Abstract
We report the characterization of two members of a gene family from Arabidopsis that encode, respectively, cytosolic (cPMSR) and plastid-targeted (pPMSR) isoforms of the oxidative-stress-repair enzyme peptide methionine sulfoxide reductase. Overexpression of these proteins in Escherichia coli confirmed that each had PMSR enzyme activity with a synthetic substrate, N-acetyl-[3H]-methionine sulfoxide, or a biological substrate, α-1 proteinase inhibitor. The pPMSR was imported into intact chloroplasts in vitro with concomitant cleavage of its approximately 5-kD N-terminal signal peptide. The two PMSR isoforms exhibited divergent pH optima, tissue localization, and responses to developmental and environmental effects. Analysis of the Arabidopsis database indicated that there are probably at least two p-pmsr-like genes and three c-pmsr-like genes in the Arabidopsis genome. Expression of the p-pmsr genes and their protein products was restricted to photosynthetic tissues and was strongly induced following illumination of etiolated seedlings. In contrast, the c-pmsr genes were expressed at moderate levels in all tissues and were only weakly affected by light. Exposure to a variety of biotic and abiotic stresses showed relatively little effect on pmsr gene expression, with the exception of leaves subjected to a long-term exposure to the cauliflower mosaic virus. These leaves showed a strong induction of the c-pmsr gene after 2 to 3 weeks of chronic pathogen infection. These data suggest novel roles for PMSR in photosynthetic tissues and in pathogen defense responses in plants.
Oxygen is essential to all aerobic organisms but can also have many harmful effects (Davies, 1995). Oxidative stress is manifested by tissue damage caused by the oxidation of compounds such as proteins, lipids, and nucleic acids. Potentially destructive reactive oxygen species (ROS) can be produced during normal metabolic processes such as photosynthesis in plants and some bacteria (Salin, 1987), or it can result from a wide range of both biotic (Berlett and Stadtman, 1997) and abiotic stresses (Dann and Pell, 1989). Despite the presence of antioxidant defenses in cells, ROS such as hydroxyl radicals and superoxide ions can readily oxidize protein amino acid residues, in particular Met, often with a consequent loss of enzymatic activity (Dann and Pell, 1989). Oxidation of Met residues has been implicated in several serious conditions in humans, including adult respiratory distress syndrome, rheumatoid arthritis, smokers' emphysema, and Alzheimer's disease (Abrams et al., 1981; Vogt, 1995; Moskovitz et al., 1996a; Gabitta et al., 1999). The best-characterized response to the oxidation of peptide residues in both plants and animals is the induction of proteases that cause the complete breakdown of the oxidized protein and its eventual replacement by a protein synthesized de novo (Davies, 1995; Grune and Davies, 1997; Grune et al., 1997).
The repair of oxidized proteins, rather than their breakdown, has generally been regarded as a minor component of the response of organisms to ROS. Nevertheless, there is now growing evidence that enzymatic repair of oxidized Met may play a key role in organisms ranging from bacteria to humans (Moskovitz et al., 1995, 1996b; Wizemann et al., 1996, 1997, 1998; El Hassouni et al., 1999). In addition to being the most common form of oxidative damage to proteins, the oxidation of Met to Met sulfoxide (MetSO) is unique in being readily reversible by the enzyme peptide-Met sulfoxide reductase (PMSR; EC 1.8.4.6), suggesting that PMSR may be able to repair oxidatively damaged proteins (Brot et al., 1982a, 1982b) in vivo. Indeed, pmsr-null mutants of yeast (Moskovitz et al., 1997) and Escherichia coli (Moskovitz et al., 1995) are significantly more susceptible to oxidative stress than wild-type controls, and this phenotype in yeast can be reversed by re-addition of further copies of the pmsr gene (Moskovitz et al., 1998). In mammals, pmsr gene expression and enzyme activity are localized in tissues such as kidneys (Moskovitz et al., 1996b), neutrophils (Fliss et al., 1983), macrophages (Moskovitz et al., 1996a), and the retina (Moskovitz et al., 1996a), where high levels of oxidative stress may be expected.
A role for PMSR in microbial pathogenicity is suggested by the reduced binding of pmsr-deficient mutants of E. coli, Neisseria gonorrhoea, and Streptococcus pneumoniae to eukaryotic host cell receptors (Wizemann, et al., 1996). More recently, it has been shown that a pmsr gene is a major virulence determinant in the plant pathogen Erwinia chrysanthemi (El Hassouni et al., 1999). Studies with the artificial substrate N-acetyl-MetSO have shown PMSR activity in plants (Ferguson and Burke, 1994), where it was particularly strong in photosynthetic tissues (Sanchez et al., 1983). We have previously isolated a pmsr gene from Brassica napus and demonstrated that the corresponding protein had PMSR activity (Sadanandom et al., 1996). In the present study we report the discovery of a complex family of at least five pmsr genes encoding two different protein isoforms in the model plant Arabidopsis. Comparison of the subcellular and tissue localization and regulation of the two plant PMSR isoforms suggests that they play different roles in response to oxidative stress caused by factors such as pathogen infection and photosynthesis.
RESULTS
Isolation and Genomic Organization of Arabidopsis pmsr Genes
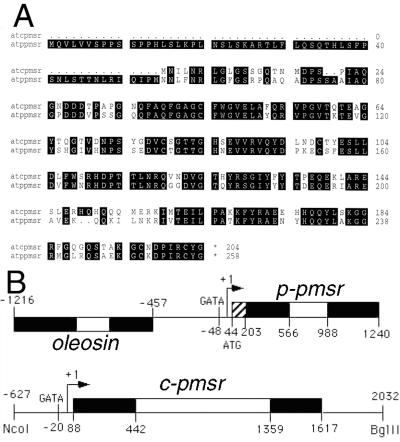
An Arabidopsis genomic fragment of 3.4 kb contained a pmsr reading frame within an oleosin gene promoter in a similar orientation to that described for Brassica napus (Sadanandom et al., 1996). A specific DNA probe based on the Arabidopsis genomic sequence was used to screen an Arabidopsis leaf cDNA library. The longest clone isolated contained a cDNA of 1,059 nt having a 40-nt poly(A+) tail and a putative polyadenylation site 130 nt from the translational stop codon. Primer extension using 5′-RACE showed that the 5′ end of the cDNA was 43 nt upstream from the start of the translation codon. Comparison of this cDNA sequence with the sequence of the pmsr-oleosin genomic clone allowed us to elucidate the structure of this entire genomic fragment. The intergenic distance (as defined by the transcriptional start sites) between the pmsr and oleosin genes was 414 nt. Arabidopsis pmsr has a single intron of 422 nt and two exons of 522 and 252 nt, respectively. There were no obvious TATA boxes near the pmsr transcriptional start site, although a GATA box, which has been shown to play an important role in regulating light-controlled genes (Teakle and Kay, 1995), was observed 48 nt upstream of the transcriptional start site. This gene encoded a 258-residue, 28,568-D protein with a putative plastidial targeting signal and was termed p-pmsr.
