Abstract
PRACTALL is a joint initiative of the American Academy of Allergy, Asthma & Immunology and the European Academy of Allergy and Clinical Immunology to provide shared evidence-based recommendations on cutting-edge topics in the field of allergy and immunology. PRACTALL 2017 is focused on what has been established regarding the role of the microbiome in patients with asthma, atopic dermatitis, and food allergy. This is complemented by outlining important knowledge gaps regarding its role in allergic disease and delineating strategies necessary to fill these gaps. In addition, a review of progress in approaches used to manipulate the microbiome will be addressed, identifying what has and has not worked to serve as a baseline for future directions to intervene in allergic disease development, progression, or both.
Keywords: Microbiome, microbiota, dysbiosis, asthma, atopic dermatitis, food allergy, systems biology, probiotic, prebiotic
There is increasing evidence that resident microbial communities in the human gastrointestinal tract, airway, and skin contribute to health and disease. This complex relationship between the microbiota (Table I) and the human host could lend itself to manipulation to benefit the host. This fits with recognition that the microbiome plays an important role in early immunologic development, raising the possibility that experimental manipulation can modulate the immune system.1 Linking the microbiota to both immune development and response heightens the potential that the microbiome plays an important role in the development and manifestations of allergic diseases. In fact, at a clinical level, this connection had its origins with observations in the 1980s that children in large families had a lower incidence of allergic rhinitis and atopic dermatitis (AD) compared with children from smaller families, observations that led to the hygiene hypothesis.2 Subsequent epidemiologic studies yielded data documenting a lower rate of allergic disease in children raised in European rural environments linked to raw milk and stable exposure, particularlyduring the first year of life.3,4 A host of additional studies established that in addition to the findings noted above, absence of early antibiotic exposure, exclusive breast-feeding for the first 4 months of life, vaginal delivery, furry pets in the home during infancy, lack of maternal antibiotic use during pregnancy, and maternal animal exposure during pregnancy all were associated with lower rates of allergic disease.5-9 Taken together, these clinical observations established a strong link between microbes and the development of allergic disease.
TABLE I.
Key definitions
| Microbiome | The sum of microbes, their genomic elements, and interactions in a given ecological niche |
| Microbiota | The sum of microbes found in a given ecological niche |
| Richness | The number of species or taxa found in a sample or niche |
| Evenness | The relative distribution of species or taxa in a sample or niche |
| Diversity | A calculated index that incorporates measures of richness and species distribution. Many different diversity measures exist:
|
| Taxon or taxa | Synonymous with operational taxonomic unit (OTU); for bacteria identified by using 16S rRNA gene-based analysis, a taxon is defined as a group of species with very similar sequence homology (eg, ≥97%). |
| Dysbiosis | Descriptive term for imbalance in a microbial ecosystem; for example, dysbiosis of the intestinal or respiratory tract associated with a disease state compared with health |
| Pathobiont | An organism normally part of the microbiota (commensal) but able to promote pathologic disease under certain host conditions; examples include Clostridium difficile, Haemophilus influenzae, and Klebsiella pneumoniae |
The science connecting the microbiome to immunologically mediated disorders, including allergic disease, has been advanced by the capacity to catalog microbiota and define imbalances in microbial communities, which are referred to as dysbiosis (Table I). These studies have used next-generation DNA sequencing to profile the total bacterial community in a particular site, including evaluation of the richness, evenness, and diversity of the species present (Table I). This approach represents an important step forward because standard culture methods typically detect only approximately 1% of the bacterial community in a particular site when compared with molecular methods. However, there remain limitations to these data, including the recognition that many studies characterizing bacterial content have not captured other microbiota, such as fungi and viruses.
Nonetheless, the potential contribution of the microbiota to the documented increase in rates of asthma, AD, and food allergy has become increasingly appealing to consider. The clinical findings noted above, together with the increased rate of allergic disease, suggest that urban living in Western society is a major contributor to this change. There are now clinical research studies and animal studies that link the microbiota of the gastrointestinal tract, as well as those of the skin and respiratory tract, to allergic disease. Two recent examples include a study that suggests that microbiota (together with bacterial metabolic products) during early infancy affects the risk of childhood asthma.10 In addition, in genetically comparable populations, environmental exposures linked to varied microbial content affect the development of asthma by shaping the innate immune response.11 These and many other studies clearly provide very strong connections between bacterial microbiota and the development of allergic disease. However, most of these studies have focused on one organ system and have used varied end points. Thus there remains a host of unanswered questions to further clarify and expand on these connections. Finally, it has been suggested that modifying the microbiome to the host’s advantage could prove to be an approach for preventing and/or treating allergic disease.
MICROBIAL ECOLOGY
Trillions of microbes colonize the skin and mucosal body surfaces. These microbes are highly adapted to survive within complex community structures, requiring nutrients from other microbes, host processes, or both (Fig 1). Many bacterial species within these communities lack genes that are essential for bacterial fitness in other environments, whereas they possess genes that benefit the host with little or no benefit to the bacterium, suggesting a symbiotic coevolution of bacterial communities within specific host niches.12 DNA-sequencing techniques have facilitated more in-depth analysis of the gastrointestinal tract, skin, genitourinary tract, and lung microbiota, revealing a microbial superorgan residing in symbiosis with host mucosal surfaces. These communities evolve within a host from birth, constantly being fine-tuned to maintain a homeostatic balance with the host’s immune system. This evolution is influenced by host factors, such as adaptive and innate immune responses13; external factors, such as diet, medication, and toxin exposure; infection; and illness. Given the potentially vast number of interactions existing between microbes, diet/nutrients, host metabolism, immune responses, and xenobiotics, it has been extremely difficult to model population dynamics experimentally.
FIG 1.
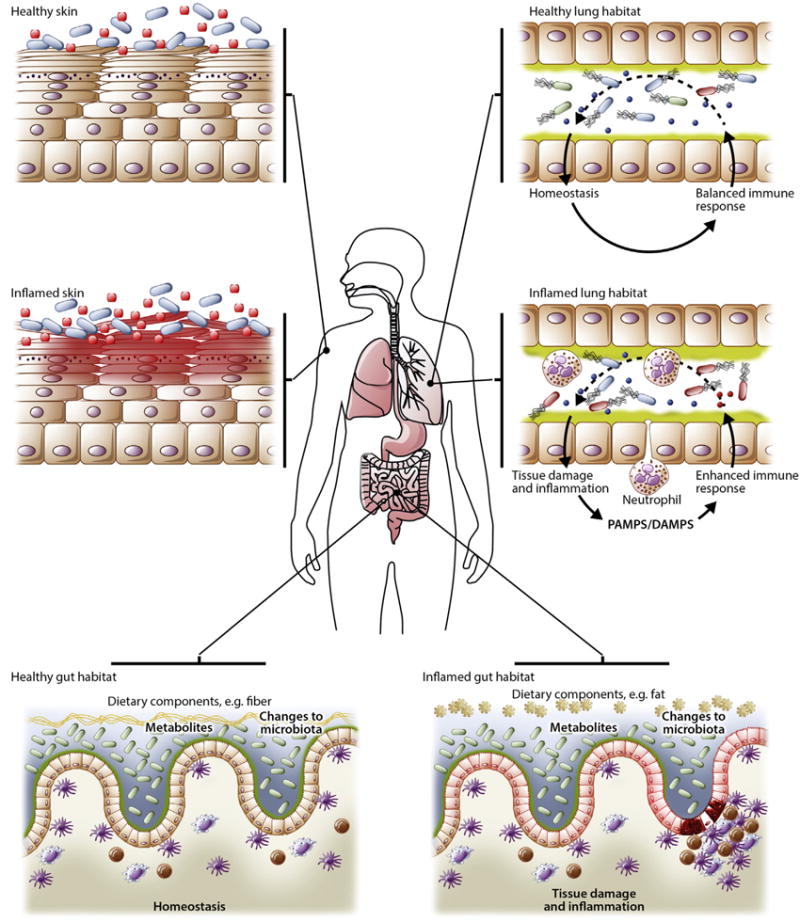
Microbial ecology. The human body provides a diverse habitat for the microbes that reside on its epithelial surfaces. Differences in oxygen levels, pH, and availability of nutrients create environments that select for the microbes specifically adapted to colonize the tissue. However, the human body ecosystem is dynamic, with changes driven by age, diet, environmental exposures, and disease. Nutrients provided from diet are key ecological influencers within the intestinal tract because they represent energy sources for bacteria that have the metabolic machinery to use them. Diet changes with both age and lifestyle matched by a coordinated change in the constituents of the microbiota. Inflammation is a powerful modifier of the local microbial habitat both directly through antimicrobial immune mechanisms and indirectly by influencing putative energy sources such as mucins. Physiologic changes, including pH or oxygen levels, change the environment, providing microbes with the toolkit to handle these microenvironments. Host-microbe cross-talk is a key determinant of the tissue habitat. Tonic signals from the microbiota set the tone of the local immune system, which, under steady-state conditions, results in a state of mutualism; however, dysregulation of this interaction, such as through exposure to an exogenous pathogen (or a bloom of a pathobiont), medical treatment (antibiotics, antifungal drugs, and immunosuppression), or inflammation/tissue damage, can perpetuate inflammation or cause exacerbations. DAMPS, Danger-associated molecular patterns; PAMPs, pathogen-associated molecular patterns.
