Abstract
The domestic goat (Capra hircus) is an important ruminant species both as a source of antibody-based reagents for research and biomedical applications and as an economically important animal for agriculture, particularly for developing nations that maintain most of the global goat population. Characterization of the loci encoding the goat immune repertoire would be highly beneficial for both vaccine and immune reagent development. However, in goat and other species whose reference genomes were generated using short-read sequencing technologies, the immune loci are poorly assembled as a result of their repetitive nature. Our recent construction of a long-read goat genome assembly (ARS1) has facilitated characterization of all three antibody loci with high confidence and comparative analysis to cattle. We observed broad similarity of goat and cattle antibody-encoding loci but with notable differences that likely influence formation of the functional antibody repertoire. The goat heavy-chain locus is restricted to only four functional and nearly identical IGHV genes, in contrast to the ten observed in cattle. Repertoire analysis indicates that light-chain usage is more balanced in goats, with greater representation of kappa light chains (~ 20–30%) compared to that in cattle (~ 5%). The present study represents the first characterization of the goat antibody loci and will help inform future investigations of their antibody responses to disease and vaccination.
Electronic supplementary material
The online version of this article (10.1007/s00251-017-1033-3) contains supplementary material, which is available to authorized users.
Keywords: B cell receptor, Kappa light chain, Lambda light chain, Heavy chain
Introduction
Antibodies are critical constituents of adaptive humoral immunity in vertebrates. In mammalian species, antibodies are encoded by three distinct, but closely related genetic loci: the immunoglobulin (IG) heavy-chain (IGH) locus and the kappa (IGK) and lambda (IGL) light-chain loci. The IGH, IGK, and IGL loci each contain multiple variable (V), diversity (D, only in IGH), and joining (J) genes, which undergo recombination during immune cell maturation to generate a highly diverse combinatorial repertoire (Lefranc 2014). The number of V, D, and J genes is variable between species, and some species also have multiple V gene subgroups as a means to increase repertoire diversity. Humans, for example, have 123 to 129 IGHV genes of which 38 to 46 are functional and belong to seven functional IGHV subgroups (Matsuda et al. 1998; Pallares et al. 1999; Watson Corey et al. 2013). In contrast, cattle have only one functional subgroup containing ten functional IGHV genes (Niku et al. 2012; Sinclair et al. 1997). Functional diversity is maintained in cattle by an increased occurrence of sequence changes during recombination, a substantially increased average length of the heavy-chain complementarity-determining region (CDR) 3, and a small number of antibodies with an ultra-long CDR3 resulting from the usage of an exceptionally long IGHD gene (Berens et al. 1997; Ma et al. 2016; Pasman et al. 2017; Wang et al. 2013).
The tandem arrays of V, D, and J genes within the antibody loci have evolved by a process of duplication and subsequent diversification, resulting in closely related repeating sequences up to a few kilobases in length. This repetitive nature makes the characterization of antibody loci notoriously challenging. Until very recently, whole genome sequencing projects have relied on short-read sequencing with read lengths of 75 bp to 1 kb. The ability to assemble genome sequences across repetitive regions is difficult or impossible if the reads are shorter than the length of the repeat. The antibody loci in most current genome assemblies are therefore highly fragmented, incorrectly assembled, contracted due to repeats piling up at the same position during automatic assembly, or expanded due to the incorrect incorporation of allelic variants between haplotypes. The use of mate-pair sequencing to span larger distances, which can help to scaffold the assemblies to some extent, has not overcome this limitation in genome assembly projects. As a result, the best characterizations of antibody loci to date have relied on the targeted sequencing of bacterial artificial chromosomes (BACs) containing antibody sequence. Even though BAC clones are relatively short (~ 100–200 kb) and represent a single haplotype, the use of short reads in sequencing these clones cannot fully surmount the difficulties in assembling the germline sequence.
The IGH, IGK, and IGL loci have been characterized in a few species within the Cetartiodactyla, a mammalian clade which includes many agriculturally important and model species. The pig (Sus scrofa) is perhaps the best characterized of these. Much of the pig IGH locus has been characterized from overlapping BAC sequences (Eguchi-Ogawa et al. 2012; Eguchi-Ogawa et al. 2010), as have the IGK and IGL loci (Guo et al. 2016; Schwartz et al. 2012a; Schwartz et al. 2012b; Schwartz and Murtaugh 2014). Results of these studies were consistent with analyses of porcine antibody repertoire and B cell development, reviewed extensively by Butler et al. (2017). Ruminants, such as cattle (Bos taurus), sheep (Ovis aries), and goats (Capra hircus), diverged from pigs approximately 60 Mya (Meredith et al. 2011) and have evolved notable differences with respect to their antibody repertoires. For example, cattle B cells almost exclusively express lambda light chains, whereas pig B cells express lambda and kappa at approximately equal frequency (Arun et al. 1996; Butler et al. 2005; Hood et al. 1967; Sinkora et al. 2001). Cattle exclusively express a completely different IGHV subgroup compared to pigs and rely on only a single IGLV subgroup instead of two or three as in pigs. It should be noted that the antibody loci of cattle have largely been characterized using the available reference genome assemblies, which are fragmented into many contigs (Ekman et al. 2009; Niku et al. 2012; Pasman et al. 2010), and only very recently was the cattle IGH characterized using overlapping BAC clones (Ma et al. 2016). In sheep, the heavy-chain locus in the genome assembly (Oar_v3.1) is heavily disrupted and mostly missing so that only the light-chain loci are reasonably characterized (Qin et al. 2015).
Goats diverged from cattle approximately 30 Mya (Hiendleder et al. 1998) and from sheep approximately 15 Mya (Nomura et al. 2013). However, despite the economic importance of goats in both agriculture and for the production of antibody-based reagents, surprisingly little is known about the goat antibody repertoire. The San Clemente goat is a highly homozygous domesticated breed that was recently used to create the most contiguous genome assembly for a non-model organism reported to date (Bickhart et al. 2017). The assembly process used a combination of technologies including long-read Pacific Biosciences (PacBio) single molecule real-time sequencing, physical mapping methods, and error correction using shorter Illumina reads. The resulting assembly had greatly improved representations of repetitive immune complexes, including the antibody loci, which were mostly intact relative to the previous goat reference assembly, CHIR_2.0 (Bickhart et al. 2017). As such, we sought to more completely characterize the three antibody loci in the domestic goat and compare them to what is known in other related species, such as cattle, sheep, and pigs.
