Abstract
As the importance of bacteriophages as novel antimicrobials and potential diagnostics comes increasingly into focus, there is a heightened interest in understanding the mechanisms of how they interact with their bacterial hosts. The first step of a bacteriophage (phage) infection is the recognition of specific moieties on the bacterial cell surface as determined by their phage receptor binding proteins (RBPs). Knowledge of RBPs and how they interact with bacteria has been driven by studies of model phages and of industrially important phages, such as those that impact the dairy industry. Therefore, data from these phage groups constitute the majority of this review. We start with a brief introduction to phages, their life cycles and known receptors. We then review the state-of-the-art knowledge of phage RBPs of Gram-positive bacteria in the context of the better understood Gram-negative bacterial RBPs. In general, more is known about the RBPs of siphoviruses than myoviruses, which is reflected here, but for both virus families, where possible, we show what RBPs are, how they are arranged within phage genomes and what is known about their structures. As RBPs are the key determinant of phage specificity, studying and characterising them is important, for downstream applications such as diagnostic and therapeutic purposes.
Keywords: Receptor binding proteins, Siphoviruses, Myoviruses, Gram-positive bacteria, Bacteria–phage interaction
Introduction
The relationship between phages and their bacterial hosts is of significant interest to microbiologists who seek to understand how phages impact the biology of their bacterial hosts. A key part of this relationship, and the focus here, is how phages initially interact with their hosts. This interaction has been widely studied for phages that target Gram-negative bacteria, but far less is known for phages that infect Gram-positive bacteria (Mahony and van Sinderen 2015). For all phages, the receptor binding proteins (RBPs), or tail fibre proteins, are the key factors that determine phage specificity. The stability, specific binding nature and affinity of RBPs to specific carbohydrate-binding proteins has meant that they have been exploited as therapeutic tools to reduce bacterial colonisation (Simpson et al. 2016, Waseh et al. 2010).
In addition, RBPs are being used, and developed for use, as diagnostic tools for several bacterial genera. For example, the phage NCTC12673 RBPs were found to selectively bind to and, thus, be used as a diagnostic test for Campylobacter, both C. jejuni and C. coli (Javed et al. 2013). The study and characterisation of RBPs and the mechanisms used by phages to adsorb to their bacterial hosts is critical to enhancing our fundamental understanding of phages, and to being able to better exploit the use of them (Bertozzi Silva et al. 2016).
RBPs differ structurally between phages, according to phage morphology, and by the specific mechanisms by which phages attach to bacteria. For example, in myoviruses such as T4, the attachment to Escherichia coli occurs via both long and short tail fibres, but in siphoviruses such as the Lactococcus lactis phage TP901-1, the RBPs are found within a large structure known as a baseplate. Alternatively, RBPs are found on tail spikes such as in Bacillus subtilis phage SPP1 (Veesler et al. 2012; Simpson et al. 2016).
Both phage receptors and RBPs have been well studied and characterised for phages that infect E. coli, particularly for T4 and lambda (Table 1). However, recently, there has been an increase in the focus to study RBPs from phages that target Gram-positive bacteria (Mahony and van Sinderen 2015). Previous excellent reviews, such as that of Mahony et al. (2016), have highlighted recent progress on the phage–host interactions for Gram-positive bacteria. The main focus of previous reviews has been on phages (mainly siphoviruses) that target lactic acid bacteria. This review aims to extend the focus and cover state-of-the-art knowledge and characterisation of the RBPs of phages that infect all Gram-positive bacteria, and, where possible, we include information for both siphoviruses and myoviruses.
Table 1.
List of well-studied phages that infect Gram-negative bacteria and Gram-positive bacteria, along with their known receptors to which phages attach
| Host | Phage | Phage family | Name of the receptors on the host | References |
|---|---|---|---|---|
| Escherichia coli | Lambda | Siphoviridae | Protein LamB | Chatterjee and Rothenberg (2012) |
| Escherichia coli | T5 | Siphoviridae | Protein FhuA | Mahony and van Sinderen (2012) |
| Salmonella | ES18 | Siphoviridae | Protein FhuA | Killmann et al. (2001) |
| Streptococcus thermophilus | OBJ | Siphoviridae | Glucosamine/ribose | Quiberoni et al. (2000) |
| S. thermophilus | CYM | Siphoviridae | Glucosamine/rhamnose | Quiberoni et al. (2000) |
| Lactococcus lactis | TP901-1 | Siphoviridae | Saccharide | Mahony and van Sinderen (2012) |
| L. lactis | Tuc2009 | Siphoviridae | Saccharide | Mahony and van Sinderen (2012) |
| Bacillus subtilis | SPP1 | Siphoviridae | Protein YueB | São-José et al. (2004) |
| Staphylococcus aureus | W | Siphoviridae | N-acetylglucosamine (GlcNAc) glycoepitope on wall teichoic acids | Xia et al. (2011) |
| S. aureus | ϕSLT | Siphoviridae | Poly(glycerophosphate) moiety of lipoteichoic acid | Kaneko et al. (2009) |
Brief bacteriophage background
To contextualise the importance of RBPs in phages, it is useful to give a little background on phage biology. Phages are bacterial viruses and are thought to be the most abundant and diverse biological entities on Earth. They can have either single- or double-stranded RNA or DNA genomes and these are surrounded by a protein capsid connected to their phage tail. This phage tail binds to the host receptor sites (Haq et al. 2012). The vast majority of phages that have been morphologically characterised belong to the order Caudovirales, which have dsDNA genomes. Within this order, there are three families that can be distinguished from one another on their overall appearance and, of particular relevance to this review, their tail morphology. Siphoviruses possess long non-contractile tails, but myoviruses have contractile tails and podoviruses have short tails (Spinelli et al. 2014a).
