Abstract
Myosin-II is required for the assembly and constriction of cytokinetic contractile rings in fungi and animals. We used electron microscopy, fluorescence recovery after photobleaching (FRAP), and fluorescence correlation spectroscopy (FCS) to characterize the physical properties of Myo2 from fission yeast Schizosaccharomyces pombe. By electron microscopy, Myo2 has two heads and a coiled-coiled tail like myosin-II from other species. The first 65 nm of the tail is a stiff rod, followed by a flexible, less-ordered region up to 30 nm long. Myo2 sediments as a 7 S molecule in high salt, but aggregates rather than forming minifilaments at lower salt concentrations; this is unaffected by heavy chain phosphorylation. We used FRAP and FCS to observe the dynamics of Myo2 in live S. pombe cells and in cell extracts at different salt concentrations; both show that Myo2 with an N-terminal mEGFP tag has a diffusion coefficient of ~ 3 μm2 s−1 in the cytoplasm of live cells during interphase and mitosis. Photon counting histogram analysis of the FCS data confirmed that Myo2 diffuses as doubled-headed molecules in the cytoplasm. FCS measurements on diluted cell extracts showed that mEGFP-Myo2 has a diffusion coefficient of ~ 30 μm2 s−1 in 50 to 400 mM KCl concentrations.
Key terms: myosin-II, contractile ring, electron microscopy
Introduction
Cytokinesis of fission yeast Schizosaccharomyces pombe (S. pombe) depends on myosin-II (Myo2) (Kitayama et al. 1997; May et al. 1997; Bezanilla et al. 1997; Motegi et al. 1997), as in other fungi (Calvert et al. 2011), amoebas (DeLozanne and Spudich 1987) and animal cells (Mabuchi and Okuno 1977). Extensive work on fission yeast cytokinesis has produced an extensive genetic inventory of components (Guertin et al. 2002; Pollard and Wu 2010), a detailed timeline of the process (Wu et al. 2003) and quantitation of many participating proteins (Wu and Pollard 2005). Detailed computer simulations of ring assembly (Vavylonis et al. 2008) and constriction (Stachowiak et al. 2014) account for the cytokinetic time course in wild type and mutant cells.
Fission yeasts have two type-II myosins formed from different heavy chains: Myo2p is the heavy chain of the “conventional” dimeric myosin-II (hereafter Myo2); and Myp2p is the heavy chain of an “unconventional” myosin-II (Bezanilla and Pollard 2000; Kitayama et al. 1997; May et al. 1997; Bezanilla et al. 1997; Motegi et al. 1997; Balasubramanian et al. 1998; Motegi et al. 2000; Lord and Pollard 2004). IQ motifs of both myosin-II heavy chains associate with regulatory light chain Rlc1p and essential light chain Cdc4p (Naqvi et al. 1999; Motegi et al. 2000). Myo2 has a long coiled-coil tail formed from the C-terminal halves of the two heavy chains, while the purified tail of Myp2 forms a much shorter rod and may be monomeric and folded back on itself (Bezanilla and Pollard 2000).
The cytokinetic contractile ring of fission yeast assembles by a two-step process. Early in mitosis, Myo2 concentrates in nodes distributed in a broad band around the cell’s equator. Later, these nodes coalesce to form the contractile ring (Bezanilla et al. 2000; Motegi et al. 2000; Wu et al. 2003; Vavylonis et al. 2008). Association of Myo2 with nodes and placement of the contractile ring in the middle of the cell depends on anillin Mid1p and the IQGAP protein Rng2p (Sohrmann et al. 1996; Motegi et al. 2004; Laporte et al. 2011; Padmanabhan et al. 2011; Saha and Pollard 2012). Super-resolution fluorescence microscopy of nodes in live cells shows that Mid1p and Rng2p anchor the C-terminal tails of about ten Myo2 molecules to the plasma membrane with the heads oriented towards the cytoplasm (Laplante et al. 2016). Myo2 contributes to both the assembly and constriction of the contractile ring (Laplante et al. 2015; Laplante and Pollard 2017; Zambon et al. 2017). Phosphorylation of S1444 near the C-terminus of Myo2 heavy chain (Motegi et al. 2004) is not required for contractile ring assembly, but promotes its constriction (Sladewski et al. 2009). Myp2 joins the contractile ring after it forms (Bezanilla et al. 2000; Wu et al. 2003) and contributes to both ring constriction and septum formation (Bezanilla et al. 1997). Myp2 also promotes the recruitment of Mto1p, a member of a protein complex that positions the microtubule-organizing center (Samejima et al. 2010).
Myo2 exchanges between the cytoplasm and contractile rings on a time-scale of tens of seconds (Pelham and Chang 2002; Sladewski et al. 2009), but our knowledge of its physical state in the cytoplasm is limited. We used electron microscopy of purified Myo2 to show that the molecule has two heads and a rod-like, 65 nm tail with a heterogeneous region at the end. Myo2 aggregates at low ionic strength, but we did not observe bipolar filaments by electron microscopy regardless of salt concentration, presence of accessory proteins, or heavy chain phosphorylation. We used fluorescence correlation spectroscopy (FCS) to characterize the diffusion of mEGFP-Myo2 in the cytoplasm of living cells and in cell extracts. Photon counting histogram analysis of the FCS data showed that mEGFP-Myo2 behaves two-headed monomeric molecule in the cytoplasm. Fluorescence recovery after photobleaching (FRAP) confirmed the behavior of Myo2 seen in live-cell FCS experiments. Readers will be interested in a complementary, independent study of the hydrodynamics, solubility and motor activity of Myo2 (Pollard et al. 2017).
RESULTS
Electron microscopy of purified Myo2 molecules
Unlike the tails of many other myosin-II heavy chains, the 711-residue tail of Myo2 contains many proline residues within the last ~ 150 residues, which disrupt coiled-coil structures. Recombinant Myo2 tail domains form dimers by hydrodynamic criteria; by electron microscopy these dimers appear as rods ~87 nm long (Bezanilla and Pollard 2000). We prepared Myo2 from yeast extracts under conditions where residue serine-1444 was largely phosphorylated or unphosphorylated when analyzed by LC/MS/MS (see Supplementary materials).