Materials and methods
Ethics statement
Peripheral blood samples from goats and cattle were collected in accordance with the UK Animal (Scientific Procedures) Act, 1986, and approved by The Pirbright Institute Animal Welfare Ethical Review Board.
Genomic sequence, annotation, and nomenclature
The long-read goat genome sequence assembly (Bickhart et al. 2017) was deposited in GenBank (BioProject accession: PRJNA290100). The IGH locus is contained on the scaffold for chromosome 21 (GenBank: NC_030828.1) and nine additional unplaced contigs (GenBank: LWLT01000331, LWLT01000361, LWLT01000364, LWLT01000390, LWLT01008200, LWLT01000862, LWLT01008318, LWLT01001482, and LWLT01010025). The IGL locus is on chromosome 17 (GenBank: NC_030824.1), and the IGK locus is on chromosome 11 (GenBank: NC_030818.1). These regions were manually annotated for antibody gene features using Artemis (Rutherford et al. 2000). Sequence similarity to the known cattle V, D, and J genes and light-chain constant regions (Ekman et al. 2009; Ma et al. 2016; Pasman et al. 2010) was used to predict the related genes in the goat using the basic local alignment search tool (BLAST) (Altschul et al. 1990). The identification of more divergent V genes was further aided by searching for immunoglobulin domains using CD-search (Marchler-Bauer and Bryant 2004) and comparing the resulting sequences to the IMGT database (Giudicelli et al. 2005). Pseudogenes were defined based on the presence of a truncation, nonsense mutation, frameshift, or defective initiation codon, any of which would prevent the production of a functional protein. All alignments were generated using CLUSTALW with default parameters (Thompson et al. 1994). Genes were named and amino acid positions were numbered according to the IMGT nomenclature (Lefranc 2007, 2011; Lefranc et al. 2003). Subgroup or clan designations were based on sequence similarity to known V genes in other species using the IMGT/V-QUEST (Brochet et al. 2008).
All goat IGHV genes were found to be most closely related to the human IGHV2, IGHV3, and IGHV4 subgroups using the IMGT/V-QUEST (number of aligned reference sequences 15; advanced parameters with allele *01 only), which is consistent with cattle and sheep. All four functional goat IGHV genes belong to the ruminant IGHV1 subgroup and are approximately 72 to 73% identical to the human IGHV4 subgroup. Thus, the existing IMGT nomenclature system for ruminants was used to classify the goat IGHV genes to the ruminant IGHV1, IGHV2, and IGHV3 subgroups. The IGHG1, IGHG2, and IGHG3 constant genes were named based in their similarity and likely orthology with those in cattle. Sheep sequences for IGHG were acquired from GenBank (IGHG1, accession: X69797; IGHG2, accession: X70983) or from the sheep reference assembly Oar_v3.1 (IGHG3, chr. 18: 68,549,650–68,551,242) using the Ensembl genome browser (Cunningham et al. 2015).
Animals and RNA isolation
Four apparently healthy Friesian cattle, aged 30 months, were selected from a herd at The Pirbright Institute. Additionally, four female Saanen goats, aged 6 months, were selected from a healthy commercial herd in the UK. Peripheral blood was collected by venipuncture into heparinized tubes, and mononuclear cells (PBMCs) were isolated by density gradient cell separation using Histopaque-1077 (Sigma-Aldrich), and contaminating erythrocytes were lysed in ammonium chloride lysis buffer (160 mM ammonium chloride, 170 mM Tris, pH 7.65). PBMCs were mixed with TRIzol (Thermo Fisher Scientific) and chloroform, and total RNA was extracted from the aqueous phase and purified using isopropanol and ethanol precipitations according to the manufacturer’s guidelines. Complementary DNA (cDNA) was synthesized using oligo(dT)12–18 primers and the SuperScript II reverse transcriptase kit (Thermo Fisher Scientific) according to the manufacturer’s protocol and quantified using a Qubit fluorometer (Thermo Fisher Scientific).
Relative quantification of light-chain transcripts
Oligonucleotide primers were designed to amplify IGKC and IGLC as well as the housekeeping genes PPIA, SDHA, and ACTB from both cattle and goats with product sizes of approximately 120 bp (Supplementary Table 1). Specific amplification for each primer pair was confirmed using Sanger sequencing on PCR products derived from multiple animals. Quantitative real-time PCR (qPCR) reactions were performed in triplicate with the isolated cDNA using the Luminaris Color HiGreen Low ROX qPCR master mix (Thermo Fisher Scientific) and the QuantStudio 5 (Applied Biosystems) platform. Reactions contained 10 ng cDNA, 1× master mix, and 300 nM of each primer in a final volume of 20 μl. Thermal cycling conditions were as follows: 50 °C for 2 min, 95 °C for 5 min, followed by 40 cycles at 95 °C for 15 s, 58 °C for 1 min, and 72 °C for 45 s. Dissociation curves were also generated for each reaction product using incremental heating steps from 58 to 95 °C to further assess specificity. The qbase+ software (Biogazelle NV, Belgium) was used for qPCR sample comparisons with PPIA, SDHA, and ACTB as reference genes and positive controls. The cycle threshold (Ct) values were used to calculate an expression ratio between IGKC and IGLC for each individual animal. These calculations were then used to perform a one-tailed Wilcoxon rank-sum test on the hypothesis that the distribution of IGKC expression is increased in goats compared to cattle. This test was chosen instead of the t test as it does not assume normality.