Since the previously reported PMSR proteins in other organisms lack organelle-targeting sequences and are therefore probably cytosolic, we re-screened the Arabidopsis leaf cDNA library for putative c-pmsr genes, but only obtained additional p-pmsr clones. However, screening of the Arabidopsis Expressed Sequence Tag (EST) database revealed several more divergent pmsr-like clones that were only 59% identical at the DNA level but 70% identical at the protein level to the p-pmsr. One of these clones, EST t125, was used in a RACE-PCR strategy to isolate an 847-nt cDNA encoding a full-length c-pmsr containing 88 nt upstream of the ATG codon, a 612-nt open reading frame, and a 147-nt downstream region extending to a poly(A+) tail. This cDNA was used to isolate a corresponding genomic clone from an Arabidopsis λGEM II library. A 7-kb clone was isolated and restricted to a 2.7-kb NcoI-BglII fragment that contained the entire c-pmsr open reading frame and flanking regions. The sequence of this fragment included a 627-nt promoter region followed by two exons of 354 and 258 nt, separated by a 917-nt intron. No TATA or CAAT boxes were found, but there was a putative GATA box (Teakle and Kay, 1995) 20 nt upstream of the transcription start site. The two exons encoded a 204-residue, PMSR-like protein of 22,705 D. These data are summarized in Figure 1A, and the full genomic sequences are filed as GenBank accession nos. x97236 (p-pmsr) and aj133753, aj133754 (c-pmsr). Comparison of the derived amino acid sequences of the p-pmsr and c-pmsr genes shows 70% identity and 82% similarity in the region of overlap (Fig. 1B). The p-PMSR sequence contains an additional 54-residue N-terminal domain that is rich in Ser, is relatively hydrophobic, and can potentially form an amphipathic β-strand, all of which are attributes of plastidial targeting signals (Von Hijne et al., 1989).
Figure 1.
A, Alignment of the protein sequences derived from the Arabidopsis p-pmsr and c-pmsr genes, showing the presence of an N-terminal plastidial targeting sequence in the p-pmsr. B, Genomic organization of the Arabidopsis p-pmsr and c-pmsr genes characterized in this study. Exons and introns are indicated by black and white boxes, respectively. The hatched box in the first p-pmsr exon indicates the coding region for the cleavable plastidial targeting sequence. The p-pmsr gene is located within the oleosin gene promoter as in B. napus (Sadanandom et al., 1996).
Database analysis confirmed that the locus corresponding the p-pmsr gene isolated in this study was located on overlapping bacterial artificial chromosome (BAC) clones F13M23 and F24A6 on chromosome 4, with seven ESTs corresponding to this sequence (H37471, AA712385, N37941, N65094, F13739, H36504, and N96968). A second p-pmsr-like sequence was found on two ESTs of unknown location (W43259 and W43286). We were unable to locate the genomic region corresponding to the c-pmsr genomic clone isolated in this study (containing a 917-nt intron), but preliminary RFLP analysis (Sadanandom, 1998) indicates that it was in an unsequenced region on the upper half of chromosome 5. This gene was highly expressed, as shown by the presence of seven corresponding ESTs (AA042165, AA005830, T45651, T44845, T44422, AI099810, and Z18051). A second c-pmsr gene (containing a 428-nt intron) was on a previously sequenced region (BAC clone K11 J9) on the lower half of chromosome 5, encoded a 77% identical, 86% similar cPMSR-like protein, and had a single corresponding EST (N97091). A third c-pmsr-like sequence was found in BAC clone T27K22 from chromosome 2 and had one corresponding EST (T45426). We conclude that there are at least two p-pmsr and three c-pmsr-like genes in the Arabidopsis genome.
Comparison with Other PMSR Sequences
The relationship between the Arabidopsis pPMSR and cPMSR sequences and some of the other reported genes encoding proteins having similar domains in other plant species was analyzed using public databases. In addition to the Arabidopsis and B. napus PMSRs shown here, there is a relatively tight cluster of other putative higher plant PMSR sequences. For example, the PMSR-like sequences from tomato (gi100204) and strawberry (gi1310665) are 61% identical and 73% similar to the Arabidopsis cPMSR and also lack plastidial targeting sequences. Database searches with the Arabidopsis PMSR also revealed significant hits (60%–80% identity over 85–150 residues) with partial sequences from alfalfa, kidney bean, rice bean, loblolly pine, and rice. The core PMSR domains that were compared are each about 150 residues in length, and the non-plant sequences are 35%–52% identical and 51%–65% similar to the overlapping Arabidopsis PMSR domain.
