Abstract
Phytophthora sojae (Kaufmann and Gerdemann) is an oomycete that causes stem and root rot on soybean (Glycine max L. Merr) plants. We have constructed three cDNA libraries using mRNA isolated from axenically grown mycelium and zoospores and from tissue isolated from plant hypocotyls 48 h after inoculation with zoospores. A total of 3,035 expressed sequence tags (ESTs) were generated from the three cDNA libraries, representing an estimated 2,189 cDNA transcripts. The ESTs were classified according to putative function based on similarity to known proteins, and were analyzed for redundancy within and among the three source libraries. Distinct expression patterns were observed for each library. By analysis of the percentage G+C content of the ESTs, we estimate that two-thirds of the ESTs from the infected plant library are derived from P. sojae cDNA transcripts. The ESTs originating from this study were also compared with a collection of Phytophthora infestans ESTs and with all other non-human ESTs to assess the similarity of the P. sojae sequences to existing EST data. This collection of cDNA libraries, ESTs, and accompanying annotation will provide a new resource for studies on oomycetes and on soybean responses to pathogen challenge.
The Stramenopiles comprise a diverse group of organisms that have recently been consolidated as a result of analysis of mitochondrial and ribosomal DNA sequences. Species that were previously referred to as heterokont algae and fungal oomycetes belong to this group of related organisms (Gunderson et al., 1987; Förster et al., 1990). Stramenopiles include autotrophic and heterotrophic species that may differ enormously in their morphology and mode of life, but most of which are vegetatively diploid and possess distinctive tinsel-like flagella at some point in their life cycle. Members of this group occupy key ecological niches in marine environments. For example, the diatoms constitute the most abundant component of marine plankton, while the brown algae (e.g. Fucus, Sargassum, and Laminaria spp.) may form extensive floating or attached communities such as the kelp forests. Stramenopiles have also succeeded in terrestrial environments as plant pathogens. Oomycetes (e.g. Peronospora, Pythium, and Phytophthora spp.) include many obligate and facultative parasites that cause disease on a wide spectrum of herbaceous and woody plants throughout the world in tropical and temperate environments (Erwin and Ribeiro, 1996).
Phytophthora sojae Kaufmann and Gerdemann (syn. Phytophthora megasperma f. sp. glycinea Kuan and Erwin) is the causal agent of stem and root rot of soybean (Glycine max [L.] Merr). Extensive outbreaks of Phytophthora root rot on soybeans were first noted over 40 years ago (Kaufmann and Gerdemann, 1958; Hildebrand, 1959), and the disease remains an endemic and serious problem in most soybean-producing areas (Schmitthenner, 1985; Doupnik, 1993; Wrather et al., 1995). Chemical methods may be used to control the disease, but the most cost-effective and widespread strategy to reduce the incidence of the disease has been through the development of resistant or tolerant soybean cultivars. Genetic analysis has shown that tolerance is a quantitative trait, whereas resistance is inherited qualitatively (Schmitthenner, 1985; Ward, 1990; Kasuga et al., 1997). Single dominant host resistance (Rps) genes and corresponding dominant avirulence (Avr) genes in the parasite have been described previously (Tyler et al., 1995; Whisson et al., 1995; Gijzen et al., 1996a). Despite breeding efforts to exploit such resistance-gene-mediated protection, resistant plants eventually become susceptible to disease due to the emergence of new virulent races (Anderson and Buzzell, 1992).
P. sojae is a diploid hemibiotroph that passes through several morphological phases to complete its life cycle. Asexual, single-celled zoospores are biflagellate, motile, and chemotactic to soybean plants (Ho et al., 1967; Morris et al., 1998). Zoospores encyst and germinate on the root or hypocotyl surface, and the resulting germ tube may swell to form an appressorium-like structure at the point of penetration into host tissues (Stossel et al., 1980). Resistance is manifested by rapid induction of host defenses, phytoalexin accumulation, and hypersensitive cell death at the infection site (Ward et al., 1989; Kamoun et al., 1999b). In susceptible interactions, large, water-soaked lesions develop as the pathogen rapidly invades the host. P. sojae spreads through the intercellular matrix of the plant and forms haustoria for intimate contact with host cells. Under moist conditions, zoospores are produced in sporangia that develop at the hyphal tips of vegetative mycelium. P. sojae is homothallic, and thus may reproduce sexually by the development of oospores through self-fertilization or by outcrosses between different strains (Förster et al., 1994). Estimations of genome size in P. sojae vary from 62 to 97 Mb (Rutherford and Ward, 1985; Mao and Tyler, 1991; Voglmayr and Greilhuber, 1998).
Despite thorough investigations into the biology of disease development, little is known concerning the pathogenic determinants of P. sojae. Molecular studies of pathogenicity and virulence of oomycetes are relatively rare compared with those on plant pathogenic fungi, bacteria, and viruses. Plant pathogenic basidiomycetes and ascomycetes, and model organisms such as Saccharomyces, Aspergillus, or Neurospora spp. are taxonomically far removed and fundamentally different in their cell wall composition, reproductive biology, and genetics from oomycete species (Judelson, 1997). Thus, expressed sequence tags (ESTs) and genome sequence information from fungal species may be of limited utility in studies of oomycetes. For these reasons, we undertook a study to profile gene expression patterns in P. sojae by analysis of ESTs. A model strain of P. sojae, P6497, was chosen for this analysis because it has been used for genome size estimations, genetic mapping of avirulence loci, and for construction of large insert bacterial artificial chromosome libraries arranged in ordered arrays. Sequence and annotation information that results from this research will also help to establish a public database to facilitate research on P. sojae and other oomycetes (Waugh et al., 2000).
We have constructed three cDNA libraries using mRNA from axenically grown mycelium, zoospores, and in planta infection sites. More than 1,000 ESTs were generated from each library to determine how patterns of gene expression change during development and pathogenesis. We show how the percentage G+C content of an EST sequence provides a measure that can help distinguish P. sojae from soybean cDNAs, and validate this with selected transcripts by hybridization analysis. Sequence data from each of the cDNA libraries were also systematically compared with EST data from Phytophthora infestans and all other non-human ESTs. Overall, our results indicate that P. sojae gene expression patterns and metabolic processes fundamentally shift during pathogenesis. We have identified transcripts encoding proteins that are likely to be involved in the recognition, attachment, penetration, and invasion of the host soybean plant.