The composition and diversity of the microbiome varies across body sites, resulting in a series of unique habitats within and between subjects that can change dramatically over time. The gastrointestinal tract has the greatest number and diversity of microbes and is dominated by facultative and strictly anaerobic bacteria of the phyla Firmicutes, Bacteroidetes, Actinobacteria, Verrucomicrobia, and Proteobacteria.14 The dominant microbial phyla of the healthy human lung also include Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria, in addition to Fusobacteria.15 Bacterial communities differ across the skin surface, and this biogeography is dependent on pH, temperature, moisture, sebum content, and topography.16 Importantly, changes in the composition, metabolic activity, or both of the gut, lung, and skin microbiomes have been associated with asthma, AD, and food allergy, respectively.
ASSESSMENT OF THE MICROBIOTA
Classically, microbiota composition and diversity were assessed with tools, such as microscopy and in vitro culture. More recently developed culture-independent approaches targeting small subunit (16S) rRNA gene sequences or metagenomic shotgun sequencing have allowed for unprecedented detailed assessments of the diversity, composition, and function of many microbial ecosystems.17 However, comparative analysis between different studies has been problematic because of differences in phenotyping, sample collection and storage, DNA isolation, amplification, and sequencing protocols. In addition, these analyses depend on the quality of available databases that allow the 16S rRNA gene sequences to be annotated with a particular bacterial taxon and the types of analysis and data interpretation tools that are used. Consensus on best practice experimental conditions and data analysis tools is urgently required to prevent potentially misleading interpretations of the data.
In addition to the assessment of changes in bacterial community composition and diversity, it is important to better understand the functional consequences of these changes. Meaningful in vitro and in vivo models are required to link changes in microbiome signatures with disease-relevant end points. This requires that target bacterial strains are isolated, cultured, and well characterized.
In addition, new and improved tools to evaluate the fungal and viral components within each niche habitat will also need to be developed to fully understand the role of the human microbiome in health and disease. Despite the technical and analytic challenges involved in studying fungal and viral communities (as discussed later), several studies have described the types of fungal and viral species detected in different body sites, including the gut, respiratory tract, and skin.18-20 Fungal and viral community differences associated with disease states, including allergic rhinitis and asthma, also have been reported.21,22 However, the clinical implications of such findings remain unclear given poor insight into the functional consequences of these microbial associations in disease or health, a focus on DNA viruses (eg, bacteriophages), and limited understanding of the dynamics of human-associated fungal and viral communities. For example, a study of sputum microbiota in patients with cystic fibrosis found greater fluctuation in fungal community than bacterial community composition, which suggests that inhaled fungal elements might be more transient in the respiratory microbiomes of some subjects.23
ASTHMA
Significant advances have been made in our understanding of the genetic, environmental, and immunologic factors that shape asthma. However, asthma is clinically heterogeneous, and underlying causes for many phenotypes remain poorly understood.24-28 Microbes have long been postulated to play a role in asthma and might also shape its heterogeneity.29,30 These roles include the following contexts: (1) the effect of early-life exposure to microbially rich environments on susceptibility to childhood asthma; (2) the role of the microbiota in immune system development; (3) the effects of acute viral infections and patterns of respiratory tract bacterial colonization on childhood asthma; and (4) the potential role of bronchial colonization by particular bacteria in phenotypes of asthma. Increasingly, further evidence in support of these roles has been derived from studies that considered the ecology of microbial communities found in different sample types or organ-specific niches (microbiota). New insights from these and other human microbiome investigations are blurring classical concepts of “colonization” and “infection,”31,32 leading researchers to consider new hypotheses about the role of microbiota-host interactions in patients with disease, including the inception, chronicity, and severity of asthma.
Recent applications of modern molecular and bioinformatic tools to understand the composition and functional potential of microbial communities has significantly expanded our knowledge of patterns in the microbiome linked to asthma.10,33-43 To date, the predominant focus has been on bacteria, and a number of studies have described asthma-associated differences in the composition of bacterial microbiota found in the respiratory (upper and lower) and gastrointestinal tracts.10,34-41,44 For example, in the lower respiratory tract a repeating signature is asthma-associated enrichment in members of Proteobacteria, a large phylum representing many species with known potential to cause respiratory illnesses.33-37,39,40,42,43 These include members of the genera Haemophilus, Moraxella, Neisseria, and Streptococcus (Box 1).
Box 1.
Bacterial genera implicated in asthma or asthma-related features (from recent microbiota studies)
| Bacterial genus | Microbiome compartment | Asthma context |
|---|---|---|
| Haemophilus | Respiratory | Increase associated with asthma in children and adults |
| Neisseria | Respiratory | Increase associated with asthma in adults |
| Moraxella | Respiratory | Increase associated with asthma in children and adults |
| Streptococcus | Respiratory | Increase associated with asthma in children |
| Lactobacillus | Gastrointestinal, respiratory | Decrease associated with asthma in children and adults |
| Bifidobacterium | Gastrointestinal | Decrease associated with risk for asthma development in childhood |
| Faecalibacterium | Gastrointestinal | Decrease associated with risk for asthma development in childhood |
| Akkermansia | Gastrointestinal | Decrease associated with risk for asthma development in childhood |
| Morganella morganii | Gastrointestinal | Increase associated with asthma in nonobese adults |
Although differences exist across studies in which specific members of the microbiota are implicated (ie, differences by organ site or asthmatic group studied), the theme of note is that imbalances in the composition of respiratory and gastrointestinal microbiota (dysbiosis) are both linked to asthma. However, it remains poorly understood how functional consequences of this dysbiosis, particularly in the respiratory tract, shape or reflect asthma susceptibility or phenotype. Specific clinical and inflammatory features of asthma have been found to associate with different patterns of lower airway microbiota composition.33-35,39,40 These phenotypic features include airway hyperresponsiveness, asthma control, obesity-associated asthma, responsiveness to corticosteroids and macrolides, and patterns of bronchial epithelial gene expression, including low type 2 airway inflammation.
Evidence for a direct effect of the microbiota on asthma has been obtained from animal models, particularly in the context of development of allergic responses and allergic airway inflammation. An exaggerated susceptibility to allergic responses that is normally seen in germ-free mice is reversed by colonization of germ-free animals with a microbiota before allergen sensitization.45 Interestingly, there appear to be key postnatal steps through which microbial colonization educates the immune system and influences disease susceptibility, the so-called window of opportunity. Neonatal mice are highly susceptible to allergic inflammation after allergen exposure, a susceptibility that diminishes after development of the airway microbiome, which occurs in the first 2 to 3 postnatal weeks.46,47
The clinical parallel is earlier landmark epidemiologic studies from Europe showing that living in a farming environment, which is linked to exposures to a diverse microbial community, is associated with a lower incidence of allergies.48 Functional evidence was recently reported showing that exposing mice to “farm dust,” which is presumably laden with bacteria or their components, could reduce the magnitude of their allergic airway responses.49 The mechanism has been linked to Toll-like receptor 4 signaling50 and activation of A2049 in airway epithelial cells. These data are supported by earlier studies showing that exposure to bacteria isolated from farm dust, such as Acinetobacter lwoffii F78 and Lactococcus lactis G121, can protect mice against airway inflammation,51 as can exposure to innocuous bacterial strains52 or their products.53 These preclinical models have elucidated different mechanisms of action that consistently show a protective role of bacteria against allergic inflammation. However, significant gaps remain in our understanding of the dynamics of lung microbial communities and how these dynamics influence host-microbe interactions. Recent evidence in mice54 and human subjects55 argues for a cross-talk between microbes and immune cells, whereby inflammation can shape the microbiota and vice versa.56
The effects of the microbiota are not limited to the local tissue environment. The gut microbiota, for example, can have powerful effects on respiratory responses. Dietary fermentable fibers can influence allergic lung inflammation in mice by altering the gut microbiota, leading to an increase in circulating short-chain fatty acids (SCFAs).57-59 SCFAs have been shown to influence the maturation state of dendritic cell progenitors in the bone marrow, rendering them less capable of instigating TH2 responses in the lungs.57 SCFAs are also known to act as histone deacetylase inhibitors; a study from Choi et al60 reported that trichostatin A, a histone deacetylase inhibitor, protected against allergic airway inflammation in mice. Indeed, recently, it was shown that SCFAs might lead to epigenetic modifications in the forkhead box P3 pathway in utero,61 thus protecting mice against allergic inflammation.