Activity of the Overexpressed PMSRs
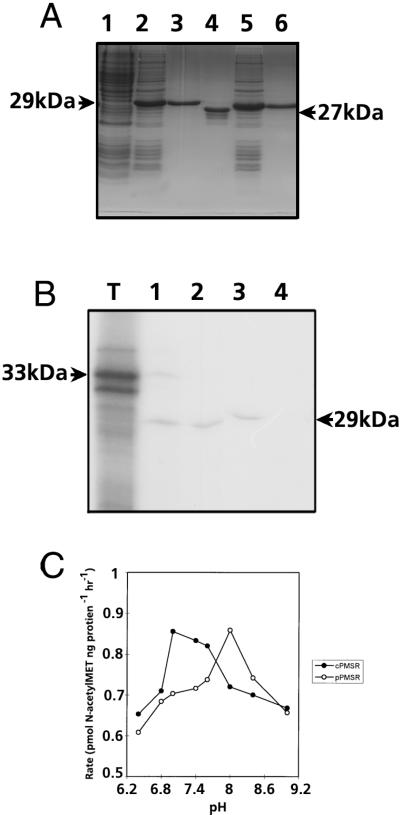
The biological activities of the putative Arabidopsis p-PMSR (minus the targeting sequence) and c-PMSR were demonstrated by overexpression and purification of the recombinant proteins in E. coli. The PMSRs were initially expressed with N-terminal poly-His tags to allow purification by affinity chromatography, as shown in Figure 2A. Both recombinant PMSR proteins exhibited apparent molecular masses on SDS-PAGE about 4 to 5 kD higher than predicted, as did the PMSR from plant tissue extracts, as detected by immunoblotting. This was probably due to the relatively acidic nature (pI = 5) of these proteins. The N-terminal His tag of the p-PMSR was removed by thrombin cleavage to generate a single polypeptide running at 28 kD. This protein was used to raise anti-PMSR antibodies in rabbits. The purified His-tagged c-PMSR, which ran at 27 kD, was enzymatically active and was used directly for assays. The activities of the purified recombinant PMSRs were assayed with either a protein substrate (oxidized α-1 protease inhibitor) or a synthetic substrate (N-acetyl-[3H]-MetSO). As shown in Table I, both substrates were efficiently reduced by the Arabidopsis PMSRs, using dithiothreitol as an electron donor. Using the synthetic substrate, it was also shown that PMSR activity was a linear function of enzyme concentration and time over the range of conditions used in this study (data not shown).
Figure 2.
A, SDS-PAGE gel showing overexpression and purification of recombinant pPMSR and cPMSR proteins in E. coli. Lane 1, Non-induced C41 cells; lane 2, C41 cells after 3 h of His-pPMSR induction; lane 3, affinity-purified His-pPMSR; lane 4, purified pPMSR after removal of His tag by thrombin; lane 5, C41 cells after 3 h of His-cPMSR induction; lane 6, affinity-purified His-cPMSR. B, Autoradiograph showing chloroplast import of the Arabidopsis PMSR polypeptide. Lane T, PMSR translation product; lane 1, PMSR translation product incubated with intact pea chloroplasts; lane 2, thermolysin-treated intact chloroplasts after the import incubation; lane 3, stromal fractions after isolation from the thermolysin-treated chloroplasts; and lane 4, thylakoid fractions after isolation from the thermolysin-treated chloroplasts. C, Variation in activity of p-PMSR (○) and c-PMSR (●) at different pH values. Data points are means of duplicate assays performed using N-acetyl-[3H]-MetSO as a substrate.
Table I.
Enzymatic activity of recombinant Arabidopsis pPMSR and cPMSR
| Dithiothreitol | Protein | Substrate 1b | Assay 1 |
|---|---|---|---|
| pmol MetSO reduced | |||
| − | − | − | 0 |
| − | − | − | 0 |
| + | − | + | 2 |
| + | pPMSR | + | 69.0 |
| − | pPMSR | + | 4.8 |
| + | cPMSR | + | 67.3 |
| − | cPMSR | + | 5.0 |
| + | BSAa | + | 2.0 |
| Dithiothreitol | Protein | Substrate 2c | Assay 2 |
|---|---|---|---|
| % of substrate | |||
| − | − | + | 0 |
| + | − | + | 3 |
| + | pPMSR | + | 75 |
| − | pPMSR | + | 4 |
| + | cPMSR | + | 75 |
| − | cPMSR | + | 5 |
| + | cPMSR, boiled | + | 3 |
BSA, Bovine serum albumin.
Substrate 1, N-Acetyl-[3H]-MetSO.
Substrate 2, Bovine α-1 proteinase inhibitor.
The targeting of the p-PMSR to the chloroplast stroma was demonstrated by the in vitro transcription-translation of the full-length p-pmsr cDNA (including targeting sequence) to yield a polypeptide with an apparent mass of 33 kD in this gel system, as shown in Figure 2B. This full-length polypeptide was imported into intact chloroplasts, with an associated cleavage of its approximately 5-kD targeting sequence. The mature 28-kD protein was shown by protease protection and fractionation assays to be located in the soluble, stromal compartment of the chloroplast. Control experiments (not shown) demonstrated that in the absence of the N-terminal signal sequence, the p-PMSR was not imported into intact chloroplasts. The c-PMSR had a pH optimum of about 7.0 to 7.2, while that of p-PMSR was 8.0 (Fig. 2C). This is consistent with the localization of p-PMSR in the stromal compartment of chloroplasts, which also has a pH of 8.0 during photosynthesis (Neumann and Jagendorf, 1964).
Tissue Localization and Regulation of PMSR Isoforms
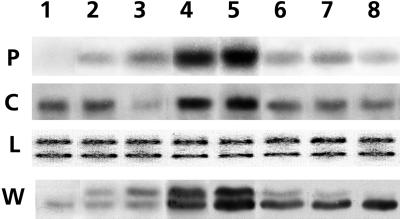
All tissues expressed the mRNA encoding c-pmsr, with the highest levels occurring in leaves and roots (Fig. 3). The c-PMSR protein was also present in all tissues, although its distribution was not exactly the same as that of the corresponding mRNA. Expression of the p-pmsr mRNA was largely confined to green tissues and was particularly prominent in the rosette and cauline leaves, which are the major sites of photosynthesis in Arabidopsis. The p-PMSR protein distribution largely, but not exactly, mirrored that of its mRNA. Subcellular fractionation of leaf homogenates using Suc density gradient centrifugation confirmed the finding in Figure 2B that the pPMSR was localized in the chloroplast fraction, while the c-PMSR was confined to the soluble, cytosolic fraction (data not shown).
Figure 3.