Results
The goat Ig heavy-chain locus
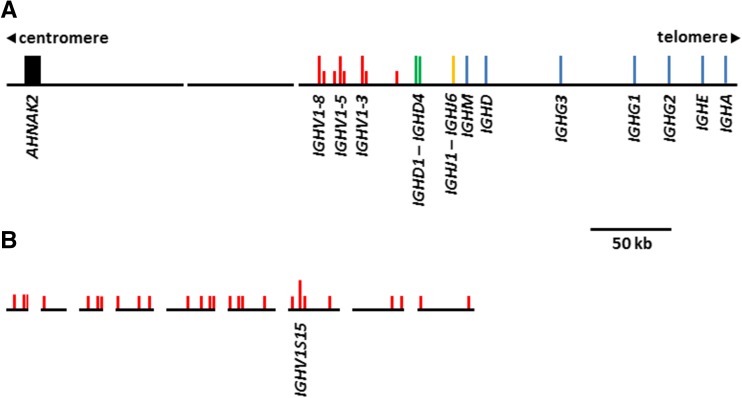
The goat heavy-chain locus is found on chromosome 21q24 (Schibler et al. 2009). Our previous analyses revealed that this region remains partially fragmented in the ARS1 assembly, possibly because the sequence data was generated from whole blood cell DNA (Bickhart et al. 2017). However, it is substantially improved compared to the previous reference assembly, CHIR_2.0. Ten contigs spanning the IGH locus were identified in the ARS1 assembly. Of these contigs, the bulk of the locus is contained on a single 275 kb contig on the chromosome 21 scaffold (Fig. 1a), whereas the remaining nine contigs comprise approximately 219 kb of unplaced sequence (Fig. 1b). Our analyses identified 34 IGHV genes across all ten contigs (Fig. 1a, b and Supplementary Table 2). Of the 34 IGHV, only four are putatively functional, and these are all very similar to each other (Supplementary Fig. 1), indicating a lack of functional germline diversity. The remaining 30 non-functional IGHV genes are typically heavily disrupted by frameshifts, premature stop codons, truncations, and disrupted promoter sequences and splice site junctions (Supplementary Table 2). Several of the unmapped IGHV genes appear to be duplicated (IGHV1S11/IGHV1S19, IGHV3S12/IGHV3S20, and IGHV2S17/IGHV2S30), although it is uncertain if this is due to the presence of nearly identical duplications in the genome or the incorporation of sequence from both chromosomes into the assembly. As in cattle, the goat IGHV genes belong to three ruminant-specific IGHV subgroups, and like cattle, the IGHV2 and IGHV3 subgroup genes are all non-functional (Niku et al. 2012). Consequently, the goat IGH repertoire is severely restricted at the germline level, suggesting that heavy-chain diversity in this species must be largely driven by post-recombinatorial mechanisms, such as somatic gene conversion and/or somatic hypermutation.
Fig. 1.
Organization of the goat antibody heavy-chain locus on the chromosome 21 scaffold (a) and unplaced contigs (b). Vertical bars indicate the position of individual genes or exons. Long vertical bars indicate putatively functional genes, whereas short bars indicate putatively non-functional genes. Only functional genes are labeled. Sequence gaps are indicated by breaks in the horizontal backbone. Red, V gene; green, D gene; orange, J gene; blue, constant region; black, other. The contigs shown in (b) are in the following order from left to right (GenBank accessions): LWLT01010025, LWLT01008318, LWLT01008200, LWLT01001482, LWLT01000862, LWLT01000390, LWLT01000364, LWLT01000361, LWLT01000331. Of these, the second, third, sixth, seventh, and ninth have been inverted to make their orientation consistent with the other contigs
The 275 kb contig placed with chromosome 21 contains the entirety of the constant region, four IGHD genes (Supplementary Fig. 2), six IGHJ genes (Supplementary Fig. 3), and the first eight IGHV genes. In contrast to cattle, there is no evidence that goats have duplicated the IGHD, IGHJ, and IGHC genes (Ma et al. 2016). As a result, goats lack the ultra-long IGHD gene as seen in cattle (Ma et al. 2016; Wang et al. 2013). We previously noted that the flanking gene AHNAK2 is found upstream from the IGHV region in the new goat ARS1 assembly, whereas in humans, this gene is downstream from the IGH constant region. This structure in the goat was supported by three radiation hybridization probes (Bickhart et al. 2017), indicating that the goat IGH locus is inverted relative to humans.
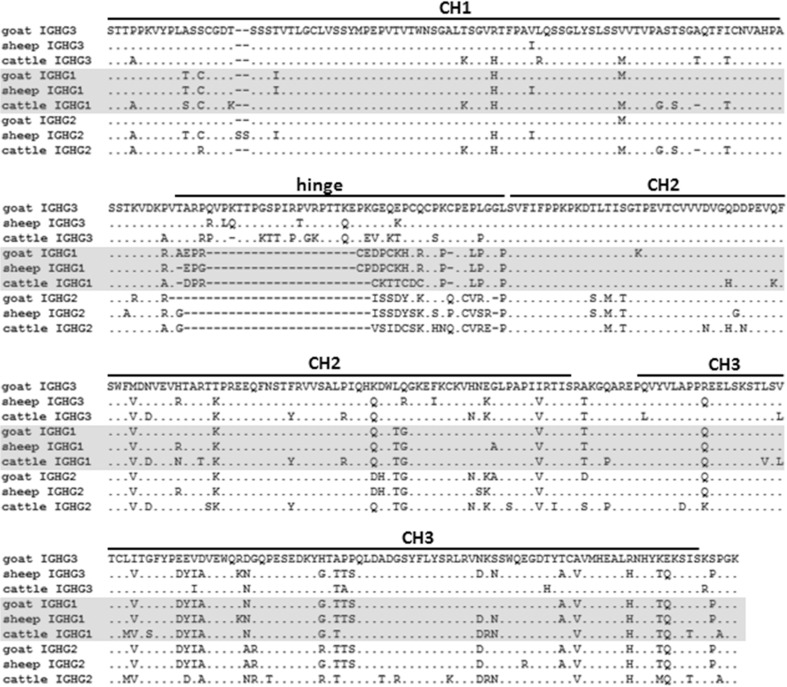
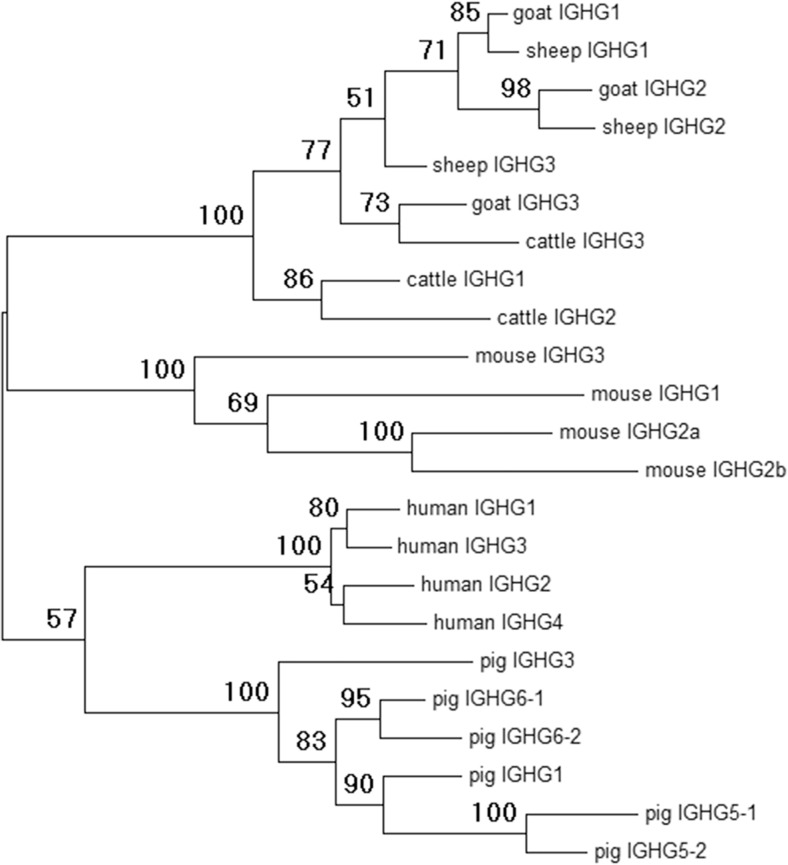
Goats have three constant region IGHG subclasses (Fig. 1a), as in cattle and sheep. Although the three immunoglobulin-like heavy-chain constant (CH) domains are all very similar to each other, the three subclasses differ greatly in the hinge region (Fig. 2), which reflects their differential abilities to interact with various Fc receptors. Phylogenetic analysis confirmed that these three ruminant subclasses also clade together when compared to pigs, mice, and humans (Fig. 3). However, among ruminants, they do not consistently clade with each other or with their respective species. This is most likely due to sequence similarity across the CH domains, which appear to have homogenized so that they appear more similar within each species (Fig. 2). Together, these data confirm that the goat IGHG subclasses share common ancestry with cattle and sheep and are likely functionally equivalent between these species.