Phages rely on their bacterial hosts to replicate, and two distinct life cycles have been observed. The first is known as the lytic life cycle, where the phages penetrate their bacterial hosts, multiply and then mature progeny are released after they lyse the bacterial cells. Lytic phages are a source of promising novel therapeutics and, indeed, have a long history of use to treat bacterial infections (Czaplewski et al. 2016). The second life cycle is termed the lysogenic life cycle and, here, the phage genome integrates into the bacteria genome, often remaining integrated for extended periods. However, if the bacteria encounters stressful conditions, such as ultraviolet radiation or antibiotics, the integrated (temperate) phage is induced and enters the lytic cycle, as described above (Haq et al. 2012).
Phage adsorption
By definition, the physical phage interaction with bacterial hosts is a sine qua non or absolutely essential for phage infection, and is, in most cases, thought to be highly specific. The infection process starts with phages attaching to one or more components or receptors on their host bacteria and then penetrating the cell membranes (Rakhuba et al. 2010). This is mediated via interactions between proteins located at the distal end of the phage, the RBPs (sometimes termed as anti-receptors or adhesins) and ligands (receptors) on the bacterial surface. This highly specific binding is termed adsorption and has two main steps (Mahony and van Sinderen 2012). The first is when phages bind reversibly to a constituent on the bacterial surface. Here, phage–bacterial binding is not complete and the phages can be desorbed from bacteria, as evidenced by the presence of viable phages within supernatant (Bertozzi Silva et al. 2016). During the second step however, phages irreversibly attach to either the same receptor as in the first step of the adsorption or to a second receptor. Following this step, the phages penetrate the host cell and insert their DNA. This binding between phages and bacteria is sometimes assisted by enzymatic cleavage that helps phage DNA to be injected (São-José et al. 2004; Bertozzi Silva et al. 2016).
Bacteriophage receptors on the surface of the bacteria
Many diverse molecular structures on the surface of bacteria can act as phage receptors, but their nature and position on the cell differs with specific bacteria–phage interactions. The receptors can be protein, polysaccharide, lipopolysaccharides (LPS) and carbohydrate moieties (Bertozzi Silva et al. 2016). In Gram-negative bacteria, LPS is a common receptor for phages. In addition, other receptors are outer membrane proteins, pili and flagella (Sørensen et al. 2011). The exact mechanisms of action have been well studied in the model E. coli myovirus T4. This phage attaches reversibly to LPS or to the outer membrane protein porin OmpC, depending on the strain of E. coli. This attachment then leads to irreversible binding of T4 with the outer core region of E. coli LPS. The model E. coli phage T7 also uses LPS as the main irreversible phage binding site (Sørensen et al. 2011; González-García et al. 2015).
For phages that infect Gram-positive bacteria, peptidoglycan is an important phage receptor, as it is a major polymer on bacterial surfaces, along with teichoic acids, that are attached covalently to the peptidoglycan layer (Bertozzi Silva et al. 2016). Polysaccharides that are exposed on the surface of the bacteria are also common receptors (Bertozzi Silva et al. 2016). In many ways, it is surprising that only a small number of phage receptors have been identified for Gram-positive bacteria; this is, in part, due to their complex outer structure and, in part, due to the scarcity of research activities on phages that target Gram-positive bacteria in general. An example of a Gram-positive bacteria phage with a known receptor is the phage 3C that attaches to the N-acetyl glucosamine moiety of teichoic acids on the surface of Staphylococcus aureus. Another example is the D-glucose chain of the teichoic acid on the B. subtilis surface, which functions as the receptor for phages SP2 and SP10 (Rakhuba et al. 2010).
RBPs for siphoviruses of Gram-positive bacteria
The vast majority of phages that infect Gram-positive bacteria and have had their RBPs characterised are siphoviruses. This has largely been driven by the fact that phages from this family are important in the dairy industry and, thus, they have been thoroughly studied (Table 2).
Table 2.