Localization of p-pmsr and c-pmsr mRNA and proteins in various tissues. P and C are northern blots probed with specific cDNA probes for the plastidial (1.1 kb) and cytosolic (0.9 kb) forms of pmsr, respectively. Lanes contain 10 μg of total RNA. L, Loading control (same RNA stained with ethidium bromide); W, western blot probed with polyclonal antibodies that recognized both the mature pPMSR protein at 28 kD and the cPMSR protein at 27 kD. Lane 1, Roots; lane 2, base of stem; lane 3, upper stem; lane 4, rosette leaves; lane 5, cauline leaves; lane 6, flower buds; lane 7, open flowers; lane 8, siliques (mid-maturation).
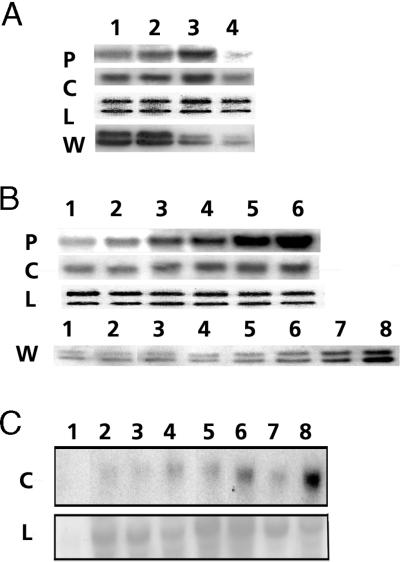
The differing responses of the two PMSR isoforms during developmental and environmental regulation are shown in Figure 4. Figure 4A shows expression of the PMSRs at the four developmental stages of leaves, ranging from immature to almost senescent. The mRNA levels of both pmsr isoforms peaked at stage 3, although the c-pmsr mRNA was consistently expressed at higher levels than that of the p-pmsr. In contrast, the protein levels of both isoforms peaked at stage 2, after which the p-PMSR protein declined rapidly to very low levels by stage 4, while the c-PMSR levels decreased more gradually.
Figure 4.
Regulation of PMSR during leaf development (A), light-induced greening (B), and pathogen infection (C). In A and B, the panels labeled P and C are northern blots probed, respectively, with specific cDNA probes for the p-pmsr and c-pmsr. All lanes contain 10 μg of total RNA. The panels labeled W are western blots probed with antibodies to PMSR that recognize both plastidial (28-kD) and cytosolic (27-kD) isoforms. Panels labeled L are the loading control (same RNA stained with ethidium bromide). A, Lane 1, immature expanding leaves still undergoing greening; lane 2, fully green and 80% expanded leaves; lane 3, mature fully expanded leaves; and lane 4, late mature leaves just prior to the onset of senescence. B, Panels P, C, and L, lane 1, 5-d-old etiolated seedlings; lanes 2, 3, 4, and 5, 5-d-old etiolated seedlings exposed to light for 20 min, 40 min, 60 min, and 3 h, respectively; lane 6, 5-d-old green seedlings. L, Loading control. Panel W, Western blot probed with polyclonal PMSR antibodies. Lane 1, 5-d-old etiolated seedlings; lanes 2, 3, 4, 5, 6, and 7, 5-d-old etiolated seedlings exposed to light for 2 h, 6 h, 15 h, 30 h, 50 h, 72 h, respectively; lane 8, 7-d-old green seedlings. C, Response to pathogen infection. Upper panel C, Northern blot probed with a specific cDNA probe for the c-pmsr; lower panel L, α-tubulin loading control. Lanes contain 20 μg of total RNA from leaves at various times after mock inoculation (1, 3, 5, and 7) or inoculation with CaMV isolate Cabb-B-JI (2, 4, 6, and 8). Lanes 1 and 2, 7 d post inoculation; lanes 3 and 4, 14 d; lanes 5 and 6, 21 d; lanes 7 and 8, 35 d.
Figure 4B shows the response of both PMSR isoforms to the illumination of dark-grown Arabidopsis seedlings. The c-pmsr mRNA was present at relatively high levels, even in etiolated leaves, and increased only moderately (about 2-fold) over 3 h of greening. The p-pmsr mRNA was relatively less abundant (but not absent) in etiolated leaves but showed a dramatic induction of expression within 20 to 40 min of greening. After 3 h of illumination the p-pmsr mRNA had almost reached levels present in light-grown leaves. Despite its relatively rapid induction by light, the p-pmsr gene was not activated by a phytochrome-mediated pathway, since it showed no response to red and far-red light treatments (data not shown). Western-blotting studies showed that p-PMSR protein levels increased more slowly than the mRNA, with only a slight increase after 6 h of greening but a more rapid and sustained increase after 15 h.
We tested the response of both p-pmsr and c-pmsr genes to a variety of stresses that have been associated with increased levels of ROS production. Treatments included high (52°C) and low (4°C) temperatures, wounding, application of jasmonic acid, and infection with the virulent pathogen Pseudomonas syringae. Northern analysis of leaves from affected plants failed to show any consistent or statistically significant effect on the levels of either p-pmsr or c-pmsr mRNAs. The only exception noted in this study was the pronounced induction of the c-pmsr gene in response to infection of Arabidopsis leaves by the pathogen CaMV, as shown in Figure 4C. Some response was seen within 7 d after inoculation, but the major response occurred at 21 to 35 d, as the symptoms of infection by this relatively slow-acting virus (Cecchini et al., 1997) became more pronounced. Note that mock inoculations of leaves with 10 mm MgC12 did not result in similar increases in c-pmsr gene expression, which rules out an effect due to tissue damage alone. Parallel blots probed with the p-pmsr cDNA showed no response to viral infection (data not shown).