RESULTS
ESTs from Infected Plant Tissues Are More Likely to Produce Highly Significant Matches
Three directional cDNA libraries were constructed from in vitro-grown mycelium, from zoospores, and from P. sojae-infected soybean tissue. The average size of the cDNA inserts for each of the libraries was 0.83, 1.5, and 1.1 kb, respectively. Over 1,000 clones from each library were randomly chosen for plasmid purification and partial DNA sequencing, resulting in a total of 3,035 robust quality ESTs. Most cDNA clones were properly oriented in the vector, since only 2.7% of the ESTs possessed poly(A+) tracts adjacent to the T3 vector site. Thus, the vast majority of the ESTs reflect 5′ end sequences of cDNA from mRNA transcripts.
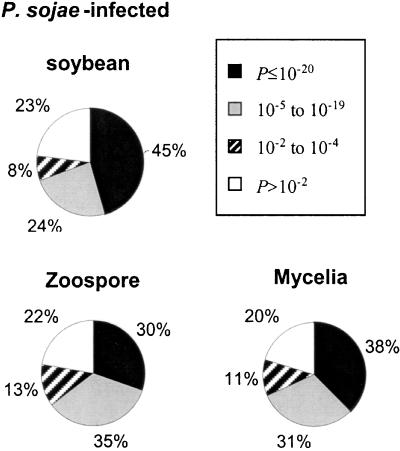
After editing, an average length of 575 bp was used in database searches. Figure 1 shows the distribution of ESTs for each of the source libraries based upon deduced amino acid sequence homology to known or hypothetical proteins. From 20% to 23% of the ESTs from all three libraries returned a P value of >10−2. Of the remaining ESTs with P values of ≤10−2, fewer than 10% of these matched proteins of no known function. Highly significant matches occurred most frequently (45%) with sequences originating from the infected plant library. This reflects the current bias of the DNA sequence databases for plant proteins and genes relative to sequences from oomycetes and related organisms. Thus, soybean EST sequences from the infected plant library are more likely to result in highly significant match scores.
Figure 1.
Frequency distributions of ESTs from each of the source cDNA libraries according to P-value scores returned from BLASTX version 1.4.11 searches.
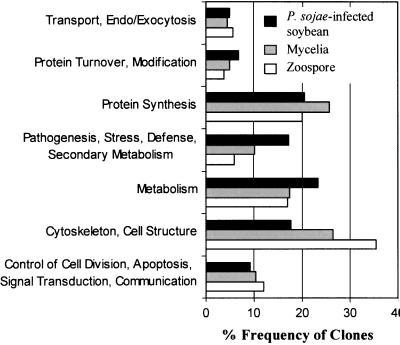
ESTs with P values of ≤10−2 were grouped into seven functional categories, as outlined in Figure 2, for tissue-specific expression profile comparisons. Matches to structural proteins comprised the most abundant class of ESTs in both the zoospore and mycelium libraries, whereas matches to metabolic enzymes were most numerous in the infected plant library. ESTs that matched ribosomal proteins and other factors required for protein synthesis were also highly represented in all three libraries.
Figure 2.
Classification of ESTs according to putative biological function. Protein matches resulting from the BLASTX searches were assigned to one of seven functional categories for life cycle comparisons. Shown is the percentage frequency of clones in each category for the total set of ESTs from each source library.
Two Populations of ESTs That Differ in Percentage G+C Content Are Present in the Infected Plant Library
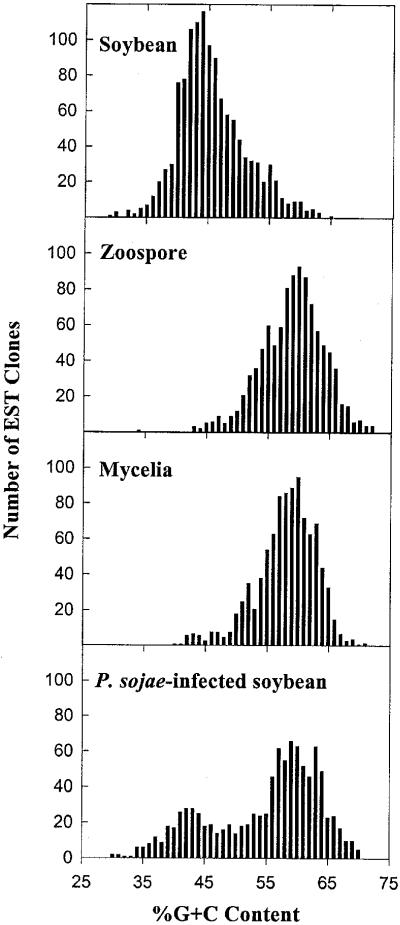
Figure 3 shows a comparison of the percentage G+C content of ESTs from each of the cDNA libraries described in this study, and 1,300 soybean ESTs derived from a seed coat cDNA library. The average G+C content of soybean ESTs was 46%, whereas P. sojae zoospore and mycelium ESTs clustered around a mean of 58% G+C. The graphs indicate that the percentage G+C content of ESTs from soybean and P. sojae produce distinct, but overlapping, normal distribution curves. When ESTs from the P. sojae-infected soybean tissue are similarly plotted, two separate peaks of percentage G+C content are evident, with each peak closely corresponding to the mean percentage G+C content of the uninfected soybean and axenically grown P. sojae ESTs. This bimodal distribution of ESTs based on percentage G+C content can be used to determine the proportion of P. sojae and soybean transcripts within the infected plant library. By fitting normal distribution curves to each of the two different populations of ESTs present in the infected plant library, and comparing the area covered by each of these curves, we estimate that 60% to 70% of the EST sequences from the infected plant library are derived from P. sojae cDNA transcripts.
Figure 3.
Distribution of ESTs on the basis of percentage G+C content. The ESTs were grouped in 1% increments of percentage G+C content and the resulting distributions plotted. Source cDNA library is indicated for each histogram.
Most ESTs Occur as Singletons
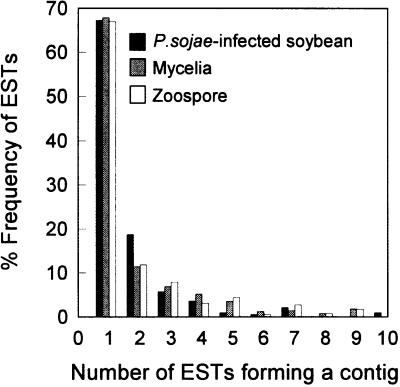
Nucleotide sequences were clustered by similarity in each of the libraries to deduce tissue-specific redundancy. Figure 4 shows that the majority of ESTs from all three libraries occurred as singletons. Thus, 680/1,002 (68%) mycelium, 691/1,031 (67%) zoospore, and 674/1,002 (67%) infected plant ESTs did not assemble into contiguous sequences with other ESTs when compared for similarity. Total redundancy was also determined by pooling EST sequences from all three libraries. By this analysis 1,894/3,035 (62%) of the ESTs are singletons. The total EST assembly further enabled us to positively identify many P. sojae transcripts in the infected plant library. Sequences from the mycelium or zoospore library that match ESTs from the infected plant library most likely originate from P. sojae mRNAs.