List of some of the phages that target Gram-positive bacteria for which their receptor binding proteins (RBPs) have been identified
| Phages | Host | RBPs | References |
|---|---|---|---|
| bIL170 | L. lactis | ORF20 | Dupont et al. (2004) |
| TP901-1 | L. lactis | BppL | Vegge et al. (2006) |
| Tuc2009 | L. lactis | ORF53 | Vegge et al. (2006) |
| SPP1 | B. subtilis | Gp21 | Vinga et al. (2012) |
| A118 | Listeria monocytogenes | Gp19 and Gp20 | Bielmann et al. (2015) |
| P35 | L. monocytogenes | Gp16 | Bielmann et al. (2015) |
| Phage φ11 | S. aureus | Gp45 | Li et al. (2016) |
RBPs are generally identified using bioinformatics analysis, followed by antibody studies, for example using polyclonal antibodies raised against overexpressed putative tail fibre proteins, to neutralise phage infection (Li et al. 2016). Molecular approaches are also used; for example, a chimeric phage was produced to identify the RBPs of the L. lactis phages TP901-1 and Tuc2009. In this work, the gene encoding the TP901-1 lower baseplate protein (BppL) was replaced with the gene (orf53) from phage Tuc2009 (Vegge et al. 2006). The results showed that the chimeric TP901-1 phage was able to infect the Tuc2009 host strain efficiently, indicating that the TP901-1 BppL and gene orf53 of phage Tuc2009 are both responsible for the phage attachment to the bacteria (Vegge et al. 2006). To gain further insights into how these phages attach to their host, structural information was determined using X-ray crystallography and morphological information from transmission electron microscopy (Mahony and van Sinderen 2012).
Genomic architecture and the position of tail fibre/baseplate sequences
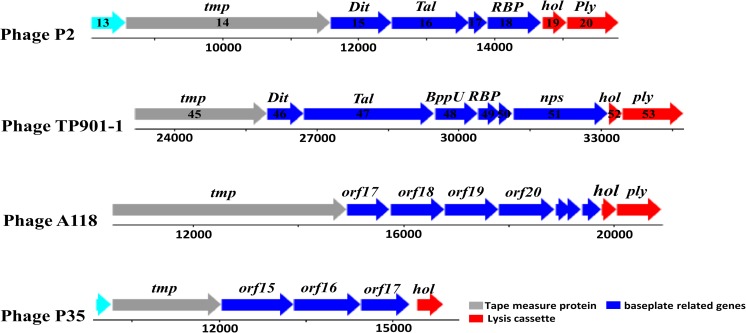
Based on the genome sequences of siphoviruses that infect the Gram-positive species such as B. subtilis, L. lactis, L. monocytogenes, S. aureus and C. difficile, it can be seen that genes that encode tail fibre proteins, or proteins that form the baseplate, are located between tmp genes encoding the tape measure protein TMP (that determines the length of the phage tail) and the genes encoding the holin and endolysin proteins (Li et al. 2016). There are usually four genes located between these anchor points, as shown in Fig. 1, one of these being the Dit gene that encodes the distal tail protein (Dit), which has been shown to be fairly conserved within siphoviruses. After Dit usually comes the Tal gene that encodes tail-associated lysin protein (Tal). Tal promotes efficient phage penetration and infection by degrading peptidoglycan (Bielmann et al. 2015; Li et al. 2016; Mahony and van Sinderen 2012; Stockdale et al. 2013).
Fig. 1.
Schematic representation of the baseplate genes location of five siphoviruses, P2, TP901-1, A118 and P35. These genes are located between tmp and holin and endolysin encoding genes. This figure was adapted from Bebeacua et al. (2013) and Bielmann et al. (2015)
The Tal gene architecture has distinct differences within different phages, consistent with the divergent structure of siphovirus tail tips (Spinelli et al. 2014b). For example, in the Lactococcus phage TP901-1 and in most siphoviruses that have been characterised, there are two genes located downstream to the Tal gene; one encodes the upper baseplate protein (BppU) and the other encodes RBPs. However, in the B. subtilis SPP1 phage, there is no BppU but, instead, this phage has a tail spike protein (Bebeacua et al. 2010). The four gene products (Dit, Tal, BppU and RBPs) are discussed in detail below.
Structural aspects of siphoviruses baseplate
Distal tail proteins (Dit)
The gene encoding the Dit protein is involved in siphoviruses baseplate formation, where it forms the central hub of the phage baseplate. Using HHpred analysis, and data from crystal structures studies, this protein can be identified in several siphoviruses, including the B. subtilis phage SPP1, L. lactis TP901-1, Tuc2009 and P2 phages, A118 and P35 that infect L. monocytogenes, φ11 phage that infects S. aureus (Bielmann et al. 2015; Li et al. 2016) and CDHS1 phage that targets C. difficile (Dowah et al., under review). This diverse set of phages with a recognisable Dit protein demonstrate the high level of proteins conservation within the siphoviruses (Veesler et al. 2012).