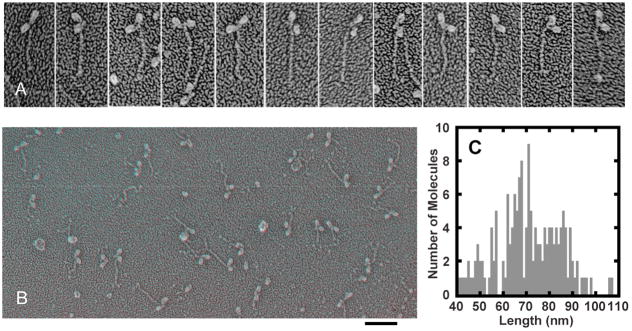
We prepared specimens of Myo2 by adsorbing purified protein to mica flakes, quick-freezing, deep etching and rotary-shadowing (Heuser 1983). Transmission electron micrographs of these specimens contain only single molecules with two heads and a tail (Figure 1AB). Pollard et al. (Pollard et al. 2017) published similar micrographs of a few Myo2 molecules under unspecified conditions. The lengths of the Myo2 tails in our specimens varied from about 40 nm to more than 100 nm (Figure 1C). These data fit a population with an average length of 70.5 nm ± 14.0 nm (mean ± SD). Higher-order models of Gaussian mixtures include subpopulations of 66.7 nm ± 4.4 nm and 82.5 nm ± 7.1 nm, which is consistent with previous work (Bezanilla and Pollard 2000). Virtually all of the tails in our preparations include a relatively straight proximal segment ~65 nm long, the length expected for a coiled-coil of 431 residues extending from residue 814 at the beginning of the predicted coiled-coil to residue 1245, the location of the first break in the predicted coiled-coil sequence. For about a third of all molecules analyzed, no tail extends beyond this 65 nm coiled-coil. Most of the longer tails bent at about 65 nm and the distal segments (here called the tailpiece) vary considerably in length, orientation and appearance. Serine 1444 is located near the end of this tailpiece. Myo2 molecules appear the same under all conditions tested: double-headed molecules with rod-shaped tails.
Figure 1.
Electron micrographs of purified Myo2 prepared by rapid freezing on mica, fracturing, deep etching and rotary shadowing. A. Electron micrographs of rotary-shadowed Myo2 prepared under dephosphorylation conditions in 500 mM KCl, 10 mM imidazole pH 7.0. The first four molecules from the left end measure around 65 nm. The 8 molecules on the right have extensions beyond 65 nm varying in shape and orientation. Panel height is 150 nm. B. Anaglyph of electron micrographs of a field of rotary-shadowed Myo2 prepared under dephosphorylation conditions in 50 mM KCl, 2 mM MgCl2, 10 mM imidazole pH 7.0. Scale bar, 100 nm. C. Distribution of Myo2p tail lengths. Myo2 purified under dephosphorylation conditions was prepared in 500 mM KCl, 10 mM imidazole pH 7.0.
Myo2 forms aggregates but not minifilaments at low ionic strength
In 200 mM KCl, Myo2 sediments at 6–7 S in sucrose or glycerol gradients (Figure S1A) like similar Type II nonmuscle myosins. Pollard et al. (Pollard et al. 2017) measured a sedimentation coefficient of 6.8 S for purified Myo2 in 50 to 500 mM KCl with or without phosphorylation of the regulatory light chain.
In 50 mM KCl >90% of Myo2 aggregates and pellets during centrifugation at 14,000 g for 20 min (Figure S1B) leaving a small fraction of soluble Myo2 that sediments at 7 S. No Myo2 sediments at 10–30 S as observed for minifilaments of other types of myosin-II (Sinard and Pollard 1989; Reisler et al. 1986). Similar results were obtained at a range of KCl concentrations; with or without 2 mM MgCl2, which aggregates myosin-II minifilaments from some species; in the presence of the fission yeast chaperone protein Rng3p (Wong et al. 2000; Lord and Pollard 2004); and with or without heavy chain phosphorylation. These results indicate that purified Myo2 exists in equilibrium between a stable 7 S molecule and an aggregated state in low-salt concentrations, but does not form minifilaments in vitro under any conditions we or Pollard et al. (Pollard et al. 2017) tested.
We used electron microscopy to examine purified Myo2 under a wide range of conditions. We tested a range of KCl concentrations from 50–200 mM, in the presence or absence of 2 mM MgCl2, and under purification conditions that favor either phosphorylation or dephosphorylation of Myo2p heavy chain (see Supplementary materials). We prepared these specimens of Myo2 for electron microscopy by deep-etching and rotary-shadowing, negative staining, or shadowing after spraying onto mica. We observed individual molecules (Figure 1B) and aggregates of protein, but no bipolar filaments. Control preparations of skeletal muscle myosin contained bipolar filaments. These experiments are consistent with independent work by Pollard et al. (Pollard et al. 2017), who found that Rlc1p phosphorylation does not affect the 7 S sedimentation coefficient or solubility of Myo2 in a range of KCl concentrations.
Photobleaching experiments show single Myo2 molecules in the cytoplasm of fission yeast
We used fluorescence recovery after photobleaching, FRAP (Axelrod et al. 1976; Edidin et al. 1976), to measure the mobile fraction and diffusion coefficient of Myo2 molecules in the cytoplasm of live S. pombe cells during mitosis and interphase. We used a strain with a mEGFP tag on the N-terminus of the Myo2 heavy chain in the genome (mEGFP-Myo2), so it was expressed from its native promoter and functioned like wild type Myo2 (S. pombe strain JW1109; Table 1)(Coffman et al. 2009). As a standard, we used freely diffusing mEGFP in the cytoplasm expressed under the control of the 3nmt1 promoter (S. pombe strain JW3290).
Table 1.