Finally, the leading hypotheses to date have been limited to the role of the bacterial microbiota in the influence of allergic responses. However, it is notable that in addition to complex bacterial communities being detected in farm dust, fungal loads have also been reported to be high.48 Within the context of allergy, fungi are typically considered within the framework of allergens or inducers of allergic inflammation.62 However, fungi could also be key regulators of inflammation and immune homeostasis,63 opening the avenue of beneficial roles of fungi in shaping immune responsiveness. Indeed, a recent report by Wheeler et al64 has reported that disruption of the “mycobiome” with antifungal drugs can predispose mice to both colitis and allergic airway inflammation.
AD
AD is a complex familial transmitted skin disease with distinct phenotypes and endotypes.65 Two major biologic pathways are responsible for AD: epidermal epithelial dysfunction and altered innate/adaptive immune responses.66,67 The increased prevalence of AD, particularly in industrialized regions, has been hypothesized to be due to excessive hygiene accompanying the Western lifestyle reducing exposure of the host’s immune system to education provided by beneficial microbes. A subset of patients with AD are prone to microbial dysbiosis with bacteria, viruses, and fungi, which can exacerbate skin inflammation.68
Staphylococcus aureus is the most common pathogen grown from AD skin. Colonization and infection with S aureus is associated with increased IgE responses, food allergy, and the severity of AD skin disease.69,70 These detrimental effects are likely mediated through production of staphylococcal virulence factors, including superantigens that stimulate type 2 immune responses and subvert the activity of regulatory T cells; cytolysins, which lyse epithelial cells; and serine proteases and lipases, which damage the skin barrier. S aureus colonization occurs as a result of skin barrier dysfunction (eg, reduced filaggrin expression) and increased IL-4 and IL-13 expression. These abnormalities can occur as a result of genetic and immune responses triggered by allergens, scratching, or both. IL-4 and IL-13 enhance S aureus binding to AD skin by inducing S aureus adhesins and reduce S aureus killing by inhibiting the production of keratinocyte derived antimicrobial peptides required for control of S aureus abundance. The observation that a humanized mAb that blocks the action of IL-4 and IL-13 leads to a dramatic reduction in AD skin severity supports the concept that IL-4 and IL-13 blockade can also reduce S aureus colonization in patients with AD.71 Using new therapies that specifically target polarized immune pathways will allow us to sort out key mechanisms of disease pathogenesis.
The use of next-generation DNA sequencing methods has provided a more complete evaluation of the bacterial composition in AD skin. These data suggest an important role for other members of the skin bacterial community beyond S aureus.72 16S rRNA gene sequencing has demonstrated that there is remarkable diversity in the healthy skin microbiome at the genus level. Importantly, exacerbation of AD is associated with loss of microbial diversity and increased abundance of S aureus. Interestingly, anti-inflammatory and emollient therapy is associated with increased microbial diversity, supporting the importance of skin epithelial function in maintaining a normal microbiome.72,73 Future studies will involve metagenomics or DNA sequencing of full-length microbial genomes to identify microbes at the strain level and permit exploration of mechanisms through which individual bacteria affect the skin barrier and their immune response.74 It is critical that such studies control for host factors, including age and body regions, because these variables have been found to alter the bacterial composition of the skin.75,76
Although antibiotics are often used in the treatment of S aureus infection, these medications have the disadvantage of not only killing S aureus but also killing beneficial bacteria and potentially selecting for antibiotic-resistant bacteria, such as methicillin-resistant S aureus. There is increasing evidence that commensal skin microbes from normal skin can improve the skin barrier and augment host defense against skin pathogens, including S aureus. Staphylococcus epidermidis and Staphylococcus hominis secrete antimicrobial activities that inhibit S aureus growth and biofilm formation.77,78 S epidermidis has also been found to stimulate Toll-like receptor 2 to induce production of keratinocyte-derived antimicrobial peptides and increased tight junctions to enhance the skin barrier.79,80 Success with fecal microbiota therapy, in which the gastrointestinal disease microbiota of patients with recurrent Clostridium difficile infections is replaced with bacteria from healthy subjects, might be applicable to patients with AD. In this regard there has been interest in microbial skin “transplant” therapy of commensal bacteria from healthy skin to replace S aureus on the skin of patients with AD (Box 2).
Box 2.
Microbes associated with AD
| Microbe | Effects reported |
|---|---|
| S aureus | Detrimental |
| Group A Streptococcus | Detrimental |
| S epidermidis | Beneficial |
| S hominis | Beneficial |
| Herpes simplex virus | Detrimental |
| Vaccinia virus | Detrimental |
Eczema herpeticum (EH) is a life-threatening complication of AD and can result in severe disfigurement and corneal blindness associated with ocular infection. These patients frequently have serum IgE levels of greater than 1000 IU/mL, are colonized with S aureus, and have severe AD.81 Interestingly, S aureus has been shown to secrete products that enhance viral replication in keratinocytes. Because the majority of patients with AD have high titers of immunoglobulin (IgG) antibodies to herpes simplex virus but less than 5% have EH, host factors, such as genetic risk variants, are thought to determine AD with EH susceptibility. Genetic effects in patients with AD with a history of EH are supported by the dramatic 10.1-fold odds ratio conferred by filaggrin-null mutations.82 Susceptibility to EH has also been associated with mutations in tight junctions, as well as thymic stromal lymphopoietin genes. Gene transcriptome studies of patients with AD prone to EH have also revealed a defect in their interferon-signaling pathways.83 Targeted deep sequencing has identified rare loss-of-function variants in IFNGR1 for the risk of AD complicated by EH.84 In experimental animal models of AD, filaggrin-deficient mice, as well as type 2 immune responses mediated by thymic stromal lymphopoietin and IL-33, promote vaccinia virus replication.85 Overall, these studies support the concept that the propensity to disseminated skin infection is caused by genetic and acquired defects in epithelial skin barrier function, reduced innate immune responses, and increased type 2 immune responses.
FOOD ALLERGY
Food allergy is thought to involve deviation from the default state of mucosal immune tolerance that can be driven by diet, commensal microbiota, and the interactions between them.86 Studies of the gut microbiota in patients with food allergy have yielded variable findings on bacteria associated with the condition. Heterogeneity in diagnostic criteria, variable food allergy subphenotypes (eg, allergy to different foods), altered diet, and differences in profiling and analytic approaches underlie the disparate findings that have been reported (Box 3).
Box 3.
Microbial orders implicated in food allergy and food allergy–related outcomes
| Order | Effects reported |
|---|---|
| Clostridiales | Mostly beneficial |
| Bacteroidales | Beneficial and detrimental |
| Enterobacteriales | Mostly detrimental |
| Lactobacillales | Mostly beneficial |
Studies of children with milk allergy have shown that infants with milk allergy have higher total bacteria and anaerobic counts compared with healthy control subjects; after 6 months of differential formula intake, the 46 infants with milk allergy studied had higher proportions of lactobacilli and lower proportions of enterobacteria and bifidobacteria observed in bacterial cultures.87 A 16S rRNA gene sequencing–based study of 39 children showed that infants with milk allergy have increased microbial diversity with an increased abundance of Ruminococcaceae and Lachnospiraceae compared with control subjects.88 In contrast to these studies of milk allergy status, a recent multi-center longitudinal study of 226 children with milk allergy examined the relationship between the gut microbiome and food allergy resolution, finding that Firmicutes, including Clostridia, were enriched in the early infant gut microbiome of patients whose milk allergy resolved by age 8 years.89
Given differences in the presentations and natural histories of specific food allergies, it is plausible that the microbiota associated with each food allergy subphenotype are also distinct. Reduced microbial richness and increased Bacteroides were observed in subjects with self-reported peanut or tree nut allergy compared with those not reporting these allergies.90 Although differences in the implicated taxa reported in this study versus the milk allergy studies might have been due to the specific food allergy targeted, additional explanations include this study’s focus on adults (vs children) and its use of self-reported food allergy, which is prone to reporting bias.