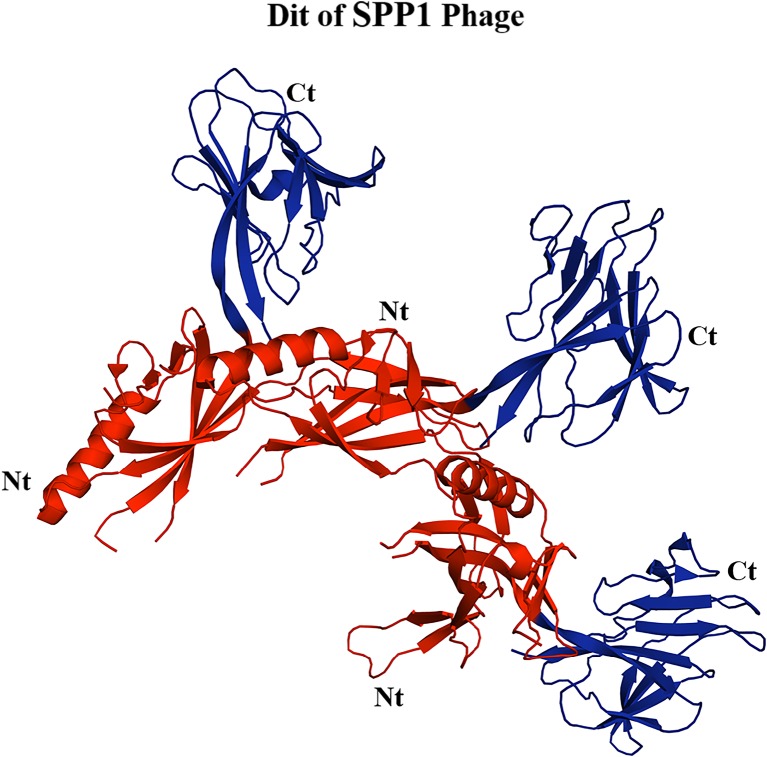
In terms of the physical structure, the Dit protein is organised as a hexametric ring that links the tail tube to the phage baseplate. The N-terminal of each Dit monomer consists of a β-sandwich, α-helix and a β-hairpin, and the C-terminal is a galectin-like β-sandwich (Veesler et al. 2012; Flayhan et al. 2014). Figure 2 shows the structure of the Dit protein for B. subtilis phage SPP1. Structural comparison reveals a striking similarity between SPP1 Dit and that of other phages, such as the Dit of TP901-1 and P2 phages (Veesler et al. 2012). Furthermore, the N-terminal region of Dit has been identified in myoviruses, including T4 and Mu, and in siphoviruses, such as T5 that infects E. coli (Koç et al. 2016). Thus, the conservation of the Dit protein extends between both the siphoviruses and the myoviruses.
Fig. 2.
Structure of the Dit protein of SPP1 phage that infects B. subtilis (Veesler et al. 2010). The N-terminal of this protein is highlighted in red and the C-terminal is highlighted in blue. This figure was generated using PyMol software (http://www.pymol.org)
Tail-associated lysin proteins (Tal)
The second gene of interest generally located next to dit is tal, which encodes the Tal protein that can be released via self-cleavage at a consensus site. The Tal protein belongs to a group of proteins known as virion-associated peptidoglycan hydrolases (VAPGHs). The main function of these proteins or enzymes is peptidoglycan layer degradation, to assist phage infection. The virion-associated muralytic activities may offer a promising tool to treat bacterial pathogens (Drulis-Kawa et al. 2015). There are four enzymes included in VAPGHs based on their enzymatic activity: lysozymes, for example, gp5 from T4 phage; lytic trans glycosylases, such as gp16 from T7 phage; protein P7 from PRD1, glucosaminidases and endopeptidases as in protein P5 from phi6 phage; and Tal2009 from Tuc2009 phage (Drulis-Kawa et al. 2015; Kenny et al. 2004). The N-terminal domain of the Tal protein is similar to a group of proteins including ORF16 of L. lactis phage p2, gp21 of phage SPP1 and ORF47 of TP901-1. In addition, the N-terminal of the Tal protein is thought to be involved in phage baseplate structure formation (Sciara et al. 2008).
Interestingly, the N-terminal domain of the Tal protein has been found to be conserved in phages that infect Gram-negative bacteria, such as the myoviruses T4 and Mu and siphoviruses such as T5 (Koç et al. 2016). However, the C-terminus of the Tal protein has proteolytic activity that is involved in the peptidoglycan cleavage, thus facilitating phage DNA injection into the bacteria. This phenomenon has also been noted in the Tal C-terminal of Lactococcus phages TP901-1 and Tuc2009. Conversely, no such activity has been established for Lactococcus phage p2. Instead, the Tal C terminal of B. subtilis SPP1 phage is found to be involved in phage attachment with YueB on the surface of the host (Bebeacua et al. 2010; Spinelli et al. 2014b).
Upper baseplate protein (BppU)
BppU protein is formed as six asymmetric trimers which link to the Dit central core and the RBPs. Each monomer of BppU contains an N-terminal domain, a linker and two helices joined by a kink and then the C-terminal domain (Veesler et al. 2012). HHpred analysis revealed that the homology of this BppU protein’s N-terminal was found in several phages, such as A118 that infects L. monocytogenes, φ11 phage that infects S. aureus (Li et al. 2016; Bielmann et al. 2015) and in CDHS1 that infects C. difficile (Dowah et al., under review). Finally, downstream to BppU are the RBPs.