Fission yeast strains used in the study
| Strain | Genotype | Source |
|---|---|---|
| TP 150 | h- leu1-32 sm902 | M. Yanagida |
| ML 386 | h- leu1-32 ura4-D18 kanR:3nmt1-myo2 | Lord and Pollard, 2004 |
| ML 387 | h- leu1-32 ura4-D18 kanR:3nmt1-myo2 | Lord and Pollard, 2004 |
| JW3290 | h- leu1-32-kanMX6-3nmt1-mEGFP ade6-M210 ura4-D18 | J.-Q. Wu lab |
| JW3291 | h- leu1-32-kanMX6-41nmt1-mEGFP ade6-M210 ura4-D18 | J.-Q. Wu lab |
| JW1109 | h+ kanMX6-Pmyo2-mEGFP-myo2 ade6-M210 leu1-32 ura4-D18 | Coffman et al., 2009 |
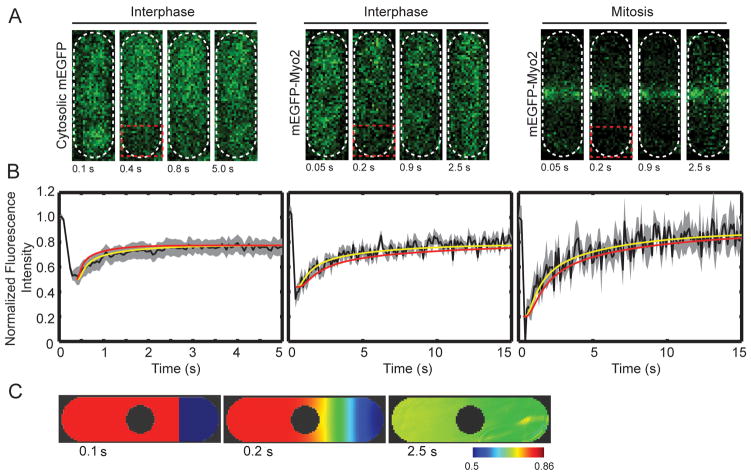
We used a laser scanning confocal microscope to bleach mEGFP-Myo2 or mEGFP, in roughly one fourth of each cell (Figure 2A). We followed the redistribution of fluorescence in the whole cell as a function of time (Figure 2B). After photobleaching, both cytosolic mEGFP and mEGFP–Myo2 fluorescence recover rapidly to levels expected after irreversible photobleaching of a large fraction of molecules. The mean half times ± SEM are 0.34 ± 0.02 s for mEGFP (n = 9 cells), 1.1 ± 0.05 s for mEGFP-Myo2 in interphase cells (n = 8 cells) and 1.0 ± 0.04 s for mEGFP-Myo2 in mitotic cells (n = 8 cells).
Figure 2.
FRAP of live cells.(A) Time series of images of cells from FRAP experiments. Each image is of 4 μm thick confocal section, including all the fluorescence in the cell. Left, cell expressing mEGFP; middle, interphase cell expressing mEGFP-Myo2; right, mitotic cell expressing mEGFP-Myo2. Red box indicates area of cells that was photobleached. (B) Averaged time courses of the recovery of fluorescence in the bleached zone (black lines): Left, mEGFP (n = 9 cells), middle, mEGFP-Myo2 in interphase (n = 8 cells) and right, mEGFP-Myo2 in mitotic phase (n = 8 cells). The grey areas show the standard error of the mean. Yellow lines are the time courses of recovery simulated by Vcell with diffusion coefficients of 22 μm2/s (mEGFP), 3.0 μm2/s (mEGFP-Myo2 in interphase) and 2.5 μm2/s (mEGFP-Myo2 in mitosis). Red lines are time courses of recovery calculated by Vcell using diffusion coefficients from FCS experiments. (C) Time course of fluorescence fluctuations in the contractile ring in a cell expressing mEGFP-Myo2, after photobleaching the tip of the cell. (D) Vcell model of time course of the recovery from photobleaching of mEGFP in the first frame (0.1 s). The second frame is time 0.2 s. The third frame is time 2.5 s. These images are sections through the mid plane of the cell. The pseudo-color scale gives the concentrations in μM used in the Vcell model based on the normalized fluorescence intensity in FRAP experiments.
We used the Virtual Cell modeling environment (Slepchenko et al. 2003) (VCell; www.vcell.org) to estimate diffusion coefficients from the FRAP data. The model assumed a single molecular species diffusing in a 3D space the size and shape of the cytoplasm of a fission yeast cell. To simulate photobleaching, we reduced the concentration of fluorescent molecules in the bleached region of the cell to experimentally observed values (Figure 2D, in blue). A spatial-deterministic model simulated the redistribution of bleached and unbleached molecules until they were evenly distributed in the cell. The diffusion coefficients and extent of bleaching were varied to match the experimental data (Figure 2B). Models with single diffusing species, i.e. Myo2 molecules, fit the fluorescence recovery after photobleaching (Figure 2B) and gave values for the diffusion coefficients (Table 2). Our value for the diffusion coefficient of cytosolic mEGFP (22 μm2/s) is close to the 26 μm2/s measured previously in live S. pombe cells (Bridges et al. 2014).
Table 2.
Diffusion coefficients
| Method, conditions | Diffusing species | D (μm2/s) Mean ±SD |
Number of cells or measurements |
|---|---|---|---|
| Cytoplasm of live cells | |||
| FRAP live interphase cells | mEGFP monomer | 22 | 9 |
| FCS live interphase cells | mEGFP monomer | 27. ± 8.5 | 20 |
| FRAP live interphase cells | mEGFP-Myo2 | 3 | 8 |
| FRAP live mitotic cells | mEGFP-Myo2 | 2.5 | 8 |
| FCS live interphase cells | mEGFP-Myo2 | 1.8 ± 0.5 | 20 |
| FCS live mitotic cells | mEGFP-Myo2 | 1.9 ± 0.5 | 20 |
| Diluted cell extracts | |||
| FCS diluted cell extract 50 mM KCl | mEGFP monomer | 76.7 ± 23.0 | 60 |
| FCS diluted cell extract 50 mM KCl | mEGFP-Myo2 | 37.8 ± 9.4 | 60 |
| FCS diluted cell extract 200 mM KCl | mEGFP-Myo2 | 29.7 ± 3.4 | 60 |
| FCS diluted cell extract 400 mM KCl | mEGFP-Myo2 | 30.0 ± 4.7 | 60 |
In FCS experiments, 10 measurements were acquired and averaged from single points in cells or cell extracts. The samples consisted of 20 cells or 20 points in each of 3 cell extracts.
