Abstract
Oncogenic rearrangements leading to targetable gene fusions are well-established cancer driver events in lung adenocarcinoma. Accurate and reliable detection of these gene fusions is crucial to select the appropriate targeted therapy for each patient. We compared the targeted next-generation-sequencing Oncomine Focus Assay (OFA; Thermo Fisher Scientific) with conventional ALK FISH and anti-Alk immunohistochemistry in a cohort of 52 lung adenocarcinomas (10 ALK rearranged, 18 non-ALK rearranged, and 24 untested cases). We found a sensitivity and specificity of 100% for detection of ALK rearrangements using the OFA panel. In addition, targeted next generation sequencing allowed us to analyze a set of 23 driver genes in a single assay. Besides EML4-ALK (11/52 cases), we detected EZR-ROS1 (1/52 cases), KIF5B-RET (1/52 cases) and MET-MET (4/52 cases) fusions. All EML4-ALK, EZR-ROS1 and KIF5B-RET fusions were confirmed by multiplexed targeted next generation sequencing assay (Oncomine Solid Tumor Fusion Transcript Kit, Thermo Fisher Scientific). All cases with EML4-ALK rearrangement were confirmed by Alk immunohistochemistry and all but one by ALK FISH. In our experience, targeted next-generation sequencing is a reliable and timesaving tool for multiplexed detection of targetable rearrangements. Therefore, targeted next-generation sequencing represents an efficient alternative to time-consuming single target assays currently used in molecular pathology.
Keywords: Next generation sequencing, Lung adenocarcinoma, Gene fusion, ALK gene
1. Introduction
Since its discovery in 2007, oncogenic EML4-ALK rearrangements have been intensively studied in lung cancer biology and therapy [[1], [2], [3], [4]]. Meanwhile, first line Alk kinase inhibitor therapy with crizotinib is the current standard of care in ALK rearranged lung adenocarcinoma (LUAD) with increased progression-free survival compared with conventional chemotherapy [5]. Therefore, a reliable and accurate detection of such ALK rearrangements is essential for the molecular pathology workflow. Approximately 3–7% of LUADs harbor ALK rearrangements in Caucasian populations. Histological morphology of ALK rearranged LUAD is typically solid with few foci of signet ring cells [3]. Other cancer driver fusion genes in LUAD are ROS1 and RET [[6], [7], [8]]. The resulting chimeric proteins also are therapeutic targets [[9], [10], [11]]. ROS1 rearrangements are found in approximately 2% of LUADs and RET rearrangements in 1%, respectively. MET splice site mutations resulting in exon 14 skipping and activation of the c-Met pathway occur in approximately 4% of LUADs [12,13]. Patients with these mutations were shown to respond to MET inhibition [14]. Multiplexed assays like targeted next-generation-sequencing (NGS) approaches allow the analysis of large set of genomic alterations compared with single target assays like fluorescence in-situ hybridization (FISH) and immunohistochemistry (IHC). We have previously demonstrated the feasibility and reliable application of DNA- and RNA- based targeted sequencing in a cohort of small tissue samples and cytological specimens [15]. The aim of the present study was to investigate the performance of a RNA-based targeted NGS assay for detection of targetable fusion genes and compare the results with corresponding FISH and IHC assays.
2. Materials and methods
2.1. Patient samples and cell lines
We tested a cohort of advanced lung adenocarcinomas (LUADs) (n = 52) in this retrospective validation study (Table 1). Formalin-fixed paraffin-embedded (FFPE) LUAD tissue blocks were collected from our archives between 2003 and 2008. All samples were processed according to National Comprehensive Cancer Network (NCCN) and Swiss Society of Pathology (SSPath) guidelines. Tumor cell content was assessed by board-certified pathologists on a multi-headed microscope (VT, AS, BV). Only unambiguous LUAD samples with tumor cell content ≥ 60% were included in the study cohort. Among included LUAD cases, 10 samples were ALK rearranged, 18 cases were non-ALK rearranged as detected by fluorescence in-situ hybridization (FISH). The remaining 24 cases had not been tested before (Table 1). The study was approved by the Cantonal Ethics Committee of Zurich (StV-No. 2009-0029 and KEK-ZH-No. 2014-0604).
Table 1.
LUAD samples included in the study cohort (n = 52).
| ALK status | Sample No. |
|---|---|
| Positive, n = 10 | 1–10 |
| Negative, n = 18 | 11–20, 32, 36, 42, 43, 45, 46, 51, 52 |
| Unknown, n = 24 | 21–31, 33–35, 37–41, 44, 47–50 |
ALK, anaplastic lymphoma kinase.
H3122 (EML4-ALK rearranged) and HCC-44 (no EML4-ALK rearrangement) were grown in RPMI 1640 medium (Thermo Fisher Scientific, Carlsbad, CA) with 10% FBS at 37 °C in humidified atmosphere with 5% CO2 to 70% confluency. The cells were harvested after rinsing with phosphate buffered saline using 0.25% trypsine (Thermo Fisher Scientific, Carlsbad, CA). Cells were washed in RPMI medium, pelleted and formalin fixated. For cell blocks, protein glycerol (Morphisto GmbH, Frankfurt) clotting followed by routine histological processing was performed.