Receptor binding proteins (RBPs)
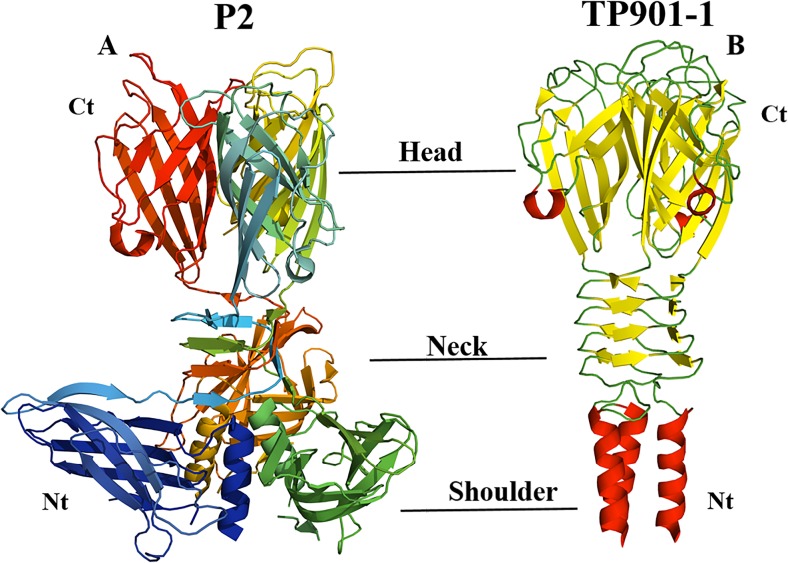
Although the RBPs from various siphoviruses that target different Gram-positive bacteria have been identified, the majority of known structures are from phages that infect L. lactis. The first three Lactococcus phages that had their RBPs structurally defined were 936, p2 and P335 TP901-1 (Fig. 3) and 936 phage bIL170 (Ricagno et al. 2006). The overall structures of RBPs for these phages are composed of three parts: the shoulder, the neck and the head. The N-terminus (shoulder) of this protein contains an α-helix bundle, while the neck is a β-prism domain connected to the C-terminus (the head), which possesses a saccharide binding site that is attached to the receptors on the surface of the organism (Sciara et al. 2008).
Fig. 3.
Structure of receptor binding proteins (RBPs) of two phages that infect L. lactis, P2 (a) (Tremblay et al. 2006) and TP901-1 (b) (Spinelli et al. 2006). This figure was generated using PyMol software (http://www.pymol.org)
The structural comparisons of RBPs between phages reveal that RBPs of phage bIL170 have a high sequence identity of 89% to 936 phage p2 at the N-terminus domain. However, they exhibit differences on the neck and head domains; this explains the difference in host range for these two phages (Ricagno et al. 2006). On the other hand, the RBPs head domain of P335 and phage TP901 are 28% identical to the RBPs head of 936 phage p2 In contrast, no similarity has been noted at the RBPs N-terminal of those phages (Spinelli et al. 2014b). Recently, the RBP structure for phage φ11 that infects S. aureus has been resolved. The structure of this protein also has three regions: the N-terminal domain that consists of a triple-helical bundle; the central part, with three β-propeller domains; and the C-terminal “tower” region (Koç et al. 2016). Compared to the RBPs structure of L. lactis phages, the RBPs of phage φ11 is distinct from the RBPs of any known phages, apart from the first 30 amino acids being similar to that of the P335 and TP901 phages (Koç et al. 2016).
RBPs shape depend on the bacterial receptors nature
The information that has been obtained from studying siphoviruses RBPs, particularly the phages that infect Gram-positive bacteria, allows for an initial prediction of the nature of the host receptor (protein or carbohydrate) to which phages bind. For example, if a phage binds to host receptor that is protein in nature, the end of the tail fibres will be sharp or spiked in form. The B. subtilis SPP1 phage, B. anthracis γ phage and c2-type phages that infect L. lactis are good examples of siphoviruses that have their RBPs as tail spikes that bind to protein receptors on bacterial surfaces (Mahony et al. 2016). In contrast, the phages that attach to a carbohydrate constituent form a larger baseplate where binding occurs. For example, P2 and TP901-1 that infect L. lactis are known to bind to a carbohydrate on the surface of the host (Mahony and van Sinderen 2012).
Two strategies found within siphoviruses binding
A comparison of structural studies of these phages has highlighted differences in phage infection mechanisms. Lactococcus lactis siphoviruses P2 and TP901 have been the most extensively studied, both structurally and morphologically (Bebeacua et al. 2013; Veesler et al. 2012). Data obtained from X-ray crystallography and electron microscopy analysis revealed that they undergo two different mechanisms to attach to their bacterial host. First, the 18 RBPs in the P2 phage are oriented toward the capsid. These RBPs then undergo Ca2+-mediated conformational changes and effectively infect the bacteria (Spinelli et al. 2014b). Some phages, on the other hand, have simple attachment mechanisms; for example, phage TP901 has its 54 RBPs displayed in the head domain and they are pointing toward the host, and, therefore, is in ready to infect form without needing a conformational change or Ca2+ for activation (Veesler et al. 2012).