After bleaching mEGFP-Myo2 in the cytoplasm of mitotic cells, the fluorescence in contractile rings declines slowly (Figure 2B). After photobleaching contractile rings, mEGFP-Myo2 fluorescence recovers with halftimes between 25–30 s as reported (Pelham and Chang 2002; Sladewski et al. 2009).
Fluorescence correlation spectroscopy show single Myo2 molecules in the cytoplasm and extracts of fission yeast
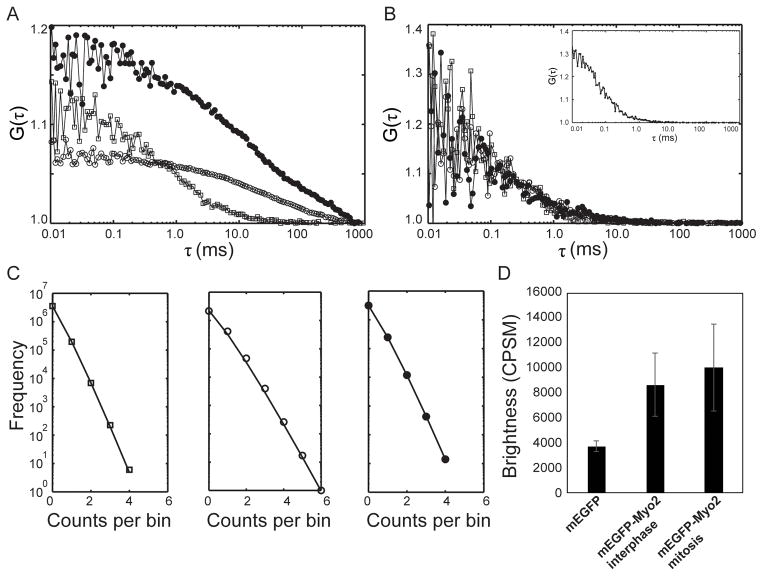
We used FCS (Magde et al. 1974) to measure the fluorescence fluctuations emitted by mEGFP and mEGFP-Myo2 molecules in live yeast and extracts of yeast cells (Figure 3). We expressed mEGFP at a moderate concentration under the control of 41nmt1 promoter (S. pombe strain JW3291). We determined the confocal volume (0.5–1 fL) using 10 nM Alexa Fluor 488 in buffer (Figure 3B, inset). FCS gave an average interphase cytoplasmic concentration of mEGFP-Myo2 of 0.47 ± 0.01 μM (Figure 3A), the same concentration measured by quantitative fluorescence microscopy (Wu 2005). In mitotic cells, the cytoplasmic concentration of mEGFP-Myo2 was 0.27 ± 0.03 μM, confirming that about half of the protein assembles in the contractile ring (Wu 2005).
Figure 3.
Fluorescence correlation spectroscopy on mEGFP and mEGFP-Myo2 in live cells and cell extracts. (A) Cytoplasm of live cells: (□) cell expressing mEGFP; (○) interphase cell expressing mEGFP-Myo2; and (●) mitotic cell expressing mEGFP-Myo2. (B) Cell extracts. Asynchronous cells were lysed by bead beating at 4°C after mixing 1:1 with 10 mM imidazole pH 7.0, 50 to 400 mM KCl, 1 mM MgCl2, 1 mM EGTA, 1% sucrose, 1 mM DTT, EDTA-free Protease Inhibitor tablet- cOmplete (catalog #11873580001, Roche) 1 tablet/50 ml buffer and 2 mM ATP. After clarification at 100,000 g for 20 min, samples of supernatant were diluted 10-fold in the same buffer; (□) mEGFP in cell lysate with 50 mM KCl; mEGFP-Myo2 in interphase cell lysates with (○) 50 mM KCl, and (●) 400mM KCl. Inset shows the autocorrelation curve for 10 nM Alexa 488 dye (Molecular Probes, Eugene) dissolved in dimethylformamide and diluted to calibrate the confocal volume. (C) Photon counting histograms of (□) a cell expressing mEGFP (left); (○) an interphase cell expressing mEGFP-Myo2 (middle); and (●) a mitotic cell expressing mEGFP-Myo2 (right). Symbols are the raw data. The black lines are curves fit to this data. (D) Cytosolic molecular brightness plotted as counts per second per molecule (CPSM) for 10 cells expressing mEGFP, 6 interphase cells expressing mEGFP-Myo2 and 7 mitotic cells expressing mEGFP-Myo2. Means ± SDs.
We used the anomalous diffusion equation with a single diffusing species to take into account molecular crowding in the yeast cytoplasm when fitting the FCS data to measure mean diffusion coefficients (Table 2). The measured diffusion coefficient of cytosolic mEGFP (27.2 ± 1.2 μm2/s) in S. pombe agrees with a previous report (Bridges et al. 2014). The diffusion coefficients of mEGFP-Myo2 in the cytoplasm of interphase and mitotic cells were indistinguishable (1.9 ± 0.1 μm2/s). These diffusion coefficients also gave good fits to the FRAP data for these molecules in the VCell model (Figure 2B, red lines).
We used photon counting histogram (PCH) analysis of the FCS photon counts to determine the number of mEGFPs associated with the diffusing species of Myo2 in S. pombe cytoplasm (Figure 3CD). PCH analysis considers the intensity distribution of the point spread function (PSF), Poisson shot noise from the detector, and fluctuations in molecule numbers to estimate the number and brightness of molecules (Chen et al. 2003; Chen et al. 1999). We compared the brightness of the molecules diffusing in the cytoplasm of yeast strains expressing mEGFP and mEGFP-Myo2. The analysis shows that mEGFP-Myo2 has twice the brightness of mEGFP in interphase and mitotic cells, indicating that the diffusing species of Myo2 in S. pombe cytoplasm is associated with two molecules of mEGFP, as expected for a myosin with two heavy chains (Figure 3CD).
To test the effect of salt concentration on the diffusion coefficient of cytoplasmic mEGFP-Myo2, we lysed S. pombe cells in buffers containing 50, 200 or 400 mM KCl, then diluted about 10 times relative to original cytoplasmic concentration. For comparison, we did FCS experiments with lysates of cells expressing mEGFP in 50 mM KCl. We fit the FCS data in cell lysates with the 3D diffusion equation instead of the anomalous diffusion equation and calculated the mean diffusion coefficients (Table 2). The diffusion coefficient of mEGFP is similar to a previous study (Terry et al. 1995). The diffusion coefficients of mEGFP-Myo2 in lysates at all three salt concentrations were around 30 μm2/s, which is about two-fold higher than measured for purified Myo2 (Lord and Pollard 2004).