Figure 4.
Assessment of EST redundancy in each of the three cDNA libraries by contiguous sequence assembly analysis. Contigs that consist of one sequence are considered singletons, whereas contigs comprised of two or more sequences are classified as redundant ESTs. Shown is a plot of the frequency of singleton and redundant ESTs for each of the source libraries.
Highly Represented Transcripts Vary among the Three cDNA Libraries
Table I shows a comparison of the most abundantly represented transcripts (excluding ribosomal proteins) from each library. This includes all contiguous sequences of four or more ESTs that resulted from the assembly analysis. Transcripts encoding ribosomal proteins were abundantly represented in all three libraries at levels up to 9-fold (0.9%) redundancy. However, other redundant ESTs were generally specific for a particular library. From the infected soybean library, we conclude that five of the eight redundant ESTs shown in Table I are P. sojae transcripts, and the remaining three represent soybean cDNAs. This was determined by hybridization analysis, or was predicted from the representation, percentage G+C content, and BLAST similarity match of the ESTs comprising the sequence contig. The three abundant soybean transcripts correspond to two different pathogenesis-related proteins, and to a soybean chalcone synthase. Abundant P. sojae transcripts from the infected soybean library include enzymes of intermediary metabolism and a mitochondrial ATP synthase subunit. In contrast, mycelium and especially zoospores showed very different patterns of gene expression. Transcripts matching structural proteins were more prevalent in the mycelium and zoospore ESTs. Almost none (1/12) of the abundant zoospore transcripts shown in Table I were represented in the mycelium or infected plant library.
Table I.
Most prevalent mRNAs as measured by EST redundancya
| cDNA Library | ESTb | Best Match | P Valuec | Redundancyd | Representatione | G + C | Source |
|---|---|---|---|---|---|---|---|
| % | |||||||
| Infected soybean | 3-10E-HA | Pathogenesis-related protein 1 | 10−119 | 10 | HA | 42 | G. maxf |
| 3-4G-HA | Formate dehydrogenase | 10−127 | 7 | HA | 67 | P. sojaef | |
| 4-5G-HA | Alcohol dehydrogenase | 10−67 | 7 | HA | 64 | P. sojaef | |
| 4-6H-HA | Stress-induced PR protein | 10−87 | 6 | HA | 45 | G. maxg | |
| 4-7B-HA | ATP synthase, β-chain | 10−157 | 4 | HA, MY | 64 | P. sojaeg | |
| 8-6C-HA | Chalcone synthase | 10−210 | 4 | HA | 47 | G. maxg | |
| 5-2H-HA | 2-Phosphoglycerate dehydratase | 10−164 | 4 | HA, MY | 56 | P. sojaeg | |
| 2-11G-HA | Glu dehydrogenase | 10−53 | 4 | HA | 64 | P. sojaeg | |
| Mycelium | 8-12D-MY | RIC1 protein | 10−32 | 8 | MY | ||
| 9-9D-MY | Unknown/hypothetical protein | 10−7 | 8 | MY | |||
| 11-9A-MY | Annexin | 10−27 | 7 | HA, MY | |||
| 7-4D-MY | Guanine nucleotide binding protein | 10−155 | 6 | HA, MY | |||
| 6-10C-MY | Unknown/hypothetical protein | 10−7 | 5 | MY | |||
| 4-9A-MY | Thioredoxin peroxidase | 10−76 | 4 | MY | |||
| 3-7A-MY | Superoxide dismutase | 10−55 | 4 | HA, MY | |||
| Zoospore | 3-10A-ZO | Elicitin | 10−5 | 8 | ZO | ||
| 11-6F-ZO | Unknown/hypothetical protein | 10−9 | 8 | ZO | |||
| 4-2B-ZO | Unknown/hypothetical protein | 10−21 | 8 | ZO | |||
| 6-5E-ZO | ABC transporter | 10−31 | 7 | ZO | |||
| 9-2D-ZO | GTP cyclohydrolase | 10−67 | 6 | ZO | |||
| 10-7B-ZO | Ribonucleotide reductase | 10−128 | 6 | ZO | |||
| 5-7D-ZO | Actin A | 10−219 | 5 | HA, MY, ZO | |||
| 4-2F-ZO | Membrane glycoprotein | 10−10 | 5 | ZO | |||
| 7-3H-ZO | HAM34 protein | 10−18 | 5 | ZO | |||
| 10-6E-ZO | Mucin | 10−14 | 4 | ZO | |||
| 3-5A-ZO | Unknown/hypothetical protein | 10−9 | 4 | ZO | |||
| 4-1H-ZO | Transcription factor protein | 10−4 | 4 | ZO |
Not including ribosomal proteins. Total no. of ESTs in each library: 1,002 infected plant (HA); 1,002 mycelium (MY); and 1,031 zoospore (ZO).
Representative EST clone from assembly contig.
P value from BLASTX search using entire contiguous sequence resulting from assembly of ESTs.
No. of ESTs from an individual source library that assembled into a contiguous sequence.
Representation of transcript in all three source libraries.
Determined by hybridization analysis.
Predicted from representation, BLASTX match, and percentage G + C content.
