Lett. 1992;91:257–264. doi: 10.1111/j.1574-6968.1992.tb05218.x. [DOI] [PubMed] [Google Scholar]
- Kamiya R, Asakura S. Helical transformations of Salmonella flagellain vitro. J Mol Biol. 1976;106:167–186. doi: 10.1016/0022-2836(76)90306-5. [DOI] [PubMed] [Google Scholar]
- Kamiya R, Asakura S. Flagellar transformations at alkaline pH. J Mol Biol. 1977;108:513–518. doi: 10.1016/S0022-2836(76)80133-7. [DOI] [PubMed] [Google Scholar]
- Kamiya R, Asakura S, Wakabayashi K, Namba K. Transition of bacterial flagella from helical to straight forms with different subunit arrangements. J Mol Biol. 1979;131:725–742. doi: 10.1016/0022-2836(79)90199-2. [DOI] [PubMed] [Google Scholar]
- Kitao A, Yonekura K, Maki-Yonekura S, Samatey FA, Imada K, Namba K, Go N. Switch interactions control energy frustration and multiple flagellar filament structures. Proc Natl Acad Sci USA. 2006;103:4894–4899. doi: 10.1073/pnas.0510285103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori T, Shimamoto N, Yamaguchi S, Namba K, Aizawa S-I. Morphological pathway of flagellar assembly in Salmonella typhimurium. J Mol Biol. 1992;226:433–446. doi: 10.1016/0022-2836(92)90958-M. [DOI] [PubMed] [Google Scholar]
- Kuo WT, Chin KH, Lo WT, Wang AH, Chou SH. Crystal structure of the C-terminal domain of a flagellar hook-capping protein from Xanthomonas campestris. J Mol Biol. 2008;381:189–199. doi: 10.1016/j.jmb.2008.05.083. [DOI] [PubMed] [Google Scholar]
- Kutsukake K, Doi H. Nucleotide sequence of the flgD gene of Salmonella typhimurium which is essential for flagellar hook formation. Biochim Biophys Acta. 1994;1218:443–446. doi: 10.1016/0167-4781(94)90202-X. [DOI] [PubMed] [Google Scholar]
- Kutsukake K, Suzuki T, Yamaguchi S, Iino T. Role of gene flaFV on flagellar hook formation in Salmonella typhimurium. J Bacteriol. 1979;140:267–275. doi: 10.1128/jb.140.1.267-275.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen SH, Reader RW, Kort EN, Tso W-W, Adler J. Change in direction of flagellar rotation is the basis of the chemotactic response in Escherichia coli. Nature. 1974;249:74–77. doi: 10.1038/249074a0. [DOI] [PubMed] [Google Scholar]
- Macnab RM. How bacteria assemble flagella. Annu Rev Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- Macnab RM. Type III flagellar protein export and flagellar assembly. Biochim Biophys Acta. 2004;1694:207–217. doi: 10.1016/j.bbamcr.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Macnab RM, Koshland DE., Jr The gradient-sensing mechanism in bacterial chemotaxis. Proc Natl Acad Sci USA. 1972;69:2509–2512. doi: 10.1073/pnas.69.9.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab RM, Ornston MK. Normal-to-curly flagellar transitions and their role in bacterial tumbling. Stabilization of an alternative quaternary structure by mechanical force. J Mol Biol. 1977;112:1–30. doi: 10.1016/S0022-2836(77)80153-8. [DOI] [PubMed] [Google Scholar]
- Maki S, Vonderviszt F, Furukawa Y, Imada K, Namba K. Plugging interactions of HAP2 pentamer into the distal end of flagellar filament revealed by electron microscopy. J Mol Biol. 1998;277:771–777. doi: 10.1006/jmbi.1998.1663. [DOI] [PubMed] [Google Scholar]
- Maki-Yonekura S, Yonekura K, Namba K. Domain movements of HAP2 in the cap-filament complex formation and growth process of the bacterial flagellum. Proc Natl Acad Sci USA. 2003;100:15528–15533. doi: 10.1073/pnas.2534343100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki-Yonekura S, Yonekura K, Namba K. Conformational change of flagellin for polymorphic supercoiling of the flagellar filament. Nat Struct Mol Biol. 2010;17:417–422. doi: 10.1038/nsmb.1774. [DOI] [PubMed] [Google Scholar]
- Manson MD, Tedesco P, Berg HC, Harold FM, Van der Drift C. A protonmotive force drives bacterial flagella. Proc Natl Acad Sci USA. 1977;4:3060–3064. doi: 10.1073/pnas.74.7.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami H, Barker CS, Yoon YH, Wolf M, Samatey FA. Complete structure of the bacterial flagellar hook reveals extensive set of stabilizing interactions. Nat Commun. 