DISCUSSION
Most of the tails of the double-headed Myo2 molecules were much shorter in our electron micrographs than the expected length for a 711-residue coiled-coil tail. Our interpretation is that the proximal tail forms a relatively rigid coiled-coil of about 65 nm, followed by a flexible region at residue 1245 and a heterogeneous distal segment of about 20 nm corresponding to residues 1246–1380, where a second large break in the coiled-coil sequence occurs (Bezanilla and Pollard 2000). The end of the tail between residues 1400–1525 is predicted to form two short coiled-coils, but this region was heterogeneous in the replicas, so very few of the tails were measured at over 100 nm long. Recombinant Myo2 tails prepared by glycerol spraying and rotary shadowing (Bezanilla and Pollard 2000) had a broad distribution of lengths, with an average of 87 nm but again with a substantial number of longer species, so distal segments also appeared heterogeneous by this method. This Myo2 tailpiece is thought to interact with IQGAP Rng2p (Laporte et al. 2011) in cytokinesis nodes in the contractile ring (Laplante et al. 2016). In low salt ± 2 mM MgCl2 most Myo2 molecules form insoluble aggregates with a few soluble 7 S molecules. We did not observe in these samples any bipolar minifilaments by electron microscopy or the corresponding 10–30 S species by sedimentation velocity.
To gain some insight into the Myo2 species that exchange between the 0.3 μM cytoplasmic pool and cytokinesis nodes and contractile rings (Pelham and Chang 2002; Sladewski et al. 2009), we used FRAP and FCS to characterize the physical state of Myo2 in the cytoplasm of living cells. Previous work did not establish whether the cytoplasmic pool of Myo2 is monomeric or assembled into larger structures.
PCH analysis of the FCS data provided the most direct measurement of the oligomeric state of cytoplasmic Myo2. This analysis revealed that the brightness of mEGFP-Myo2 is twice that of monomeric mEGFP (Figure 3CD). This is strong evidence that two molecules of mEGFP are fused to the species of Myo2 diffusing in the cytoplasm, as expected for single molecules of a myosin-II with two heavy chains (along with four light chains).
Measurements of diffusion coefficients by FRAP and FCS are consistent with this conclusion but with added insights. According to the Stokes-Einstein equation, the diffusion coefficient is inversely proportional to the molecular radius, so measurement of the diffusion coefficient of a fluorophore-tagged protein provides information about its oligomeric state (Youker and Teng 2014). Since the diffusion coefficient is also inversely proportional to the viscosity, diffusion coefficients are expected to be much lower in the crowded cytoplasm than dilute solution. For example, the diffusion coefficient of mEGFP was three-fold lower in the cytoplasm of our live cells (D = 28 μm2/s) than in dilute solution (D = 76 μm2/s).
FRAP and FCS gave similar diffusion coefficients of 2–3 μm2/s for mEGFP-Myo2 in the cytoplasm of living yeast cells, about 5-fold less than a Myo2 molecule with a Stokes’ radius of 13.4 nm in dilute solution (Lord and Pollard 2004). These diffusion coefficients are consistent with the conclusion of the PCH analysis that cytoplasmic Myo2 diffuses as single molecules with two heavy chains. Thus, conditions in the cytoplasm must favor the solubility of Myo2 molecules with two heads. These single molecules are the most likely species exchanging between the cytoplasm and cytokinetic nodes, where the end of the Myo2 tail is tethered to the plasma membrane (Laplante et al. 2016).
One surprise is that the diffusion coefficients of mEGFP-Myo2 in cell extracts diluted with 50 to 500 mM KCl are higher (~30 μm2/s) than the purified protein in dilution solution. Given these high diffusion coefficients, conditions in the cytoplasmic extract may favor a more compact conformation than the purified Myo2 with an extended tail as observed by electron microscopy.
FCS experiments confirmed that about half of mEGFP-Myo2 is incorporated into the contractile ring in mitotic cells (Wu and Pollard 2005). We verified that the halftime for fluorescence recovery of mEGFP-Myo2 in the contractile ring is about 25–30 s (Clifford et al. 2008; Pelham and Chang 2002). Super-resolution microscopy will be required to determine if nodes exchange individual Myo2 molecules or whole nodes turn over.
Materials and methods
We used standard procedures for S. pombe growth and genetic manipulation (Moreno et al. 1991). Table 1 lists the strains used in this work.
Myo2 purification
We isolated Myo2 from S. pombe strains over expressing the Myo2 heavy chain (ML 386 or 387) and transformed with pGST-Rlc1 or pGST-Cdc4 plasmids to over-express Rlc1p and Cdc4p light chains tagged with GST (Lord and Pollard 2004). We lysed the cells by vortexing with glass beads in an equal volume of 750 mM KCl, 25 mM Tris-HCl, 4 mM MgCl2, 20 mM sodium phosphate, 2 mM EGTA 0.1% Triton X-100 (pH 7.4) (Lord and Pollard 2004) and added 4 mM ATP and 3 mM phenylmethylsulfonyl fluoride to the homogenate before high-speed centrifugation. “Dephosphorylated Myo2” was prepared after incubating the clarified cell lysate 45 min at 4 °C. “Phosphorylated Myo2” was isolated in lysis buffer with 20 mM sodium phosphate, 50 mM NaF, 100 μM NaVO4 and 20 mM β-glycerophosphate to reduce dephosphorylation. “Phosphorylated Myo2” was confirmed by mass spectrometry to be phosphorylated on S1444 (Figure S2) and reacted more strongly with an antibody specific for this site than “unphosphorylated Myo2” (Figure S3).
Sedimentation velocity ultracentrifugation
Samples of 200 μL of 200 μg/ml Myo2 or protein standards were applied to 5 mL linear gradients of 5–25% glycerol or 10–50% sucrose made in 10 mM imidazole pH 7.0, KCl from 50–500 mM, 1 mM DTT and 4 mM MgCl2. These samples were centrifuged in Beckman swinging-bucket rotors at 35,000 rpm for 3 h in a SW41 rotor or 48,000 rpm for 18 h in a SW55Ti rotor. Fractions of 250 μL were collected from the bottoms of the tubes with a peristaltic pump and analyzed by SDS-PAGE and silver staining.