Fig. 2.
Alignment of putative amino acid sequences for the IGHG subclasses in ruminants. IGHG subclasses are separated by gray shading. Cattle sequences are derived from a genomic BAC construct (GenBank: KT723008) as recently published (Ma et al. 2016). Sheep IGHG1 and IGHG2 are based on the mRNA GenBank accession sequences X69797 and X70983, respectively. Sheep IGHG3 is based on genomic sequence found in the Oar_v3.1 genome assembly contig AMGL01043778, acquired from the Ensembl genome browser and manually annotated
Fig. 3.
Phylogenetic analysis of putative IGHG amino acid sequences from goats, sheep, cattle, mice, humans, and pigs. Amino acid sequences for goats, sheep, and cattle were acquired as described in Fig. 2. Pig sequences were acquired from either of two genomic BAC constructs (GenBank accession: AB699686 for IGHG1, IGHG3, IGHG5-1, and IGHG5-2; AB699687 for IGHG6-1 and IGHG6-2) as previously described (Eguchi-Ogawa et al. 2012). Mouse sequences were acquired from a genomic contig (GenBank: D78344) as previously published (Akahori and Kurosawa 1997). Sequence for human IGHG1 was acquired from the genome assembly for chromosome 14 (GenBank: NC_000014.9). The remaining human sequences were acquired from cDNA (IGHG2, AJ250170; IGHG3, AJ390284; and IGHG4, AJ294733). The tree was generated using maximum likelihood based on the Jones-Taylor-Thornton model (Jones et al. 1992) and 100 bootstrap iterations within MEGA6 (Tamura et al. 2013). Nodal bootstrap values are indicated
The goat Ig lambda light-chain locus
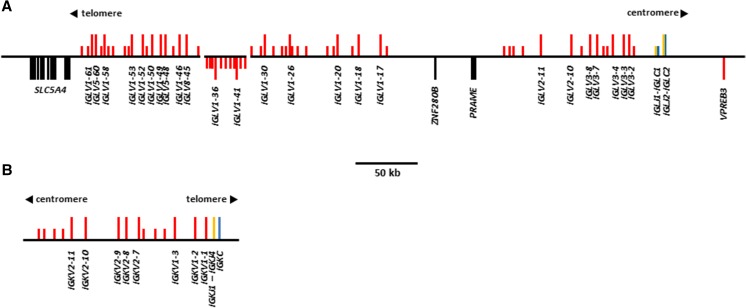
In ARS1, the IGL locus spans approximately 460 kb and contains two sequence gaps on a single scaffold representing chromosome 17. A contig located in the middle of the IGLV region is most likely erroneously inverted relative to the rest of the locus (Fig. 4a) (Bickhart et al. 2017). Our previous analysis of the goat IGLV region revealed that a substantial amount of sequence was absent from the CHIR_2.0 reference assembly—a result of the highly repetitive nature of these genes (Bickhart et al. 2017). In ARS1, the IGLV genes are separated into two distinct clusters by an 89 kb region containing the genes ZNF280B and PRAME and flanked on the 5′ end by SLC5A4. This organization is consistent with other described species, including cattle and pigs (Pasman et al. 2010; Schwartz et al. 2012b).
Fig. 4.
Organization of the antibody light-chain lambda (a) and kappa (b) loci in the domestic goat. Genes are labeled as in Fig. 1. Orientation of vertical bars relative to the horizontal backbone indicates transcriptional orientation. The regions displayed represent the ARS1 genomic scaffolds for chromosome 17: 125,000–725,000 (IGL; GenBank: NC_030824.1) and chromosome 11: 46,750,000–46,925,000 (IGK; GenBank: NC_030818.1)
The constant region is comprised of IGLJ-IGLC cassettes, of which there are two in the goat. However, goat IGLC1 is truncated by a nonsense mutation at IMGT position 41 (Supplementary Fig. 4). This likely results in a non-functional constant region, as it would prevent both the formation of the disulfide bond between the light and heavy chains as well as the internal disulfide bond between C23 and C104. This observation is consistent with sheep, which possess the same nonsense mutation (Qin et al. 2015). Likewise, in both goats and sheep, the associated IGLJ gene contains a non-canonical LGGG motif, despite having an intact recombination signal (RS) (Supplementary Fig. 5). Thus, both goats and sheep are functionally restricted to the single remaining IGLJ-IGLC cassette. Consequently, IGL-CDR3 diversity in the goat is predicted to be nearly entirely derived from the rearrangeable IGLV genes.