Murine models of food allergy have provided experimental dimensions to the study of microbiota and food allergy. Food allergy–prone mice with a gain-of-function mutation in the IL-4 receptor α chain demonstrated differential abundance of Lach-nospiraceae, Lactobacillaceae, Rikenallaceae, and Porphyromo-nonadaceae compared with wild-type mice.91 Interestingly, the food allergy phenotype could be transmitted by transferring the gut microbiota from affected to wild-type germ-free mice.91 Other murine studies have shown that Clostridia strains in particular modulate allergy.92 Investigators identified 17 strains of bacteria from human stool that enhanced regulatory T-cell abundance, with genome sequencing revealing that all 17 strains were within the Clostridia.92 Oral administration of these 17 Clostridia strains into mice attenuated disease in models of colitis and allergic diarrhea.92 Colonization of germ-free mice with Clostridium species clusters protected against sensitization to peanut/cholera toxin, with reduced levels of peanut-specific and total IgE compared with those in germ-free control mice and no temperature decrease on allergen challenge.93 The functional effects of Clostridia in food allergy can occur through bacterial metabolites that regulate epithelial integrity and immune responses in the gut.94 The clostridial families Lachnospiraceae and Ruminococcaceae ferment dietary fiber to produce SCFAs, which ultimately promote epithelial integrity, repair, and homeostasis.95
Findings that specific bacterial taxa modulate allergy suggest potential therapeutic utility from the manipulation of gut microbiota in patients with food allergy. Only a few trials of probiotics for the prevention or treatment of challenge-proved food allergies have been published. Trials of probiotic supplementation with Lactobacillus casei and Bifidobacterium lactis for 12 months showed no effect on milk allergy resolution, although Lactobacillus rhamnosus combined with extensively hydrolyzed casein formula increased rates of milk allergy resolution compared with a control group receiving formula alone.96,97 The probiotic Lactobacillus rhamnosus GG administered with peanut oral immunotherapy for 18 months induced desensitization compared with placebo.98 However, because there was no oral immuno-therapy–only or probiotic-only group, the efficacy of the probiotic itself is unclear. The effects of probiotic treatment are likely strain specific, and the data are currently inconclusive to support probiotic supplementation with specific taxa for food allergy.
Early infancy could be the key window of opportunity for intervention given age-dependent associations between the gut microbiome and food allergy outcomes. Gut microbial richness at age 3 months is associated with increased likelihood of food sensitization by age 1 year, whereas there is no association at age 12 months.99 Gut microbiome composition at age 3 to 6 months, but not 7 to 12 or 13 to 16 months, is associated with milk allergy resolution.89 Murine models also support age-sensitive interactions with microbiota.47 Colonization of germ-free mice with a diverse microbial population early but not late in life suppresses IgE and prevents mice from having food allergy.100 These collective findings support the notion that microbial effects on early immune system development play a role in subsequent food allergy development.
STRATEGIES TO MANIPULATE THE MICROBIOME
Probiotic and prebiotic trials in patients with food allergy, AD, and asthma represent attempts to deliberately modify microbiota and their metabolism.101 Probiotics can be defined as live micro-organisms that, when administered in adequate amounts, have the potential to confer a health benefit on the host. Notably, the definition of a probiotic does not differentiate between the wide range of potential health benefits, and it is clear that not all probiotics will influence the immune system in the same way. Even strains within the same species display different host effects, such as induction of forkhead box P3–positive regulatory T cells,102 showing that health claims relating to one probiotic cannot be extrapolated to other bacterial strains within that species.
Prebiotics can be defined as selectively fermented ingredients that allow specific changes in the composition and/or activity in the gastrointestinal microbiota that confer benefits on host wellbeing and health. As with the probiotic definition, not all prebiotics will have the same effect on immunologic functions, and their effect could vary widely depending on the composition of the host’s gut microbiota.
The combination of probiotics and prebiotics is termed synbiotics. The probiotic and prebiotic human studies to date have focused on administration to the gut, but lung or skin application can provide more potent therapeutic responses in patients with asthma or AD, respectively.
Diet is a critical regulator of the gut microbiota composition and function, as demonstrated by a large number of observational and intervention studies.103 Short-term dramatic changes in diet can rapidly alter the gut microbiota, and long-term dietary habits seem to be a dominant force shaping the composition and activity of the microbiota. Dietary fermentable fibers seem particularly important because microbial fermentation of these fibers generates SCFAs that can influence the severity of allergic inflammation.57 Dietary practices early in life influence later risk of atopy and asthma,104 which might be mediated indirectly by dietary-associated changes in the microbiota. Dietary modification in combination with appropriate microbiota manipulation (eg, with probiotics) can provide the most effective preventive and therapeutic effects and form part of a precision medicine approach.
In addition to treating patients with a single or a small number of microbes, fecal microbiota transplantation is an efficient method for transferring entire communities of organisms from a healthy subject to a patient. Fecal microbiota transplantation has been successfully used in patients with C difficile colitis,105,106 and studies are ongoing in other patient groups. However, there are significant questions concerning this approach relating to donor selection, mechanism of action, and the “ideal” microbiota for transplantation. To address these issues, significant research is ongoing to engineer a microbial cocktail that can effectively establish itself in the human gut and replace the damaging elements of the microbiota. However, these engineered microbial consortia have yet to make it to the clinic, and their exclusion of other members of the microbiome (eg, fungi, viruses, and phages) might lead to unexpected hurdles in the future.
Delivery mode alters microbial diversity and composition, and cesarean section delivery has been associated with increased risk for immune and metabolic disorders.107 Recently, innovative efforts have begun to try altering the microbiota of cesarean section–delivered infants to be more like those of vaginally born infants. By using swabs, babies were exposed to their maternal vaginal contents, thus transferring the vaginal microbes to the infant.108 This pilot study showed that bacterial communities can be partially restored in newborns delivered by means of cesarean section. Although larger cohorts and longitudinal observation are required to determine whether this procedure modifies disease risk in later life, this is an interesting proof of concept. A summary of strategies to mitigate the risk of allergic disease is presented in Box 4.
Box 4.
Current opportunities for practical clinical intervention to mitigate risk of allergic disease
| Clinical intervention | Rationale |
|---|---|
| Reduce elective cesarean sections | Microbial dysbiosis can be mitigated to reduce the risk of allergic immune responses and inflammation. |
| Reduce indiscriminateuse of antibiotics during infancy/perinatal period | |
| Encourage breast-feeding, when possible | |
| Increase dietary intake of fermentable fiber | Microbial fermentation produces SCFAs, which in vitro and in vivo data suggest can mitigate allergic responses, including in the lung. |
| Topical anti-inflammatory medication | Inflammation reduces host antimicrobial responses, and use of anti-inflammatory agents reduces S aureus colonization. |
CHALLENGES AND RESEARCH OPPORTUNITIES
Analytics
One limitation associated with many current studies is that they focus exclusively on the bacterial microbiota and omit the likely important contribution of the mycobiome and virome. Approaches to characterize the mycobiome composition are developing but still difficult. In particular, isolation of fungal DNA requires specific processing that is typically not used in classical bacterial microbiota analysis. In addition, fungal sequence databases, which allow the DNA sequence fragments to be assigned to specific fungi, are still in development. For viruses, there is no parallel to 16S rRNA sequencing in bacteria, which means that a deep-sequencing approach and powerful bioinformatic tools are required to better understand changes in virome composition. However, analysis of specific viruses by using classical techniques, such as PCR, could be an interim step toward understanding the importance of subclinical viral communities that might provide tonic signals to the immune system and members of the microbiome.
Although there is strong evidence that very early-life factors shape allergy and asthma risk in childhood, the heterogeneity of allergic diseases and asthma presents both challenges and opportunities to understand how the microbiome can shape disease phenotypes. For example, it is now recognized that a significant proportion of asthma in adults is not driven by type 2 airway inflammation.28 Emerging evidence suggests that asthma-associated alterations in the respiratory microbiome are linked to diminished type 2 airway inflammation and diminished response to corticosteroids.39,40 Whether this respiratory dysbiosis is simply a reflection of the underlying host state or actively shapes the phenotypic features of asthma is difficult to dissect from clinical studies. Nonetheless, the microbiota-phenotypic feature link is striking in some cases, such as different patterns of airway microbiota enrichment associated with obesity or stability of asthma control in patients with severe asthma.34
Thus further studies in diverse groups of patients with asthma and allergic disorders are merited. However, it is important to emphasize that such studies require thoughtful prospective considerations in study design and execution. These include attention to clinical phenotyping and sample collection and processing procedures for microbiome-specific analyses. Otherwise, the results might be difficult to interpret or compare across studies. Moreover, acquiring and analyzing (in vitro or in vivo) host cells from the same tissue site as the microbiota is an important step toward testing causality and understanding how host-microbe interactions could be modulated to prevent or treat asthmatic patients.
To understand ultimately how the microbiome shapes allergic diseases and asthma in patients will require a combination of both discovery- and hypothesis-driven research. Clinical insights can drive relevant questions regarding the microbiome for which a variety of analytic tools are now available and continue to be developed. Prespecified hypotheses, informed by clinical knowledge or basic science insights, can be formulated to interrogate microbiome-related data (here referring to a variety of possible omic techniques, including 16S rRNA, metagenomics, metatranscriptomics, and metabolomics). A recent perspective article on “where next for microbiome research” described areas for attention,109 several of which are highly relevant to the focus and intent of this PRACTALL report. These include understanding interactions among microbiota and between microbiota and the host, systems biology for integrative analysis of the human microbiome, and manipulation of the microbiota as a therapeutic approach.
For asthma, extensions of these topics are important to consider. First, there is a dearth of knowledge on the stability of the microbiome, particularly in adults and the lower respiratory tract. Longitudinal observational studies to address this are a challenge but important to establish foundational knowledge that allows interpretation of how observed perturbations to the microbiome shape or predict respiratory outcomes.