Electron microscopy; quick frozen samples
Purified Myo2 at concentrations of 50–150 nM in a range of buffers was adsorbed to mica flakes, rapidly frozen, fractured, deep etched and rotary shadowed (Heuser 1983). Conditions included 10 mM imidazole pH 7.0 or 10 mM Tris pH 8.5, KCl concentrations from 50–200 mM, the presence or absence of 2 mM MgCl2, and the presence or absence of 500 nM Rng3p (Lord and Pollard 2004).
Electron microscopy; negative stain
Myo2 or Myo2p tail fragments were dialyzed into various low-salt conditions in 10 mM imidazole pH 7.0 or 10 mM Tris pH 8.5 and adsorbed to glow-discharged Formvar/carbon films on copper grids for 1 min, washed twice in buffer, drained, stained with two drops of 1% uranyl acetate, drained and dried.
Electron microscopy; spray and rotary shadow
Small volumes of Myo2 at 100–500 nM in 10 mM imidazole pH 7.0 or 20 mM Tris pH 7.5 or 8.0, KCl concentrations from 50–200 mM, with or without 2 mM MgCl2 were sprayed onto 1 cm × 1 cm squares of freshly cleaved mica. After drying, the mica was rotary-shadowed with platinum and stabilized with carbon. Films were floated on distilled water and picked up on copper grids.
Electron microscopy; microscopy and quantitation
Samples were observed with a JEOL-1100 electron microscope equipped with a digital camera. We measured lengths of Myo2 tails in rotary shadowed preparations using a custom best-fit algorithm (Kuhn and Pollard 2005) in ImageJ (National Institutes of Health, rsb.info.nih.gov).
Fluorescence recovery after photobleaching (FRAP)
We carried out FRAP experiments on a Zeiss LSM-880 confocal microscope (Jena, Germany), equipped with 63X, 1.4 NA Plan-Apochromatic oil immersion objective at 25 ºC. The pinhole was opened 4 μm wide to collect all the fluorescence from entire cell. Excitation for image acquisition (λ = 488 nm) was set at 0.8 – 1% of the maximal laser intensity. Photobleaching was achieved with a 100% laser power with 5–10 iterations (over 0.17–0.3 s). In each FRAP experiment one-fourth of the volume of the cell from one of the cell tip was photobleached. Images of the cells were captured before and immediately after bleaching, following the recovery phase at intervals of 50–100 ms for up to 8 s. Zeiss LSM software was used to remove background fluorescence and correct for photobleaching during the acquisition of recovery data.
Analysis of photobleaching recovery
A Virtual Cell simulation (VCell; www.vcell.org) was used to model the FRAP results and to determine the 3D diffusion coefficients. S. pombe cells were modeled as cylinders with a diameter of 3.5 μm and length of 9–12 μm to approximate their physical dimensions. The nucleus is the largest organelle and can hinder the diffusion of molecules, so it was included as a sphere with diameter 2.2 μm at the cell center. The photobleaching condition in FRAP experiment was used in the model as the initial condition with a lower concentration of the molecules in the bleached region and higher concentration of the molecules in the unbleached region of the cell based on the experimental FRAP recovery data. Molecules diffused according to defined diffusion coefficients and spatial constraints. Simulations used 50–100 ms time steps with spatial simulation mesh of 0.1 μm in x, y and z axes.
Fluorescence correlation spectroscopy (FCS)
FCS measurements were carried out with a Zeiss LSM-880 Confocal microscope outfitted with a ConfoCor 3 module and a 40X, 1.2 NA C-Apochromatic water immersion objective at 25 ºC. The laser power was kept ~ 0.5 μW or 0.15% for cells and ~ 2.3 μW or 0.7% for cell extracts of maximum intensity at the back aperture of the objective.
Cells expressing mEGFP or mEGFP-Myo2 were excited with 488 nm laser line using a MBS488 dichroic mirror. A BP 499–535 nm filter was used to collect the fluorescence emission. The pinhole was set to 1.0 airy unit. A fluorescent image of a field of cells expressing mEGFP-Myo2 was acquired before each FCS experiment. FCS measurements were carried out for 5 s and repeated ten times at single point in the cytoplasm, near the tip of each cell. The Zeiss software calculated the autocorrelation curve from the fluorescence trace using the standard formula,
where δF(t) = F(t) − 〈F〉.
The molecular crowding in the cytoplasm obstructed the diffusion of mEGFP and mEGFP-Myo2 molecules in the cytoplasm. Hence, correlation curves from cells expressing mEGFP were found to best fit to an anomalous diffusion equation instead normal 3D diffusion equation. Custom-built software in MATLAB was used to fit the correlation curves to the standard equation of anomalous diffusion,
where G(∞) represents the level of background autocorrelation at long time scales and s is the aspect ratio, height to width, of the confocal volume. N is the average number of fluorophores and τdiff, their characteristic residence time in the confocal volume; τdiff, is related to the diffusion coefficient D of the fluorophores and to the radial half width ω0 of the confocal volume:
FCS measurements were first carried out with 10 nM Alexa Fluor 488 dye in solution, which has a known diffusion coefficient of 414 μm2/s at 25 ºC (Petrov and Schwille 2008). The residence time τdiff was calculated from fits of the autocorrelation curves of Alexa Fluor 488 to determine radial dimension ω0 from the above equation.
FCS in cell extracts
The S. pombe strain expressing mEGFP-Myo2 was grown in 300 mL of YE5S media overnight to OD595 = 0.4. Cells were centrifuged at 3000 rpm and washed twice with water. An equal amount of volume buffer (10 mM imidazole pH 7.0, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 1% sucrose, 1 mM DTT, EDTA free protease inhibitor cocktail tablet–cOmplete (catalog #11873580001, Roche) (1 tablet/50 ml buffer) and 2 mM ATP) was added to the cell pellet and the cells were lysed by vortexing with glass beads in a cell homogenizer (FastPrep-24 Instrument, MP Biomedical). The lysate was centrifuged at 100,000 × g in a tabletop ultracentrifuge for 20 min at 4 ºC. The supernatant was diluted 10 times, transferred to a Lab-Tek 8-chamber borosilicate coverglass system and used for the FCS measurements. Due to lack of molecular crowding in the diluted supernatant, mEGFP and mEGFP-Myo2 molecules would move freely. Therefore, correlation curves obtained in the FCS experiments of cell extracts were fit with the normal 3D diffusion equation. To increase the KCl concentration from 50–400 mM, we added extra KCl in the buffer.