Comparative Analyses Indicate Varied Patterns of Similarity to Other ESTs
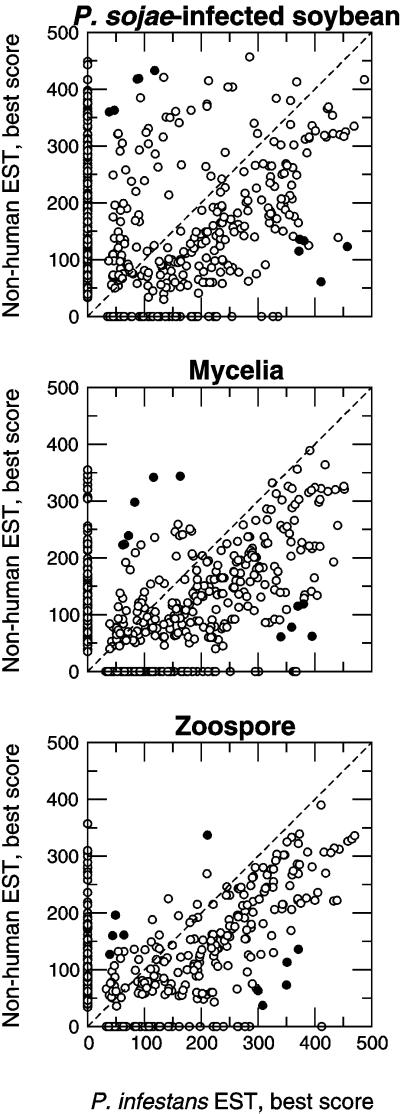
Raw scores from the best TBLASTX match found in each of two targets are plotted in Figure 5, for each of the three cDNA libraries. Several patterns indicate that sequences from P. sojae resemble sequences obtained from P. infestans mycelium to varying degrees. In general, some P. sojae sequences match one target but have no corresponding matches in the other. Those sequences that match both targets tend to resemble the P. infestans sequences more closely than the non-human target, as a majority of points lie below the identity function (dotted line). This pattern is least prominent in the infected soybean library, likely indicating sequences originating from soybean. This is consistent with the observation that highly significant matches occurred most frequently in sequences originating from the infected plant library. The comparative results also indicate cases in which the best score was relatively high in one target but low in the other. Table II summarizes the best examples of the difference between scores from either target being the greatest but neither score being zero, with the best match against the “nr” amino acid target.
Figure 5.
Comparative analysis of ESTs derived from each of the source cDNA libraries using TBLASTX, version 2.0.6. P. sojae and soybean EST sequences generated in this study were compared with two distinct data sets, one consisting of P. infestans mycelium ESTs and another, larger data set of non-human ESTs from all taxa, as described in “Materials and Methods.” Raw scores from the best match to each target are shown from infected soybean (top), mycelium (middle), and zoospore (bottom) libraries. The black circles indicate the 10 sequences from each library that the had the greatest difference between scores, but where neither score was zero, as shown in Table II.
Table II.
P. sojae ESTs having the greatest difference in raw scores from comparative TBLASTX analyses
| Name | P. infestans Score | Non-Human EST Score | E Value | Best Match |
|---|---|---|---|---|
| 6-1E-HA | 411 | 61 | 10−31 | Ser/Thr protein kinase |
| 8-2C-HA | 457 | 123 | 10−48 | Yeast YGR163 gene |
| 8-4A-HA | 372 | 115 | 10−60 | DNA damage checkpoint protein RAD24 |
| 11-9H-HA | 381 | 133 | 10−60 | DNA damage checkpoint protein RAD24 |
| 5-2D-HA | 373 | 136 | 10−45 | 60S ribosomal protein L9 |
| 10-4C-HA | 87 | 418 | 10−22 | Elongation factor 1-α |
| 6-11B-HA | 90 | 419 | 10−90 | Ubiquitin-protein ligase-like protein |
| 10-11B-HA | 38 | 360 | 10−26 | High mobility group protein HMG2A |
| 2-12F-HA | 47 | 363 | 10−101 | Alcohol dehydrogenase |
| 11-1H-HA | 118 | 433 | 10−101 | ADP-ribosylation factor |
| 3-7A-MY | 395 | 62 | 10−51 | Superoxide dismutase precursor (Mn) |
| 10-11B-MY | 359 | 78 | 10−26 | High mobility group protein HMG2A |
| 5-12G-MY | 340 | 61 | 10−07 | GRR1 protein |
| 1-4F-MY | 380 | 119 | 10−57 | 14-3-3-Like protein |
| 11-5H-MY | 370 | 115 | 10−49 | Triose-P isomerase |
| 4-3G-MY | 116 | 342 | 10−83 | ADP-ribosylation factor 1 |
| 5-8A-MY | 83 | 298 | 10−88 | Heat shock protein 80 |
| 1-12F-MY | 163 | 344 | 10−80 | Calmodulin |
| 10-7C-MY | 72 | 239 | 10−71 | 2-Oxoglutarate dehydrogenase |
| 5-5A-MY | 62 | 223 | 10−50 | 40S Ribosomal protein S2 |
| 6-9D-ZO | 350 | 73 | 10−8 | Hypothetical 17.7-kD protein |
| 3-6B-ZO | 308 | 37 | 10−7 | Plasma membrane ATPase 1 (proton pump) |
| 5-9H-ZO | 351 | 113 | 10−46 | Ribosomal protein L17 |
| 10-2G-ZO | 300 | 63 | 10−14 | Regulator of acetyl-CoA synthetase activity |
| 10-8E-ZO | 371 | 136 | 10−45 | Ribosomal protein L9 |
| 8-1H-ZO | 49 | 196 | 10−65 | 60s Ribosomal protein L3 |
| 9-2B-ZO | 211 | 337 | 10−83 | Protein kinase ADK1-like protein |
| 7-11G-ZO | 44 | 160 | 10−71 | Casein kinase II, α-chain |
| 6-8B-ZO | 64 | 161 | 10−44 | Plasma membrane proton ATPase |
| 9-7D-ZO | 39 | 127 | 10−53 | Alkyl-DHAP synthase precursor |
DISCUSSION
We analyzed gene expression patterns in P. sojae by comparing populations of ESTs generated from three different cDNA libraries. We chose to harvest diseased soybean tissue for library construction 48 h after challenge to increase the probability of cloning P. sojae transcripts involved in pathogenesis.
Determining the percentage G+C content of these partially resolved cDNA sequences is a useful indicator in the analysis of ESTs from the infected plant library, since soybean and P. sojae transcripts are different in their average G+C content. This measure provides a means to estimate the total proportion of soybean and P. sojae cDNA transcripts present in the mixed-source library. Individual soybean and P. sojae ESTs may also be distinguished from one another by comparison with other sequence databases. As sequence information accumulates and ESTs approach saturation coverage for soybean and P. sojae, determining the origin of a particular EST from a mixed library may simply be accomplished by a comparative representation analysis.
The high representation (60%–70%) of P. sojae cDNAs present in the infected plant library was surprising, despite the fact that this pathogen is aggressive and fast growing. P. sojae is a considered a hemibiotroph, and therefore it first establishes itself in host tissues as a biotroph but then switches to a more necrotrophic type of growth, rapidly invading and killing host cells. The high proportion of P. sojae ESTs from the infected library may result from this pathogenic strategy, since by 48-h post infection, P. sojae had ramified throughout the hypocotyl tissues and caused large, water-soaked lesions. Further EST analyses at different stages of the infection process and of other host-pathogen interactions would help to put these results in context.