2016;7:13425. doi: 10.1038/ncomms13425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori Y, Yamashita I, Murata K, Fujiyoshi Y, Yonekura K, Toyoshima C, Namba K. The structure of the R-type straight flagellar filament of Salmonella at 9 Å resolution by electron cryomicroscopy. J Mol Biol. 1995;249:69–87. doi: 10.1006/jmbi.1995.0281. [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Vonderviszt FC, Namba K. Location of terminal segments of flagellin in the filament structure and their roles in polymerization and polymorphism. J Mol Biol. 1997;270:222–237. doi: 10.1006/jmbi.1997.1111. [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Vonderviszt F, Yamashita I, Fujiyoshi Y, Namba K. Direct interaction of flagellin termini essential for polymorphic ability of flagellar filament. Proc Natl Acad Sci USA. 1996;93:15108–15113. doi: 10.1073/pnas.93.26.15108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Imada K, Namba K. Mechanisms of type III protein export for bacterial flagellar assembly. Mol BioSyst. 2008;4:1105–1115. doi: 10.1039/b808065h. [DOI] [PubMed] [Google Scholar]
- Morgan DG, Owen C, Melanson LA, DeRosier DJ. Structure of bacterial flagellar filaments at 11 Å resolution: packing of the α-helices. J Mol Biol. 1995;249:88–110. doi: 10.1006/jmbi.1995.0282. [DOI] [PubMed] [Google Scholar]
- Moriya N, Minamino T, Hughes KT, Macnab RM, Namba K. The type III flagellar export specificity switch is dependent on FliK ruler and a molecular clock. J Mol Biol. 2006;359:466–477. doi: 10.1016/j.jmb.2006.03.025. [DOI] [PubMed] [Google Scholar]
- Moriya N, Minamino T, Imada K, Namba K. Genetic analysis of the bacterial hook-capping protein FlgD responsible for hook assembly. Microbiology. 2011;157(Pt 5):1354–1362. doi: 10.1099/mic.0.047100-0. [DOI] [PubMed] [Google Scholar]
- Nambu T, Minamino T, Macnab RM, Kutsukake K. Peptidoglycan-hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium. J Bacteriol. 1999;181:1555–1561. doi: 10.1128/jb.181.5.1555-1561.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nithichanon A, Rinchai D, Gori A, Lassaux P, Peri C, Conchillio-Solé O, Ferrer-Navarro M, Gourlay LJ, Nardini M, Vila J, Daura X, Colombo G, Bolognesi M, Lertmemonkolchai G. Sequence- and structure-based Immunoreactive epitope discovery for Burkholderia pseudomallei Flagellin. PLoS Negl Trop Dis. 2015;9:e0003917. doi: 10.1371/journal.pntd.0003917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien EJ, Bennett PM. Structure of straight flagella from a mutant Salmonella. J Mol Biol. 1972;73:133–152. doi: 10.1016/0022-2836(72)90168-4. [DOI] [PubMed] [Google Scholar]
- Ohnishi K, Ohto Y, Aizawa S-I, Macnab RM, Iino T. FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J Bacteriol. 1994;176:2272–2281. doi: 10.1128/jb.176.8.2272-2281.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postel S, Deredge D, Bonsor DA, Yu X, Diederichs K, Helmsing S, Vromen A, Friedler A, Hust M, Egelman EH, Beckett D, Wintrode PL, Sundberg EJ (2016) Bacterial flagellar capping proteins adopt diverse oligomeric states. Elife 5. 10.7554/eLife.18857 [DOI] [PMC free article] [PubMed]
- Pulić I, Cendron L, Salamina M, Polverino de Laureto P, Matković-Čalogović D, Zanotti G. Crystal structure of truncated FlgD from the human pathogen helicobacter pylori. J Struct Biol. 2016;194:147–155. doi: 10.1016/j.jsb.2016.02.003. [DOI] [PubMed] [Google Scholar]
- Saijo-Hamano Y, Matsunami H, Namba K, Imada K. Expression, purification, crystallization and preliminary X-ray diffraction analysis of a core fragment of FlgG, a bacterial flagellar rod protein. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69(Pt 5):547–550. doi: 10.1107/S1744309113008075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo-Hamano Y, Uchida N, Namba K, Oosawa K. In vitro characterization of FlgB, FlgC, FlgF, FlgG, and FliE, flagellar basal body proteins of Salmonella. J Mol Biol. 2004;339:423–435. doi: 10.1016/j.jmb.2004.03.070. [DOI] [PubMed] [Google Scholar]
- Samatey FA, Imada K, Nagashima S, Vonderviszt F, Kumasaka T, Yamamoto M, Namba K. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature. 2001;410:331–337. doi: 10.1038/35066504. [DOI] [PubMed] [Google Scholar]
- Samatey FA, Matsunami H, Imada K, Nagashima S, Shaikh TR, Thomas DR, Chen JZ, DeRosier DJ, Kitao A, Namba K. Structure of the bacterial flagellar hook and implication for the molecular universal joint mechanism. Nature. 2004;431:1062–1068. doi: 10.1038/nature02997. [DOI] [PubMed] [Google Scholar]