Photon counting histogram (PCH) analysis
Photon counts from FCS experiments were plotted as histograms to determine the photon count variance. The ratio of the photon count variance to the average intensity gave an estimate of the molecular brightness in terms of counts per second per molecule (CPSM) (Youker and Teng 2014). The molecular brightness of mEGFP and mEGFP-Myo2 were compared to determine the oligomerization state of Myo2 in S. pombe cytoplasm. PCH analysis was carried out with a freely available ImageJ plugin created by Jay Unruh at Stowers Institute for Medical Research in Kansas City, Missouri (Unruh 2014).
Supplementary Material
Acknowledgments
Research reported in this publication was supported by National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM026338. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors thank Brian Slaughter and Joseph Wolenski for help with florescence correlation spectroscopy, Ann Cowen for advice on modeling photobleaching recovery, Caroline Laplante, Eugene Davidov, Kyle Friend, Jeff Kuhn, Michael Snyder and Janine Mok for assistance with the project and Pollard lab members for helpful discussions.
References
- Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys J. 1976;16:1055–69. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian MK, McCollum D, Chang L, Wong KC, Naqvi NI, He X, Sazer S, Gould KL. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics. 1998;149(3):1265–75. doi: 10.1093/genetics/149.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M, Pollard TD. Myosin-II tails confer unique functions in Schizosaccharomyces pombe: characterization of a novel myosin-II tail. Mol Biol Cell. 2000;11(1):79–91. doi: 10.1091/mbc.11.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M, Forsburg SL, Pollard TD. Identification of a second myosin-II in Schizosaccharomyces pombe: Myp2p is conditionally required for cytokinesis. Mol Biol Cell. 1997;8(12):2693–705. doi: 10.1091/mbc.8.12.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M, Wilson JM, Pollard TD. Fission yeast myosin-II isoforms assemble into contractile rings at distinct times during mitosis. Curr Biol. 2000;10(7):397–400. doi: 10.1016/s0960-9822(00)00420-6. [DOI] [PubMed] [Google Scholar]
- Bridges AA, Zhang H, Mehta SB, Occhipinti P, Tani T, Gladfelter AS. Septin assemblies form by diffusion-driven annealing on membranes. Proc Natl Acad Sci U S A. 2014;111:2146–51. doi: 10.1073/pnas.1314138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert ME, Wright GD, Leong FY, Chiam KH, Chen Y, Jedd G, Balasubramanian MK. Myosin concentration underlies cell size-dependent scalability of actomyosin ring constriction. J Cell Biol. 2011;195:799–813. doi: 10.1083/jcb.201101055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wei LN, Muller JD. Probing protein oligomerization in living cells with fluorescence fluctuation spectroscopy. Proc Natl Acad Sci U S A. 2003;100(26):15492–7. doi: 10.1073/pnas.2533045100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Muller JD, So PT, Gratton E. The photon counting histogram in fluorescence fluctuation spectroscopy. Biophys J. 1999;77(1):553–67. doi: 10.1016/S0006-3495(99)76912-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford D, Wolfe B, Roberts-Galbraith R, McDonald W, Yates, Gould K. The Clp1/Cdc14 phosphatase contributes to the robustness of cytokinesis by association with anillin-related Mid1. The Journal of cell biology. 2008;181:79–88. doi: 10.1083/jcb.200709060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman VC, Nile AH, Lee IJ, Liu H, Wu JQ. Roles of formin nodes and myosin motor activity in Mid1p-dependent contractile-ring assembly during fission yeast cytokinesis. Mol Biol Cell. 2009;20(24):5195–210. doi: 10.1091/mbc.E09-05-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLozanne A, Spudich JA. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 1987;236:1086–91. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- Edidin M, Zagyansky Y, Lardner TJ. Measurement of membrane protein lateral diffusion in single cells. Science. 1976;191:466–68. doi: 10.1126/science.1246629. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Trautmann S, McCollum D. Cytokinesis in eukaryotes. Microbiol Mol Biol Rev. 2002;66(2):155–78. doi: 10.1128/MMBR.66.2.155-178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser JE. Procedure for freeze-drying molecules adsorbed to mica flakes. Journal of Molecular Biology. 1983;169:155–95. doi: 10.1016/s0022-2836(83)80179-x. [DOI] [PubMed] [Google Scholar]
- Kitayama C, Sugimoto A, Yamamoto M. Type II myosin heavy chain encoded by the myo2 gene composes the contractile ring during cytokinesis in Schizosaccharomyces pombe. The Journal of cell biology. 1997;137(6):1309–19. doi: 10.1083/jcb.137.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J. 2005;88(2):1387–402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante C, Pollard TD. Response to Zambon et al. Curr Biol. 2017;27:R101–R02. doi: 10.1016/j.cub.2016.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante C, Huang F, Tebbs IR, Bewersdorf J, Pollard TD. Molecular organization of cytokinesis nodes and contractile rings by super-resolution fluorescence microscopy of live fission yeast. Proc Natl Acad Sci U S A. 2016;113:E5876–E85. doi: 10.1073/pnas.1608252113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante C, Berro J, Karatekin E, Lee R, Hernandez-Leyva A, Pollard TD. Three myosins contribute uniquely to the assembly and constriction of the cytokinetic contractile ring in fission yeast. Curr Biol. 2015;25:1955–65. doi: 10.1016/j.cub.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Coffman V, Lee I, Wu J. Assembly and architecture of precursor nodes during fission yeast cytokinesis. The Journal of cell biology. 2011;192:1005–21. doi: 10.1083/jcb.201008171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord M, Pollard TD. UCS protein Rng3p activates actin filament gliding by fission yeast myosin-II. The Journal of cell biology. 