RBPs for myoviruses of Gram-positive bacteria
Fewer myovirus RBPs have been identified and characterised for myoviruses of Gram-positive bacteria compared to siphoviruses (Spinelli et al. 2014a). Indeed, the phage A511 that infects L. monocytogenes is one of very few myoviruses whose RBPs have been identified (Habann et al. 2014).
A511 belongs to a group of SPO1-related phages, and is strictly lytic with low G + C content, like all phages within this group (Klumpp et al. 2010). When A511 is compared to other SPO1-related phages, such as Bacillus phage SPO1 and the Staphylococcus phages Twort and ISP, it can be seen that the genes located between the tmp gene and helicase are responsible for encoding the baseplate proteins that contains RBPs. Taking this into account, five gene products (gp98, gp99, gp104, gp106 and gp108) of phage A511 were identified between TMP and helicase. They are thought to be involved in baseplate formation. Polyclonal antibodies were raised against those proteins and the results showed that two of these proteins (gp98 and gp108) are, indeed, involved in phage attachment (Habann et al. 2014). Furthermore, the baseplate is a big complex device attached to long and short tail fibres, and, during the infection, the baseplate undergoes conformational change and rearranges into a double-ringed shape, accompanied by the contraction of the phage tail (Habann et al. 2014).
Conclusions
Studying phage–bacterial interactions is essential if we are to improve our understanding and downstream exploitation of phages. The key proteins responsible for phage–bacterial interactions are the receptor binding proteins (RBPs). In this review, we have highlighted the current knowledge on the RBPs of phages that infect Gram-positive bacteria. Although we have focused on the siphoviruses, where known, we have included information on myoviruses. We include information from molecular, bioinformatics and structural studies of these viruses. We have summarised the known genes that encode proteins involved in the phage baseplate formation and shown how the genomic architecture of these genes within different siphoviruses is conserved. We have also highlighted the known RBPs structures.
To date, very few myoviruses that infect Gram-positive bacteria have had their RBPs characterised. In this review, phage A511 from the group of SPO1-related phages was used as a reference phage as is a well-characterised myovirus. It is clear that further study of RBPs of myoviruses that infect Gram-positive bacteria is much needed to enhance our understanding of how these viruses interact with their hosts, and how this interaction mechanism differs within different myoviruses. Significant efforts have been exerted in trying to identify and characterise the RBPs for phages that infect Gram-positive bacteria, although the focus has mainly been on phages that infect Lactococcus lactis. In summary, we have presented an overview of what is known about RBPs for phages of Gram-positive bacteria but emphasise that this exciting field is rich with undiscovered structures and details of interactions which are likely to unravel new and exciting methods in which phages and their hosts interact.
Acknowledgements
We are grateful to the Libyan Ministry of Higher Education for funding the PhD studies of ASAD.
Compliance with ethical standards
Conflict of interest
Ahmed S.A. Dowah declares that he has no conflict of interest. Martha R. J. Clokie declares that she has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is part of a Special Issue on ‘Biomolecules to Bio-nanomachines - Fumio Arisaka 70th Birthday’ edited by Damien Hall, Junichi Takagi and Haruki Nakamura.
References
- Bebeacua C, Bron P, Lai L, Vegge CS, Brøndsted L, Spinelli S, Campanacci V, Veesler D, van Heel M, Cambillau C. Structure and molecular assignment of lactococcal phage TP901-1 baseplate. J Biol Chem. 2010;285:39079–39086. doi: 10.1074/jbc.M110.175646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebeacua C, Tremblay D, Farenc C, Chapot-Chartier MP, Sadovskaya I, van Heel M, Veesler D, Moineau S, Cambillau C. Structure, adsorption to host, and infection mechanism of virulent lactococcal phage p2. J Virol. 2013;87:12302–12312. doi: 10.1128/JVI.02033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertozzi Silva J, Storms Z, Sauvageau D. Host receptors for bacteriophage adsorption. FEMS Microbiol Lett. 2016;363:fnw002. doi: 10.1093/femsle/fnw002. [DOI] [PubMed] [Google Scholar]