DISCUSSION
The data presented here show that the genome of Arabidopsis contains at least five pmsr-like genes. This contrasts with all animal and microbial genomes described to date, which only contain a single pmsr gene (Kuschel et al., 1999). The relatively large number of pmsr genes in Arabidopsis is particularly significant, since it has one of the smallest genomes (120 Mb) of any higher eukaryote (Meinke et al., 1998). Our analysis of the Arabidopsis EST database indicates that all five of the identified pmsr genes are indeed expressed, and the relative abundance of such ESTs (totaling 18) indicates that pmsr genes are expressed at high levels in plants. Higher plants are also unique in having two different PMSR isoforms that we have shown to be localized in separate subcellular compartments. The two isoforms are differentially expressed in various plant tissues and in response to both development and stress, indicating that PMSR may play additional and perhaps more complex roles in plants than in other organisms. Such roles may be necessary because of the increased exposure of plants to oxidative stress due to oxygenic photosynthesis (Salin, 1987) and their lack of motility, which precludes their avoidance of many environmental sources of oxidative stress such as drought, salt, high light and temperature (Demmig-Adams and Adams III, 1992; Prasad, 1996; Foyer et al., 1997), and pathogen attack (Wojtaszek, 1997).
A role for PMSR in the alleviation of oxidative stress is strongly indicated by studies of pmsr-null mutants of E. coli (Moskovitz et al., 1995) and yeast (Moskovitz et al., 1997) and the greatly enhanced resistance to oxidative stress of yeast or human T cells transfected with additional copies of pmsr genes (Moskovitz et al., 1998). Several protein substrates for PMSR have been investigated in animal and microbial systems. These include the α-1-proteinase inhibitor (Abrams et al., 1981), calmodulin (Sun et al., 1999), apolipoprotein A1 (Sigalov and Stern, 1998), and α/β-spore proteins (Hayes et al., 1998). We found that both plant PMSR isoforms were able to repair the bovine oxidized α-1-proteinase inhibitor protein at rates comparable to those reported in the literature for non-plant PMSRs. This is interesting in view of our observation of a strong but gradual dramatic induction of c-pmsr gene expression in response to infection with the slow-acting viral pathogen CaMV. One of the most highly expressed classes of pathogen-induced genes in many plants encodes proteinase inhibitor (pin) proteins (Johnson et al., 1989; Cordero et al., 1994), and it is possible that PMSR can also repair oxidized pin proteins from plants.
Plants can also respond to pathogen attack by inducing a relatively rapid hypersensitive response (HR), which involves localized apoptotic cell death in the affected region and is accompanied by a release of ROS (Doke, 1983a, 1983b; for reveiws, see Lamb and Dixon, 1997; Wojtaszek, 1997). This phenomenon, termed the “oxidative burst,” may be analogous to the oxygenic response of mammalian neutrophils to microbial infection, in which PMSR may repair proteins damaged by the ROS (Moskovitz et al., 1996a). The relative slowness of c-pmsr induction by CaMV and the lack of strong induction by more virulent pathogens indicates that c-pmsr is probably not involved in the HR. Some particularly virulent plant pathogens, such as Erwinia chrysanthemi, do not elicit HR and their major virulence determinants were formerly believed to encode extracellular lytic enzymes that allow invasion of the host tissue. However, a recent report showed that a hitherto unrecognized virulence gene in E. chrysanthemi encodes a PMSR, and that pmsr-null mutants were non-virulent on plants and extremely susceptible to oxidative stress (El Hassouni et al., 1999). This surprising finding indicates that, even in the absence of HR, plants attempt to mount a defense against virulent pathogens by generating ROS. The more successful pathogens, such as E. chrysanthemi, can overcome this oxidative defense thanks to their PMSR-based repair system. Our data on the gradual induction of c-pmsr expression following infection by a slow-acting viral pathogen (i.e. one that does not induce an HR) imply that plants may also protect themselves from the long-term consequences of their release of anti-pathogenic ROS. Therefore, both the plant host and the microbial pathogen are able to deploy the PMSR protein repair system to mitigate oxidative damage during pathogenesis.
Other roles for cPMSR can be inferred from results obtained in different plant species. The putative c-pmsr homologs in tomato (Cordes et al., 1989) and strawberry (GenBank accession no. gi1310664) are both strongly induced during fruit ripening. This process involves the production of large amounts of ROS, and the PMSR may be required to protect key proteins from oxidative damage. In this study we found relatively high levels of cPMSR (but not pPMSR) in roots and maturing seed (siliques) of Arabidopsis. Differential screening studies have also revealed c-pmsr homologs that are highly up-regulated during root nodulation in alfalfa (GenBank accession no. gi1310665), seed maturation in rice (GenBank accession no. gi3760326), and xylem formation in loblolly pine (Allona et al., 1998). These processes all involve an increase in ROS, even during normal metabolism levels, and demonstrate the possible manifold functions of the cPMSR isoform in plants.
We have shown that expression of the p-pmsr gene in Arabidopsis is largely restricted to actively photosynthesizing tissues. The chloroplasts of such tissues are the most oxygenic sites of any biological system and are therefore uniquely exposed to damage by ROS, even during normal daytime metabolism. It is believed that the principal defense mechanism of plants and animals involves scavenging of ROS by antioxidants or via enzymes such as catalase or glutathione reductase (Davies, 1995). Oxidative damage to proteins has previously been regarded as largely irrepairable, resulting in their turnover by proteolysis (Mehta et al., 1992; Davies, 1995; Grune and Davies, 1997; Grune et al., 1997; Pell et al., 1997). However, the presence of two light-inducible p-pmsr genes in Arabidopsis strongly indicates that PMSR performs an important repair function for chloroplast proteins that are oxidized during normal photosynthesis. It is noteworthy that the levels of the plastidial PMSR protein declined significantly immediately prior to leaf senescence. During this pre-senescent stage, there are elevated levels of proteolysis of key enzymes such as the CO2-fixing protein Rubisco (He et al., 1997; Oh et al., 1997; Desimone et al., 1998), followed by a buildup of ROS levels leading to the more visible symptoms of leaf senescence such as chlorophyll loss and yellowing. It is therefore tempting to speculate about a link between the sudden decline in pPMSR levels and the onset of senescence in plants. In some plants, ethylene modulates the activation of senescence-related genes in tissues such as fruits and leaves (Buchanan-Wollaston, 1997; Oh et al., 1997). The tomato cPMSR homolog E4 is strongly induced by ethylene during fruit ripening, and it would be interesting to investigate its regulation during leaf senescence.