2.2. RNA extraction
We extracted RNA from three tissue cores (diameter 0.6 mm) punched from the formalin-fixed paraffin-embedded (FFPE) tissue blocks or from FFPE sections of the cell blocks from cell lines. Normal tissue was not analyzed. Tumor tissue cylinders were deparaffinized with 1000 μl xylene and washed twice in 800 μl ethanol. After drying at 37 °C the samples were digested with proteinase K at 56 °C overnight. To avoid genomic DNA contamination, samples were treated with DNase1 for 15 min at room temperature (RT). RNA extraction was performed applying the Maxwell 16 LEV RNA FFPE Purification Kit (Promega). RNA was quantified with Qubit 2.0 using the RNA HS Assay Kit (Thermo Fisher Scientific). We assessed RNA quality with the Agilent 2100 Bioanalyzer (Agilent Technologies, Basel, Switzerland).
2.3. Targeted NGS
Targeted RNA-based NGS of LUADs has already been performed on a cohort of small biopsies and cytology smears at the Institute of Pathology and Molecular Pathology [15]. Targeted RNA-based NGS was conducted with the Oncomine Focus Assay (OFA) panel (Thermo Fisher Scientific, Carlsbad, CA) which is designed to detect gene fusions involving 23 fusion drivers (ABL1, ALK, AKT3, AXL, BRAF, EGFR, ERBB2, ERG, ETV1, ETV4, ETV5, FGFR1, FGFR2, FGFR3, MET, NTRK1, NTRK2, NTRK3, PDGFRA, PPARG, RAF1, RET, ROS1).
We validated the OFA results with the Oncomine Solid Tumor Fusion Transcript Kit (Thermo Fisher Scientific), another targeted NGS assay to detect relevant gene fusions (ALK, ROS1, RET, NTRK). All libraries were prepared using 10 ng of starting RNA. RNA was first reverse transcribed with the Invitrogen SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific). The resulting cDNA was used as input for the targeted amplification. After targeted amplification with the corresponding panel, all libraries were labeled with Ion Xpress™ Barcode Adapters (Thermo Fisher Scientific). Libraries were quantified and mixed according to manufacturer’s recommendations. The Ion Hi-Q Chef Kit and the Ion Chef System were used for template preparation and enrichment. Enriched libraries were then loaded on the Ion 318 Select Chip and sequenced on the Ion PGM System (Thermo Fisher Scientific) using the Ion PGM Hi-Q Sequencing Kit (Thermo Fisher Scientific). The detection of a rearrangement in a sample was judged as true positive following the manufacturer’s recommendations (Thermo Fisher Scientific). Statistics and sequencing data analysis were performed as described [15]. Visualization of detected fusion events was made using Integrative Genomics Viewer (IGV; Broad Institute, Cambridge, MA) demonstrating the alignment of sequenced reads to the reads of known fusion breakpoints and the reference human genome hg19.
2.4. Fluorescence in-situ hybridization (FISH)
For testing of ALK, ROS1, and RET rearrangements, the Abbott Molecular/Vysis LSI ALK Break Apart Rearrangement Probe (Abbott Molecular, Baar, Switzerland), ZytoLight SPEC ROS1 Dual Color Break Apart Probe (Zytovision GmbH, Bremerhaven, Germany), and ZytoLight SPEC RET Dual Break Apart Probe (Zytovision GmbH, Bremerhaven, Germany) were applied. FISH testing was performed on whole sections of LUAD specimens. For each particular case, a board certified pathologist analyzed 100 tumor nuclei. A sample was called true positive if ≥15% of tumor nuclei showed split signals according to the manufacturer’s evaluation guidelines (Abbott Molecular, Des Plaines, IL). A second pathologist independently counted borderline cases.
2.5. Immunohistochemistry (IHC)
Alk and Ros1 IHC was performed on 0.6 mm tissue cylinders as previously described [16,17]. For Alk IHC, the mouse anti-human ALK monoclonal antibody was applied (clone 5A4, Leica Biosystems). Ros1 IHC was conducted using a rabbit anti-human ROS1 monoclonal antibody (clone D4D6, Cell Signaling Technology). For c-Met IHC a monoclonal rabbit anti-human Met antibody was used (clone SP44, Spring Biosciences). All buffers, including pretreatment CC1 standard incubation buffer, secondary antibody (UltraMap anti-Rabbit HRP) and detection system Discovery ChromoMap DAB, were purchased from Roche Ventana (Tucson, AZ). Immunostainings were performed on the automated immunostainer DiscoveryUltra (Roche Ventana).
3. Results
3.1. RNA metrics
We examined 52 LUAD cases, of which 10 had ALK rearrangement confirmed by FISH and immunohistochemistry (Table 1). Eighteen cases were confirmed non-ALK rearranged samples by FISH and immunohistochemistry (Table 1). The remaining 24 cases had an unknown ALK status (Table 1). The extracted RNA of the three FFPE tissue cores per case showed an average RNA concentration of 48 ng/μl (median 42.2 ng/μl, standard deviation ± 29.5 ng/μl, range 11.2–152 ng/μl) and a low RNA integrity number (RIN) of 1.1–2.6 (13 cases RIN not available, Table 2). The fragment length >150 bp ranged from 22 to 90% (13 cases not available, Table 2). All RNA samples were processed using two assays, the Oncomine Focus Assay (OFA) and the Oncomine Solid Tumor Fusion Transcript Kit, respectively. For the OFA panel, the total mapped fusion panel reads ranged from 25,705–269,517 (Table 2). Amplification of the 5 control genes was successful in all cases. The number of total mapped fusion panel reads (TMFPR) was not correlated with the RIN (correlation coefficient 0.108, p-value 0.5139) or with the percentage of fragments >150 bp (correlation coefficient 0.268, p-value 0.09).