By considering the spectrum and abundance of ESTs represented in the infected library, the present study provides much information about the mechanisms of pathogenesis and of the host defense response. P. sojae genes that appear to be differentially expressed in planta are predominantly associated with intermediary metabolism. The abundant ESTs that match formate dehydrogenase, alcohol dehydrogenase, and glycolytic enzymes such as 2-phosphoglycerate dehydratase suggest that P. sojae relies on glycolysis and mixed alcohol/formic acid fermentation for substrate catabolism and energy generation during growth in host tissues. The oxidative deamination of Glu to α-ketoglutarate also seems to be an important route in the degradation and assimilation of amino acid carbon skeletons during pathogenesis, since at least six different Glu dehydrogenase transcripts were represented by 10 ESTs from the infected library. This enzyme plays a key role in nitrogen metabolism in many organisms, because free ammonia is released as a product of catalysis (Garnier et al., 1997).
The high proportion of infection site ESTs that encode enzymes of intermediary metabolism (most of which appear to be P. sojae transcripts) indicates that rapid growth and invasion of host tissues puts a massive demand on central metabolic processes to furnish simple precursors and ATP. Thus, critical steps within these pathways could provide new targets to control the growth of P. sojae and reduce its virulence on soybean plants. In this regard, many other ESTs from the infected library were also potentially significant. For example, several different glucanases and proteinases were also identified from the infection site and zoospore libraries, as shown in Table III. Secreted hydrolytic enzymes are important components that aid in the processes of physical ingress and nutrient solubilization, and thus may constitute quantitative factors that contribute to the overall virulence of the pathogen. Furthermore, it has been well documented that Phytophthora spp. lack a complete sterol synthesis pathway and require an exogenous source of these lipids for normal growth and development (Nes, 1987). A putative progesterone receptor and an estradiol dehydrogenase homolog are thus noteworthy for their possible role in sterol metabolism.
Table III.
Sample of ESTs and their known or predicted function
| EST | Best Match | P Valuea |
|---|---|---|
| 7-2e-ZO | Aminopeptidase | 10−20 |
| 7-10a-HA | Endo-1,3-1,4-β-d-glucanase | 10−8 |
| 8-9d-ZO | Endocellulase | 10−6 |
| 8-2d-HA | Estradiol dehydrogenase | 10−14 |
| 2-5g-HA | 2′-Hydroxydihydrodaidzein reductase | 10−32 |
| 2-1h-ZO | Iron transporter | 10−23 |
| 9-2h-ZO | Kinesin motor protein | 10−23 |
| 3-10h-ZO | Myotubulin | 10−30 |
| 10-11h-HA | Neprilysin | 10−20 |
| 9-9d-HA | Peroxidase | 10−62 |
| 8-8c-HA | Polyphenol oxidase | 10−41 |
| 6-6e-HA | Preprocathepsin D | 10−26 |
| 8-11h-HA | Progesterone receptor | 10−6 |
| 4-6e-ZO | Thrombospondin-related adhesive | 10−13 |
| 2-10c-HA | Tripeptidyl-peptidase II | 10−30 |
P value from BLASTX search.
The ESTs from the infection site also offer insight into host plant responses, since an estimated 300 to 400 of these sequences originate from soybean cDNA transcripts. Matches to pathogenesis-related and other defense-related proteins (e.g. peroxidase and polyphenol oxidase) were prevalent among these ESTs, as were matches to enzymes in the phenylpropanoid pathway (e.g. cinnamyl alcohol dehydrogenase, chalcone synthase/reductase, isoflavone reductase). These enzymes produce antimicrobial compounds such as phytoalexins and quinones, and are also involved in the polymerization of phenolic compounds to impede pathogen spread. The isoflavonoid glyceollins are the main phytoalexins produced by soybean via a metabolic pathway of at least 11 different enzyme-catalyzed steps from the precursor coumaroyl-CoA. Molecular characterization of the enzymes and corresponding genes of many of these steps remain to be accomplished. Nonetheless, at least five of the 11 steps in glyceollin synthesis were represented by EST matches, including a cDNA encoding a P450 enzyme (4-1B-HA) that was subsequently characterized and shown to catalyze the aryl migration reaction of isoflavonoid biosynthesis (Steele et al., 1999). Clearly, the infection site ESTs are a rich source of genes involved in host defense responses and offer good opportunities for identifying new enzymes and proteins that participate in these processes.
The collection of zoospore ESTs will also be useful for studies on infection processes and pathogenicity, since zoospores are considered to be the primary infectious propagules of most Phytophthora spp. (Hardham et al., 1991; Erwin and Ribeiro, 1996). Highly represented in the zoospore ESTs were matches to cell structural proteins, including transcripts that may be important for adhesion. Also abundant was a transcript encoding a protein similar to the Bremia lactucae HAM34 protein (Judelson and Michelmore, 1990). The HAM34 mRNA is an abundant transcript in B. lactucea spores, and promoter sequences from this gene have been used in the construction of oomycete transformation vectors (Judelson et al., 1993). Other zoospore ESTs, such as matches to myotubulin and kinesin motor protein, may be involved in motility as components of the prominent tinsel and whiplash flagella.
Mycelium ESTs were more likely to match sequences from the infected plant library than were zoospore ESTs, and were therefore useful to identify many P. sojae transcripts from the mixed infection site. Among the most abundant transcripts in the mycelium library was a match to RIC1, a putative membrane protein from P. infestans that is also similar to stress-induced proteins from yeast (van West et al., 1999). A mycelium superoxide dismutase transcript also present in the infected-plant library is potentially significant because H2O2 and other reactive oxygen species generated at infection sites are part of the plant defense response.
The EST data generated in this study represent an important complement to ongoing mapping studies that focus on the genetic localization of avirulence genes by molecular cloning strategies (Tyler et al., 1995; Whisson et al., 1995; Gijzen et al., 1996a). Linkages found between avirulence genes, and sequencing of contiguous stretches of P. sojae genomic DNA have raised the possibility of pathogenicity islands in P. sojae. Thus, positioning ESTs onto physical maps could also provide useful information about gene organization and function in this species.
P. sojae ESTs could also be used for developing new transformation methods. Transient expression assays have shown that vectors constructed using promoters derived from ascomycetes and basidiomycetes were nonfunctional in P. sojae (Judelson et al., 1993). Current P. sojae transformation vectors rely on promoter sequences from Bremia lactucae that may be less than optimal for expression in P. sojae. Promoters from tissue-specific or constitutively present ESTs identified in this study can be isolated from genomic DNA and used for construction of Phytophthora specific transformation vectors.