In conclusion, we have demonstrated that the two PMSR isoforms investigated in this study are involved in various developmentally and environmentally regulated processes in Arabidopsis, some of which are unique to plants. It will now be important to clarify the roles of PMSRs in more detail and in particular to elucidate its various protein substrates. Since plant PMSRs are encoded by a multigene family and are only one component of a highly redundant system for dealing with oxidative stress, it will not necessarily be easy to assess their physiological importance in vivo. Nevertheless, the clear impact of pmsr-null mutations on the oxidative stress response of yeast (Moskovitz et al., 1997), E. coli (Moskovitz et al., 1995), and E. chrysanthemi (El Hassouni et al., 1999) demonstrates that PMSR does have a significant function in these organisms. We are therefore using a pmsr gene-knockout strategy based on transposon mutagenesis in order to elucidate more fully the biological roles of PMSR in higher plants.
MATERIALS AND METHODS
Overexpression and Purification of Arabidopsis pmsr Gene Products
The plastidial PMSR protein (pPMSR) was overexpressed without its N-terminal targeting sequence, i.e. from Met-54. Ligation of the DNA containing the truncated p-pmsr coding region to the pET vector resulted in the fusion of six His residues to the N terminus of the mature pPMSR protein. This construct was then under the control of isopropylthio-β-galactoside-inducible lac promoter linked to the T7 polymerase. Cultures of E. coli C41 were transformed with the pET expression vector, and the size of the fusion protein was determined by SDS-PAGE. The fusion protein was affinity-purified on a nickel-agarose column and the His tag removed by thrombin cleavage, as described in the pET system manual (Novagen, Madison, WI). For production of the cytosolic PMSR protein (cPMSR), the cDNA corresponding to the Arabidopsis c-pmsr gene was cloned into the pQE expression vector. The construct was transformed into either XL1B or C41 strains of E. coli, and overexpression of the His-tagged cPMSR was induced by isopropylthio-β-galactoside. The cPMSR was affinity-purified as described above, and both pPMSR and cPMSR were dialyzed and concentrated to about 1 mg mL−1. Long-term storage was at −80°C, and both PMSR isoforms retained >80% specific activity for at least 6 months.
Assays
PMSR assays were performed using a bovine α-1-proteinase inhibitor substrate (0.044 unit), which was oxidized with 30 μmol of chloramine-T in 500 μL of buffer (160 mm Tris-HC1, pH 8.0) at 20°C for 120 min, and dialyzed overnight against 1.5 L of 20 mm Tris-HC1, pH 7.5, at 4°C to remove the chloramine-T. After the PMSR reaction, the amount of α-1-proteinase inhibitory activity was measured with an elastase assay. To 100 μL of reaction medium containing α-1-proteinase inhibitor, porcine pancreatic elastase (0.003 unit) in 400 μL of buffer (Tris-HC1, pH 8.0, 0.1% [v/v] Triton X-100) was added and mixed. Alternatively, the synthetic substrate MetSO was used in PMSR assays (Brot et al., 1982b). Chloroplast import assays were performed by translating p-pmsr mRNA in 200 μL of wheat germ cell-free lysate with [35S] Met. Along with the translation product, the import sample contained 200 mm MgC12, 100 mm ATP, and 60 μL (1 mg ml−1 chlorophyll) of intact pea chloroplasts in sorbitol resuspension medium (250 mm 4-[2-hydroxyethyl]-1-piperazineethanesulfonic acid [HEPES] and 1.65 m sorbitol, pH 8.0). Protease treatment and lysis of chloroplasts was carried out essentially as stated in Robinson and Ellis (1984). Samples were analyzed on SDS-PAGE gels.
Plant Growth and Treatments
Arabidopsis ecotype Columbia plants were grown in a climate chamber with a 6-h photo period, a photon flux density (PFD) of 200 μmol photons m−2 s−1, a temperature of 22°C, and a relative humidity of 75%. For the light treatment, plants were grown from seed on Murashige and Skoog medium in darkness for 5 d, and then illuminated as above. For viral treatments, fully expanded rosette leaves from 6-week-old plants were inoculated with a purified cauliflower mosaic virus (CaMV) (Gardner and Shepherd, 1980) isolate, Cabb-B-JI. The inoculum (1 μL) was applied with a trace of celite abrasive to the surface of leaves 2, 3, or 4 followed by gentle rubbing with a Pasteur pipette tip flattened into a spatula shape. Control mock inoculations were performed with 10 mm MgCl2 and celite in the same manner.
Protein and RNA Analyses
All operations to prepare protein extracts from Arabidopsis plants were performed on ice. Various tissues were ground in liquid nitrogen and extracted as in Sadanandom et al. (1996). Protein amounts were determined as in Bradford (1976), analyzed by SDS-PAGE on acrylamide gels (Laemmli, 1970), and transferred to PVDF membranes according to the instructions of the manufacturer (electrophoresis manual, Bio-Rad Laboratories, Hercules, CA). Membranes were incubated with anti-PMSR primary antibodies at 1:500 dilution and developed using a western-blotting detection system (ECL PLUS, Amersham, Buckinghamshire, UK). Total RNA from various plant tissues was extracted (Prescott and Martin, 1987), and 10-μg aliquots were separated on 1.6% (v/v) agarose gels and probed with gene-specific c-pmsr or p-pmsr cDNAs as in Sadanandom et al. (1996).
ACKNOWLEDGMENTS
We thank Uli Bechtold, Kanu Patel, Jo Ross, and Steve Rawsthorne for advice and help.
Footnotes
This work was supported by the Biotechnology and Biological Science Research Council grant (to John Innes Centre), by a John Innes Foundation studentship (to A.S.), and by Vavilov Frankel and Royal Society fellowships (to Z.P.).