Second, there is a need to understand the functional consequences of asthma-associated respiratory dysbiosis and how these relate to disease pathogenesis or phenotype. Understanding how inhaled exposures (pollutants, particulates, microbes, and drugs) affect the respiratory microbiome adds another layer of complexity. Current capabilities to collect a massive amount of information and variety of data types will require novel integrative approaches for analysis, such as through systems biology or machine-learning methods.
Third, how and when to intervene with potential approaches to therapeutically modify the microbiome remains a difficult question. Current evidence indicates a very early window of opportunity to minimize the risk of childhood asthma or atopy. Are such approaches viable in adults to “reverse” asthma or other atopic diseases? Beyond allergic diseases, leveraging microbiota manipulation to treat nonallergic diseases continues to present challenges and is an active area of investigation.
Systems biology
Systems biology is an approach to understanding living systems focused on modeling diverse types of high-dimensional interactions to develop a more comprehensive understanding of biology at multiple scales: molecular, cellular, tissue, organ, organism, and community.110 System-wide multiscale profiles of populations are coupled with computational techniques to build network models.110 Networks provide a mathematic framework for exploring the context in which genes, gene products, microbes, metabolites, and other variables operate, connect, and interact.110 Networks can place microbiota of interest in the context of biologic pathways and molecular interactions, and the best nodes from a network can be used to assess the disease and serve as targets for therapeutic intervention.110
Systems biology approaches, such as probabilistic causal network analysis, can be used to move beyond associations between the microbiota and allergy outcomes to inferring causal directionality. In such instances Bayesian methods can be used to infer causal relationships by considering thousands of molecular, microbial, and clinical variables and using statistical techniques to select a consensus model that best fits the data and identifies directionality of relationships between the variables.110,111 The connectivity structure of networks in disease helps us understand how biological processes are defined at the molecular level, how they are disrupted in disease, and how we can assess disease risk and potentially intervene.110,111 Causal network approaches have been applied to several disease areas outside of allergy, resulting in new mechanistic understandings for metabolic disease,112 cardiovascular disease,113 and Alzheimer disease.114 The implementation of such systems biology approaches to better understand the role of the microbiome in patients with asthma, AD, and food allergy represents a challenge to be addressed in the future.
To further our understanding of the microbiome in patients with asthma and allergic diseases, the objective should be to build on the level of data integration thus far (Fig 2).110 To that end, we need to generate multiple dimensions of data on each subject to lend power and provide the raw materials needed for a systems biology approach to examine asthma, AD, and food allergy at a more holistic level.110 The resulting network can then be used to predict the behavior of these conditions and generate novel and biologically relevant information.
FIG 2.
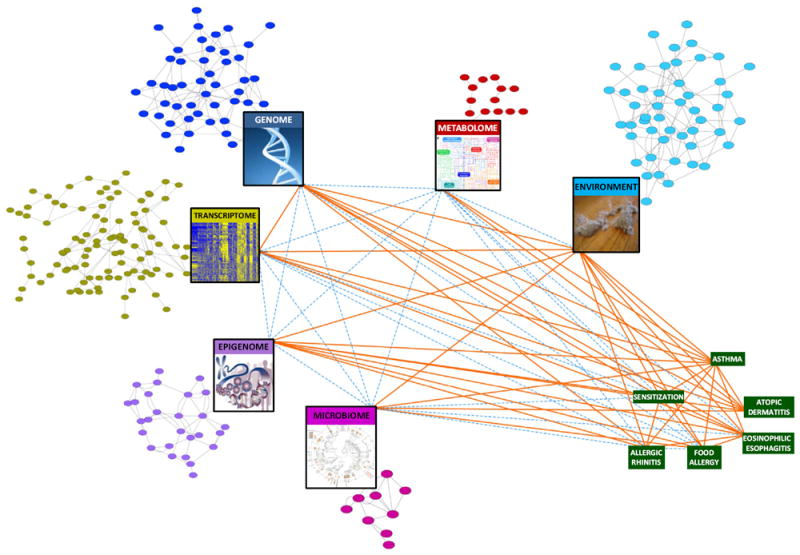
Moving toward a systems biology view of the microbiome in asthma and allergy. The colored circular nodes represent microbial, genetic, regulatory, metabolite, and environmental entities associated with asthma and allergic disorders. Their identification and potential connectivity can be assessed by the profiling represented in the large rectangular nodes. The green rectangular nodes represent diseases of interest. Orange lines (edges) denote evidence for associations between the implicated nodes in asthma and allergy. Dashed blue edges denote relationships that are currently less well studied. Examination of the network’s collective nodes and edges or a substantial subset thereof would move us toward a systems biology understanding of allergic disease. Figure adapted from Bunyavanich and Schadt.110
CONCLUSION
Because asthma, AD, and food allergy are complex and heterogeneous diseases, it is unlikely that the microbiota implicated in these diseases or even the microbiome in its entirety can fully capture the interdependent dynamics of the molecular networks involved in these diseases (Fig 2). Our understanding of allergic diseases has been advanced not only by studies of the microbiome but also by data generated through genome-wide association, transcriptomic, epigenomic, and metabolomic studies.110 Integrating these types of system-wide data will facilitate construction of models that are predictive of complex biological interactions and systems, a necessary step to developing a more complete understanding of asthma, AD, and food allergy.110 It is likely that bacterial, viral, and fungal biomes interact with the human genome in complex ways to influence these disorders.110 Systems biology approaches have been used to examine relationships between the microbiota and host genomic profiles in other disease areas, such as inflammatory bowel disease, suggesting that similar approaches can be applied to allergic disease.115,116 The ultimate objective is to better understand causal relationships among molecular features within a given cell or tissue type, between different tissues,117 and between the host and environment, including the microbiome.110
Developing a clear understanding of the role played by the microbiota in the development and evolution of allergic disease will afford the opportunity for targeted and rational means to manipulate the microbiota to the advantage of the host. Such an approach will contribute to and build on precision medicine for allergy care.65 Studies to date have yet to delineate clear approaches for the clinical manipulation of the microbiota with regard to asthma and allergy. However, the increasing availability of data in these areas should lead to carefully controlled clinical trials of microbiota-directed therapy. Such trials will clarify the degree to which microbial manipulation is a useful option for the prevention, management, or both of allergic diseases. Primary prevention through microbiota-directed therapy is particularly appealing to potentially decrease the incidence of asthma and allergy. Ultimately, clinical efficacy is unlikely to be achieved with a single approach, and a variety of strategies customized to different patient phenotypes/endotypes or perhaps individual patients will emerge.
In conclusion, our understanding of the role of the microbiota in the development and progress of allergic disease continues to evolve. Advances in this field will be driven by progress in our ability to address the gaps in approach, analysis, and interpretation that we have identified and to integrate knowledge of the microbiome into a systems biology context of the human host and environment. By moving forward research on the microbiome in patients with asthma and allergy, we can further our understanding of these disorders and derive new approaches for their prevention and management.
Acknowledgments
Supported in part by the National Institutes of Health (NIH) Intramural Program of the NIH Clinical Center.
Abbreviations used
- AD
Atopic dermatitis
- EH
Eczema herpeticum
- SCFA
Short-chain fatty acid
Footnotes
Disclosure of potential conflict of interest: Y. J. Huang has received travel support and payment for lectures from the American Academy of Allergy, Asthma & Immunology and has received travel support from the National Academy of Science, Engineering, and Medicine; the National Institutes of Health (NIH), the American College of Chest Physicians, the European Respiratory Society, and the Massachusetts Institute of Technology. S. Bunyavanich has received a grant from the NIH/National Institute of Allergy and Infectious Diseases. L. O’Mahony has consultant arrangements with Alimentary Health Ltd and has received grants from GlaxoSmithKline. D. Y. M. Leung has received a grant from MedImmune; has received consulting fees or honoraria from Novartis, Regeneron, and Sanofi-Aventis; and has received payment for writing or reviewing this manuscript from Omnia-Prova Education Collaborative. A. Muraro has consultant arrangements with Meda, Novartis, and Menarini; is employed by Padua University Hospital; and has received payment for lectures from Meda and Menarini. T. A. Fleisher is President of the American Academy of Allergy, Asthma & Immunology; has received payment for lectures from the Boston City Wide Allergy Meeting, the Louisiana Society of Allergy, Asthma, and Immunology, and the Alaska Society of Allergy, Asthma, and Immunology; and has received royalties as coeditor of Clinical Immunology: Principles and Practice. B. J. Marsland declares that he has no relevant conflicts of interest.