A total of 63 IGLV genes were identified upstream from the constant region (Fig. 4a). This is equivalent to the number previously reported in the cattle Btau_3.1 assembly (Ekman et al. 2009), yet more than the 43 IGLV in sheep (Qin et al. 2015), and more than the 23 IGLV in pigs (Schwartz et al. 2012b; Schwartz and Murtaugh 2014). The IGLV subgroups are relatively consistent between these species, with those of the IGLC-proximal cluster being most similar to the IGLV2 and IGLV3 subgroups of humans, and those of the IGLC-distal cluster most similar to the IGLV1, IGLV5, and IGLV8 subgroups. Of the 63 IGLV genes in the goat, 25 are putatively functional and spread across these five subgroups, although 15 of these belong to IGLV1 and five belong to IGLV3 (Supplementary Table 3 and Supplementary Fig. 6).
The goat Ig kappa light-chain locus
The goat IGK locus on chromosome 11 is contained on a single contig (Bickhart et al. 2017). The 5′ end of the locus is flanked by a gene desert with no additional immunoglobulin domains identified within 200 kb upstream of the first IGKV gene, indicating that the entire locus is present. The IGK locus is considerably reduced in size compared to the IGL and contains only 15 IGKV genes (Fig. 4b), which is also fewer than the 24 IGKV reported in cattle (Ekman et al. 2009). However, both goats and cattle possess eight putatively functional IGKV (Supplementary Table 4 and Supplementary Fig. 7). Of these functional IGKV in the goat, five belong to the IGKV2 subgroup and the other three to the IGKV1. Two of the three IGKV1 genes possess a non-canonical heptamer in their RS, likely rendering V-J recombination highly inefficient. In contrast, all five IGKV2 genes possess a canonical RS, suggesting that these are likely represented in the majority of expressed kappa chains.
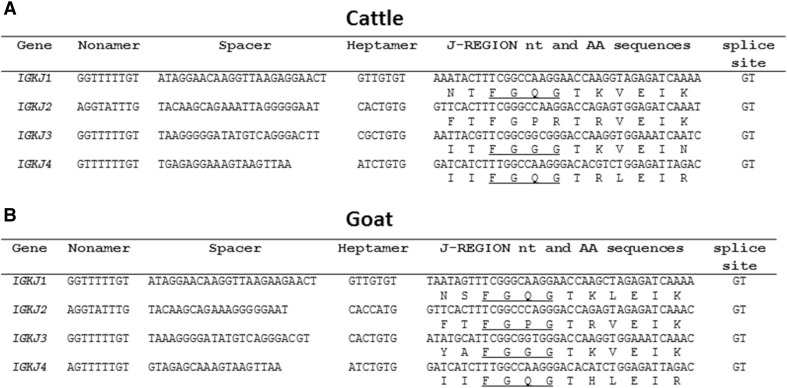
There are four IGKJ genes downstream from the IGKV region, one more than previously reported in cattle (Ekman et al. 2009). However, the current cattle reference genome contains a sequence homologous to the fourth goat IGKJ in the position implied by conserved synteny, indicating that the IGKJ repertoire is highly similar between these two species (Fig. 5a, b). Of the four goat IGKJ genes, IGKJ3 is the only one associated with both a canonical heptamer (i.e. CACTGTG) and a canonical 23-bp spacer within the RS. This strongly suggests that only IGKJ3 can be efficiently rearranged in the goat (Fig. 5b). We additionally observed from the three published cattle IGKJ sequences (as well as the unpublished IGKJ4) that only cattle IGKJ2 possesses a canonical heptamer (Fig. 5a). However, cattle IGKJ2 is rendered non-functional due to a mutated W/F-G-X-G motif in its framework region (Ekman et al. 2009). As a result, cattle most likely rely on IGKJ3 which is likely inefficiently rearranged due to its mutated heptamer.
Fig. 5.
Genomic features of the cattle (a) and goat (b) IGKJ genes. The canonical W/F-G-X-G framework motif is underlined where intact
IGK is more abundantly expressed in goats compared to cattle
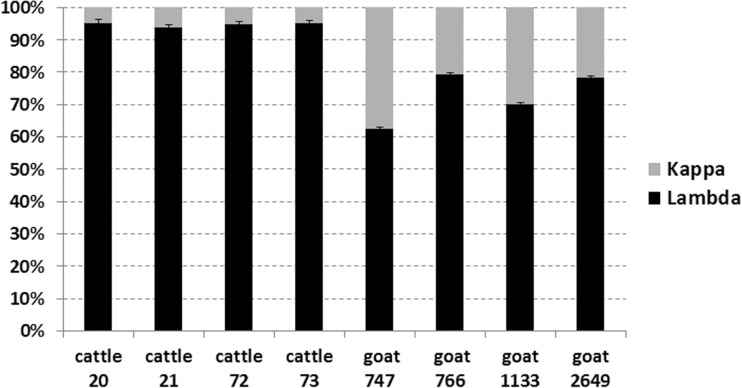
Because we observed that all of the cattle IGKJ sequences possess either a non-canonical RS or a non-functional framework motif and that goats possess a fully functional IGKJ3 with an intact RS, we hypothesized that the proportional contribution of kappa light chains in the expressed antibody repertoire is higher in goats relative to cattle. We therefore tested the relative contribution of IGLC and IGKC in both goat and cattle PBMCs using qPCR. Our results confirmed that IGK usage in the cattle antibody repertoire is approximately 5% (Fig. 6), which is consistent with previous reports (Arun et al. 1996; Butler et al. 2005; Hood et al. 1967; Sinkora et al. 2001), and approximately 20–35% in goats (Fig. 6). This difference between cattle and goat light-chain usage was statistically significant (P = 0.014286) using a Wilcoxon rank-sum test. The single mutation in the heptamer of IGKJ3 (i.e. CACTGTG > CGCTGTG) likely drives the restriction of IGK in cattle.
Fig. 6.
Relative quantification of IGKC (gray) and IGLC (black) transcripts in cattle and goat PBMCs from four animals each. Results represent the average of three replicates, and standard error between replicates is shown. The greater expression of IGKC in goats is statistically significant (P = 0.014286) using a one-tailed Wilcoxon rank-sum test
Discussion
The goat antibody loci and light-chain usage relative to other related species
We have presently characterized the three antibody loci in the domestic goat and, where possible, compared their germline repertoire to related species. Although the IGHV region remains disrupted in the new long-read ARS1 assembly, our identification of numerous IGHV genes confirms that the goat utilizes the same subgroup of closely related genes as cattle. However, only four nearly identical putatively functional IGHV were identified in the goat compared to ten identified previously in cattle (Niku et al. 2012). It is possible given the degree of fragmentation in this region that there are more IGHV genes in goats not identified here; although given the completeness of the other antibody loci and the use of both PacBio and Illumina reads to generate and error correct the assembly, it seems unlikely that many would be missing. The degree of fragmentation observed in the IGH locus is quite possibly due to the use of DNA extracted from blood as the sequencing material, since much of this would be from circulating B cells that have rearranged their antibody loci. Furthermore, unlike cattle, goats have not duplicated the IGHD or IGHJ regions or the IGHC region (specifically IGHD and IGHM) (Ma et al. 2016; Wang et al. 2013). Because of this, goats lack the IGHD gene that enables cattle to generate ultra-long heavy-chain CDR3s. The paucity of germline heavy-chain diversity in the goat suggests that this and other species are heavily reliant on junctional diversity, somatic hypermutation, and/or somatic gene conversion for the diversification of its primary heavy-chain repertoire, although detailed studies of repertoire formation are needed to understand these processes.