LITERATURE CITED
- Abrams WR, Weinbaum G, Weissbach L, Weissbach H, Brot N. Enzymatic reduction of oxidized alpha-1-proteinase inhibitor restores biological activity. Proc Natl Acad Sci USA. 1981;78:7483–7486. doi: 10.1073/pnas.78.12.7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allona I, Quinn M, Shoop E, Swope K, Cyr SS, Carlis J, Riedl J, Retzel E, Campbell MM, Sederoff R, Whetten RW. Analysis of xylem formation in pine by cDNA sequencing. Proc Natl Acad Sci USA. 1998;95:9693–9698. doi: 10.1073/pnas.95.16.9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brot N, Weissbach L, Werth J, Weissbach H. The biochemistry of methionine sulfoxide residues in proteins. Biofactor. 1982a;3:91–96. [PubMed] [Google Scholar]
- Brot N, Werth J, Koster D, Weissbach H. Reduction of N-acetyl methionine sulfoxide: a simple assay for peptide methionine sulfoxide reductase. Anal Biochem. 1982b;122:291–294. doi: 10.1016/0003-2697(82)90283-4. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V. The molecular biology of leaf senescence. J Exp Bot. 1997;48:181–199. [Google Scholar]
- Cecchini E, Gong ZH, Geri C, Covey SN, Milner JJ. Transgenic Arabidopsis lines expressing gene VI from cauliflower mosaic virus variants exhibit a range of symptom-like phenotypes and accumulate inclusion bodies. Mol Plant-Microbe Interact. 1997;10:1094–1101. doi: 10.1094/MPMI.1997.10.9.1094. [DOI] [PubMed] [Google Scholar]
- Cordero MJ, Raventos D, San Segundo B. Expression of a maize proteinase inhibitor gene is induced in response to wounding and fungal infection: systemic wound-response of a monocot gene. Plant J. 1994;6:141–150. doi: 10.1046/j.1365-313x.1994.6020141.x. [DOI] [PubMed] [Google Scholar]
- Cordes S, Deikman J, Margossian LJ, Fischer RL. Interaction of a developmentally regulated DNA-binding factor with sites flanking 2 different fruit-ripening genes from tomato. Plant Cell. 1989;1:1025–1034. doi: 10.1105/tpc.1.10.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann M, Pell E. Decline of activity and quantity of ribulose bisphosphate carboxylase/oxygenase and net photosynthesis in ozone-treated foliage. Plant Physiol. 1989;91:427–432. doi: 10.1104/pp.91.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KJA. Oxidative stress: the paradox of aerobic life. Biochem Soc Symp. 1995;61:1–31. doi: 10.1042/bss0610001. [DOI] [PubMed] [Google Scholar]
- Demmig-Adams B, Adams WW., III Photoprotection and other responses of plants to high light stress. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:599–626. [Google Scholar]
- Desimone M, Wagner E, Johanningmeier U. Degradation of active-oxygen-modified ribulose-1,5-bisphosphate carboxylase/oxygenase by chloroplastic proteases requires ATP-hydrolysis. Planta. 1998;205:459–466. [Google Scholar]
- Doke N. Generation of superoxide anion by potato tuber protoplasts during the hypersensitive response to hyphal cell wall components of Phytophthora infestans and specific inhibition of reaction by suppressors of hypersensitivity. Physiol Plant Pathol. 1983a;23:359–367. [Google Scholar]
- Doke N. Involvement of superoxide anion generation in hypersensitive response of potato tuber tissues to infection with an incompatible race of Phytophthora infestans. Physiol Plant Pathol. 1983b;23:345–358. [Google Scholar]
- El Hassouni M, Chambost JP, Expert D, VanGijsegem F, Barras F. The minimal gene set member msrA, encoding peptide methionine sulfoxide reductase, is a virulence determinant of the plant pathogen Erwinia chrysanthemi. Proc Natl Acad Sci USA. 1999;96:887–892. doi: 10.1073/pnas.96.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson DL, Burke JJ. Methionyl sulfoxide content and protein-methionine-S-oxide reductase activity in response to water deficits or high temperature. Physiol Plant. 1994;90:253–258. [Google Scholar]
- Fliss H, Weissbach H, Brot N. Oxidation of methionine residues in proteins of activated human neutorphils. Proc Natl Acad Sci USA. 1983;80:7160–7164. doi: 10.1073/pnas.80.23.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, LopezDelgado H, Dat JF, Scott IM. Hydrogen peroxide- and glutathione-associated mechanisms of acclimatory stress tolerance and signaling. Physiol Plant. 1997;100:241–254. [Google Scholar]
- Gabitta SP, Aksenov MY, Lovell MA, Markesbery WR. Decrease in peptide methionine sulfoxide reductase in Alzheimer's disease brain. J Neurochem. 1999;73:1660–1666. doi: 10.1046/j.1471-4159.1999.0731660.x. [DOI] [PubMed] [Google Scholar]
- Gardner R, Shepherd R. A procedure for rapid isolation and analysis of cauliflower mosaic virus DNA. Virology. 1980;106:159–161. doi: 10.1016/0042-6822(80)90234-2. [DOI] [PubMed] [Google Scholar]
- Grune T, Davies KJ. Breakdown of oxidized proteins as a part of secondary antioxidant defenses in mammalian cells. Biofactors. 1997;6:165–172. doi: 10.1002/biof.5520060210. [DOI] [PubMed] [Google Scholar]
- Hayes CS, Illades Aguiar B, Casillas Martinez L, Setlow P. In vitro and in vivo oxidation of methionine residues in small, acid-soluble spore proteins from Bacillus species. J Bacteriol. 1998;180:2694–2700. doi: 10.1128/jb.180.10.2694-2700.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, von Caemmerer S, Hudson G, Price G, Badger M, Andrews T. Ribulose-1,5-bisphosphate carboxylase/oxygenase activase deficiency delays senescence of ribulose-1,5-bisphosphate carboxylase/oxygenase but progressively impairs its catalysis during tobacco leaf development. Plant Physiol. 1997;115:1569–1580. doi: 10.1104/pp.115.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Narvaez J, An G, Ryan CA. Expression of proteinase inhibitors I and II in transgenic tobacco plants: effects on natural defense against Manduca sexta larvae. Proc Natl Acad Sci USA. 1989;88:9871–9875. doi: 10.1073/pnas.86.24.9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuschel L, Hansel A, Schonherr R, Weissbach H, Brot N, Hoshi T, Heinemann SH. Molecular cloning and functional expression of a human peptide methionine sulfoxide reductase (hMsrA) FEBS Lett. 1999;456:17–21. doi: 10.1016/s0014-5793(99)00917-5. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamb C, Dixon RA. The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:251–275. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- Mehta RA, Fawcett TW, Porath D, Mattoo AK. Oxidative stress causes rapid membrane translocation and in vivo degradation of ribulose-1,5-bisphosphate carboxylase/oxygenase. J Biol Chem. 1992;267:2810–2816. [PubMed] [Google Scholar]