2004;167(2):315–25. doi: 10.1083/jcb.200404045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuchi I, Okuno M. The effect of myosin antibody on the division of starfish blastomeres. The Journal of cell biology. 1977;74(1):251–63. doi: 10.1083/jcb.74.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magde D, Elson EL, Webb WW. Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers. 1974;13:29–61. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- May KM, Watts FZ, Jones N, Hyams JS. Type II myosin involved in cytokinesis in the fission yeast, Schizosaccharomyces pombe. Cell Motil Cytoskel. 1997;38(4):385–96. doi: 10.1002/(SICI)1097-0169(1997)38:4<385::AID-CM8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods in enzymology. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Motegi F, Nakano K, Mabuchi I. Molecular mechanism of myosin-II assembly at the division site in Schizosaccharomyces pombe. J Cell Sci. 2000;113(Pt 10):1813–25. doi: 10.1242/jcs.113.10.1813. [DOI] [PubMed] [Google Scholar]
- Motegi F, Mishra M, Balasubramanian MK, Mabuchi I. Myosin-II reorganization during mitosis is controlled temporally by its dephosphorylation and spatially by Mid1 in fission yeast. The Journal of cell biology. 2004;165(5):685–95. doi: 10.1083/jcb.200402097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi F, Nakano K, Kitayama C, Yamamoto M, Mabuchi I. Identification of Myo3, a second type-II myosin heavy chain in the fission yeast Schizosaccharomyces pombe. FEBS Lett. 1997;420(2–3):161–6. doi: 10.1016/s0014-5793(97)01510-x. [DOI] [PubMed] [Google Scholar]
- Naqvi NI, Eng K, Gould KL, Balasubramanian MK. Evidence for F-actin-dependent and -independent mechanisms involved in assembly and stability of the medial actomyosin ring in fission yeast. Embo J. 1999;18(4):854–62. doi: 10.1093/emboj/18.4.854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan A, Bakka K, Sevugan M, Naqvi N, D’souza V, Tang X, Mishra M, Balasubramanian M. IQGAP-related Rng2p organizes cortical nodes and ensures position of cell division in fission yeast. Curr Biol. 2011;21:467–72. doi: 10.1016/j.cub.2011.01.059. [DOI] [PubMed] [Google Scholar]
- Pelham RJ, Chang F. Actin dynamics in the contractile ring during cytokinesis in fission yeast. Nature. 2002;419:82–86. doi: 10.1038/nature00999. [DOI] [PubMed] [Google Scholar]
- Petrov EP, Schwille P. State of the Art and Novel Trends in Fluorescence Correlation Spectroscopy. Standardization and Quality Assurance in Fluorescence Measurements II. 2008;6:145–97. [Google Scholar]
- Pollard LW, Bookwalter CS, Tang Q, Krementsova EB, Trybus KM, Lowey S. Fission yeast myosin Myo2 is down-regulated in actin affinity by light chain phosphorylation. Proc Natl Acad Sci U S A. 2017 doi: 10.1073/pnas.1703161114. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Wu J-Q. Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol. 2010;11:149–55. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisler E, Cheung P, Borochov N. Macromolecular assemblies of myosin. Biophys J. 1986;49:335–42. doi: 10.1016/S0006-3495(86)83646-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Pollard TD. Anillin-related protein Mid1p coordinates the assembly of the cytokinetic contractile ring in fission yeast. Molec Biol Cell. 2012;23:3982–92. doi: 10.1091/mbc.E12-07-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samejima I, Miller V, Rincon S, Sawin K. Fission Yeast Mto1 Regulates Diversity of Cytoplasmic Microtubule Organizing Centers. Curr Biol. 2010;20:1959–65. doi: 10.1016/j.cub.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinard JH, Pollard TD. The effect of heavy-chain phosphorylation and solution conditions on the assembly of Acanthamoeba myosin-II. J Cell Biol. 1989;109:1529–35. doi: 10.1083/jcb.109.4.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladewski T, Previs M, Lord M. Regulation of fission yeast myosin-II function and contractile ring dynamics by regulatory light-chain and heavy-chain phosphorylation. Mol Biol Cell. 2009;20:3941–52. doi: 10.1091/mbc.E09-04-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slepchenko BM, Schaff JC, Macara I, Loew LM. Quantitative cell biology with the Virtual Cell. Trends Cell Biol. 2003;13(11):570–6. doi: 10.1016/j.tcb.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Sohrmann M, Fankhauser C, Brodbeck C, Simanis V. The dmf1/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes Dev. 1996;10(21):2707–19. doi: 10.1101/gad.10.21.2707. [DOI] [PubMed] [Google Scholar]
- Stachowiak MR, Laplante C, Chin HF, Guirao B, Karatekin E, Pollard TD, O’Shaughnessy B. Mechanism of cytokinetic contractile ring constriction in fission yeast. Dev Cell. 2014;29:547–61. doi: 10.1016/j.devcel.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry BR, Matthews EK, Haseloff J. Molecular characterisation of recombinant green fluorescent protein by fluorescence correlation microscopy. Biochem Biophys Res Commun. 1995;217(1):21–7. doi: 10.1006/bbrc.1995.2740. [DOI] [PubMed] [Google Scholar]
- Unruh JR. Stowers ImageJ Plugins. 2014 http://researchstowersorg/imagejplugins/zipped_pluginshtml.
- Vavylonis D, Wu J-Q, Hao S, O’Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- Wong KC, Naqvi NI, Iino Y, Yamamoto M, Balasubramanian MK. Fission yeast Rng3p: an UCS-domain protein that mediates myosin II assembly during cytokinesis. Journal of cell science. 2000;113(Pt 13):2421–32. doi: 10.1242/jcs.113.13.2421. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310(5746):310–4. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Kuhn JR, Kovar DR, Pollard TD. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev Cell. 2003;5(5):723–34. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- Youker RT, Teng H. Measuring protein dynamics in live cells: protocols and practical considerations for fluorescence fluctuation microscopy. J Biomed Opt. 2014;19(9):90801. doi: 10.1117/1.JBO.19.9.090801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon P, Palani S, Kamnev A, Balasubramanian MK. Myo2p is the major motor involved in actomyosin ring contraction in fission yeast. Curr Biol. 2017;27:R99–R100. doi: 10.1016/j.cub.2016.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.