We identified the same number of IGLV genes in the goat (n = 63) as previously identified in cattle (Ekman et al. 2009), which is also more than found in sheep (n = 43) (Qin et al. 2015). However, both the sheep and cattle IGL characterizations were based on the current reference genome assemblies which are heavily fragmented across the IGLV region (Ekman et al. 2009; Qin et al. 2015). It is therefore quite likely that there remain unidentified genes in these species. Indeed, our preliminary analysis of a recent long-read assembly in cattle indicates that there are many additional unreported IGLV genes in that species (unpublished).
The goat IGK locus is substantially reduced in size and complexity relative to the IGL. Although a similar arrangement exists in cattle, we found that goats utilize IGK more than cattle (~ 20–30 vs ~ 5%, respectively) but less than pigs which use IGK in approximately half of all antibodies (Arun et al. 1996; Butler 1983; Butler et al. 2005; Hood et al. 1967; Sinkora et al. 2001). Nevertheless, the ruminant IGK locus appears to be no less complex than in pigs which possess ≥ 9 functional IGKV and five functional IGKJ, although only one of these IGKJ has a canonical RS (Schwartz et al. 2012a). Goats, cattle, and pigs all possess intact kappa enhancers, a kappa-deleting element, and an IGKJ-IGKC intronic RS heptamer (Das et al. 2009). This, combined with the lack of canonical RS heptamers in the functional cattle IGKJ genes suggests that in cattle, recombination may favor either non-functional IGK rearrangements or IGK ablation via recombination with the intronic heptamer and/or the kappa-deleting element downstream from the IGKC. In either case, this results in a substantially reduced expressed IGK repertoire in cattle. The abundance of goat light-chain genes, the relative lack of functional IGH genes, and the greater usage of IGK in goats indicate that the potential light-chain diversity in the available repertoire is greater in goats than in cattle, while germline IGH diversity is reduced. The comparison of the expressed (i.e. mRNA) antibody repertoire with the genomic sequence is beyond the scope of the present study. Future work is therefore necessary to confirm gene usage and investigate the mechanisms of post-recombinatorial diversification of goat B cells.
IGHG subclasses are predicted to be functionally equivalent between cattle and goats
The IGHG constant region subclasses are functionally responsible for a diverse range of responses, which are often subclass-specific. Except between closely related species, such as ruminants, the IGHG subclasses have independently expanded and diversified following speciation (Butler and Wertz 2006; Butler et al. 2009; Eguchi-Ogawa et al. 2012). In contrast, early investigations indicated that cattle, sheep, and goat IgG subclasses were very similar to each other based on the cross-reactivity of subclass-specific antisera (Butler 1983). We identified apparent gene conversion-mediated homogenization between the IGHG subclasses within the Ig domains, perhaps indicative of selective pressure to maintain similar Ig constant domains between the IGHG subclasses. However, the hinge region remained unique between the subclasses and very similar between cattle, goats, and sheep. Because the CH2-proximal portion of the hinge is known to be directly involved in FcγR binding (Chappel et al. 1991; Duncan et al. 1988; Radaev and Sun 2002), the relative conservation of the hinge region of these species’ IGHG subclasses strongly suggests that the subclasses are both functionally equivalent between these species and descended from common ancestral genes. Cattle IGHG2 is known to have a unique Fc receptor (FcR) encoded by FCG2R (Zhang et al. 1995) and which appears to play a major role in pathogen opsonization and phagocytosis (Howard 1984). Peculiarly, this receptor is poorly related to the other FcγRs but very closely related to the leukocyte immunoglobulin-like receptors (LILR) encoded within the leukocyte receptor complex (LRC) (Morton et al. 1999; Zhang et al. 1995). The conservation of the hinge region in goat IGHG2 is further consistent with our observations that FCG2R is conserved in the goat LRC yet absent from the pig LRC (J.C. Schwartz and J.A. Hammond, unpublished). The current results will enable more detailed studies into goat humoral immunity and B cell development and fill an important evolutionary link in our understanding of the B cell repertoire.
Electronic supplementary material
(PDF 1194 kb)
Acknowledgements
We kindly thank Dr. Karin Darpel and Sophia Hodgson (The Pirbright Institute) for providing peripheral blood from the commercial goats. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the United State Department of Agriculture (USDA).
Funding information
JCS and JAH were supported by the United Kingdom Biotechnology and Biological Sciences Research Council (BBSRC) through projects BBS/E/I/00001710 and BBS/E/I/00007015. RLP was supported by pre-doctoral studentship funding from The Pirbright Institute and from the University of Glasgow West of Scotland Bioscience (WestBio) Doctoral Training Partnership funded by the BBSRC grant BB/J013854/1. DMB was supported in part by appropriated project 1265-31000-096-00, “Improving Genetic Predictions in Dairy Animals Using Phenotypic and Genomic Information”, of the USDA Agricultural Research Service (ARS). DMB and TPLS were also supported by the Agricultural Food Research Initiative (AFRI) competitive grant number 2015-67015-22970 from the USDA National Institute of Food and Agriculture (NIFA) Animal Health Program.
Compliance with ethical standards
Ethics approval
Peripheral blood samples from goats and cattle were collected in accordance with the UK Animal (Scientific Procedures) Act, 1986, and approved by The Pirbright Institute Animal Welfare Ethical Review Board.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s00251-017-1033-3) contains supplementary material, which is available to authorized users.