- Meinke DW, Cherry JM, Dean C, Rounsley SD, Koornneef M. Arabidopsis thaliana: a model plant for genome analysis. Science. 1998;282:679–682. doi: 10.1126/science.282.5389.662. [DOI] [PubMed] [Google Scholar]
- Moskovitz J, Berlett BS, Barbara S, Poston JM, Stadtman ER. The yeast peptide-methionine sulfoxide reductase functions as an antioxidant in vivo. Proc Natl Acad Sci USA. 1997;94:9585–9589. doi: 10.1073/pnas.94.18.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz J, Flescher E, Berlett BS, Azares J, Poston JM, Stadtman ER. Overexpression of peptide-methionine sulfoxide reductase in Saccharomyces cerevisiae and human T cells provides them with high resistance to oxidative stress. Proc Natl Acad Sci USA. 1998;95:14071–14075. doi: 10.1073/pnas.95.24.14071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz J, Jenkins NA, Gilbert DJ, Copeland NG, Jursky F, Weissbach H, Brot N. Chromosomal localization of the mammalian peptide-methionine sulfoxide reductase gene and its differential expression in various tissues. Proc Natl Acad Sci USA. 1996a;93:3205–3208. doi: 10.1073/pnas.93.8.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz J, Rahman MA, Strassman J, Yancey SO, Kushner SR, Brot N, Weissbach H. Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. J Bacteriol. 1995;177:502–507. doi: 10.1128/jb.177.3.502-507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz J, Weissbach H, Brot N. Cloning and expression of a mammalian gene involved in the reduction of methionine sulfoxide residues in proteins. Proc Natl Acad Sci USA. 1996b;93:2095–2099. doi: 10.1073/pnas.93.5.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann JS, Jagendorf AT. Light-induced pH changes related to photophosphorylation by chloroplasts. Arch Biochem Biophys. 1964;107:109–117. doi: 10.1016/0003-9861(64)90276-0. [DOI] [PubMed] [Google Scholar]
- Oh SA, Park JH, Lee GI, Paek KH, Park SK, Nam HG. Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. Plant J. 1997;12:527–535. doi: 10.1046/j.1365-313x.1997.00527.x. [DOI] [PubMed] [Google Scholar]
- Pell EJ, Schlagnhaufer CD, Arteca RN. Ozone-induced oxidative stress: mechanisms of action and reaction. Physiol Plant. 1997;100:264–273. [Google Scholar]
- Prasad TK. Mechanisms of chilling-induced oxidative stress injury and tolerance in developing maize seedlings: change in antioxidant system, oxidation of proteins and lipids, and protease activities. Plant J. 1996;10:1017–1026. [Google Scholar]
- Prescott A, Martin C. A rapid method for the quantitative assessment of levels of specific mRNAs in plants. Plant Mol Biol Rep. 1987;4:219–224. [Google Scholar]
- Robinson C, Ellis RJ. Transport of proteins into chloroplasts: partial purification of a chloroplast protease involved in the processing of imported precursor polypeptides. Eur J Biochem. 1984;142:337–342. doi: 10.1111/j.1432-1033.1984.tb08291.x. [DOI] [PubMed] [Google Scholar]
- Sadanandom A. Molecular regulation of protein in plants. PhD thesis. Norwich, UK: University of East Anglia and John Innes Centre; 1998. [Google Scholar]
- Sadanandom A, Piffanelli P, Knott T, Robinson C, Sharpe A, Lydiate D, Murphy D, Fairbairn DJ. Identification of a peptide methionine sulphoxide reductase gene in an oleosin promoter from Brassica napus. Plant J. 1996;10:235–242. doi: 10.1046/j.1365-313x.1996.10020235.x. [DOI] [PubMed] [Google Scholar]
- Salin ML. Toxic oxygen species and protective systems of the chloroplast. Physiol Plant. 1987;72:681–689. [Google Scholar]
- Sanchez J, Nikolau BJ, Stumpf PK. Reduction of N-acetyl methionine sulfoxide in plants. Plant Physiol. 1983;73:619–623. doi: 10.1104/pp.73.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigalov AB, Stern LJ. Enzymatic repair of oxidative damage to human apolipoprotein A-I. FEBS Lett. 1998;433:196–200. doi: 10.1016/s0014-5793(98)00908-9. [DOI] [PubMed] [Google Scholar]
- Sun H, Gao J, Ferrington DA, Biesiada H, Williams TD, Squier TC. Repair of oxidized calmodulin by methionine sulfoxide reductase restores ability to activate the plasma membrane Ca-ATPase. Biochemistry. 1999;38:105–112. doi: 10.1021/bi981295k. [DOI] [PubMed] [Google Scholar]
- Teakle GR, Kay SA. The GATA-binding protein CGF-1 is closely related to GT-1. Plant Mol Biol. 1995;29:1253–1266. doi: 10.1007/BF00020466. [DOI] [PubMed] [Google Scholar]
- Vogt W. Oxidation of methionyl residues in proteins-tools, targets and reversal. Free Radic Biol Med. 1995;18:93–105. doi: 10.1016/0891-5849(94)00158-g. [DOI] [PubMed] [Google Scholar]
- Von Hijne G, Steppuhn J, Herrmann RG. Domain-structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- Wizemann TM, Moskovitz J, Pearce BJ, Cundell D, Arvidson CG, So M, Weissbach H, Brot N, Masure HR. Peptide methionine sulfoxide reductase contributes to the maintenance of adhesions in three major pathogens. Proc Natl Acad Sci USA. 1996;93:7985–7990. doi: 10.1073/pnas.93.15.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtaszek P. Oxidative burst: an early plant response to pathogen infection. Biochem J. 1997;322:681–692. doi: 10.1042/bj3220681. [DOI] [PMC free article] [PubMed] [Google Scholar]