In summary, the comparative EST-based study presented here represents a resource for genetic and biochemical studies on P. sojae and soybean. We have successfully produced an inventory of cDNA clones and corresponding sequences that will help unravel the underlying mechanisms defining virulence, pathogenicity, and host specificity of P. sojae and defense responses of the host plant.
MATERIALS AND METHODS
Culture and Growth Conditions
Phytophthora sojae strain P6497 is a race 2 phenotype originally isolated in Mississippi and obtained from the Phytophthora culture collection at the University of California, Riverside (Förster et al., 1994). Working stocks of the organism were routinely maintained on vegetable juice agar at 25°C in the dark. Sporangial development was induced by repeatedly flooding 5- to 7-d-old mycelium colonies with sterile distilled water. Zoospores were collected by centrifugation at 2,000g (Ward et al., 1979). To obtain axenically prepared mycelium, P6497 was grown for 7 d on synthetic agar media selective for Phytophthora growth (Hoitink and Schmitthenner, 1969). Mycelium discs cut from the growing edge of each colony were transferred into flasks containing 50 mL of liquid synthetic medium and grown for 3 weeks at 25°C in the dark. Liquid cultures were vacuum decanted onto filter paper, and mycelium tissue was collected, frozen in liquid N2, lyophilized, and stored at −80°C.
Soybean (Glycine max [L.] Merr) cv Harosoy seeds were from the collection at Agriculture and Agri-Food Canada. This cultivar possesses the Rps7 gene for resistance to P. sojae, but it is susceptible to infection by P6497 and most other strains of P. sojae. Etiolated seedlings of cv Harosoy were placed in trays and each hypocotyl inoculated with 10 drops of a zoospore suspension, with each 10-μL drop containing approximately 103 zoospores (Ward et al., 1979; Gijzen et al., 1996b). Challenged seedlings were incubated for 48 h at 25°C in the dark. Infected, water-soaked tissue was excised, frozen in liquid N2, lyophilized, and stored at −80°C.
Nucleic Acid Extraction
Freshly collected zoospores, lyophilized mycelium, or infected plant tissues were pulverized to a fine powder in liquid N2 using a mortar and pestle. Total RNA from zoospores and mycelium was extracted in a solution of phenol-guanidine isothiocyanate (TRIZOL, Life Technologies/Gibco-BRL, Cleveland) according to instructions provided by the manufacturer. Total RNA was isolated from inoculated soybean hypocotyls following the procedure of Wang and Vodkin (1994), because these tissues contained interfering phenolic compounds. P. sojae and soybean genomic DNA was prepared according to the method of Dellaporta et al. (1983) with modifications. Following phenol-chloroform extraction, genomic DNA was incubated with RNAse A for 30 min at 37°C prior to isopropanol precipitation. To remove residual protein and polysaccharide complexes from soybean genomic DNA, samples were further purified by repeated chloroform-isoamyl alcohol (24:1, v/v) extractions in the presence of 0.8 m NaCl and 0.1 volume of 10% (w/v) cetyl-trimethyl-ammonium bromide in 0.7 m NaCl.
cDNA Library Construction
Poly(A+) RNA was purified from total RNA by oligo(dT)-cellulose chromatography. The synthesis of cDNA, size selection, addition of linkers, insertional ligation, and packaging into λ vector (λZAP Express, Stratagene, La Jolla, CA) followed the manufacturer's instructions and did not deviate significantly from standard methods (Sambrook et al., 1989). The total primary titer of each library in recombinant plaque forming units was: mycelium, 1.1 × 106; zoospore, 7.1 × 106; and infected plant, 9.0 × 105. After amplification, samples of each of the cDNA libraries were used to subclone inserts by mass excision for conversion of λ to phagemid vector. The resulting phagemid libraries were plated at low density on Luria Bertani agar plates containing kanamycin (25 mg L−1). Over 1,000 randomly selected bacterial colonies from each of the three cDNA libraries were cultured for plasmid isolation and long-term storage in microtiter plates. Identification codes for ESTs were derived from plate and well numbers and from the cDNA library source (mycelium, MY; zoospore, ZO; cv Harosoy-infected tissue, HA). Sequence data from a seed coat cDNA library (Gijzen, 1997) was used to determine the percentage G+C content of soybean ESTs.
Nucleotide Sequencing
Plasmid DNA was purified from Escherichia coli cultures by alkaline lysis, vacuum filtration, and anion-exchange chromatography using a high-throughput, 96-well format system (Qiagen, Mississauga, Ontario, Canada). Automated cycle sequencing of DNA was carried out using T3 primer and dye-labeled terminators, and products were resolved by electrophoresis through acrylamide gels (model 377, Applied Biosystems, Foster City, CA). All EST data are publicly available through the National Center for Genome Resources (NCGR), coordinating site of the Phytophthora Genome Initiative Project (http://www.ncgr.org/pgi/).
Sequence Data Analysis
Raw DNA sequence data were edited to remove vector sequences and poor quality data, and the percentage G+C content was determined using a computer program (LASERGENE software, DNASTAR, Inc., Madison, WI). Computer-processed sequences were checked manually, compared with electropherograms, and further edited if necessary to improve the quality and reliability of the data. The six deduced amino acid translations of the partial cDNAs were searched against a non-redundant database available through the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) using the BLASTX 1.4.11 program for comparative identification (Altschul et al., 1990). The probability (P) value was rounded to an order of magnitude and used to classify ESTs as highly significant (P < 10−19), moderately significant (P values from 10−5–10−19), or weakly significant (P values from 10−2–10−4) matches, where P represents the smallest sum of probability in a Poisson distribution (Adams et al., 1991). ESTs that returned P values greater than 10−2 were deemed not statistically significant matches.
Assembly of ESTs into Sequence Contigs
The edited EST sequences were arranged into nucleotide-matched clusters using an alignment program (Seqman, LASERGENE, DNASTAR, Inc.) installed in a personal computer (Dell OptiPlex GX1, Pentium II 350 MHz processor, 128 MB RAM) to determine the frequency of sampling redundancy. A pre-pass function optimizes assembly order by constructing a matrix of 16-mer content for each sequence and grouping candidate overlapping ESTs with the greatest number of 16-mers in common. From this data set, sequences were processed into contigs based on the strategy of pair-wise alignment. This approach involves implementation of the Martinez algorithm (Martinez, 1983), which assesses regions of a perfect match between paired sequences, followed by the Needleman-Wunch method, which aligns regions between these matched sites (Needleman and Wunch, 1970). Default parameters (including a 75% minimum match threshold) were used to construct the alignments for each of the databases derived from EST sequences from mycelium, zoospores, or infected plant tissue. For the final assembly, EST sequences from all three libraries were assembled using a minimum match threshold of 65%.