Table 2.
Metrics of extracted RNA and NGS parameters of the Oncomine Focus Assay panel.
| Sample No. | Quantification | Quality |
NGS metrics |
|||
|---|---|---|---|---|---|---|
| Qubit (ng/μl) | RIN | RNA fragments >150 base pairs (%) | TMFPR | Gene (Exons) | Read Counts | |
| 1 | 27 | 2.2 | 67 | 194460 | EML4(6) – ALK(20) | 527 |
| 2 | 29.8 | 2.5 | 46 | 106905 | EML4(6) – ALK(20) | 518 |
| 3 | 152 | NA | NA | 38816 | EML4(6) – ALK(20) | 632 |
| 4 | 11.2 | 2.5 | 51 | 199487 | EML4(6) – ALK(20) | 2277 |
| 5 | 11.3 | 2.4 | 59 | 194261 | EML4(6) – ALK(20) | 370 |
| 6 | 12.4 | 2.5 | 38 | 137627 | EML4(13) – ALK(20) | 3108 |
| 7 | 40 | NA | NA | 49725 | EML4(6) – ALK(20) | 574 |
| 8 | 18.3 | NA | NA | 68458 | EML4(13) – ALK(20) | 3294 |
| 9 | 26 | NA | NA | 42774 | EML4(6) – ALK(20) | 1790 |
| 10 | 20.6 | NA | NA | 27414 | EML4(6) – ALK(20) | 533 |
| 11 | 14.7 | 2.3 | 67 | 179102 | EZR(10) – ROS1(34) | 8981 |
| 12 | 44.2 | 2.4 | 24 | 148025 | no fusion | – |
| 13 | 42.6 | 1.1 | 22 | 151081 | no fusion | – |
| 14 | 49.8 | NA | NA | 41152 | no fusion | – |
| 15 | 24.4 | NA | NA | 63798 | no fusion | – |
| 16 | 52.2 | NA | NA | 196200 | no fusion | – |
| 17 | 35.4 | NA | NA | 230033 | no fusion | – |
| 18 | 47 | NA | NA | 131974 | no fusion | – |
| 19 | 35.4 | 2.3 | 60 | 269517 | no fusion | – |
| 20 | 34.8 | 2.5 | 47 | 99363 | no fusion | – |
| 21 | 64.4 | 2.4 | 68 | 114466 | no fusion | – |
| 22 | 76.6 | 2.3 | 73 | 186135 | no fusion | – |
| 23 | 66.4 | 2.4 | 49 | 84206 | no fusion | – |
| 24 | 30.6 | 2.5 | 46 | 135882 | no fusion | – |
| 25 | 23.2 | 2.5 | 42 | 38390 | no fusion | – |
| 26 | 37.2 | 2.4 | 52 | 150894 | no fusion | – |
| 27 | 22.2 | 2.5 | 46 | 172565 | no fusion | – |
| 28 | 21 | 2.5 | 43 | 192538 | no fusion | – |
| 29 | 40.6 | 2.5 | 40 | 114472 | no fusion | – |
| 30 | 63 | 2.3 | 63 | 253671 | EML4(13) – ALK(20) | 2626 |
| 31 | 87.2 | 2.3 | 64 | 183584 | no fusion | – |
| 32 | 50.8 | 2.4 | 51 | 234307 | no fusion | – |
| 33 | 19.6 | 1.6 | 76 | 35200 | no fusion | – |
| 34 | 55.8 | 2.4 | 57 | 215167 | no fusion | – |
| 35 | 83.6 | 2.3 | 58 | 196873 | no fusion | – |
| 36 | 74.8 | 2.4 | 57 | 237336 | KIF5B(15) – RET(12) | 14257 |
| 37 | 50.8 | 2.4 | 65 | 169419 | no fusion | – |
| 38 | 73 | 2.4 | 54 | 131217 | no fusion | – |
| 39 | 61.6 | 2.3 | 65 | 183111 | no fusion | – |
| 40 | 130 | 2 | 74 | 114982 | no fusion | – |
| 41 | 26.6 | 2.2 | 38 | 25705 | no fusion | – |
| 42 | 26.4 | 2.4 | 52 | 188899 | no fusion | – |
| 43 | 50.4 | 2.4 | 62 | 206721 | no fusion | – |
| 44 | 50.8 | 2.5 | 43 | 169713 | no fusion | – |
| 45 | 25.6 | 2.6 | 37 | 102602 | no fusion | – |
| 46 | 80 | 2.4 | 50 | 119869 | no fusion | – |
| 47 | 56.4 | 2.2 | 58 | 206807 | no fusion | – |
| 48 | 100 | 2.1 | 69 | 175943 | no fusion | – |
| 49 | 30.8 | 2.3 | 90 | 181785 | EZR(10) – ROS1(34) | 76 |
| 50 | 41.8 | NA | NA | 95416 | no fusion | – |
| 51 | 100 | NA | NA | 143052 | no fusion | – |
| 52 | 43.6 | NA | NA | 59579 | no fusion | – |
Legend: RIN, RNA integrity number; NGS, next-generation sequencing; TMFPR, total mapped fusion panel reads; NA, not assessed; EML4-ALK, echinoderm microtubule-associate protein-like 4-anaplastic lymphoma kinase; EZR-ROS1, ezrin gene-proto-oncogene tyrosine-protein kinase 1; KIF5B-RET, the kinesin family 5 B gene-ret proto-oncogene.