- Bielmann R, Habann M, Eugster MR, Lurz R, Calendar R, Klumpp J, Loessner MJ. Receptor binding proteins of Listeria monocytogenes bacteriophages A118 and P35 recognize serovar-specific teichoic acids. Virology. 2015;477:110–118. doi: 10.1016/j.virol.2014.12.035. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Rothenberg E. Interaction of bacteriophage l with its E. coli receptor, LamB. Viruses. 2012;4:3162–3178. doi: 10.3390/v4113162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplewski L, Bax R, Clokie M, Dawson M, Fairhead H, Fischetti VA, Foster S, Gilmore BF, Hancock REW, Harper D, Henderson IR, Hilpert K, Jones BV, Kadioglu A, Knowles D, Ólafsdóttir S, Payne D, Projan S, Shaunak S, Silverman J, Thomas CM, Trust TJ, Warn P, Rex JH. Alternatives to antibiotics—a pipeline portfolio review. Lancet Infect Dis. 2016;16:239–251. doi: 10.1016/S1473-3099(15)00466-1. [DOI] [PubMed] [Google Scholar]
- Drulis-Kawa Z, Majkowska-Skrobek G, Maciejewska B. Bacteriophages and phage-derived proteins—application approaches. Curr Med Chem. 2015;22:1757–1773. doi: 10.2174/0929867322666150209152851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont K, Vogensen FK, Neve H, Bresciani J, Josephsen J. Identification of the receptor-binding protein in 936-species lactococcal bacteriophages. Appl Environ Microbiol. 2004;70:5818–5824. doi: 10.1128/AEM.70.10.5818-5824.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flayhan A, Vellieux FM, Lurz R, Maury O, Contreras-Martel C, Girard E, Boulanger P, Breyton C. Crystal structure of pb9, the distal tail protein of bacteriophage T5: a conserved structural motif among all siphophages. J Virol. 2014;88:820–828. doi: 10.1128/JVI.02135-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-García VA, Pulido-Cid M, Garcia-Doval C, Bocanegra R, van Raaij MJ, Martín-Benito J, Cuervo A, Carrascosa JL. Conformational changes leading to T7 DNA delivery upon interaction with the bacterial receptor. J Biol Chem. 2015;290:10038–10044. doi: 10.1074/jbc.M114.614222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habann M, Leiman PG, Vandersteegen K, Van den Bossche A, Lavigne R, Shneider MM, Bielmann R, Eugster MR, Loessner MJ, Klumpp J. Listeria phage A511, a model for the contractile tail machineries of SPO1-related bacteriophages. Mol Microbiol. 2014;92:84–99. doi: 10.1111/mmi.12539. [DOI] [PubMed] [Google Scholar]
- Haq IU, Chaudhry WN, Akhtar MN, Andleeb S, Qadri I. Bacteriophages and their implications on future biotechnology: a review. Virol J. 2012;9:9. doi: 10.1186/1743-422X-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed MA, Poshtiban S, Arutyunov D, Evoy S, Szymanski CM. Bacteriophage receptor binding protein based assays for the simultaneous detection of Campylobacter jejuni and Campylobacter coli. PLoS One. 2013;8:e69770. doi: 10.1371/journal.pone.0069770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko J, Narita-Yamada S, Wakabayashi Y, Kamio Y. Identification of ORF636 in phage phiSLT carrying Panton–Valentine leukocidin genes, acting as an adhesion protein for a poly(glycerophosphate) chain of lipoteichoic acid on the cell surface of Staphylococcus aureus. J Bacteriol. 2009;191:4674–4680. doi: 10.1128/JB.01793-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny JG, McGrath S, Fitzgerald GF, van Sinderen D. Bacteriophage Tuc2009 encodes a tail-associated cell wall-degrading activity. J Bacteriol. 2004;186:3480–3491. doi: 10.1128/JB.186.11.3480-3491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killmann H, Braun M, Herrmann C, Braun V. FhuA barrel-cork hybrids are active transporters and receptors. J Bacteriol. 2001;183:3476–3487. doi: 10.1128/JB.183.11.3476-3487.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp J, Lavigne R, Loessner MJ, Ackermann HW. The SPO1-related bacteriophages. Arch Virol. 2010;155:1547–1561. doi: 10.1007/s00705-010-0783-0. [DOI] [PubMed] [Google Scholar]
- Koç C, Xia G, Kühner P, Spinelli S, Roussel A, Cambillau C, Stehle T. Structure of the host-recognition device of Staphylococcus aureus phage varphi11. Sci Rep. 2016;6:27581. doi: 10.1038/srep27581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Koç C, Kühner P, Stierhof Y-D, Krismer B, Enright MC, Penadés JR, Wolz C, Stehle T, Cambillau C, Peschel A, Xia G. An essential role for the baseplate protein Gp45 in phage adsorption to Staphylococcus aureus. Sci Rep. 2016;6:26455. doi: 10.1038/srep26455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony J, van Sinderen D. Structural aspects of the interaction of dairy phages with their host bacteria. Viruses. 2012;4:1410–1424. doi: 10.3390/v4091410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony J, van Sinderen D. Gram-positive phage–host interactions. Front Microbiol. 2015;6:61. doi: 10.3389/fmicb.2015.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony J, Stockdale SR, Collins B, Spinelli S, Douillard FP, Cambillau C, van Sinderen D. Lactococcus lactis phage TP901-1 as a model for Siphoviridae virion assembly. Bacteriophage. 2016;6:e1123795. doi: 10.1080/21597081.2015.1123795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiberoni A, Stiefel JI, Reinheimer JA. Characterization of phage receptors in Streptococcus thermophilus using purified cell walls obtained by a simple protocol. J Appl Microbiol. 2000;89:1059–1065. doi: 10.1046/j.1365-2672.2000.01214.x. [DOI] [PubMed] [Google Scholar]