References
- 1.Rooks MG, Garrett WS. Gut microbiota:metabolites and host immunity. Nat Rev Immunol. 2016;16:341–52. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carpenter L, Beral V, Stachan D, Ebi-Kryston KL, Inskip H. Respiratory symptoms as predictors of 27 year mortality in a representative sample of British adults. BMJ. 1989;299:357–61. doi: 10.1136/bmj.299.6695.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Neerven RJ, Knol EF, Heck JM, Savelkoul HF. Which factors in raw cow’s milk contribute to protection against allergies? J Allergy Clin Immunol. 2012;130:853–8. doi: 10.1016/j.jaci.2012.06.050. [DOI] [PubMed] [Google Scholar]
- 4.Von Mutius E, Radon K. Living on a farm:impact on asthma induction and clinical course. Immunol Allergy Clin North Am. 2008;28:631–47. doi: 10.1016/j.iac.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Raciborski F, Tomaszewska A, Komorowski J, Samel-Kowalik P, Bialoszewski AZ, Walkiewicz A, et al. The relationship between antibiotic therapy in early childhood and the symptoms of allergy in children aged 6-8 years—the questionnaire study results. Int J Occup Med Environ Health. 2012;25:470–80. doi: 10.2478/S13382-012-0056-0. [DOI] [PubMed] [Google Scholar]
- 6.Silvers KM, Frampton CM, Wickens K, Pattemore PK, Ingham T, Fishwick D, et al. Breastfeeding protects against current asthma up to 6 years of age. J Pediatr. 2012;160:991–6. doi: 10.1016/j.jpeds.2011.11.055. [DOI] [PubMed] [Google Scholar]
- 7.Roduit C, Scholtens S, de Jongste JC, Wijga AH, Gerritsen J, Postma DS, et al. Asthma and 8 years of age in children born by ceasarian section. Thorax. 2009;64:107–13. doi: 10.1136/thx.2008.100875. [DOI] [PubMed] [Google Scholar]
- 8.Fujimura KE, Johnson CC, Ownby DR, Cox MJ, Brodie EL, Havastad SL, et al. Man’s best friend? The effect of pet ownership on house dust microbial communities. J Allergy Clin Immunol. 2010;126:410–2. doi: 10.1016/j.jaci.2010.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynch SV, Wood RA, Boushey H, Bacharier LB, Bloomberg GR, Kattan M, et al. Effects of early-life exposure to allergens and bacteria on recurrent wheeze and atopy in urban children. J Allergy Clin Immunol. 2014;134:593–601.e12. doi: 10.1016/j.jaci.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7:1–14. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 11.Stein MM, Hrusch CL, Gozdz J, Igartua C, Pivniouk V, Murray SE, et al. Innate immunity and asthma risk in Amish and Hutterite farm children. N Engl J Med. 2016;375:411–21. doi: 10.1056/NEJMoa1508749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2011;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 13.Goodrich GK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, et al. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe. 2016;11:731–43. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clavel T, Desmarchelier C, Haller D, Gérard P, Rohn S, Lepage P, et al. Intestinal microbiota in metabolic diseases:from bacterial community structure and functions to species of pathophysiological relevance. Gut Microbes. 2014;5:544–51. doi: 10.4161/gmic.29331. [DOI] [PubMed] [Google Scholar]
- 15.Noval Rivas M, Crother TR, Arditi M. The microbiome in asthma. Curr Opin Pediatr. 2016;6:764–71. doi: 10.1097/MOP.0000000000000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oh J, Byrd AL, Deming C, Conlan S. NISC Comparative Sequencing Program, Kong HH, Segre JA. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514:59–64. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinstock GM. Genomic approaches to studying the human microbiota. Nature. 2012;489:250–6. doi: 10.1038/nature11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui L, Morris A, Ghedin E. The human mycobiome in health and disease. Genome Med. 2013;5:63. doi: 10.1186/gm467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med. 2015;21:1228–34. doi: 10.1038/nm.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hannigan GD, Meisel JS, Tyldsley AS, Zheng Q, Hodkinson BP, SanMiguel AJ, et al. The human skin double-stranded DNA virome:topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome. MBio. 2015;6:e01578–615. doi: 10.1128/mBio.01578-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung WH, Croll D, Cho JH, Kim YR, Lee YW. Analysis of the nasal vestibule mycobiome in patients with allergic rhinitis. Mycoses. 2015;58:167–72. doi: 10.1111/myc.12296. [DOI] [PubMed] [Google Scholar]
- 22.van Woerden HC, Gregory C, Brown R, Marchesi JR, Hoogendoorn B, Matthews IP. Differences infungi present ininduced sputum samples from asthma patientsand non-atopic controls:a community based case control study. BMC Infect Dis. 2013;13:69. doi: 10.1186/1471-2334-13-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kramer R, Sauer-Heilborn A, Welte T, Guzman CA, Abraham WR, Höfle MG. Cohort study of airway mycobiome in adult cystic fibrosis patients:differences in community structure between fungi and bacteria reveal predominance of transient fungal elements. J Clin Microbiol. 2015;53:2900–7. doi: 10.1128/JCM.01094-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haldar P, Pavord ID, Shaw DE, Berry MA, Thomas M, Brightling CE, et al. Cluster analysis and clinical asthma phenotypes. Am J Respir Crit Care Med. 2008;178:218–24. doi: 10.1164/rccm.200711-1754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loza MJ, Adcock I, Auffray C, Chung KF, Djukanovic R, Sterk PJ, et al. Longitudinally stable, clinically defined clusters of patients with asthma independently identified in the ADEPT and U-BIOPRED asthma studies. Ann Am Thorac Soc. 2016;13(suppl 1):S102–3. doi: 10.1513/AnnalsATS.201508-519MG. [DOI] [PubMed] [Google Scholar]
- 26.Modena BD, Tedrow JR, Milosevic J, Bleecker ER, Meyers DA, Wu W, et al. Gene expression in relation to exhaled nitric oxide identifies novel asthma phenotypes with unique biomolecular pathways. Am J Respir Crit Care Med. 2014;190:1363–72. doi: 10.1164/rccm.201406-1099OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore WC, Fitzpatrick AM, Li X, Hastie AT, Li H, Meyers DA, et al. Clinical heterogeneity in the severe asthma research program. Ann Am Thorac Soc. 2013;10(suppl):S118–24. doi: 10.1513/AnnalsATS.201309-307AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fahy JV. Type 2 inflammation in asthma—present in most, absent in many. Nat Rev Immunol. 2015;15:57–65. doi: 10.1038/nri3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smits HH, Hiemstra PS, Prazeres da Costa C, Ege M, Edwards M, Garn H, et al. Microbes and asthma:opportunities for intervention. J Allergy Clin Immunol. 2016;137:690–7. doi: 10.1016/j.jaci.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 30.Sutherland ER, Martin RJ. Asthma and atypical bacterial infection. Chest. 2007;132:1962–6. doi: 10.1378/chest.06-2415. [DOI] [PubMed] [Google Scholar]
- 31.Casadevall A, Pirofski LA. Microbiology:ditch the term pathogen. Nature. 2014;516:165–6. doi: 10.1038/516165a. [DOI] [PubMed] [Google Scholar]
- 32.Byrd AL, Segre JA. Infectious disease. Adapting Koch’s postulates. Science. 2016;351:224–6. doi: 10.1126/science.aad6753. [DOI] [PubMed] [Google Scholar]
- 33.Denner DR, Sangwan N, Becker JB, Hogarth DK, Oldham J, Castillo J, et al. Corticosteroid therapy and airflow obstruction influence the bronchial microbiome, which is distinct from that of bronchoalveolar lavage in asthmatic airways. J Allergy Clin Immunol. 2016;137:1398–405.e3. doi: 10.1016/j.jaci.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang YJ, Nariya S, Harris JM, Lynch SV, Choy DF, Arron JR, et al. The airway microbiome in patients with severe asthma:Associations with disease features and severity. J Allergy Clin Immunol. 2015;136:874–84. doi: 10.1016/j.jaci.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. 2011;127:372–81, e1-3. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marri PR, Stern DA, Wright AL, Billheimer D, Martinez FD. Asthma-associated differences in microbial composition of induced sputum. J Allergy Clin Immunol. 2013;131:346–52, e1-3. doi: 10.1016/j.jaci.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simpson JL, Daly J, Baines KJ, Yang IA, Upham JW, Reynolds PN, et al. Airway dysbiosis:Haemophilus influenzae and Tropheryma in poorly controlled asthma. Eur Respir J. 2016;47:792–800. doi: 10.1183/13993003.00405-2015. [DOI] [PubMed] [Google Scholar]