3.2. Detected gene fusions and performance of NGS based assays
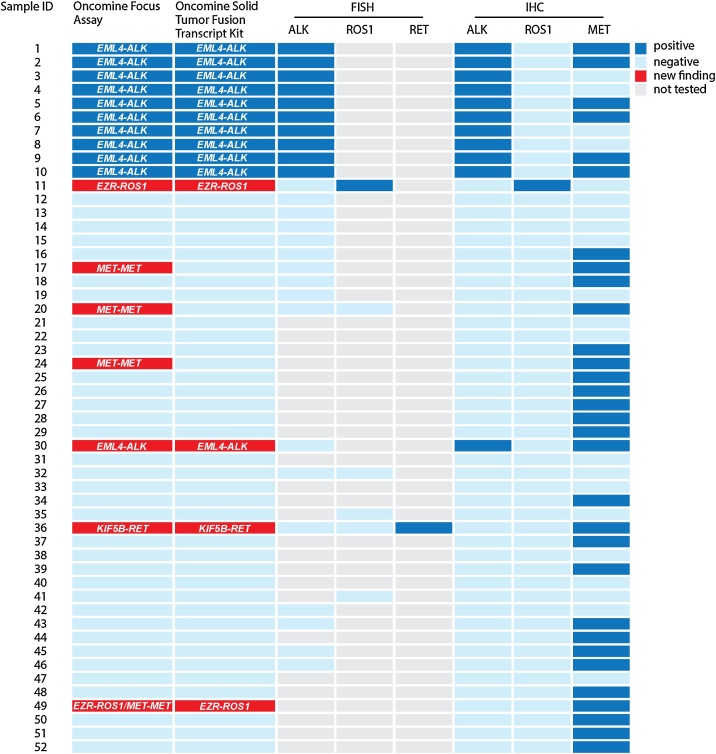
EML4-ALK fusion gene variant 1 with EML4 exon 13 being fused to ALK exon 20 was found in 3/11 (27%) cases and variants 3a/b with EML4 exon 6 being fused to ALK exon 20 were detected in 8/11 (72%) cases (Table 2). Variant 2 with EML4 exon 20 being fused to ALK exon 20 was not detected in our cohort. Other fusion genes than EML4-ALK, EZR-ROS1, KIF5B-RET and MET-MET were not detected in our LUAD cohort. We detected one additional EML4-ALK fusion, one EZR-ROS1 fusion and a single KIF5B-RET fusion in our LUAD cohort (Fig. 1). Another EZR-ROS1 fusion (case 49) was detected at a very low read number. However, Ros1 IHC was negative, so that the genomic event could not be confirmed on expression level. The Oncomine Solid Tumor Fusion Transcript Kit confirmed all EML4-ALK, EZR-ROS1 and KIF5B-RET events (Fig. 1). In addition, 4 cases of MET-MET fusion, resulting in MET exon 14 skipping were detected by the OFA and confirmed by c-Met overexpression (Fig. 1). One of the MET-MET fusion genes co-occurred with an EZR-ROS1 fusion (case 49, Fig. 1). For determination of the sensitivity of RNA-based targeted sequencing we compared different ratios of the EML4-ALK rearranged cell line H3122 on a background of the non-EML4-ALK rearranged cell line HCC-44 (Fig. 2). The lowest detectable ratio was 0.01 with 159 gene specific reads (Table 4).
Fig. 1.
Genetic rearrangements in study cohort of LUADs. Overview of NGS results compared to FISH and IHC. LUAD lung adenocarcinoma; NGS, next-generation-sequencing; FISH, fluorescence in-situ hybridization; IHC, immunohistochemistry; EML4-ALK, echinoderm microtubule-associate protein-like 4 − anaplastic lymphoma kinase; EZR-ROS1, ezrin gene − proto-oncogene tyrosine-protein kinase 1; KIF5B-RET, the kinesin family 5 B gene-ret proto-oncogene.
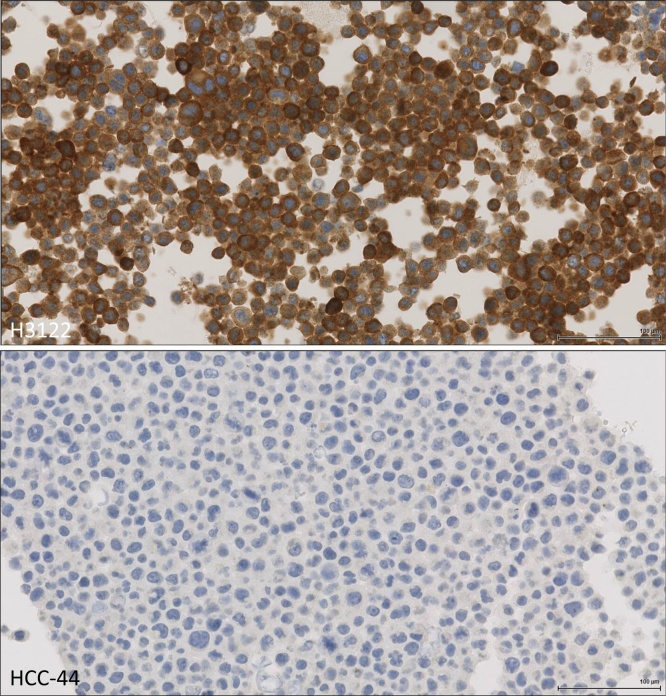
Fig. 2.