Comparative TBLASTX Analyses
To investigate further the similarity of P. sojae ESTs to other sequences, we performed a comparative analysis against two distinct sets of target sequences: one consisting of 1,490 Phytophthora infestans ESTs from a mycelium library (Kamoun et al., 1999a), and another that comprised 1,429,415 EST sequences from all taxa, excluding humans. Human sequences were excluded because these data comprise more than one-half (55%) of all ESTs and are of limited use for the purposes of a comparative analysis across taxa. Both target sets are accessible publicly at http://seqsim.ncgr.org. We used the TBLASTX algorithm, version 2.0.6 (Altschul et al., 1997), to compare all six reading frames of both query and target nucleotide sequences as amino acid translations. This program returns Expect (E) values in place of P values. Matches with E values < 10−6 were compared as raw scores from both target sets. Raw scores were the basis for comparison rather than E values, because E values are scaled according to target size, but the two targets differ in size by three orders of magnitude. In this approach, a discrepancy between scores from either target indicates that the P. sojae sequence resembles a sequence from one target more closely than from the other. For the 10 sequences from each library that had the greatest difference between scores, we ran a BLASTX search against the “nr” amino acid library to infer the function of these sequences.
Hybridization Analysis of ESTs to P. sojae and Soybean Genomic DNA
For Southern analysis, 2 μg of P. sojae or 30 μg of soybean genomic DNA was digested with EcoRI endonuclease. Restriction fragments were separated by electrophoresis on a 0.7% (w/v) agarose gel, transferred onto nylon filters, and fixed by UV cross-linkage. The membranes were pre-hybridized in 0.25 m Na2HPO4 (pH 7.2), 1% (w/v) bovine serum albumin (BSA), 1 mm EDTA, and 7% (w/v) SDS at 65°C for 5 h. Probes were 32P-labeled by random primer extension according to the manufacturer's instructions (Pharmacia, Uppsala). After an 18-h hybridization at 65°C, the filters were washed in 20 mm Na2HPO4 (pH 7.2), 1 mm EDTA, 1% (w/v) SDS at 68°C for four 20-min incubations, followed by three 5-min washes in the same buffer at room temperature. The blots were exposed to Kodak X-AR film (Eastman Kodak, Rochester, NY) with an intensifying screen at −70°C.
ACKNOWLEDGMENTS
We thank Pearl Campbell, Sandra Millar, Heather Schneider, and Ida van Grinsven for DNA sequencing; Ralph Chapman for assistance in automated DNA sequence operations; Aldona Gaidauskas-Scott, Mark Waugh, and Yihe Wu for technical assistance; B. Patrick Chapman for help with the sequence assemblies; and Mary Anne Nelson and Aaron Halpern for advice on interpretation of comparative sequence analyses.
Footnotes
This work was supported in part by a grant from the Ontario Soybean Growers.
LITERATURE CITED
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, Kerlavage AR, McCombie WR, Venter JC. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TR, Buzzell RI. Diversity and frequency of races of Phytophthora megasperma f. sp. glycinea in soybean fields in Essex County, Ontario, 1980–1989. Plant Dis. 1992;76:587–589. [Google Scholar]
- Dellaporta S, Woods H, Hicks J. A plant DNA mini-preparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Doupnik B. Soybean production and disease loss estimates for north central United States from 1989 to 1991. Plant Dis. 1993;77:1170–1171. [Google Scholar]
- Erwin DC, Ribeiro OK. Phytophthora Diseases Worldwide. St. Paul: APS Press; 1996. [Google Scholar]
- Förster H, Coffey MD, Elwood H, Sogin ML. Sequence analysis of the small subunit ribosomal RNAs of three zoosporic fungi and implications for fungal evolution. Mycologia. 1990;82:306–312. [Google Scholar]
- Förster H, Tyler BM, Coffey MD. Phytophthora sojae races have arisen by clonal evolution and by rare outcrosses. Mol Plant-Microbe Interact. 1994;7:780–791. [Google Scholar]
- Garnier A, Berredjem A, Botton B. Purification and characterization of the NAD-dependent glutamate dehydrogenase in the ectomycorrhizal fungus Laccaria bicolor (Maire) Orton. Fungal Genet Biol. 1997;22:168–176. doi: 10.1006/fgbi.1997.1004. [DOI] [PubMed] [Google Scholar]
- Gijzen M. A deletion mutation at the ep locus causes low seed coat peroxidase activity in soybean. Plant J. 1997;12:991–998. doi: 10.1046/j.1365-313x.1997.12050991.x. [DOI] [PubMed] [Google Scholar]
- Gijzen M, Förster H, Coffey MD, Tyler B. Cosegregation of Avr4 and Avr6 in Phytophthora sojae. Can J Bot. 1996a;74:800–802. [Google Scholar]
- Gijzen M, MacGregor T, Bhattacharyya M, Buzzell R. Temperature induced susceptibility to Phytophthora sojae in soybean isolines carrying different Rps genes. Physiol Mol Plant Pathol. 1996b;48:209–215. [Google Scholar]
- Gunderson JH, Elwood H, Ingold A, Kindle K, Sogin ML. Phylogenic relationships between chlorophytes, chrysophytes, and oomycetes. Proc Natl Acad Sci USA. 1987;84:5823–5827. doi: 10.1073/pnas.84.16.5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardham AR, Gubler F, Duniec J. Ultrastructural and immunological studies of zoospores of Phytophthora. In: Lucas JA, Shattock RC, Shaw DS, Cooke LR, editors. Phytophthora. Cambridge, UK: Cambridge University Press; 1991. pp. 50–59. [Google Scholar]
- Hildebrand AA. A root and stalk rot of soybeans caused by Phytophthora megasperma Drechsler var. sojae var. nov. Can J Bot. 1959;37:927–957. [Google Scholar]
- Ho HH, Zachariah K, Hickman CJ. The ultrastructure of zoospores of Phytophthora megasperma var. sojae. Can J Bot. 1967;46:37–41. [Google Scholar]
- Hoitink HA, Schmitthenner AF. Rhododendron wilt caused by Phytophthora citricola. Phytopathology. 1969;59:708–709. [Google Scholar]