- 38.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17:704–15. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goleva E, Jackson LP, Harris JK, Robertson CE, Sutherland ER, Hall CF, et al. The effects of airway microbiome on corticosteroid responsiveness in asthma. Am J Respir Crit Care Med. 2013;188:1193–201. doi: 10.1164/rccm.201304-0775OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Durack J, Lynch SV, Nariya S, Bhakta NR, Beigelman A, Castro M, et al. Features of the bronchial bacterial microbiome associated with atopy, asthma and responsiveness to inhaled corticosteroid treatment. J Allergy Clin Immunol. 2016 doi: 10.1016/j.jaci.2016.08.055. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barcik W, Pugin B, Westernann P, Perez NR, Ferst R, Wawrzyniak M, et al. Histamine-secreting microbes are increased in the gut of adult asthma patients. J Allergy Clin Immunol. 2016;138:1491–4. doi: 10.1016/j.jaci.2016.05.049. [DOI] [PubMed] [Google Scholar]
- 42.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010:5e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Green BJ, Wiriyachaiporn S, Grainge C, Rogers GB, Kehagia V, Lau L, et al. Potentially pathogenic airway bacteria and neutrophilic inflammation in treatment resistant severe asthma. PLoS One. 2014:9e100645. doi: 10.1371/journal.pone.0100645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016;22:1187–91. doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herbst T, Sichelstiel A, Schar C, Yadava K, Burki K, Cahenzli J, et al. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med. 2011;184:198–205. doi: 10.1164/rccm.201010-1574OC. [DOI] [PubMed] [Google Scholar]
- 46.Gollwitzer ES, Saglani S, Trompette A, Yadava K, Sherburn R, McCoy KD, et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat Med. 2014;20:642–7. doi: 10.1038/nm.3568. [DOI] [PubMed] [Google Scholar]
- 47.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–93. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun-Fahrlander C, et al. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364:701–9. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 49.Schuijs MJ, Willart MA, Vergote K, Gras D, Deswarte K, Ege MJ, et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science. 2015;349:1106–10. doi: 10.1126/science.aac6623. [DOI] [PubMed] [Google Scholar]
- 50.Hammad H, Chieppa M, Perros F, Willart MA, Germain RN, Lambrecht BN. House dust mite allergen induces asthma via Toll-like receptor 4 triggering of airway structural cells. Nat Med. 2009;15:410–6. doi: 10.1038/nm.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Debarry J, Garn H, Hanuszkiewicz A, Dickgreber N, Blumer N, von Mutius E, et al. Acinetobacter lwoffii and Lactococcus lactis strains isolated from farm cowsheds possess strong allergy-protective properties. J Allergy Clin Immunol. 2007;119:1514–21. doi: 10.1016/j.jaci.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 52.Nembrini C, Sichelstiel A, Kisielow J, Kurrer M, Kopf M, Marsland BJ. Bacterial-induced protection against allergic inflammation through a multicomponent immunoregulatory mechanism. Thorax. 2011;66:755–63. doi: 10.1136/thx.2010.152512. [DOI] [PubMed] [Google Scholar]
- 53.Hollingsworth JW, Whitehead GS, Lin KL, Nakano H, Gunn MD, Schwartz DA, et al. TLR4 signaling attenuates ongoing allergic inflammation. J Immunol. 2006;176:5856–62. doi: 10.4049/jimmunol.176.10.5856. [DOI] [PubMed] [Google Scholar]
- 54.Yadava K, Pattaroni C, Sichelstiel AK, Trompette A, Gollwitzer ES, Salami O, et al. Microbiota promotes chronic pulmonary inflammation by enhancing IL-17A and autoantibodies. Am J Respir Crit Care Med. 2016;193:975–87. doi: 10.1164/rccm.201504-0779OC. [DOI] [PubMed] [Google Scholar]
- 55.Bernasconi E, Pattaroni C, Koutsokera A, Pison C, Kessler R, Benden C, et al. Airway microbiota determines innate cell inflammatory or tissue remodeling profiles in lung transplantation. Am J Respir Crit Care Med. 2016;194:1252–63. doi: 10.1164/rccm.201512-2424OC. [DOI] [PubMed] [Google Scholar]
- 56.Marsland BJ, Gollwitzer ES. Host-microorganism interactions in lung diseases. Nat Rev Immunol. 2014;14:827–35. doi: 10.1038/nri3769. [DOI] [PubMed] [Google Scholar]
- 57.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–66. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 58.Vos AP, M’Rabet L, Stahl B, Boehm G, Garssen J. Immune-modulatory effects and potential working mechanisms of orally applied nondigestible carbohydrates. Crit Rev Immunol. 2007;27:97–140. doi: 10.1615/critrevimmunol.v27.i2.10. [DOI] [PubMed] [Google Scholar]
- 59.Verheijden KA, Willemsen LE, Braber S, Leusink-Muis T, Delsing DJ, Garssen J, et al. Dietary galacto-oligosaccharides prevent airway eosinophilia and hyperresponsiveness in a murine house dust mite-induced asthma model. Respir Res. 2015;16:17. doi: 10.1186/s12931-015-0171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi JH, Oh SW, Kang MS, Kwon HJ, Oh GT, Kim DY. Trichostatin A attenuates airway inflammation in mouse asthma model. Clin Exp Allergy. 2005;35:89–96. doi: 10.1111/j.1365-2222.2004.02006.x. [DOI] [PubMed] [Google Scholar]
- 61.Thorburn AN, McKenzie CI, Shen S, Stanley D, Macia L, Mason LJ, et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun. 2015;6:7320. doi: 10.1038/ncomms8320. [DOI] [PubMed] [Google Scholar]
- 62.Denning DW, O’Driscoll BR, Hogaboam CM, Bowyer P, Niven RM. The link between fungi and severe asthma:a summary of the evidence. Eur Respir J. 2006;27:615–26. doi: 10.1183/09031936.06.00074705. [DOI] [PubMed] [Google Scholar]
- 63.Zelante T, Pieraccini G, Scaringi L, Aversa F, Romani L. Learning from other diseases:protection and pathology in chronic fungal infections. Semin Immunopathol. 2016;38:239–48. doi: 10.1007/s00281-015-0523-3. [DOI] [PubMed] [Google Scholar]
- 64.Wheeler ML, Limon JJ, Bar AS, Leal CA, Gargus M, Tang J, et al. Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe. 2016;19:865–73. doi: 10.1016/j.chom.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muraro A, Lemanske RF, Jr, Hellings PW, Torres MJ, Khan D, Simon HU, et al. Precision medicine in patients with allergic diseases:airway disease and atopic dermatitis— PRACTALL document of the European Academy of Allergy and Clinical Immunology and the American Academy of Allergy, Asthma & Immunology. J Allergy Clin Immunol. 2016;137:1347–58. doi: 10.1016/j.jaci.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 66.Werfel T, Allam JP, Biedermann T, Eyerich K, Gilles S, Guttman-Yassky E, et al. Cellular and molecular immunologic mechanisms in patients with atopic dermatitis. J Allergy Clin Immunol. 2016;138:336–49. doi: 10.1016/j.jaci.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 67.Egawa G, Kabashima K. Multifactorial skin barrier deficiency and atopic dermatitis:Essential topics to prevent the atopic march. J Allergy Clin Immunol. 2016;138:350–85.e1. doi: 10.1016/j.jaci.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 68.Williams MR, Gallo RL. The role of the skin microbiome in atopic dermatitis. Curr Allergy Asthma Rep. 2015;15:65. doi: 10.1007/s11882-015-0567-4. [DOI] [PubMed] [Google Scholar]
- 69.Jones AL, Curran-Everett D, Leung DYM. Food allergy is associated with Staphylococcus aureus colonization in children with atopic dermatitis. J Allergy Clin Immunol. 2016;137:1247–8. doi: 10.1016/j.jaci.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 70.Tauber M, Balica S, Hsu CY, Jean-Decoster C, Lauze C, Redoules D, et al. Staphylococcus aureus density on lesional and nonlesional skin is strongly associated with disease severity in atopic dermatitis. J Allergy Clin Immunol. 2016;137:1272–4. doi: 10.1016/j.jaci.2015.07.052. [DOI] [PubMed] [Google Scholar]
- 71.Beck LA, Thaçi D, Hamilton JD, Graham NM, Bieber T, Rocklin R, et al. Dupilumab treatment in adults with moderate-to-severe atopic dermatitis. N Engl J Med. 2014;371:130–9. doi: 10.1056/NEJMoa1314768. [DOI] [PubMed] [Google Scholar]
- 72.Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22:850–9. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lynde CW, Andriessen A, Bertucci V, McCuaig C, Skotnicki S, Weinstein M, et al. The skin microbiome in atopic dermatitis and its relationship to emollients. J Cutan Med Surg. 2016;20:21–8. doi: 10.1177/1203475415605498. [DOI] [PubMed] [Google Scholar]