Immunohistochemical staining for Alk, upper tile H3122, lower tile HCC-44.
Table 4.
NGS metrics of different ratios of EML4-ALK to non-EML4-ALK rearrangement containing RNA.
| Ratio RNA H3122/HCC-44 | NGS metrics |
|||
|---|---|---|---|---|
| TMFPR | Gene (Exons) | Read Counts | Ratio Read Counts/TMFPR | |
| 1 | 236702 | EML4(6) – ALK(20) | 42452 | 0.179347872 |
| 0.1 | 238830 | EML4(6) – ALK(20) | 3067 | 0.01284177 |
| 0.05 | 197984 | EML4(6) – ALK(20) | 1999 | 0.010096775 |
| 0.01 | 155527 | EML4(6) – ALK(20) | 159 | 0.001022331 |
| 0.005 | 270342 | EML4(6) – ALK(20) | 0 | 0 |
| 0.001 | 234415 | EML4(6) – ALK(20) | 0 | 0 |
| 0.0005 | 214911 | EML4(6) – ALK(20) | 0 | 0 |
| 0 | 167480 | EML4(6) – ALK(20) | 0 | 0 |
3.3. Comparison of targeted NGS with FISH and IHC
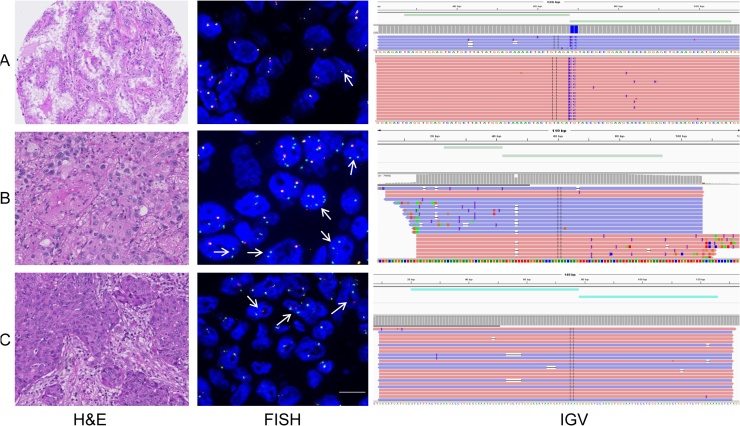
All 10 known ALK fusions and 18 non-rearranged cases were detected by OFA and Oncomine Solid Tumor Fusion Transcript Kit assay (sensitivity 100%, specificity 100%, positive predictive value 100%, negative predictive value 100%, Table 3). In addition, we found one EML4-ALK rearranged case (ID 30) within the set of the 24 previously non-tested cases (Fig. 3A). Interestingly, retrospective Alk immunohistochemistry of the same case was positive whereas ALK FISH analysis by an investigator blinded for the NGS results revealed 14% of nuclei with break-apart signals, slightly below the threshold of 15% break-apart signals required to confirm ALK rearranged LUAD. Reanalysis by an independent second pathologist confirmed the borderline nature of the case.
Table 3.
Accuracy of NGS-based targeted RNA-sequencing in detection of ALK and ROS1 rearrangements.
| ALK rearrangements | ||
| Condition positive | Condition negative | |
| Test positive | 11 (TP) | 0 (FP) |
| Test negative | 0 (FN) | 18 (TN) |
| Sensitivity 100% | Specificity 100% | |
| ROS1 rearrangements | ||
| Condition positive | Condition negative | |
| Test positive | 1 (TP) | 1 (FP) |
| Test negative | 0 (FN) | 51 (TN) |
| Sensitivity 100% | Specificity 98% | |
ALK, anaplastic lymphoma kinase; TP, true positive; FP, false positive; FN, false negative; TN, true negative; ROS1, proto-oncogene tyrosine-protein kinase 1.
Fig. 3.
3A Shows a newly identified LUAD case with EML4-ALK fusion found by OFA and Oncomine Solid Tumor Fusion Transcript Kit assay and confirmed by ALK immunohistochemistry. The FISH assay did not reveal enough break-apart signals required for classification as ALK rearrangend. 3B Newly detected EZR-ROS1 fusion in a case tested negative for ALK FISH and ALK IHC. Both OFA and Oncomine Solid Tumor Fusion Transcript Kit assay as well as retrospective ROS1 FISH and ROS1 IHC confirmed the alteration. 3C shows a RET fusion confirmed by OFA, Oncomine Solid Tumor Fusion Transcript Kit assay and RET FISH.
To confirm ROS1 rearrangement detected by OFA and the Oncomine Solid Tumor Fusion Transcript Kit assay all ROS1 positive cases were further referred to FISH and IHC. Case no. 11 was ROS1 positive when tested by FISH and IHC (Fig. 3B). The second case (ID 49) was negative for ROS1 IHC. Unfortunately, due to lack of tissue resources, ROS1 FISH analysis could not be performed. We therefore classified this result as false positive (Table 3). One case with KIF5B-RET fusion identified by both assays, the OFA and the Oncomine Solid Tumor Fusion Transcript Kit assay (Fig. 3C), showed a break-apart signal in a retrospective RET FISH.
All four samples with MET exon 14 skipping stained positive with c-Met IHC. In addition, we found c-Met overexpression in 27 of the remaining cases, indicating additional other mechanisms resulting in c-Met overexpression. All MET(13)-MET(15) fusion cases had high numbers ( > 200) of specific MET reads and sufficient total mapped fusion panel reads.