References
- Akahori Y, Kurosawa Y. Nucleotide sequences of all the gamma gene loci of murine immunoglobulin heavy chains. Genomics. 1997;41:100–104. doi: 10.1006/geno.1997.4606. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arun SS, Breuer W, Hermanns W. Immunohistochemical examination of light-chain expression (lambda/kappa ratio) in canine, feline, equine, bovine and porcine plasma cells. Zentralbl Veterinarmed A. 1996;43:573–576. doi: 10.1111/j.1439-0442.1996.tb00489.x. [DOI] [PubMed] [Google Scholar]
- Berens SJ, Wylie DE, Lopez OJ. Use of a single VH family and long CDR3s in the variable region of cattle Ig heavy chains. Int Immunol. 1997;9:189–199. doi: 10.1093/intimm/9.1.189. [DOI] [PubMed] [Google Scholar]
- Bickhart DM, Rosen BD, Koren S, Sayre BL, Hastie AR, Chan S, Lee J, Lam ET, Liachko I, Sullivan ST, Burton JN, Huson HJ, Nystrom JC, Kelley CM, Hutchison JL, Zhou Y, Sun J, Crisa A, Ponce de Leon FA, Schwartz JC, Hammond JA, Waldbieser GC, Schroeder SG, Liu GE, Dunham MJ, Shendure J, Sonstegard TS, Phillippy AM, Van Tassell CP, Smith TPL. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat Genet. 2017;49:643–650. doi: 10.1038/ng.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JE. Bovine immunoglobulins: an augmented review. Vet Immunol Immunopathol. 1983;4:43–152. doi: 10.1016/0165-2427(83)90056-9. [DOI] [PubMed] [Google Scholar]
- Butler JE, Wertz N. Antibody repertoire development in fetal and neonatal piglets. XVII. IgG subclass transcription revisited with emphasis on new IgG3. J Immunol. 2006;177:5480–5489. doi: 10.4049/jimmunol.177.8.5480. [DOI] [PubMed] [Google Scholar]
- Butler JE, Wertz N, Sun J, Wang H, Lemke C, Chardon P, Piumi F, Wells K. The pre-immune variable kappa repertoire of swine is selectively generated from certain subfamilies of Vkappa2 and one Jkappa gene. Vet Immunol Immunopathol. 2005;108:127–137. doi: 10.1016/j.vetimm.2005.07.016. [DOI] [PubMed] [Google Scholar]
- Butler JE, Wertz N, Deschacht N, Kacskovics I. Porcine IgG: structure, genetics, and evolution. Immunogenetics. 2009;61:209–230. doi: 10.1007/s00251-008-0336-9. [DOI] [PubMed] [Google Scholar]
- Butler JE, Wertz N, Sinkora M. Antibody repertoire development in swine. Annu Rev Anim Biosci. 2017;5:255–279. doi: 10.1146/annurev-animal-022516-022818. [DOI] [PubMed] [Google Scholar]
- Chappel MS, Isenman DE, Everett M, Xu YY, Dorrington KJ, Klein MH. Identification of the Fc gamma receptor class I binding site in human IgG through the use of recombinant IgG1/IgG2 hybrid and point-mutated antibodies. Proc Natl Acad Sci U S A. 1991;88:9036–9040. doi: 10.1073/pnas.88.20.9036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Giron CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Kahari AK, Keenan S, Martin FJ, Maurel T, McLaren W, Murphy DN, Nag R, Overduin B, Parker A, Patricio M, Perry E, Pignatelli M, Riat HS, Sheppard D, Taylor K, Thormann A, Vullo A, Wilder SP, Zadissa A, Aken BL, Birney E, Harrow J, Kinsella R, Muffato M, Ruffier M, Searle SM, Spudich G, Trevanion SJ, Yates A, Zerbino DR, Flicek P. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Nikolaidis N, Nei M. Genomic organization and evolution of immunoglobulin kappa gene enhancers and kappa deleting element in mammals. Mol Immunol. 2009;46:3171–3177. doi: 10.1016/j.molimm.2009.05.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AR, Woof JM, Partridge LJ, Burton DR, Winter G. Localization of the binding site for the human high-affinity Fc receptor on IgG. Nature. 1988;332:563–564. doi: 10.1038/332563a0. [DOI] [PubMed] [Google Scholar]
- Eguchi-Ogawa T, Wertz N, Sun XZ, Piumi F, Uenishi H, Wells K, Chardon P, Tobin G. Antibody repertoire development in fetal and neonatal piglets. XI. The relationship of variable heavy chain gene usage and the genomic organization of the variable heavy chain locus. J Immunol. 2010;184:3734–3742. doi: 10.4049/jimmunol.0903616. [DOI] [PubMed] [Google Scholar]
- Eguchi-Ogawa T, Toki D, Wertz N, Butler JE, Uenishi H. Structure of the genomic sequence comprising the immunoglobulin heavy constant (IGHC) genes from Sus scrofa. Mol Immunol. 2012;52:97–107. doi: 10.1016/j.molimm.2012.05.010. [DOI] [PubMed] [Google Scholar]
- Ekman A, Niku M, Liljavirta J, Iivanainen A. Bos taurus genome sequence reveals the assortment of immunoglobulin and surrogate light chain genes in domestic cattle. BMC Immunol. 2009;10:22. doi: 10.1186/1471-2172-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudicelli V, Chaume D, Lefranc MP. IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 2005;33:D256–D261. doi: 10.1093/nar/gki010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Schwartz JC, Murtaugh MP (2016) Genomic variation in the porcine immunoglobulin lambda variable region. Immunogenetics:1–9. 10.1007/s00251-016-0899-9 [DOI] [PMC free article] [PubMed]
- Hiendleder S, Lewalski H, Wassmuth R, Janke A. The complete mitochondrial DNA sequence of the domestic sheep (Ovis aries) and comparison with the other major ovine haplotype. J Mol Evol. 1998;47:441–448. doi: 10.1007/PL00006401. [DOI] [PubMed] [Google Scholar]
- Hood L, Gray W, Sanders B, Dreyer W. Light chain evolution. In: Cairns J, editor. Cold Spring Harb Symp Quant Biol. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1967. [Google Scholar]
- Howard CJ. Comparison of bovine IgG1, IgG2 and IgM for ability to promote killing of Mycoplasma bovis by bovine alveolar macrophages and neutrophils. Vet Immunol Immunopathol. 1984;6:321–326. doi: 10.1016/0165-2427(84)90057-6. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Lefranc MP. WHO-IUIS nomenclature subcommittee for immunoglobulins and T cell receptors report. Immunogenetics. 2007;59:899–902. doi: 10.1007/s00251-007-0260-4. [DOI] [PubMed] [Google Scholar]
- Lefranc MP. IMGT unique numbering for the variable (V), constant (C), and groove (G) domains of IG, TR, MH, IgSF, and MhSF. Cold Spring Harb Protoc. 2011;2011:633–642. doi: 10.1101/pdb.ip85. [DOI] [PubMed] [Google Scholar]