- 74.Grice EA. The intersection of microbiome and host at the skin interface:genomic- and metagenomic-based insights. Genome Res. 2015;25:1514–20. doi: 10.1101/gr.191320.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perez GI, Gao Z, Jourdain R, Ramirez J, Gany F, Clavaud C, et al. Body site is a more determinant factor than human population diversity in the healthy skin microbiome. PLoS One. 2016;11:e0151990. doi: 10.1371/journal.pone.0151990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi B, Bangayan NJ, Curd E, Taylor PA, Gallo RL, Leung DY, et al. The skin microbiome is different in pediatric versus adult atopic dermatitis. J Allergy Clin Immunol. 2016;138:1233–6. doi: 10.1016/j.jaci.2016.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sugimoto S, Iwamoto T, Takada K, Okuda K, Taijma A, Iwase T, et al. Staphylococcus epidermidis especially degrades specific proteins associated with Staphylococcus aureus biofilm formation and host-pathogen interaction. J Bacteriol. 2013;195:1645–55. doi: 10.1128/JB.01672-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med. 2017;9(378) doi: 10.1126/scitranslmed.aah4680. http://dx.doi.org/10.1126/scitranslmed.aah4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Otto M. Staphylococcus colonization of the skin and antimicrobial peptides. Expert Rev Dermatol. 2010;5:183–95. doi: 10.1586/edm.10.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yuki T, Yoshida H, Akazawa Y, Komiya A, Sugiyama Y, Inoue S. Activation of TLR2 enhances tight junction barrier in epidermal keratinocytes. J Immunol. 2011;187:3230–7. doi: 10.4049/jimmunol.1100058. [DOI] [PubMed] [Google Scholar]
- 81.Beck LA, Boguniewicz M, Hata TR, Schneider LC, Hanifin JM, Gallo RL, et al. Phenotype of atopic dermatitis:subjects with a history of eczema herpeticum. J Allergy Clin Immunol. 2009;124:260–9. doi: 10.1016/j.jaci.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Leung DYM. Why is eczema herpeticum unexpectedly rare? Antiviral Res. 2013;98:153–7. doi: 10.1016/j.antiviral.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bin L, Edwards MG, Heiser R, Streib JE, Richers B, Hall CF, et al. Identification of novel gene signatures in patients with atopic dermatitis complicated by eczema herpeticum. J Allergy Clin Immunol. 2014;134:848–55. doi: 10.1016/j.jaci.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gao L, Bin L, Rafaels NM, Huang L, Potee J, Ruczinski I, et al. Targeted deep sequencing identifies rare loss of-function variants in IFNGR1 for risk of atopic dermatitis complicated by eczema herpeticum. J Allergy Clin Immunol. 2015;136:1591–600. doi: 10.1016/j.jaci.2015.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oyoshi MK, Venturelli N, Geha RS. Thymic stromal lymphopoietin and IL-33 promote skin inflammation and vaccinia virus replication in a mouse model of atopic dermatitis. J Allergy Clin Immunol. 2016;138:283–6. doi: 10.1016/j.jaci.2015.12.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Berin MC, Sampson HA. Mucosal immunology of food allergy. Curr Biol. 2013;23:R389–400. doi: 10.1016/j.cub.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thompson-Chagoyan OC, Vieites JM, Maldonado J, Edwards C, Gil A. Changes in faecal microbiota of infants with cow’s milk protein allergy—a Spanish prospective case-control 6-month follow-up study. Pediatr Allergy Immunol. 2010;21:e394–400. doi: 10.1111/j.1399-3038.2009.00961.x. [DOI] [PubMed] [Google Scholar]
- 88.Berni Canani R, Sangwan N, Stefka AT, Nocerino R, Paparo L, Aitoro R, et al. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. 2016;10:742–50. doi: 10.1038/ismej.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bunyavanich S, Shen N, Grishin A, Wood R, Burks W, Dawson P, et al. Early-life gut microbiome composition and milk allergy resolution. J Allergy Clin Immunol. 2016;138:1122–30. doi: 10.1016/j.jaci.2016.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hua X, Goedert JJ, Pu A, Yu G, Shi J. Allergy associations with the adult fecal microbiota:analysis of the American Gut Project. EBioMedicine. 2016;3:172–9. doi: 10.1016/j.ebiom.2015.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Noval Rivas M, Burton OT, Wise P, Zhang Y-q, Hobson SA, Garcia Lloret M, et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J Allergy Clin Immunol. 2013;131:201–12. doi: 10.1016/j.jaci.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–6. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 93.Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, et al. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci U S A. 2014;111:13145–50. doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chinthrajah RS, Hernandez JD, Boyd SD, Galli SJ, Nadeau KC. Molecular and cellular mechanisms of food allergy and food tolerance. J Allergy Clin Immunol. 2016;137:984–97. doi: 10.1016/j.jaci.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun. 2015;6:6734. doi: 10.1038/ncomms7734. [DOI] [PubMed] [Google Scholar]
- 96.Hol J, van Leer EH, Elink Schuurman BE, de Ruiter LF, Samsom JN, Hop W, et al. The acquisition of tolerance toward cow’s milk through probiotic supplementation:a randomized, controlled trial. J Allergy Clin Immunol. 2008;121:1448–54. doi: 10.1016/j.jaci.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 97.Berni Canani R, Nocerino R, Terrin G, Coruzzo A, Cosenza L, Leone L, et al. Effect of Lactobacillus GG on tolerance acquisition in infants with cow’s milk allergy:a randomized trial. J Allergy Clin Immunol. 2012;129:580–2, e1-5. doi: 10.1016/j.jaci.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 98.Tang ML, Ponsonby AL, Orsini F, Tey D, Robinson M, Su EL, et al. Administration of a probiotic with peanut oral immunotherapy:a randomized trial. J Allergy Clin Immunol. 2015;135:737–44.e8. doi: 10.1016/j.jaci.2014.11.034. [DOI] [PubMed] [Google Scholar]
- 99.Azad MB, Konya T, Guttman DS, Field CJ, Sears MR, HayGlass KT, et al. Infant gut microbiota and food sensitization:associations in the first year of life. Clin Exp Allergy. 2015;45:632–43. doi: 10.1111/cea.12487. [DOI] [PubMed] [Google Scholar]
- 100.Cahenzli J, Koller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 2013;14:559–70. doi: 10.1016/j.chom.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Frei R, Akdis M, O’Mahony L. Prebiotics, probiotics, synbiotics, and the immune system:experimental data and clinical evidence. Curr Opin Gastroenterol. 2015;31:153–8. doi: 10.1097/MOG.0000000000000151. [DOI] [PubMed] [Google Scholar]
- 102.Lyons A, O’Mahony D, O’Brien F, MacSharry J, Sheil B, Ceddia M, et al. Bacterial strain-specific induction of Foxp3+ T regulatory cells is protective in murine allergy models. Clin Exp Allergy. 2010;40:811–9. doi: 10.1111/j.1365-2222.2009.03437.x. [DOI] [PubMed] [Google Scholar]
- 103.Sonnenburg JL, Bäckhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Roduit C, Frei R, Depner M, Schaub B, Loss G, Genuneit J, et al. Increased food diversity in the first year of life is inversely associated with allergic diseases. J Allergy Clin Immunol. 2014;133:1056–64. doi: 10.1016/j.jaci.2013.12.1044. [DOI] [PubMed] [Google Scholar]
- 105.van Nood E, Dijkgraaf MG, Keller JJ. Duodenal infusion of feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:2145. doi: 10.1056/NEJMc1303919. [DOI] [PubMed] [Google Scholar]
- 106.Youngster I, Russell GH, Pindar C, Ziv-Baran T, Sauk J, Hohmann EL. Oral, capsulized frozen fecal mcrobiolota transplantation for relapsing Clostridium difficile infection. JAMA. 2014;312:1772–8. doi: 10.1001/jama.2014.13875. [DOI] [PubMed] [Google Scholar]
- 107.Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8:343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, Cox LM, Amir A, Gonzalez A, et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med. 2016;22:250–3. doi: 10.1038/nm.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Waldor MK, Tyson G, Borenstein E, Ochman H, Moeller A, Finlay BB, et al. Where next for microbiome research? PLoS Biol. 2015;13:e1002050. doi: 10.1371/journal.pbio.1002050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bunyavanich S, Schadt EE. Systems biology of asthma and allergic diseases:a multiscale approach. J Allergy Clin Immunol. 2015;135:31–42. doi: 10.1016/j.jaci.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schadt EE, Bjorkegren JL. NEW:network-enabled wisdom in biology, medicine, and health care. Sci Transl Med. 2012;4:115rv1. doi: 10.1126/scitranslmed.3002132. [DOI] [PubMed] [Google Scholar]
- 112.Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–35. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Miller CL, Pjanic M, Wang T, Nguyen T, Cohain A, Lee JD, et al. Integrative functional genomics identifies regulatory mechanisms at coronary artery disease loci. Nat Commun. 2016;7:12092. doi: 10.1038/ncomms12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, et al. Integrated systems approach identifies genetic nodes and network. Cell. 2013;153:707–20. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Virgin HW, Todd JA. Metagenomics and personalized medicine. Cell. 2011;147:44–56. doi: 10.1016/j.cell.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dobrin R, Zhu J, Molony C, Argman C, Parrish ML, Carlson S, et al. Multi-tissue coexpression networks reveal unexpected subnetworks associated with disease. Genome Biol. 2009;10:R55. doi: 10.1186/gb-2009-10-5-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]