- Rakhuba DV, Kolomiets EI, Dey ES, Novik GI. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol J Microbiol. 2010;59:145–155. [PubMed] [Google Scholar]
- Ricagno S, Campanacci V, Blangy S, Spinelli S, Tremblay D, Moineau S, Tegoni M, Cambillau C. Crystal structure of the receptor-binding protein head domain from Lactococcus lactis phage bIL170. J Virol. 2006;80:9331–9335. doi: 10.1128/JVI.01160-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- São-José C, Baptista C, Santos MA. Bacillus subtilis operon encoding a membrane receptor for bacteriophage SPP1. J Bacteriol. 2004;186:8337–8346. doi: 10.1128/JB.186.24.8337-8346.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciara G, Blangy S, Siponen M, Mc Grath S, van Sinderen D, Tegoni M, Cambillau C, Campanacci V. A topological model of the baseplate of lactococcal phage Tuc2009. J Biol Chem. 2008;283:2716–2723. doi: 10.1074/jbc.M707533200. [DOI] [PubMed] [Google Scholar]
- Simpson DJ, Sacher JC, Szymanski CM. Development of an assay for the identification of receptor binding proteins from bacteriophages. Viruses. 2016;8:17. doi: 10.3390/v8010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen MC, van Alphen LB, Harboe A, Li J, Christensen BB, Szymanski CM, Brøndsted L. Bacteriophage F336 recognizes the capsular phosphoramidate modification of Campylobacter jejuni NCTC11168. J Bacteriol. 2011;193:6742–6749. doi: 10.1128/JB.05276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli S, Campanacci V, Blangy S, Moineau S, Tegoni M, Cambillau C. Modular structure of the receptor binding proteins of Lactococcus lactis phages. The RBP structure of the temperate phage TP901-1. J Biol Chem. 2006;281:14256–14262. doi: 10.1074/jbc.M600666200. [DOI] [PubMed] [Google Scholar]
- Spinelli S, Bebeacua C, Orlov I, Tremblay D, Klaholz BP, Moineau S, Cambillau C. Cryo-electron microscopy structure of lactococcal siphophage 1358 virion. J Virol. 2014;88:8900–8910. doi: 10.1128/JVI.01040-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli S, Veesler D, Bebeacua C, Cambillau C. Structures and host-adhesion mechanisms of lactococcal siphophages. Front Microbiol. 2014;5:3. doi: 10.3389/fmicb.2014.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockdale SR, Mahony J, Courtin P, Chapot-Chartier MP, van Pijkeren JP, Britton RA, Neve H, Heller KJ, Aideh B, Vogensen FK, van Sinderen D. The lactococcal phages Tuc2009 and TP901-1 incorporate two alternate forms of their tail fiber into their virions for infection specialization. J Biol Chem. 2013;288:5581–5590. doi: 10.1074/jbc.M112.444901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay DM, Tegoni M, Spinelli S, Campanacci V, Blangy S, Huyghe C, Desmyter A, Labrie S, Moineau S, Cambillau C. Receptor-binding protein of Lactococcus lactis phages: identification and characterization of the saccharide receptor-binding site. J Bacteriol. 2006;188:2400–2410. doi: 10.1128/JB.188.7.2400-2410.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veesler D, Robin G, Lichière J, Auzat I, Tavares P, Bron P, Campanacci V, Cambillau C. Crystal structure of bacteriophage SPP1 distal tail protein (gp19.1). A baseplate hub paradigm in gram-positive infecting phages. J Biol Chem. 2010;285:36666–36673. doi: 10.1074/jbc.M110.157529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veesler D, Spinelli S, Mahony J, Lichière J, Blangy S, Bricogne G, Legrand P, Ortiz-Lombardia M, Campanacci V, van Sinderen D, Cambillau C. Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism. Proc Natl Acad Sci U S A. 2012;109:8954–8958. doi: 10.1073/pnas.1200966109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegge CS, Vogensen FK, Mc Grath S, Neve H, van Sinderen D, Brøndsted L. Identification of the lower baseplate protein as the antireceptor of the temperate lactococcal bacteriophages TP901-1 and Tuc2009. J Bacteriol. 2006;188:55–63. doi: 10.1128/JB.188.1.55-63.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinga I, Baptista C, Auzat I, Petipas I, Lurz R, Tavares P, Santos MA, São-José C. Role of bacteriophage SPP1 tail spike protein gp21 on host cell receptor binding and trigger of phage DNA ejection. Mol Microbiol. 2012;83:289–303. doi: 10.1111/j.1365-2958.2011.07931.x. [DOI] [PubMed] [Google Scholar]
- Waseh S, Hanifi-Moghaddam P, Coleman R, Masotti M, Ryan S, Foss M, MacKenzie R, Henry M, Szymanski CM, Tanha J. Orally administered P22 phage tailspike protein reduces Salmonella colonization in chickens: prospects of a novel therapy against bacterial infections. PLoS One. 2010;5:e13904. doi: 10.1371/journal.pone.0013904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia G, Corrigan RM, Winstel V, Goerke C, Gründling A, Peschel A. Wall teichoic acid-dependent adsorption of staphylococcal siphovirus and myovirus. J Bacteriol. 2011;193:4006–4009. doi: 10.1128/JB.01412-10. [DOI] [PMC free article] [PubMed] [Google Scholar]