- Lefranc M-P. Immunoglobulin and T cell receptor genes: IMGT(®) and the birth and rise of immunoinformatics. Front Immunol. 2014;5:22. doi: 10.3389/fimmu.2014.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc MP, Pommie C, Ruiz M, Giudicelli V, Foulquier E, Truong L, Thouvenin-Contet V, Lefranc G. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev Comp Immunol. 2003;27:55–77. doi: 10.1016/S0145-305X(02)00039-3. [DOI] [PubMed] [Google Scholar]
- Ma L, Qin T, Chu D, Cheng X, Wang J, Wang X, Wang P, Han H, Ren L, Aitken R, Hammarstrom L, Li N, Zhao Y. Internal duplications of DH, JH, and C region genes create an unusual IgH gene locus in cattle. J Immunol. 2016;196:4358–4366. doi: 10.4049/jimmunol.1600158. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Ishii K, Bourvagnet P, K-i K, Hayashida H, Miyata T, Honjo T. The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J Exp Med. 1998;188:2151–2162. doi: 10.1084/jem.188.11.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith RW, Janečka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simão TLL, Stadler T, Rabosky DL, Honeycutt RL, Flynn JJ, Ingram CM, Steiner C, Williams TL, Robinson TJ, Burk-Herrick A, Westerman M, Ayoub NA, Springer MS, Murphy WJ. Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science. 2011;334:521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- Morton HC, van Zandbergen G, van Kooten C, Howard CJ, van de Winkel JG, Brandtzaeg P. Immunoglobulin-binding sites of human FcalphaRI (CD89) and bovine Fcgamma2R are located in their membrane-distal extracellular domains. J Exp Med. 1999;189:1715–1722. doi: 10.1084/jem.189.11.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niku M, Liljavirta J, Durkin K, Schroderus E, Iivanainen A. The bovine genomic DNA sequence data reveal three IGHV subgroups, only one of which is functionally expressed. Dev Comp Immunol. 2012;37:457–461. doi: 10.1016/j.dci.2012.02.006. [DOI] [PubMed] [Google Scholar]
- Nomura K, Yonezawa T, Mano S, Kawakami S, Shedlock AM, Hasegawa M, Amano T. Domestication process of the goat revealed by an analysis of the nearly complete mitochondrial protein-encoding genes. PLoS One. 2013;8:e67775. doi: 10.1371/journal.pone.0067775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallares N, Lefebvre S, Contet V, Matsuda F, Lefranc MP. The human immunoglobulin heavy variable genes. Exp Clin Immunogenet. 1999;16:36–60. doi: 10.1159/000019095. [DOI] [PubMed] [Google Scholar]
- Pasman Y, Saini SS, Smith E, Kaushik AK. Organization and genomic complexity of bovine lambda-light chain gene locus. Vet Immunol Immunopathol. 2010;135:306–313. doi: 10.1016/j.vetimm.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Pasman Y, Merico D, Kaushik AK. Preferential expression of IGHV and IGHD encoding antibodies with exceptionally long CDR3H and a rapid global shift in transcriptome characterizes development of bovine neonatal immunity. Dev Comp Immunol. 2017;67:495–507. doi: 10.1016/j.dci.2016.08.020. [DOI] [PubMed] [Google Scholar]
- Qin T, Liu Z, Zhao H. Organization and genomic complexity of sheep immunoglobulin light chain gene loci. Immunol Lett. 2015;168:313–318. doi: 10.1016/j.imlet.2015.10.010. [DOI] [PubMed] [Google Scholar]
- Radaev S, Sun P. Recognition of immunoglobulins by Fcγ receptors. Mol Immunol. 2002;38:1073–1083. doi: 10.1016/S0161-5890(02)00036-6. [DOI] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Schibler L, Di Meo GP, Cribiu EP, Iannuzzi L. Molecular cytogenetics and comparative mapping in goats (Capra hircus, 2n = 60) Cytogenet Genome Res. 2009;126:77–85. doi: 10.1159/000245908. [DOI] [PubMed] [Google Scholar]
- Schwartz JC, Murtaugh MP. Characterization of a polymorphic IGLV gene in pigs (Sus scrofa) Immunogenetics. 2014;66:507–511. doi: 10.1007/s00251-014-0785-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Lefranc MP, Murtaugh MP. Evolution of the porcine (Sus scrofa domestica) immunoglobulin kappa locus through germline gene conversion. Immunogenetics. 2012;64:303–311. doi: 10.1007/s00251-011-0589-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Lefranc MP, Murtaugh MP. Organization, complexity and allelic diversity of the porcine (Sus scrofa domestica) immunoglobulin lambda locus. Immunogenetics. 2012;64:399–407. doi: 10.1007/s00251-011-0594-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair MC, Gilchrist J, Aitken R. Bovine IgG repertoire is dominated by a single diversified VH gene family. J Immunol. 1997;159:3883–3889. [PubMed] [Google Scholar]
- Sinkora J, Rehakova Z, Samankova L, Haverson K, Butler JE, Zwart R, Boersma W. Characterization of monoclonal antibodies recognizing immunoglobulin kappa and lambda chains in pigs by flow cytometry. Vet Immunol Immunopathol. 2001;80:79–91. doi: 10.1016/S0165-2427(01)00277-X. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22 (22):4673-4680 [DOI] [PMC free article] [PubMed]
- Wang F, Ekiert DC, Ahmad I, Yu W, Zhang Y, Bazirgan O, Torkamani A, Raudsepp T, Mwangi W, Criscitiello MF, Wilson IA, Schultz PG, Smider VV. Reshaping antibody diversity. Cell. 2013;153:1379–1393. doi: 10.1016/j.cell.2013.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson Corey T, Steinberg Karyn M, Huddleston J, Warren Rene L, Malig M, Schein J, Willsey AJ, Joy Jeffrey B, Scott Jamie K, Graves TA, Wilson Richard K, Holt Robert A, Eichler Evan E, Breden F. Complete haplotype sequence of the human immunoglobulin heavy-chain variable, diversity, and joining genes and characterization of allelic and copy-number variation. Am J Hum Genet. 2013;92:530–546. doi: 10.1016/j.ajhg.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Young JR, Tregaskes CA, Sopp P, Howard CJ. Identification of a novel class of mammalian Fc gamma receptor. J Immunol. 1995;155:1534–1541. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1194 kb)