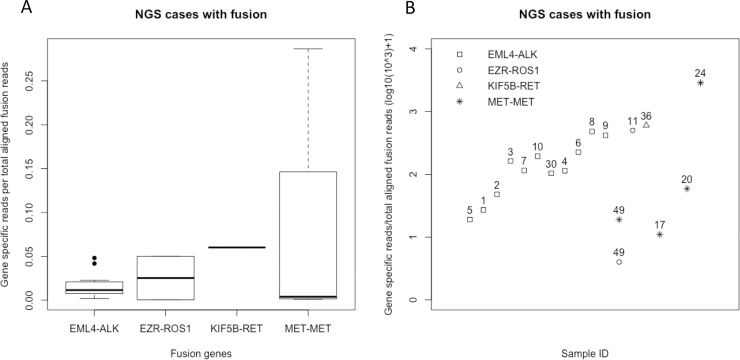
3.4. Variation of fusion read counts
We observed gene specific read counts ranging from 76 to 38946 for all detected gene fusions. The ratio of gene specific read counts and total mapped fusion panel reads ranged from 0.0004–0.2866. Fig. 4A shows boxplots of read counts for all detected fusion genes. Sample no. 49 had low read counts of 76 for ROS1. EZR-ROS1 rearrangement was confirmed with Oncomine Solid Tumor Fusion Transcript Kit assay. However, Ros1 IHC was repeatedly negative, indicating that the low number of fusion transcripts did not result in a detectable amount of endogenous Ros1 protein with standard Ros1 IHC protocol. Fig. 4 B summarizes the ratios of gene specific reads per total aligned fusion reads.
Fig. 4.
4A Boxplots of gene specific reads per total aligned fusion reads for each identified fusion genes (absolute values). 4B. Plot showing the Sample ID, genomic alteration and gene specific reads per total aligned fusion reads (log10(10^3) + 1).
4. Discussion
We studied the performance of a RNA-based targeted sequencing approach versus the gold standard FISH analysis for detection of ALK fusions in a retrospective cohort of LUADs. All previously known ALK fusions could be detected by targeted NGS. In addition, we found an additional EZR-ROS1 fusion in one of the ALK non-rearranged cases and another potential EZR-ROS1 fusion in the untested cohort. We also found one additional EML4-ALK fusion in the untested cohort.
Our results are concordant with findings by other groups. Pfarr et al. also reported a sensitivity and specificity of 100% of a targeted massively parallel sequencing approach for the detection of known targetable gene fusions [18]. Paasinen-Sohns et al. compared in their study the overall performance of DNA and RNA-based OFA panels and could confirm all detected EML4-ALK, EZR-ROS1 and MET-MET fusions with IHC in their cohort [19]. Another study investigating FISH, IHC and NGS regarding EML4-ALK rearrangements found 4 tumors positive by FISH and 8 positive by IHC in a cohort of 51 LUAD patients [20]. Of these, only 3 cases were positive by both FISH and IHC [20]. Moreover, 4 of 5 IHC positive and FISH negative patient tumors could be confirmed being EML4-ALK rearranged by NGS [20]. Two of these patients with EML4-ALK rearranged tumors confirmed by NGS received crizotinib treatment with durable progression-free survival suggesting that especially for borderline cases NGS can tip the scales [20]. A further study was focused at solving discordant ALK testing results determined by IHC and FISH with NGS [21]. NGS testing revealed three ALK FISH positive Alk IHC negative cases as ALK non-rearranged [21]. Interestingly, Alk immunohistochemistry with ALK1 clone was positive in 12 ALK FISH negative cases but turned out to be false positive when Alk IHC was repeated with clone D5F3 which we also used for our study [21].
The recommended cut-offs for FISH analyses remain controversial and might explain the false positive and negative results when comparing FISH with other assays such as NGS. Seventy four per cent of patients with advanced ALK rearranged LUAD as determined by FISH responded to first line crizotinib treatment [5]. The median progression free survival was 10.9 months [5]. False negative test results, however, make it impossible to identify patients who might benefit from targeted therapy. False positive results will exclude the patient from further molecular testings in hierarchical single target assays. In addition, false positive results lead to false hope and assignment to wrong therapeutic strategies. The ALK reads per total aligned fusion reads of our NGS positive ALK FISH negative case no. 30 match the median of all detected EML4-ALK cases (0.01) so that a low ALK read ratio reflecting a very low transcript number can be ruled out. NGS results for this particular case also fit to the observation that Alk IHC was positive. To further rule out a lack of sensitivity we performed a dilution series of ALK rearranged cell line RNA on a non-ALK rearranged RNA background. Ratios of 1, 0.1, 0.05, and 0.01 could be detected with RNA-based targeted sequencing. This means that even 1% of EML4-ALK rearranged RNA or 1% of EML4-ALK rearranged cells can be detected on a background without EML4-ALK rearrangement. This also explains the discrepancy of the case being negative according to EML4-ALK FISH evaluation criteria but positive by Alk IHC and RNA-based targeted sequencing. Case 30 could harbor a heterogeneous EML4-ALK rearrangement so that the rearrangement detection can be difficult by FISH. In addition, the ALK FISH scoring criteria could fail for heterogeneous cases aggravated by the fact that tissue sections are used where nuclei are typically clipped.