- Shaikh TR, Thomas DR, Chen JZ, Samatey FA, Matsunami H, Imada K, Namba K, DeRosier DJ. A partial atomic structure for the flagellar hook of Salmonella typhimurium. Proc Natl Acad Sci USA. 2005;102:1023–1028. doi: 10.1073/pnas.0409020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WS, Cho SY, Hong HJ, Park SC, Yoon SI. Self-Oligomerizing structure of the flagellar cap protein FliD and its implication in filament assembly. J Mol Biol. 2017;429:847–857. doi: 10.1016/j.jmb.2017.02.001. [DOI] [PubMed] [Google Scholar]
- Song WS, Jeon YJ, Namgung B, Hong M, Yoon SI. A conserved TLR5 binding and activation hot spot on flagellin. Sci Rep. 2017;7:40878. doi: 10.1038/srep40878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WS, Yoon SI. Crystal structure of FliC flagellin from Pseudomonas aeruginosa and its implication in TLR5 binding and formation of the flagellar filament. Biochem Biophys Res Commun. 2014;444:109–115. doi: 10.1016/j.bbrc.2014.01.008. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Iino T. Role of the flaR gene in flagellar hook formation in Salmonella spp. J Bacteriol. 1981;148:973–979. doi: 10.1128/jb.148.3.973-979.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Yonekura K, Namba K. Structure of the rotor of the bacterial flagellar motor revealed by electron cryomicroscopy and single-particle image analysis. J Mol Biol. 2004;337:105–113. doi: 10.1016/j.jmb.2004.01.034. [DOI] [PubMed] [Google Scholar]
- Thomas DR, Francis NR, Xu C, DeRosier DJ. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar typhimurium. J Bacteriol. 2006;188:7039–7048. doi: 10.1128/JB.00552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner L, Ryu WS, Berg HC. Real-time imaging of fluorescent flagellar filaments. J Bacteriol. 2000;182:2793–2801. doi: 10.1128/JB.182.10.2793-2801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonderviszt F, Aizawa S, Namba K. Role of the disordered terminal regions of flagellin in filament formation and stability. J Mol Biol. 1991;221:1461–1474. doi: 10.1016/0022-2836(91)90946-4. [DOI] [PubMed] [Google Scholar]
- Vonderviszt F, Imada K, Furukawa Y, Uedaira H, Taniguchi H, Namba K. Mechanism of self-association and filament capping by flagellar HAP2. J Mol Biol. 1998;284:1399–1416. doi: 10.1006/jmbi.1998.2274. [DOI] [PubMed] [Google Scholar]
- Vonderviszt F, Ishima R, Akasaka K, Aizawa S. Terminal disorder: a common structural feature of the axial proteins of bacterial flagellum? J Mol Biol. 1992;226:575–579. doi: 10.1016/0022-2836(92)90616-R. [DOI] [PubMed] [Google Scholar]
- Wagenknecht T, DeRosier DJ, Aizawa S-I, Macnab RM. Flagellar hook structures of Caulobacter and Salmonella and their relationship to filament structure. J Mol Biol. 1982;162:69–87. doi: 10.1016/0022-2836(82)90162-0. [DOI] [PubMed] [Google Scholar]
- Wagenknecht T, DeRosier DJ, Shapiro L, Weissborn A. Three-dimensional reconstruction of the flagellar hook from Caulobacter crescentus. J Mol Biol. 1981;151:439–465. doi: 10.1016/0022-2836(81)90005-X. [DOI] [PubMed] [Google Scholar]
- Williams AW, Yamaguchi S, Togashi F, Aizawa S-I, Kawagishi I, Macnab RM. Mutations in FliK and FlhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J Bacteriol. 1996;178:2960–2970. doi: 10.1128/jb.178.10.2960-2970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita I, Hasegawa K, Suzuki H, Vonderviszt F, Mimori-Kiyosue Y, Namba K. Structure and switching of bacterial flagellar filament studied by X-ray fiber diffraction. Nat Struct Biol. 1998;5:125–132. doi: 10.1038/nsb0298-125. [DOI] [PubMed] [Google Scholar]
- Yonekura K, Maki S, Morgan DG, DeRosier DJ, Vonderviszt F, Imada K, Namba K. The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science. 2000;290:2148–2152. doi: 10.1126/science.290.5499.2148. [DOI] [PubMed] [Google Scholar]
- Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- Yoon SI, Kurnasov O, Natarajan V, Hong M, Gudkov AV, Osterman AL, Wilson IA. Structural basis of TLR5-flagellin recognition and signaling. Science. 2012;335:859–864. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaloba P, Bailey-Elkin BA, Derksen M, Mark BL. Structural and biochemical insights into the peptidoglycan hydrolase domain of FlgJ from Salmonella typhimurium. PLoS One. 2016;11:e0149204. doi: 10.1371/journal.pone.0149204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Luo M, Cai X, Tang J, Niu S, Zhang W, Hu Y, Yin Y, Huang A, Wang D, Wang D. Crystal structure of a novel dimer form of FlgD from P. aeruginosa PAO1. Proteins. 2011;2011(79):2346–2351. doi: 10.1002/prot.23058. [DOI] [PubMed] [Google Scholar]