- Judelson HS. The genetics and biology of Phytophthora infestans: modern approaches to a historical challenge. Fungal Genet Biol. 1997;22:65–76. doi: 10.1006/fgbi.1997.1006. [DOI] [PubMed] [Google Scholar]
- Judelson HS, Dudler R, Pieterse CMJ, Unkles SE, Michelmore RW. Expression and antisense inhibition of transgenes in Phytophthora infestans is modulated by choice of promoter and position effects. Gene. 1993;133:63–69. doi: 10.1016/0378-1119(93)90225-r. [DOI] [PubMed] [Google Scholar]
- Judelson HS, Michelmore RW. Highly abundant and stage-specific mRNAs in the obligate pathogen Bremia lactucae. Mol Plant-Microbe Interact. 1990;3:225–232. doi: 10.1094/mpmi-3-225. [DOI] [PubMed] [Google Scholar]
- Kamoun S, Hraber PT, Sobral BWS, Nuss D, Govers F. Initial assessment of gene diversity for the oomycete pathogen Phytophthora infestans based on expressed sequences. Fungal Genet Biol. 1999a;28:94–106. doi: 10.1006/fgbi.1999.1166. [DOI] [PubMed] [Google Scholar]
- Kamoun S, Huitema E, Vleeshouwers VGAA. Resistance to oomycetes: a general role for the hypersensitive response? Trends Plant Sci. 1999b;4:196–200. doi: 10.1016/s1360-1385(99)01404-1. [DOI] [PubMed] [Google Scholar]
- Kasuga T, Salimath SS, Shi J, Gijzen M, Buzzell RI, Bhattacharyya MK. High resolution genetic and physical mapping of molecular markers linked to the Phytophthora resistance gene Rps1-k in soybean. Mol Plant-Microbe Interact. 1997;9:1035–1044. [Google Scholar]
- Kaufmann MJ, Gerdemann JW. Root and stem rot of soybeans caused by Phytophthora sojae n.sp. Phytopathology. 1958;48:201–208. [Google Scholar]
- Mao Y, Tyler BM. Genome organization of Phytophthora megasperma f.sp. glycinea. Exp Mycol. 1991;15:283–291. [Google Scholar]
- Martinez HM. An efficient method for finding repeats in molecular sequences. Nucleic Acids Res. 1983;11:4629–4634. doi: 10.1093/nar/11.13.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris PF, Bone E, Tyler BM. Chemotropic and contact responses of Phytophthora sojae hyphae to soybean isoflavonoids and artificial substrates. Plant Physiol. 1998;117:1171–1178. doi: 10.1104/pp.117.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman SB, Wunch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Nes WD. Biosynthesis and requirement of sterols in the growth and reproduction of the oomycetes. In: Fuller G, Nes WD, editors. Ecology and Metabolism of Plant Lipids. ACS Symposium Series 325. Washington, DC: American Chemical Society; 1987. pp. 304–328. [Google Scholar]
- Rutherford FS, Ward EWB. Estimation of relative DNA content in nuclei of races of Phytophthora megasperma f. sp. glycinea by quantitative fluorescence microscopy. Can J Genet Cytol. 1985;27:614–616. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schmitthenner AF. Problems and progress in control of Phytophthora root rot of soybean. Plant Dis. 1985;69:362–368. [Google Scholar]
- Steele CL, Gijzen M, Qutob D, Dixon RA. Molecular characterization of the enzyme catalyzing the aryl migration reaction of isoflavonoid biosynthesis in soybean. Arch Biochem Biophys. 1999;367:146–150. doi: 10.1006/abbi.1999.1238. [DOI] [PubMed] [Google Scholar]
- Stossel P, Lazarovits G, Ward EWB. Penetration and growth of compatible and incompatible races of Phytophthora megasperma var. sojae in soybean hypocotyl tissues differing in age. Can J Bot. 1980;58:2594–2601. [Google Scholar]
- Tyler BM, Förster H, Coffey MD. Inheritance of avirulence factors and restriction fragment length polymorphism markers in outcrosses of the Öomycete Phytophthora sojae. Mol Plant-Microbe Interact. 1995;8:515–523. [Google Scholar]
- van West P, Kamoun S, van 't Klooster JW, Govers F. Ric1, a stress induced gene of the potato late blight pathogen Phytophthora infestans. Curr Genet. 1999;36:310–315. doi: 10.1007/s002940050505. [DOI] [PubMed] [Google Scholar]
- Voglmayr H, Greilhuber J. Genome size determination in Peronosporales (Oomycota) by Feulgen image analysis. Fungal Genet Biol. 1998;25:181–195. doi: 10.1006/fgbi.1998.1097. [DOI] [PubMed] [Google Scholar]
- Wang CS, Vodkin LO. Extraction of RNA from tissues containing high levels of procyanidins that bind RNA. Plant Mol Biol. 1994;12:132–145. [Google Scholar]
- Ward EWB. The interaction of soya beans with Phytophthora megasperma f. sp. glycinea: pathogenicity. In: Hornby B, editor. Biological Control of Soilborne Plant Pathogens. Wallingford, UK: CAB International; 1990. pp. 311–327. [Google Scholar]
- Ward EWB, Cahill DM, Bhattacharyya MK. Early cytological differences between compatible and incompatible interactions of soybeans with Phytophthora megasperma f.sp. glycinea. Physiol Mol Plant Pathol. 1989;34:267–283. [Google Scholar]
- Ward EWB, Lazarovits G, Unwin CH, Buzzell RI. Hypocotyl reactions and glyceollin in soybeans inoculated with zoospores of Phytophthora megasperma var. sojae. Phytopathology. 1979;69:951–955. [Google Scholar]
- Waugh M, Hraber P, Weller J, Wu Y, Chen G, Inman J, Kiphart D, Sobral B. The Phytophthora Genome Initiative Database: informatics and analysis for distributed pathogenomic research. Nucleic Acids Res. 2000;28:87–90. doi: 10.1093/nar/28.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson SC, Drenth A, Maclean DJ, Irwin JAG. Phytophthora sojae avirulence genes, RAPD, and RFLP markers used to construct a detailed genetic linkage map. Mol Plant-Microbe Interact. 1995;8:988–995. doi: 10.1094/mpmi-8-0988. [DOI] [PubMed] [Google Scholar]
- Wrather JA, Chambers AY, Fox JA, Moore WF, Sciumbato GL. Soybean disease loss estimates for the southern United States, 1974 to 1994. Plant Dis. 1995;79:1076–1079. doi: 10.1094/PDIS.1998.82.1.114. [DOI] [PubMed] [Google Scholar]