Case 49 with potential detection of EZR-ROS1 rearrangement but negative Ros1 IHC could be most likely false positive. However, since we did not perform a dilution series of RNA of a ROS1rearranged cell line and tissue for FISH testing was not available, we cannot completely solve this case.
We observed RNA concentrations from 11.2 to 152 ng/μl although three cores had been taken and RNA was extracted from each core. The varying RNA amounts are most likely due to differences in the thickness of tissue blocks and cellularity. Although only LUADs with tumor content ≥ 60% were selected, some cores probably contained less cells in deeper areas of the core.
In summary, targeted NGS for fusion detection is a more robust and reliable method in detection of ALK and other targetable rearrangements compared to single target assays such as FISH. In addition, turnaround time, reproducibility and superior cost efficiency favor the use of targeted NGS compared with conventional single target assays for fusion detection.
5. Conclusion
Detection of targetable fusion genes is crucial for appropriate clinical action. We experienced the RNA-based OFA panel as a reliable and accurate tool in comparison with standard single target assays like FISH and IHC. Especially borderline cases can benefit from the added value of NGS testing.
Conflict of interest
The authors declare that no conflict of interest exists.
Acknowledgements
We thank Susanne Dettwiler, Fabiola Prutek and Peter Schraml from the Tissue Biobank at USZ and Ursula Rommerscheid-Fuss, Department of Pathology, University Hospital Cologne, Germany, for their excellent technical support. Cell lines were a kind gift of Prof. Dr. Roman Thomas, Cologne, Germany. This project was funded in part by Innovation Pool Grants provided by the University Hospital Zurich to V.T. and P.J.W., respectively. V. T. is the recipient of a joint ERS/EMBO Long-Term Research fellowship n° LTRF 2014-2951 and a Swiss Cancer League postdoctoral research fellowship (BIL KFS-3402-02-2014).
References
- 1.Soda M., Choi Y.L., Enomoto M., Takada S., Yamashita Y., Ishikawa S., Fujiwara S., Watanabe H., Kurashina K., Hatanaka H., Bando M., Ohno S., Ishikawa Y., Aburatani H., Niki T., Sohara Y., Sugiyama Y., Mano H. Identification of the transforming EML4?ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 2.Choi Y.L., Takeuchi K., Soda M., Inamura K., Togashi Y., Hatano S., Enomoto M., Hamada T., Haruta H., Watanabe H., Kurashina K., Hatanaka H., Ueno T., Takada S., Yamashita Y., Sugiyama Y., Ishikawa Y., Mano H. Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Res. 2008;68:4971–4976. doi: 10.1158/0008-5472.CAN-07-6158. [DOI] [PubMed] [Google Scholar]
- 3.Sasaki T., Rodig S.J., Chirieac L.R., Jänne P.A. The biology and treatment of EML4-ALK non-small cell lung cancer. Eur. J. Cancer. 2010;46:1773–1780. doi: 10.1016/j.ejca.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw A.T., Yeap B.Y., Mino-Kenudson M., Digumarthy S.R., Costa D.B., Heist R.S., Solomon B., Stubbs H., Admane S., McDermott U., Settleman J., Kobayashi S., Mark E.J., Rodig S.J., Chirieac L.R., Kwak E.L., Lynch T.J., Iafrate A.J. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J. Clin. Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solomon B.J., Mok T., Kim D.-W., Wu Y.-L., Nakagawa K., Mekhail T., Felip E., Cappuzzo F., Paolini J., Usari T., Iyer S., Reisman A., Wilner K.D., Tursi J., Blackhall F. First-line crizotinib versus chemotherapy in ALK–positive lung cancer. N. Engl. J. Med. 2014;371:2167–2177. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 6.Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., MacNeill J., Ren J.M., Yuan J., Bakalarski C.E., Villen J., Kornhauser J.M., Smith B., Li D., Zhou X., Gygi S.P., Gu T.L., Polakiewicz R.D., Rush J., Comb M.J. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 7.Takeuchi K., Soda M., Togashi Y., Suzuki R., Sakata S., Hatano S., Asaka R., Hamanaka W., Ninomiya H., Uehara H., Lim Choi Y., Satoh Y., Okumura S., Nakagawa K., Mano H., Ishikawa Y. RET, ROS1 and ALK fusions in lung cancer. Nat. Med. 2012;18:378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 8.Kohno T., Ichikawa H., Totoki Y., Yasuda K., Hiramoto M., Nammo T., Sakamoto H., Tsuta K., Furuta K., Shimada Y., Iwakawa R., Ogiwara H., Oike T., Enari M., Schetter A.J., Okayama H., Haugen A., Skaug V., Chiku S., Yamanaka I., Arai Y., Watanabe S., Sekine I., Ogawa S., Harris C.C., Tsuda H., Yoshida T., Yokota J., Shibata T. KIF5B-RET fusions in lung adenocarcinoma. Nat. Med. 2012;18:375–377. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergethon K., Shaw A.T., Ou S.H.I., Katayama R., Lovly C.M., McDonald N.T., Massion P.P., Siwak-Tapp C., Gonzalez A., Fang R., Mark E.J., Batten J.M., Chen H., Wilner K.D., Kwak E.L., Clark J.W., Carbone D.P., Ji H., Engelman J.A., Mino-Kenudson M., Pao W., Iafrate A.J. ROS1 rearrangements define a unique molecular class of lung cancers. J. Clin. Oncol. 2012;30:863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gautschi O., Milia J., Filleron T., Wolf J., Carbone D.P., Owen D., Camidge R., Narayanan V., Doebele R.C., Besse B., Remon-Masip J., Janne P.A., Awad M.M., Peled N., Byoung C.C., Karp D.D., Van Den Heuvel M., Wakelee H.A., Neal J.W., Mok T.S.K., Yang J.C.H., Ou S.H.I., Pall G., Froesch P., Zalcman G., Gandara D.R., Riess J.W., Velcheti V., Zeidler K., Diebold J., Früh M., Michels S., Monnet I., Popat S., Rosell R., Karachaliou N., Rothschild S.I., Shih J.Y., Warth A., Muley T., Cabillic F., Mazières J., Drilon A. Targeting RET in patients with RET-rearranged lung cancers: results from the global, multicenter RET registry. J. Clin. Oncol. 2017;35:1403–1410. doi: 10.1200/JCO.2016.70.9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drilon A., Wang L., Hasanovic A., Suehara Y., Lipson D., Stephens P., Ross J., Miller V., Ginsberg M., Zakowski M.F., Kris M.G., Ladanyi M., Rizvi N. Response to cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov. 2013;3:630–635. doi: 10.1158/2159-8290.CD-13-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salgia R. MET in lung cancer: biomarker selection based on scientific rationale. Mol. Cancer Ther. 2017;16:555–565. doi: 10.1158/1535-7163.MCT-16-0472. [DOI] [PubMed] [Google Scholar]
- 13.Cortot A.B., Kherrouche Z., Descarpentries C., Wislez M., Baldacci S., Furlan A., Tulasne D. Exon 14 deleted MET receptor as a new biomarker and target in cancers. J. Natl. Cancer Inst. 2017;109 doi: 10.1093/jnci/djw262. [DOI] [PubMed] [Google Scholar]
- 14.Paik P.K., Drilon A., Fan P.D., Yu H., Rekhtman N., Ginsberg M.S., Borsu L., Schultz N., Berger M.F., Rudin C.M., Ladanyi M. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring met mutations causing exon 14 skipping. Cancer Discov. 2015;5:842–850. doi: 10.1158/2159-8290.CD-14-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Velizheva N.P., Rechsteiner M.P., Wong C.E., Zhong Q., Rössle M., Bode B., Moch H., Soltermann A., Wild P.J., Tischler V. Cytology smears as excellent starting material for next-generation sequencing-based molecular testing of patients with adenocarcinoma of the lung. Cancer Cytopathol. 2017;125:30–40. doi: 10.1002/cncy.21771. [DOI] [PubMed] [Google Scholar]
- 16.Blackhall F.H., Peters S., Bubendorf L., Dafni U., Kerr K.M., Hager H., Soltermann A., O’Byrne K.J., Dooms C., Sejda A., Hernández-Losa J., Marchetti A., Savic S., Tan Q., Thunnissen E., Speel E.J.M., Cheney R., Nonaka D., De Jong J., Martorell M., Letovanec I., Rosell R., Stahel R.A. Prevalence and clinical outcomes for patients with ALK-positive resected stage I to III adenocarcinoma: results from the European Thoracic Oncology Platform Lungscape project. J. Clin. Oncol. 2014;32:2780–2787. doi: 10.1200/JCO.2013.54.5921. [DOI] [PubMed] [Google Scholar]
- 17.Rogers T.-M., Russell P.A., Wright G., Wainer Z., Pang J.-M., Henricksen L.A., Singh S., Stanislaw S., Grille J., Roberts E., Solomon B., Fox S.B. Comparison of methods in the detection of ALK and ROS1 rearrangements in lung cancer. J. Thorac. Oncol. 2015;10:611–618. doi: 10.1097/JTO.0000000000000465. [DOI] [PubMed] [Google Scholar]
- 18.Pfarr N., Stenzinger A., Penzel R., Warth A., Dienemann H., Schirmacher P., Weichert W., Endris V. High-throughput diagnostic profiling of clinically actionable gene fusions in lung cancer. Genes Chromosom. Cancer. 2016;55:30–44. doi: 10.1002/gcc.22297. [DOI] [PubMed] [Google Scholar]
- 19.Paasinen-Sohns A., Koelzer V.H., Frank A., Schafroth J., Gisler A., Sachs M., Graber A., Rothschild S.I., Wicki A., Cathomas G., Mertz K.D. Single-center experience with a targeted next generation sequencing assay for assessment of relevant somatic alterations in solid tumors. Neoplasia (United States) 2017;19:196–206. doi: 10.1016/j.neo.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pekar-Zlotin M., Hirsch F.R., Soussan-Gutman L., Ilouze M., Dvir A., Boyle T., Wynes M., Miller V.A., Lipson D., Palmer G.A., Ali S.M., Dekel S., Brenner R., Bunn P.A., Peled N. Fluorescence in situ hybridization, immunohistochemistry, and next-generation sequencing for detection of EML4-ALK rearrangement in lung cancer. Oncologist. 2015;20:316–322. doi: 10.1634/theoncologist.2014-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jang J.S., Wang X., Vedell P.T., Wen J., Zhang J., Ellison D.W., Evans J.M., Johnson S.H., Yang P., Sukov W.R., Oliveir A.M., Vasmatzis G., Sun Z., Jen J., Yi E.S. Custom gene capture and next-generation sequencing to resolve discordant ALK status by FISH and IHC in lung adenocarcinoma. J. Thorac. Oncol. 2016;11:1891–1900. doi: 10.1016/j.jtho.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]